Abstract
Atrophic gastritis is considered to be an antecedent to intestinal metaplasia and gastric cancer. A previous study identified that Helicobacter pylori was absent at the severe atrophic gastritis stage, and alterations in the gastric microbial composition resembled those in gastric cancer. To explore the role of the bacteria absence of H. pylori in gastric carcinogenesis, in the current study, we compared the microbiota of clinically collected H. pylori-free gastric fluids from 30 patients with non-atrophic gastritis (N) and 22 patients with severe atrophic gastritis (S). We estimated the bacterial loads in the N and S groups by colony counting in culture agar as well as by measuring the concentration of the extracted DNA. The results showed a significant increase in bacterial load in patients with atrophic gastritis in comparison to non-atrophic gastritis. Then, we analyzed the microbial communities of the gastric fluids from all 52 patients using high-throughput sequencing of 16S rRNA amplicons. The Chao 1, Shannon and Simpson diversity indexes demonstrated that the bacterial richness and diversity were not significantly different between the N and S groups. Moreover, principal component analysis illustrated that the microbiomes from the S group were more scattered. Microbiota composition analysis showed that the entire dataset was clustered into 27 phyla, 61 classes, 106 orders, 177 families, 292 genera and 121 species. At the genus level, only the abundance of Prevotella was significantly different between the N and S groups. Further analysis showed that all the higher taxonomic categories were significantly different between the N and S groups. To assess the effects of the metabolic products of Prevotella spp. on gastric cell physiology, we treated the human gastric epithelial cell line AGS with acetic acid and monitored nitric oxide (NO) production. The results showed that acetic acid at low concentrations (0.5 and 5 µM) significantly inhibited AGS cells to secrete NO compared to phosphate buffer saline-treated control cells. These results suggest that the microbiota in non-atrophic gastritis may influence gastric epithelial cell physiology.
Keywords: gastritis, microbiota, prevotella, nitric oxide
Introduction
Gastric cancer is one of the most common malignant tumors. It accounts for about 10% of all invasive cancers and is the second leading cause of cancer deaths worldwide (1). Although the incidence of gastric cancer in the west had declined, it remains the most common type of cancer in Asia. In China, the total number of cases and deaths from gastric cancer have increased concomitant with huge ongoing demographic changes (2,3), resulting in an urgent need to find more effective diagnostic methods and treatments.
The human stomach harbors a large number of bacteria including Helicobacter pylori (4,5); these microbial organisms collectively compose the microbial community called the microbiota. Previous studies have shown that bacterial factors play an important role in the development of gastric cancer (5,6). Gastric cancer develops from a multifactorial, multistep inflammatory process, progressing through the stages of superficial gastritis, atrophic gastritis, intestinal metaplasia, and dysplasia before the development of gastric cancer (7). At the atrophic gastritis stage, the secretion of essential substances such as hydrochloric acid, pepsin, and intrinsic factor are impaired, resulting in a neutralized stomach environment. In turn, many other bacterial species can colonize the new environment leading to gastric microbiota overgrowth as the gastritis progress and low acid production (8,9). Atrophic gastritis is considered to be an antecedent to intestinal metaplasia and gastric cancer (10,11), thus, exploring the bacterial factors that propel the progress of atrophic gastritis may be helpful for elucidating the mechanisms underlying gastric cancer development.
In previous studies, infection with H. pylori was reported to closely correlate with the other bacteria present in stomach (12), and clinical observations have found that atrophic gastritis without H. pylori is more likely to develop into gastric cancer than cases with H. pylori, an observation that has been supported by animal studies. Lee et al also showed that intervention with antimicrobial therapies delayed the onset of gastric cancer in transgenic mice irrespective of H. pylori infection status (8). Moreover, other research has shown that feeding germ-free transgenic mice with artificial microbiota accelerated the occurrence of cancer (6). Noticeably, a study by Engstrand and Lindberg found that the changes in the microbiota in individuals with atrophic gastritis resemble those seen in gastric cancer (5). Thus, the absence of H. pylori in atrophic gastritis may play a crucial role in gastric carcinogenesis; thus, the alterations in the composition of gastric microbiota and its interaction with host cells require further studies.
However, there has been little study on the characterization of the absence of H. pylori in the progress of gastritis. In the current study, we investigated the variation in gastric microbiota using gastric fluids from 30 patients with non-atrophic gastritis (N) and 22 patients with chronic atrophic gastritis (S). Our results suggest that the microbiota in gastric fluids might influence gastric epithelial cell physiology and gastric carcinogenesis.
