Abstract
MicroRNA (miR) are short non-coding RNA that bind to the 3′-untranslational region of their target genes, inhibiting translation and causing mRNA degradation. miR deregulation has been implicated in human cancer; however, the detailed regulatory mechanism of miR-137 in osteosarcoma (OS) remains largely unknown. In the present study, miR-137 and enhancer of zeste homologue 2 (EZH2) mRNA and protein expression levels were analyzed using reverse transcription-quantitative polymerase chain reaction and western blotting, respectively. MTT and transwell assays were performed to evaluate cell viability and invasion capacities and a luciferase reporter gene assay was used to determine the targeting relationship. The results of the current study indicated that miR-137 expression was significantly downregulated in OS tissues and cell lines (P<0.01). Moreover, it was observed that low miR-137 expression levels were significantly associated with lung metastasis and advanced TMN stage (P<0.05), but not associated with age, gender, tumor size, location, serum lactate dehydrogenase or serum alkaline phosphatase. Increasing levels of miR-137 significantly inhibited U2OS cell viability and invasion (P<0.01). By contrast, knockdown of miR-137 markedly increased U2OS cell viability and invasion. EZH2 was identified as a direct target gene of miR-137 in U2OS cells by luciferase reporter assay and EZH2 expression was found to be significantly increased in OS tissues and cell lines (P<0.01). EZH2 was significantly downregulated following miR-137 overexpression (P<0.01), and was upregulated following miR-137 knockdown in U2OS cells. Furthermore, EZH2 overexpression significantly attenuated the suppressive effects of miR-137 on U2OS cell viability and invasion (P<0.01), suggesting that miR-137 inhibits the viability and invasion of OS cells by targeting EZH2. Therefore, the results of the current study suggest that the miR-137/EZH2 axis may be a potential target for novel potential therapeutic strategies to treat OS.
Keywords: osteosarcoma, microRNA-137, enhancer of zeste homolog 2, tumor suppressor
Introduction
Osteosarcoma (OS) primarily occurs in regions with active bone growth and repair, and is the most common type of malignant tumor observed in the bone (1,2). Over the past few decades, the effectiveness of cancer treatments have improved, however the prognosis of patients with advanced OS remains poor, mainly due to high rates of recurrence and metastasis (1,3). It has been demonstrated that a number of oncogenes and tumor suppressors serve key roles in the development and progression of OS, and studying the molecular mechanism underlying OS may enable the development of effective therapeutic strategies (4,5).
MicroRNA (miR) are short non-coding RNA that can bind to the 3′-untranslational region (UTR) of their target genes, degrading mRNA or inhibiting translation (1). They are key regulators of various biological processes, including cell proliferation, differentiation, migration, apoptosis, angiogenesis and tumorigenesis (6,7). It has been demonstrated that various types of miR are deregulated in different types of human cancer, including OS (8–10). For example, it has been suggested that decreased levels of miR-133b and miR-503 are efficient diagnostic and prognostic factors in patients with OS (2). Furthermore, it has been demonstrated that in OS, certain miR directly target tumor suppressors or oncogenes (11,12), and it was found that miR-133a inhibits OS cell proliferation and invasion by targeting insulin-like growth factor 1 receptor (3). miR-503 is downregulated in OS tissues and cell lines, and inhibits OS cell proliferation, migration and invasion via the L1 cell adhesion molecule (13). miR-199a-3p is also downregulated in OS and serves a suppressive role in OS cell proliferation and migration (14). Therefore, miR may be potential therapeutic agents or targets for OS treatment.
It has been identified that miR-137 participates in certain types of cancer (15,16). It inhibits the proliferation of glioblastoma multiforme cell and induces the differentiation of brain tumor stem cells (15). Additionally, it can inhibit tumor growth and induce sensitivity to paclitaxel and cisplatin in lung cancer (16). Recently, miR-137 expression was found to be significantly downregulated in OS tissues and cell lines (4). Additionally, miR-137 was observed to suppress the proliferation and migration of OS 143B and Saos-2 cells, as well as tumor growth in vivo, by directly targeting FXYD6 (4). However, the underlying mechanism of miR-137 in the regulation of OS cell proliferation and invasion remains unknown.
The present study aimed to investigate the clinical significance of miR-137 in OS and to identif y the underlying mechanism of miR-137 in the regulation of OS growth and metastasis. In addition, enhancer of zeste homologue 2 (EZH2) was studied as a putative target gene of miR-137 in OS.
Materials and methods
Tissue collection
A total of 58 primary OS tissues and matched adjacent non-tumor tissues were collected by surgical resection from 58 patients who presented at the Department of Orthopedics (Second Affiliated Hospital of Nanchang University, Nanchang, China) between January 2013 and March 2015. Table I presents the clinicopathological information of all patients involved. Tissues were immediately snap-frozen in liquid nitrogen following surgical resection and stored at −80°C in liquid nitrogen before use. The present study was approved by the Ethics Committee of Second Affiliated Hospital of Nanchang University (Jiangxi, China) and informed consent was obtained from all patients.
Table I.
Association between miR-137 expression and clinicopathological characteristics in osteosarcoma.
| Variables | Total cases (n=58) | Low miR-137 (n=31) | High miR-137 (n=27) | P-value |
|---|---|---|---|---|
| Age, years | 0.599 | |||
| <20 | 24 | 14 | 10 | |
| ≥20 | 34 | 17 | 17 | |
| Gender | 0.426 | |||
| Male | 35 | 17 | 18 | |
| Female | 23 | 14 | 9 | |
| Tumor size, cm | 0.428 | |||
| <8 cm | 26 | 12 | 14 | |
| ≥8 cm | 32 | 19 | 13 | |
| Location | 0.403 | |||
| Femur or Tibia | 39 | 19 | 20 | |
| Others | 19 | 12 | 7 | |
| Lung metastasis | 0.022a | |||
| No | 40 | 17 | 23 | |
| Yes | 18 | 14 | 4 | |
| TNM stage | 0.029a | |||
| I/IIA | 21 | 7 | 14 | |
| IIB/III | 37 | 24 | 13 | |
| Serum lactate dehydrogenase | 0.788 | |||
| Normal | 22 | 11 | 11 | |
| Elevated | 36 | 20 | 16 | |
| Serum alkaline phosphatase | 0.79 | |||
| Normal | 24 | 12 | 12 | |
| Elevated | 34 | 19 | 15 |
P<0.05, representing a statistically significant difference. TNM, tumor-node-metastasis.
Cell culture
The human osteoblast cell line hFOB, and the human OS cell lines U2OS, Saos-2, MG63 and SW1353, were all purchased from the cell bank at the Chinese Academy of Sciences (Shanghai, China). All cell lines were cultured in Dulbecco's modified Eagle medium (DMEM; Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS; Thermo Fisher Scientific, Inc.) in a 37°C, 95% humidified atmosphere of 5% CO2.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA from tissues and cell lines was extracted using TRIzol reagent (Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions. Total RNA was reverse transcribed into cDNA using PrimeScript 1st Strand cDNA Synthesis kit (Takara Bio, Inc., Otsu, Japan), according to the manufacturer's instructions. To analyze levels of miR expression, qPCR was performed using an miRNA Q-PCR Detection kit (GeneCopoeia, Inc., Rockville, MD, USA) on an ABI 7500 Thermocycler (Thermo Fisher Scientific, Inc.), according to the manufacturer's instructions. The U6 gene was used as an internal control. Primers were purchased from GeneCopoeia, Inc., and the sequences are not available. For analysis of mRNA expression, a High Capacity cDNA Reverse Transcription Kit (Applied Biosystems; Thermo Fisher Scientific, Inc.) was used for reverse transcription. In brief, 5 µl total RNA was mixed with 0.15 µl of 100 Mm dNTPs (with dTTP), 1 µl (50 U) MultiScribe reverse transcriptase, 1.5 µl of reverse transcription buffer (10X), 0.19 µl RNase inhibitor (20 U/µl) and 3 µl gene-specific primers (1X). Nuclease-free H2O was added to obtain a final volume of 15 µl. Reverse transcription was performed at 16°C for 30 min, followed by an incubation step at 42°C for 30 min and enzyme inactivation at 85°C for 5 min. qPCR was performed using a SYBR-Green I Real-Time PCR kit (Biomics Biotechnologies Co., Ltd., Nantong, China), according to the manufacturer's instructions. GAPDH was used as an internal control. The specific primer pairs were as follows: Enhancer of zeste homologue 2 (EZH2), forward, 5′-AATCAGAGTACATGCGACTGAGA-3′ and reverse, 5′-GCTGTATCCTTCGCTGTTTCC-3′; GAPDH, forward, 5′-CTGGGCTACACTGAGCACC-3′ and reverse, 5′-AAGTGGTCGTTGAGGGCAATG-3′. The reaction conditions were 95°C for 5 min, followed by 40 cycles of denaturation at 95°C for 15 sec and an annealing/elongation step at 60°C for 30 sec. Relative expression was analyzed using the 2−ΔΔCq method (5).