Materials and methods
Patient samples collection and cell culture
Gastric fluids newly extracted from 30 non-atrophic gastritis and 22 atrophic gastritis patients by endoscopy were obtained from Qilu Hospital of Shandong University (Jinan, China). All patients gave their informed consent prior to their inclusion in the study. The gastric fluids were kept at −80°C and used for subsequent analysis. All studies were reviewed and approved by the Ethic Committee of Shandong University (Jinan, China). Gastric adenocarcinoma cell line AGS cells were maintained in our laboratory. AGS cells were cultured in Ham's F-12 medium (HyClone, Logan, UT, USA) supplemented with 10% FCS and 1% penicillin-streptomycin. BGC-823 cells were cultured in RPMI-1640 (Life Technologies, Carlsbad, CA, USA) supplemented with 10% FCS (Tianhang Co., Ltd., Hangzhou, China) and 1% penicillin-streptomycin.
Sequencing method
Extraction of genome DNA
Total genome DNA from samples was extracted using CTAB/SDS method. DNA concentration and purity was monitored on 1% agarose gels. According to the concentration, DNA was diluted to 1 ng/µl using sterile water.
Amplicon generation
Primer: 16S V4: 515F-806R, 18S V4: 528F-706R, 18S V9: 1380F-1510R, ITS1: ITS1FITS2. 16S/18S rRNA genes were amplified used the specific primer with the barcode. All PCR reactions were carried out in 30 µl reactions with 15 µl of Phusion® High-Fidelity PCR Master Mix (New England Biolabs, Ipswich, MA, USA); 0.2 µM of forward and reverse primers, and about 10 ng templates DNA. Thermal cycling consisted of initial denaturation at 98°C for 1 min, followed by 30 cycles of denaturation at 98°C for 10 sec, annealing at 50°C for 30 sec, and elongation at 72°C for 30 sec. Finally 72°C for 5 min.
PCR products quantification and qualification
Mix same volume of 1X loading buffer (contained SYBR-Green) with PCR products and operate electrophoresis on 2% agarose gel for detection. Samples with bright main strip between 400–450 bp were chosen for further experiments.
PCR products mixing and purification
PCR products were mixed in equidensity ratios. Then, mixture PCR products was purified with GeneJET Gel Extraction Kit (Thermo Scientific).
Library preparation and sequencing
Sequencing libraries were generated using NEB Next® Ultra™ DNA Library Prep kit for Illumina (New England Biolabs) following manufacturer's recommendations and index codes were added. The library quality was assessed on the Qubit@2.0 Fluorometer (Thermo Scientific) and Agilent Bioanalyzer 2100 system. At last, the library was sequenced on an Illumina MiSeq platform and 250/300 bp paired-end reads were generated.
Data analysis
Paired-end reads from the original DNA fragments are merged by using FLASH-a very fast and accurate analysis tool which is designed to merge paired-end reads when there are overlaps between reads1 and reads2. Paired-end reads was assigned to each sample according to the unique barcodes. Sequences were analyzed using QIIME software package (Quantitative Insights Into Microbial Ecology), and in-house Perl scripts were used to analyze alpha-(within samples) and beta-(among samples) diversity. First, reads were filtered by QIIME quality filters. Then we use pick_de_novo_otus.py to pick operational taxonomic units (OTUs) by making OTU table. Sequences with ≥97% similarity were assigned to the same OTUs. We pick a representative sequences for each OTU and use the RDP classifier to annotate taxonomic information for each representative sequence. In order to compute Alpha Divesity, we rarify the OTU table and calculate three metrics: Chao1 estimates the species abundance; Observed Species is estimates the amount of unique OTUs found in each sample, and Shannon index. Rarefaction curves were generated based on these three metrics. QIIME calculates both weighted and unweighted unifrac, which are phylogenetic measures of beta diversity.
RNA extraction and quantitative real-time PCR (QRT-PCR)
Total cellular RNA was extracted with TRIzol (Life Technologies) according to the protocol provided by the manufacturer. First-strand cDNA was synthesized from 1 µg total cellular or tissue RNA using the RevertAid™ First Strand cDNA Synthesis kit (Thermo Fisher Scientific, Waltham, MA, USA) with random primers. Then cDNA was amplified for quantitative real-time PCR, the specific primers used were as follows: for human nitric oxide synthase 2 (NOS2) forward primer 5′-GTTCTCAAGGCACAGGTCTC-3′ and reverse primer 5′-GCAGGTCACTTATGTCACTTATC-3′; for β-actin, forward primer 5′-AGTTGCGTTACACCCTTTCTTG-3′ and reverse primer 5′-CACCTTCACCGTTCCAGTTTT-3′. The real-time PCR reactions were performed at: 95°C, 10 sec (denaturation); 55°C, 30 sec (annealing); 72°C, 30 sec (extension) for 35 cycles. The real-time PCR reactions were performed on the ABI7000 Fast Real-Time PCR System with SYBR Premix Ex Taq™ according to the procedures.
Acetic acid treatment and NO detection
Cells were seeded in 6-well plates and treated with different concentrations of acetic acid for 72 h. NO was detected with Nitric Oxide Colorimetric Assay kit (Amersco, LLC, Solon, OH, USA) according to procedures the manual instructed.