Cell transfection
U2OS cells were transfected with scramble miR mimic (miR-NC), miR-137 mimic, negative control (NC) inhibitor, miR-137 inhibitor, or co-transfected with miR-137 mimic and pcDNA3.1-EZH2 expression plasmid (Yearthbio, Qingdao, China) using Lipofectamine® 2000 (Thermo Fisher Scientific, Inc.), according to the manufacturer's instructions.
MTT assay
U2OS cell suspension (5×104 cells/well) was plated in a 96-well plate, and cultured for 0, 24, 48 or 72 h. Then, 10 µl MTT (5 mg/ml) was added to each well, and cells were incubated at 37°C for 4 h. The supernatant was removed and 100 µl dimethylsulfoxide was added to each well. Absorbance was measured at 570 nm using the Model 680 Microplate Reader (Bio-Rad Laboratories, Inc., Hercules, CA, USA).
Transwell assay
To determine U2OS cell invasive capacity, 24-well transwell chambers (Chemicon; EMD Millipore, Billerica, MA, USA), which have a layer of Matrigel, were used to perform the transwell assay. U2OS cell suspension was added to the upper chamber and DMEM containing 10% FBS was added to the lower chamber. After 24 h incubation at 37°C, non-invading cells on the interior of the inserts were removed using a cotton-tipped swab. Cells on the lower surface of the membrane were stained with gentian violet, rinsed in water and then left to dry. Five fields were randomly selected and cell numbers were counted under a fluorescent inverted microscope (IX71; Olympus Corp., Tokyo, Japan).
Western blot analysis
OS cell lines were lysed with ice-cold lysis buffer and proteins were separated using 10% SDS-PAGE and transferred onto a polyvinylidene difluoride membrane (Thermo Fisher Scientific, Inc.). The membrane was then incubated with phosphate-buffered saline (PBS) containing 5% non-fat milk at room temperature for 3 h for blocking, and was subsequently incubated with rabbit polyclonal anti-human EZH2 antibody (1:100; cat. no. ab3748; Abcam, Cambridge, MA, USA) or rabbit polyclonal anti-human GAPDH (1:50; cat. no. ab9485; Abcam) at room temperature for 3 h. Following three washes with PBS, the membrane was incubated with goat polyclonal secondary antibody to rabbit immunoglobulin (Ig)G (1:5,000; cat. no. ab150077; Abcam) at room temperature for 40 min. After three washes with PBS, Super Signal West Pico Chemiluminescent Substrate kit (Pierce; Thermo Fisher Scientific, Inc.) was used to detect signals, according to the manufacture's instructions. Relative protein expression was represented as the density ratio vs. GAPDH.
Bioinformatics analysis
Targetscan online software (http://www.targetscan.org) was used to predicate the putative targets of miR-137, according to the manufacture's instructions.
Luciferase reporter assay
The mutant type (MT) of EZH2 3′UTR lacking complementarity with the miR-137 seed sequence was generated using the QuikChange Site-Directed Mutagenesis kit (Stratagene; Agilent Technologies, Inc., Santa Clara, CA, USA), according to the manufacturer's instructions. The wild-type (WT) or MT of EZH2 3′UTR was cloned downstream of the firefly luciferase coding region of the pMIR-GLOTM Luciferase vector (Promega Corporation, Madison, WI, USA). U2OS cells were co-transfected with the WT- or MT-EZH2-3′UTR luciferase reporter plasmid, and miR-NC or miR-137 mimic, respectively. Luciferase activity was detected 48 h after transfection using the Dual Luciferase Reporter Assay system (Promega Corporation), according to the manufacturer's instructions.
Statistical analysis
Data in the current study are expressed as the mean ± standard deviation of three independent experiments. SPSS version 19.0 (IBM SPSS, Inc., Armonk, NY, USA) was used to perform statistical analysis. The difference between two groups was analyzed using Student's t-test. The association between miR-137 expression and clinical characteristics of patients with OS were analyzed using the χ2 test. P<0.05 was considered to indicate a statistically significant difference.