Statistical analyses
All experiments were repeated at least three times and the data were expressed as mean ± standard deviation (SD). The differences between the three groups were compared using the Student's t-test and P<0.05 (*) was considered statistically significant.
Results
Characteristics of the study subjects
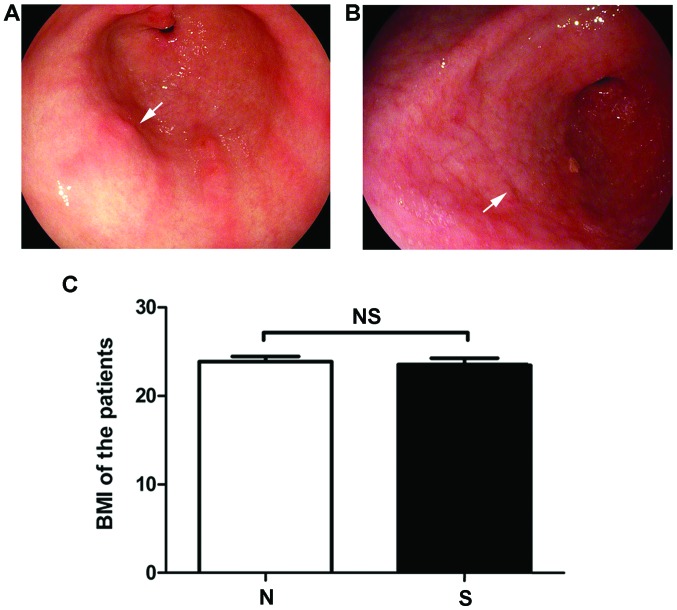
From the data obtained, the baseline characteristics of the patients in the N and S groups were compared (Table I). In terms of the degree of atrophy, there was an obvious difference in the degree of inflammation and damage in endoscopy between the N and S groups (Fig. 1A and B). However, there was no significant difference in body mass index between these two groups (Fig. 1C, P=0.73).
Table I.
Baseline characteristics of N and S patients.
| Characteristics | N | S |
|---|---|---|
| Age (mean ± SD, year) | 50.1±13.2 | 62.5±10.4 |
| Male (n, %) | 9, 30% | 10, 45.5% |
| Smoker (n, %) | 3, 10% | 5, 22.7% |
| H. pylori (n, %) (Clinical detection) | 0 | 0 |
| H. pylori (n, %) (Sequencing >0.5%) | 2, 6.7% | 4, 13.3% |
| Body mass index (mean ± SD) | 24.0±3.1 | 23.6±3.3 |
| Endoscopic results | Non-atrophic gastritis | Chronic atrophic gastritis |
Figure 1.
Characteristics of study subjects. There was an obvious difference in degree of inflammation and damage in endoscopy of stomach in (A) N and (B) S groups. Arrows: Inflammations and damage in stomach. The stomach is thinner and intestinal metaplasia occurs in atropic gastritis. However, there was no significant difference in (C) body mass index between the two groups.
Increased bacterial load in chronic atrophic gastritis
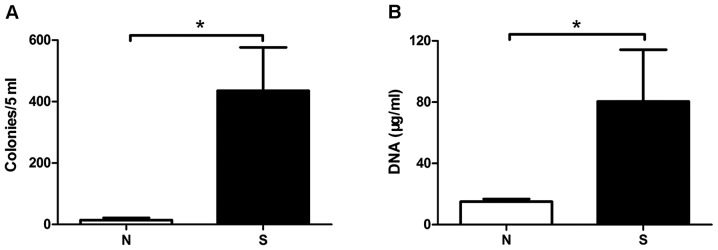
To compare the total bacterial number in gastric fluids, 5 ml of each sample fluid was centrifuged and the precipitate was used for spreading plate and microaerobic culture in blood agar. The amount of cultivable bacteria of N and S patients was calculated as 13.7 and 435 colonies per 5 ml gastric fluid, respectively, Student's t-test showed significant difference (Fig. 2A, P<0.05). This result was verified by the amount of bacterial DNA extracted from the gastric fluids (Fig. 2B, P=0.0418). Compared with non-chronic gastritis, the bacterial load in chronic atrophic gastritis was significantly increased, suggesting that there was an association between the amount of bacteria and the severity of the gastritis.
Figure 2.
The bacterial load in gastric fluids from the non- and atrophic gastritis patients. A total of 5 ml gastric fluid was centrifuged and the precipitate was used for spreading blood agar, then microaerobically cultured for 4–7 days. There was a significant difference between N and S groups in the amount of (A) cultivable bacteria. Significant difference was also existed in the amounts of (B) bacterial DNA extracted from sample gastric fluids. The bacterial DNA was determined with UV-Vis at OD260, the amounts was calculated according to formula 1OD=50 µg/ml. *P<0.05.