Results
miR-137 expression is downregulated in OS
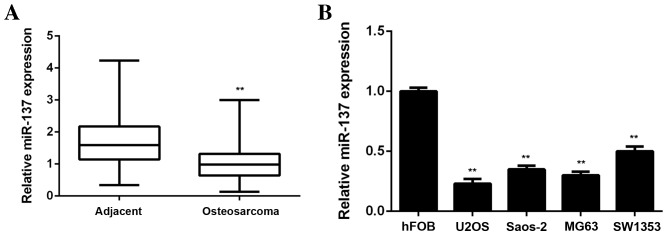
In the present study, RT-qPCR was performed to examine miR-137 expression in OS tissues and paired adjacent non-tumor tissues. miR-137 levels were observed to be significantly lower in OS tissues compared with adjacent non-tumor tissues (P<0.01; Fig. 1A). Subsequently miR-137 levels in four common human OS cell lines, including Saos-2, MG63, U2OS and SW1353, were examined. The normal human osteoblast cell line hFOB was used as a control group. It was observed that the expression of miR-137 was significantly decreased in all four OS cell lines compared with hFOB cells (P<0.01; Fig. 1B). Therefore, the results from RT-qPCR demonstrated that miR-137 expression is downregulated in OS.
Figure 1.
(A) RT-qPCR was performed to examine miR-137 levels in osteosarcoma tissues compared with adjacent non-tumor tissues. **P<0.01 vs. adjacent tissue. (B) RT-qPCR was performed to examine miR-137 levels in four osteosarcoma cell lines (U2OS, Saos-2; MG63; SW1353) compared with the hFOB cell line. **P<0.01 vs. hFOB. RT-qPCR, reverse transcription-quantitative polymerase chain reaction; hFOB, human foetal osteoblastic cells. miR, microRNA.
miR-137 downregulation is associated with OS progression
The clinical significance of miR-137 expression in OS was investigated. The mean value (1.05) of miR-137 expression in patients was used as a cutoff and patients with OS were divided into high and low miR-137 expression groups. Low miR-137 levels were significantly associated with lung metastasis and advanced TMN stage in OS (P<0.05; Table I). However, no associations between miR-137 expression and age, gender, tumor size, location, serum lactate dehydrogenase or serum alkaline phosphatase were identified (Table I). Therefore, the results of the current study suggest that reduced expression of miR-137 may stimulate OS progression.
miR-137 inhibits OS cell viability and invasion
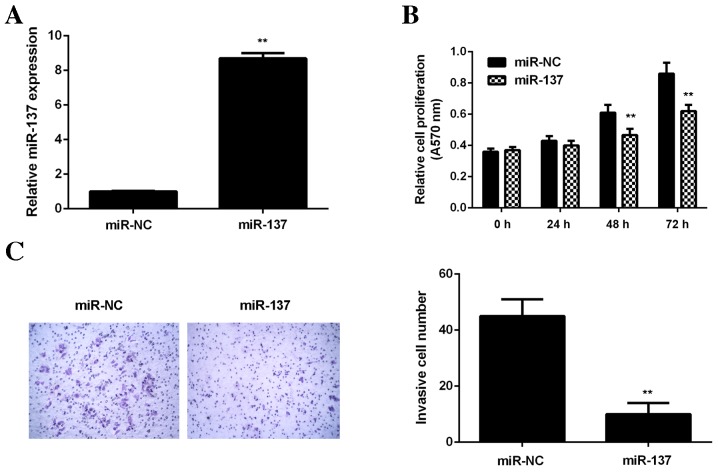
As U2OS cells exhibited the most significant decrease in miR-137 expression, this cell line was used in subsequent in vitro studies. To restore miR-137 levels, U2OS cells were transfected with a miR-137 mimic. Cells in the control group were transfected with scramble miR (miR-NC). Transfection with an miR-137 mimic significantly increased miR-137 levels compared with the miR-NC group (P<0.01; Fig. 2A). MTT (Fig. 2B) and transwell assays (Fig. 2C) were performed to determine cell viability and invasion, respectively. It was demonstrated that restoration of miR-137 expression significantly decreased the viability and invasion of U2OS cells compared with the miR-NC group (P<0.01; Fig. 2B and C). These data suggest that miR-137 has suppressive effects on OS cell viability and invasion.
Figure 2.
U2OS cells were transfected with miR-137 mimic or miR-NC. (A) Reverse transcription-quantitative polymerase chain reaction was performed to examine miR-137 levels. (B) MTT assay was used to examine cell viability and (C) transwell assay was used to examine cell invasion. **P<0.01 vs. miR-NC. A570 nm, absorbance at 570 nm; miR, microRNA; miR-NC, scramble miR.