Diversity of gastric microbiota
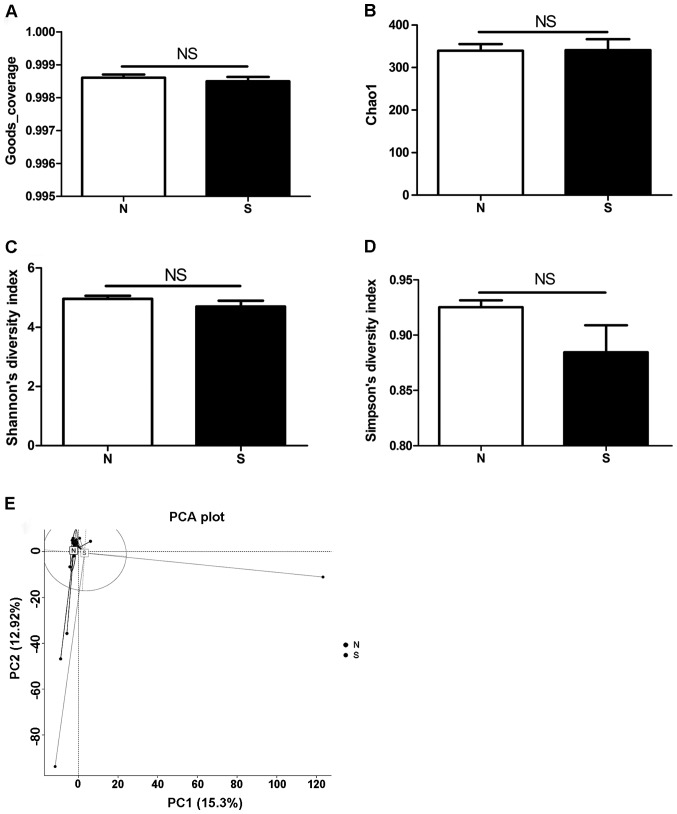
The microbial communities of the gastric fluids from the 52 patients were analyzed using high-throughput sequencing of 16S rRNA amplicons. Goods coverage, an index representing read coverage or depth was calculated, and the results showed that the coverage was more than 0.99 in both groups with no statistical significance between the two groups (P=0.5403) (Fig. 3A). To evaluate the bacterial diversity between the two groups, the Chao 1-estimated richness as well as Shannon's and Simpson's diversity indices were compared. As shown in Fig. 3B-D, the Chao 1 value of the microbiota in atrophic gastritis was similar to that in non-atrophic gastritis (N group=339.82, S group=340.72; P=0.9749) (Fig. 3B). Shannon's and Simpson's diversity indices in atrophic gastritis (4.70 and 0.88, respectively) were slightly lower than those in non-atrophic gastritis (4.96 and 0.93, respectively). However, these differences were not statistically significant (P=0.2112 for Shannon's and 0.0709 for Simpson's diversity index) (Fig. 3C and D). These results demonstrated that the diversity indices were not significantly different between the two groups.
Figure 3.
Diversity of gastric microbiota. (A) Results showed that the Goods coverage was both more than 0.99 and no statistical significance between the two groups. (B) The Chao 1, (C and D) Shannon's and Simpson's diversity indices in atrophic gastritis were not statistically significant different to that in non-atrophic gastritis. The PCA results showed the microbiota from the N group tended to cluster together, whereas samples from S group were scattered in (E) the PCA plot. All the analysis of microbiota were performed as described in Materials and methods.
The structure of the microbiota communities was explored by PCA. The results showed that the microbiota from the N group tended to cluster together, whereas samples from S group were scattered in the PCA plot (Fig. 3E). This suggests that the structure of the microbial communities in atrophic gastritis was phylogenetically diversified.
Compositional analysis of gastric microbiota
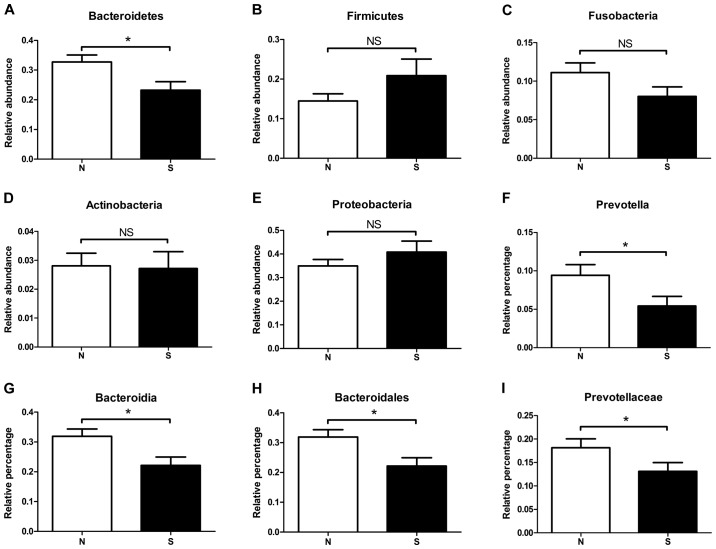
Compositional analysis showed that the entire dataset was clustered into 27 phyla, 61 classes, 106 orders, 177 families and 292 genera, respectively. The most representative phylum was Proteobacteria (representing an average of 34.9% for the N group and 40.8% for the S group of all phyla), followed by Bacteroidetes (32.8 vs. 23.3%, respectively), Firmicutes (14.4 vs. 20.9%, respectively), Fusobacteria (11.1 vs. 8.8%, respectively) and Actinobacteria (2.81 vs. 2.71%, respectively). Although the most predominant phylum varied between individuals, only one phylum, Bacteroidetes, was enriched in the non-atrophic gastritis group and had significant difference to the atrophic gastritis groups (P=0.013, Fig. 4A). The proportion of Firmicutes, Fusobacteria, Actinobacteria and Proteobacteria were varied in the two groups without statistical significances (Fig. 4B-E).
Figure 4.
Comparison of gastric microbiota in N and S groups at various taxonomic levels. (A) Phylum Bacteroidetes was enriched in the non-atrophic gastritis group and had significant difference to the atrophic gastritis groups. (B-E) The proportion of Firmicutes, Fusobacteria, Actinobacteria and Proteobacteria were varied in the two groups without statistical significances. Prevotella was accumulated in the N gastritis group at the genus level (F, P=0.05). (G) The higher taxonomic categories Bacteroidia (P=0.012), (H) Bacteroidales (P=0.012) and (I) Prevotellaceae (P=0.049) showed significant differences between the N and S groups. *P<0.05.
Similar to the phyla level, only one genus of bacteria, Prevotella, was accumulated in the N gastritis group at the genus level, and its abundance was significantly different from that of the S group (Fig. 4F). Prevotella was the major genus of the phylum Bacteroidetes, the class Bacteroidia, the order Bacteroidales, and the family Prevotellaceae. All the higher taxonomic categories besides the phylum showed significant differences between the N and S groups (Fig. 4G-I). In an analysis at the species level, we found that the genus Prevotella of the stomach microbiota was composed of P. melaninogenica (making up an average of 9.17% for the N group and 6.95% for the S group of all species); P. nanceiensis (2.55 vs. 1.48%, respectively); P. pallens (2.27 vs. 1.60%, respectively); P. tannerae (1.59 vs. 1.06%, respectively); P. intermedia (0.92 vs. 0.84%, respectively); P. nigrescens (0.27 vs. 0.18%, respectively) and P. copri (0.038 vs. 0.011%, respectively). Collectively, these species represented more than 92.4% of the species in the Prevotella genus in the tested gastric fluids.
Acetic acid, the major metabolic product of prevotella, decreases NO secretion in AGS cells
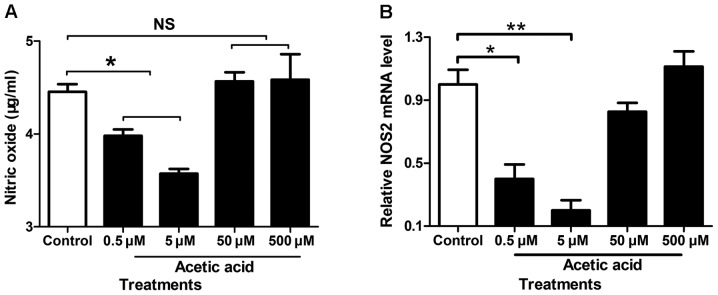
The major metabolic products of Prevotella spp. are acetic acid, butane diacid and small amount of isobutyric acid, isovaleric acid and lactic acid. To assess effects of these metabolic products of Prevotella spp. in gastric fluids on stomach epithelial cell physiology, we treated the human gastric epithelial cell line AGS with acetic acid and monitored NO production. As shown in Fig. 5A, the AGS cells incubated with low concentrations of acetic acid (0.5 and 5 µM) had significantly decreased NO secretion in culture media compared to phosphate buffered saline-treated control cells. This result was verified by QRT-PCR of NOS2 (Fig. 5B). These results suggested that Prevotella spp. in gastric fluids might impact human cell physiology.
Figure 5.
Acetic acid increases NO secretion in AGS cells. (A) The AGS cells incubated with low concentrations of acetic acid (0.5 and 5 µM) had significantly increased NO secretion in culture media compared to phosphate buffered saline-treated control cells. (B) The result was verified by QRT-PCR of NOS2. QRT-PCR was performed as described in Materials and methods. *P<0.05.