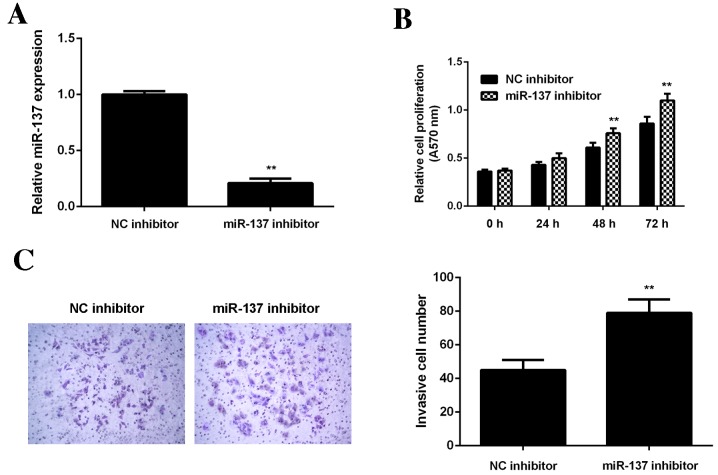
To confirm these findings, U2OS cells were transfected with miR-137 inhibitor or NC inhibitor, respectively. Transfection with an miR-137 inhibitor significantly decreased miR-137 levels in U2OS cells compared with the NC inhibitor group (P<0.01; Fig. 3A). Data from the MTT assay (Fig. 3B) and transwell assay (Fig. 3C) further indicated that inhibition of miR-137 significantly enhanced the viability and invasion of U2OS cells (P<0.01).
Figure 3.
U2OS cells were transfected with miR-137 inhibitor or a NC. (A) Reverse transcription-quantitative polymerase chain reaction was performed to assess miR-137 levels. (B) MTT assay was used to examine cell viability and (C) transwell assay was used to examine cell invasion. **P<0.01 vs. NC inhibitor. RT-qPCR, reverse transcription-quantitative polymerase chain reaction. NC, negative control; miR-NC, scramble miR; A570 nm, absorbance at 570 nm; miR, microRNA.
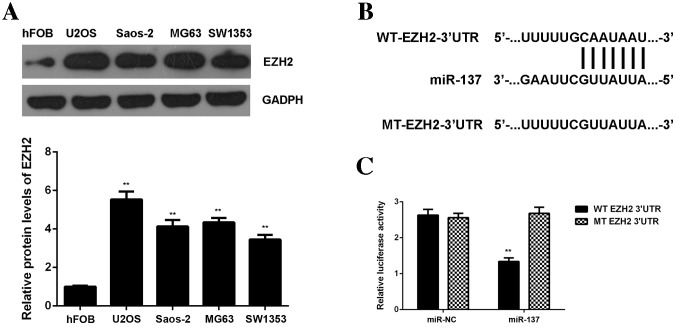
EZH2, upregulated in OS, is a direct target of miR-137
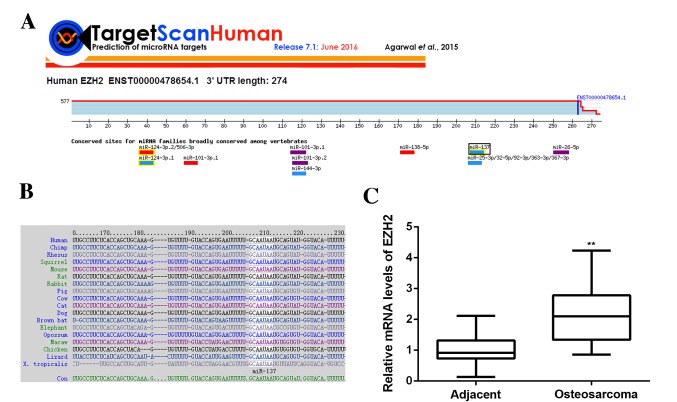
miR function by inhibiting the expression of their target genes; therefore, the potential targets of miR-137 were predicated using Targetscan software. EZH2 was found to be a putative target of miR-137 and this relationship was evolutionally conserved (Fig. 4A and B). To confirm this predication, EZH2 expression in OS tissues and cell lines was examined. Data from RT-qPCR demonstrated that EZH2 levels were significantly increased in OS tissues compared with adjacent non-tumor tissues (P<0.01; Fig. 4C). Additionally, levels of EZH2 protein were significantly increased in OS cell lines compared with hFOB cells (P<0.01; Fig. 5A), showing that EZH2 expression is upregulated in OS. To further confirm their targeting relationship, the WT or MT of EZH2 3′UTR was cloned into the pMIR-GLOTM Luciferase vector downstream of the firefly luciferase coding region, generating a WT-EZH2-3′UTR or MT-EZH2-3′UTR luciferase reporter plasmid (Fig. 5B). A luciferase reporter assay was then performed. U2OS cells were co-transfected with WT- or MT-EZH2-3′UTR luciferase reporter plasmid, and then a miR-NC or miR-137 mimic, respectively. The results demonstrated that luciferase activity was significantly lower in U2OS cells co-transfected with miR-137 mimic and WT-EZH2-3′UTR luciferase reporter plasmid compared with the control group (P<0.01), which was eliminated by transfection with MT-EZH2-3′UTR luciferase reporter plasmid (Fig. 5C). These findings indicate that miR-137 can directly bind to the 3′UTR of EZH2 mRNA in U2OS cells.