Discussion
The human microbiota consists of approximately 100 trillion microbial cells, that outnumber the human cells by 10 to 1. In healthy individuals, the gut microbiome functions as a symbiont that can offer protection from invading pathogens and prevent tumorigenesis (13). However, this commensal community exists in a careful balance that, if disrupted, enters dysbiosis and can contribute to host disease processes, including the development of cancer (14–18). The stomach harbors a large number of bacteria, and especially at the atrophic gastritis stage, the neutralized stomach can be colonized by many other bacterial species in addition to H. pylori (4). A study by Engstrand and Lindberg found that gastric microbiota overgrowth with the progress of gastritis to severe atrophic gastritis which had low acid production and no H. pylori present. They also discovered that the changes in the microbiota in individuals with atrophic gastritis resembled those seen in gastric cancer (5). Thus, it has been postulated that some microbial species colonizing the stomach may further promote gastritis, genotoxic damage and the progression to gastric cancer (6,19,20), though there is considerable variation in the most abundant bacterial species between individuals.
The stomach plays an important role in maintaining gastrointestinal health, acting as a barrier against ingested infectious disease agents of the lower gastrointestinal tract (21). A previous study found that there was no statistical significant difference between the operational taxonomic units (OTUs) of gastric fluid and mucosa, and the mean of the OTUs was higher in gastric fluid than that in mucosa. Generally, the gastric fluid samples showed higher diversity compared to gastric mucosa samples (22). Moreover, gastric fluid is easy to obtain and the detection of H. pylori is more accurate than in gastric mucosa, in which the amount of H. pylori detected may vary as the sampling location changes. Thus, in the current study, we used gastric fluids from H. pylori-negative gastritis patients for microbiota analysis.
H. pylori is the best known bacterial species that thrives in the acidic environment of the normal stomach. Inflammatory responses to H. pylori are thought to result in the destruction of cells that secrete acid into the stomach, leading to a condition known as atrophic gastritis. At the atrophic gastritis stage, gastric microbiota overgrowth and severe atrophic gastritis with low acid production occurs without H. pylori present. Clinical parameters for estimating the severity of atrophic gastritis include the degree and range of inflammation, the pH of the gastric fluids and H. pylori infection status. In fact, the existence of H. pylori is also related to the degree of inflammation and the pH of gastric fluids. However, the composition and function of microbiota absence of H. pylori remain poorly understood. Given that gastric microbes other than H. pylori are potentially linked to the development of gastric cancer. In the present study, we investigated the variation of gastric microbiota using gastric fluids from 30 patients with non-atrophic gastritis and 22 patients with chronic atrophic gastritis.
Prevotella spp. is a strictly anaerobic, Gram-negative bacterium that is non-spore forming. It does not reduce urease or nitrogen. When this organism grows on blood-containing media, it produces acid in the medium and resists acidic environments. Prevotella spp., i.e., P. melaninogenica, enrichment may contribute to the pH of the stomach becoming more acidic than in non-atrophic gastritis. Additionally, Prevotella spp. growth is inhibited by the presence of iron. The gastric mucosa is damaged at the atrophic gastritis stage, resulting in the leakage of tissue fluid and blood into the gastric fluids, which in turn, may result in higher iron content than in non-atrophic gastritis. Therefore, Prevotella spp. growth is suppressed in atrophic gastritis group.
NO is a short-lived, endogenously produced gas that acts as a signaling molecule in the body. It is synthesized by NOS and serves as a key signaling molecule in various physiological processes. Excessive and unregulated NO synthesis has been linked to contribute pathological conditions including cancer. Overexpression of NOS2 with high levels of NO generation provides a plausible link between inflammation and cancer initiation, progression, and promotion. Indeed, there is substantial evidence implicating NO in cancer as an endogenous mutagen (23), an angiogenesis factor (24), an enhancer of protooncogene expression (25), and an inhibitor of apoptosis (26). In a study by Feng et al, when lesions progressed from normal to chronic gastritis and finally to gastric cancer, positive immunostaining for p53, NOS2, and vascular endothelial growth factor (VEGF) was found to be significantly increased (27). That study also demonstrated that the positive immunostaining rates of NOS2 were correlated well with gastric cancer lymph node metastasis. NOS2 gene expression and NO production are regulated by the transcription factors NFκB, the Janus kinase (JAK)-signal transducer and activator of transcription (STAT) pathway, and c-Jun NH2-terminal kinase (JNK)-activator protein-1 (AP-1) pathways (28), whereas they are downregulated by steroids, transforming growth factor (TGF)-β, the heat shock response element, and p53 (29). A previous report showed that Short-Chain Fatty Acids (SCFAs) decreased immunoreactivity and the number of COX-2, NFκB and iNOS positive cells in colon tissue (30). Moreover, another report demonstrated that SCFAs suppressed NFκB and the production of inflammatory cytokines such as IL-6 and TNF-α, but increased IL-10 secretion from macrophages (31). All these findings suggest a role of NO in the initiation and progression of gastric cancer.
16S rRNA sequencing method has limitation, because it is unable to distinguish between living and dead bacteria. Although conventional culture could provide a complementary result, most of the gastrointestinal bacteria is uncultivable, and that bacteria with rapid growth rate would cover the plate over the slow growing bacteria, making culture method unsuitable for the present study. Due to 16S rRNA sequencing provides a complete view of the overall microbiota, our study is valuable for assessing the differences in microbiota composition from various aspects using a large number of samples. Further studies for functional role of gastric microbiota on gastric carcinogenesis such as metabolite profiling of Prevotella spp, mechanism studie using gnotobiotic mice are warranted.
Acknowledgements
We gratefully acknowledge the financial support from The National Natural Science Foundation of China (81272351), The Fundamental Research Funds of Shandong University (2015JC010), Project of Medical and Health Technology Development Program in Shandong province (Grant no. 2014ws0349) and Open Research Fund of State Key Laboratory of Environmental Chemistry and Ecotoxicology (KF2014-08).
References
- 1.Roder DM. The epidemiology of gastric cancer. Gastric Cancer. 2002;5(Suppl 1):S5–S11. doi: 10.1007/s10120-002-0203-6. [DOI] [PubMed] [Google Scholar]
- 2.Yang L, Parkin DM, Ferlay J, Li L, Chen Y. Estimates of cancer incidence in China for 2000 and projections for 2005. Cancer Epidemiol Biomarkers Prev. 2005;14:243–250. [PubMed] [Google Scholar]
- 3.Yang L, Parkin DM, Li LD, Chen YD, Bray F. Estimation and projection of the national profile of cancer mortality in China: 1991–2005. Br J Cancer. 2004;90:2157–2166. doi: 10.1038/sj.bjc.6601813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang LL, Yu XJ, Zhan SH, Jia SJ, Tian ZB, Dong QJ. Participation of microbiota in the development of gastric cancer. World J Gastroenterol. 2014;20:4948–4952. doi: 10.3748/wjg.v20.i17.4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Engstrand L, Lindberg M. Helicobacter pylori and the gastric microbiota. Best Pract Res Clin Gastroenterol. 2013;27:39–45. doi: 10.1016/j.bpg.2013.03.016. [DOI] [PubMed] [Google Scholar]
- 6.Lertpiriyapong K, Whary MT, Muthupalani S, Lofgren JL, Gamazon ER, Feng Y, Ge Z, Wang TC, Fox JG. Gastric colonisation with a restricted commensal microbiota replicates the promotion of neoplastic lesions by diverse intestinal microbiota in the helicobacter pylori INS-GAS mouse model of gastric carcinogenesis. Gut. 2014;63:54–63. doi: 10.1136/gutjnl-2013-305178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Correa P. Human gastric carcinogenesis: A multistep and multifactorial process-first american cancer society award lecture on cancer epidemiology and prevention. Cancer Res. 1992;52:6735–6740. [PubMed] [Google Scholar]
- 8.Lee CW, Rickman B, Rogers AB, Muthupalani S, Takaishi S, Yang P, Wang TC, Fox JG. Combination of sulindac and antimicrobial eradication of helicobacter pylori prevents progression of gastric cancer in hypergastrinemic INS-GAS mice. Cancer Res. 2009;69:8166–8174. doi: 10.1158/0008-5472.CAN-08-3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mowat C, Williams C, Gillen D, Hossack M, Gilmour D, Carswell A, Wirz A, Preston T, McColl KE. Omeprazole, helicobacter pylori status, and alterations in the intragastric milieu facilitating bacterial N-nitrosation. Gastroenterology. 2000;119:339–347. doi: 10.1053/gast.2000.9367. [DOI] [PubMed] [Google Scholar]
- 10.Joo YE, Park HK, Myung DS, Baik GH, Shin JE, Seo GS, Kim GH, Kim HU, Kim HY, Cho SI, Kim N. Prevalence and risk factors of atrophic gastritis and intestinal metaplasia: A nationwide multicenter prospective study in Korea. Gut Liver. 2013;7:303–310. doi: 10.5009/gnl.2013.7.3.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Park YH, Kim N. Review of atrophic gastritis and intestinal metaplasia as a premalignant lesion of gastric cancer. J Cancer Prev. 2015;20:25–40. doi: 10.15430/JCP.2015.20.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cao L, Yu J. Effect of helicobacter pylori infection on the composition of gastric microbiota in the development of gastric cancer. Gastrointest Tumors. 2015;2:14–25. doi: 10.1159/000380893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang Y, Wang X, Huycke T, Moore DR, Lightfoot SA, Huycke MM. Colon macrophages polarized by commensal bacteria cause colitis and cancer through the bystander effect. Transl Oncol. 2013;6:596–606. doi: 10.1593/tlo.13412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Candela M, Turroni S, Biagi E, Carbonero F, Rampelli S, Fiorentini C, Brigidi P. Inflammation and colorectal cancer, when microbiota-host mutualism breaks. World J Gastroenterol. 2014;20:908–922. doi: 10.3748/wjg.v20.i4.908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dicksved J, Lindberg M, Rosenquist M, Enroth H, Jansson JK, Engstrand L. Molecular characterization of the stomach microbiota in patients with gastric cancer and in controls. J Med Microbiol. 2009;58:509–516. doi: 10.1099/jmm.0.007302-0. [DOI] [PubMed] [Google Scholar]
- 16.Jobin C. Colorectal cancer: Looking for answers in the microbiota. Cancer Discov. 2013;3:384–387. doi: 10.1158/2159-8290.CD-13-0042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schwabe RF, Jobin C. The microbiome and cancer. Nat Rev Cancer. 2013;13:800–812. doi: 10.1038/nrc3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sears CL, Garrett WS. Microbes, microbiota and colon cancer. Cell Host Microbe. 2014;15:317–328. doi: 10.1016/j.chom.2014.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bik EM, Eckburg PB, Gill SR, Nelson KE, Purdom EA, Francois F, Perez-Perez G, Blaser MJ, Relman DA. Molecular analysis of the bacterial microbiota in the human stomach; Proc Natl Acad Sci USA; 2006; pp. 732–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martin ME, Bhatnagar S, George MD, Paster BJ, Canfield DR, Eisen JA, Solnick JV. The impact of helicobacter pylori infection on the gastric microbiota of the rhesus macaque. PLoS One. 2013;8:e76375. doi: 10.1371/journal.pone.0076375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martinsen TC, Bergh K, Waldum HL. Gastric juice: A barrier against infectious diseases. Basic Clin Pharmacol Toxicol. 2005;96:94–102. doi: 10.1111/j.1742-7843.2005.pto960202.x. [DOI] [PubMed] [Google Scholar]
- 22.Sung J, Kim N, Kim J, Jo HJ, Park JH, Nam RH, Seok YJ, Kim YR, Lee DH, Jung HC. Comparison of gastric microbiota between gastric juice and mucosa by next generation sequencing method. J Cancer Prev. 2016;21:60–65. doi: 10.15430/JCP.2016.21.1.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wink DA, Kasprzak KS, Maragosd CM, Elespru RK, Misra M, Dunams TM, Cebula TA, Koch WN, Andrews AW, Aleen JS, et al. DNA deaminating ability and genotoxicity of nitric oxide and its progenitors. Science. 1991;254:1001–1003. doi: 10.1126/science.1948068. [DOI] [PubMed] [Google Scholar]
- 24.Jenkins DC, Charles IG, Thomsen LL, Moss DW, Holmes LS, Baylis SA, Rhodes P, Westmore K, Emson PC, Moncada S. Role of nitric oxide in tumor growth; Proc Natl Acad Sci USA; 1995; pp. 4392–4396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ambs S, Ogunfusika MO, Merriam WG, Bennett WP, Billiar TM, Harris CC. Up-regulation of inducible nitric oxide synthase expression in cancer-prone p53 knockout mice; Proc Natl Acad Sci USA; 1998; pp. 8823–8828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li J, Billiar TR, Talanian RV, Kim YM. Nitric oxide reversibly inhibits seven members of the caspase family via S-nitrosylation. Biochem Biophys Res Commun. 1997;240:419–424. doi: 10.1006/bbrc.1997.7672. [DOI] [PubMed] [Google Scholar]
- 27.Feng CW, Wang LD, Jiao LH, Liu B, Zheng S, Xie XJ. Expression of p53, inducible nitric oxide synthase and vascular endothelial growth factor in gastric precancerous and cancerous lesions: Correlation with clinical features. BMC Cancer. 2002;2:8. doi: 10.1186/1471-2407-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Starke GR, Kerr IM, Williams BR, Silverman RH, Schreiber RD. How cells respond to interferons. Annu Rev Biochem. 1998;67:227–264. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- 29.Taylor BS, Alarcon LH, Billiar TR. Inducible nitric oxide synthase in the liver: Regulation and function. Biochemistry (Mosc) 1998;63:766–781. [PubMed] [Google Scholar]
- 30.Hijova E, Szabadosova V, Strojny L, Bomba A. Changes chemopreventive markers in colorectal cancer development after inulin supplementation. Bratisl Lek Listy. 2014;115:76–79. doi: 10.4149/bll_2014_016. [DOI] [PubMed] [Google Scholar]
- 31.Park JS, Lee EJ, Lee JC, Kim WK, Kim HS. Anti-inflammatory effects of short chain fatty acids in IFN-gamma-stimulated RAW 264.7 murine macrophage cells: Involvement of NF-kappaB and ERK signaling pathways. Int Immunopharmacol. 2007;7:70–77. doi: 10.1016/j.intimp.2006.08.015. [DOI] [PubMed] [Google Scholar]