Figure 4.
(A and B) EZH2 was shown to be a putative target of miR-137 and their relationship was evolutionally conserved. (C) Reverse transcription-quantitative polymerase chain reaction was performed to examine levels of EZH2 mRNA in osteosarcoma tissues compared with adjacent non-tumor tissues. **P<0.01 vs. adjacent tissues. EZH2, enhancer of zeste homologue 2.
Figure 5.
(A) Western blotting was performed to examine levels of EZH2 protein in osteosarcoma cell lines compared with the normal human osteoblast cell line hFOB. **P<0.01 vs. hFOB. (B) WT or MT of EZH2 3′UTR was then cloned into the downstream of the firefly luciferase coding region of pMIR-GLOTM Luciferase vector. (C) U2OS cells were co-transfected with WT- or MT-EZH2-3′UTR luciferase reporter plasmid, and miR-NC or miR-137 mimic, respectively. The luciferase activity was examined. **P<0.01 vs. miR-NC. miR, microRNA; EZH2, enhancer of zeste homologue 2; UTR, untranslated region; NC, negative control; WT, wild-type; MT, mutant type; hFOB, human foetal osteoblastic cells.
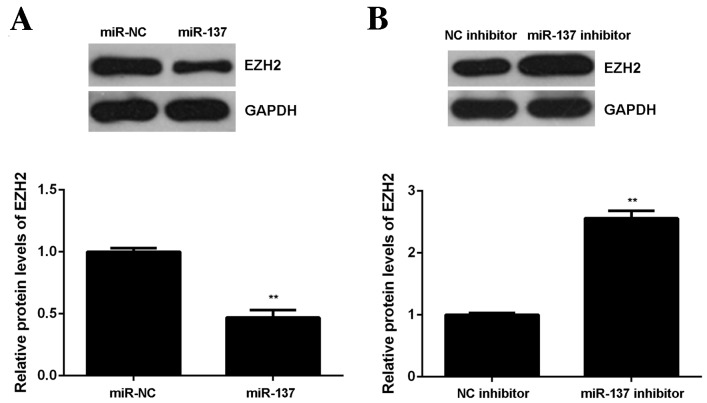
miR-137 negatively regulates EZH2 expression in OS cells
miRs generally inhibit the expression of their target genes at the post-transcriptional level, therefore expression of EZH2 protein in U2OS cells following miR-137 overexpression or downregulation was measured. Results from the western blot analysis demonstrated that overexpression of miR-137 significantly decreased levels of EZH2 protein in U2OS cells (P<0.01; Fig. 6A). By contrast, miR-137 knockdown significantly enhanced the expression of EZH2 protein in U2OS cells (P<0.01; Fig. 6B). The results demonstrate that EZH2 is negatively regulated by miR-137 in U2OS cells.
Figure 6.
(A) Western blotting was performed to examine the levels of EZH2 protein in U2OS cells transfected with miR-137 mimic or miR-NC. **P<0.01 vs. miR-NC. (B) Western blotting was performed to examine the levels of EZH2 protein in U2OS cells transfected with miR-137 inhibitor or a NC. **P<0.01, vs. NC inhibitor. miR, microRNA; EZH2, enhancer of zeste homologue 2; UTR, untranslated region; NC, negative control; miR-NC, scramble miR.
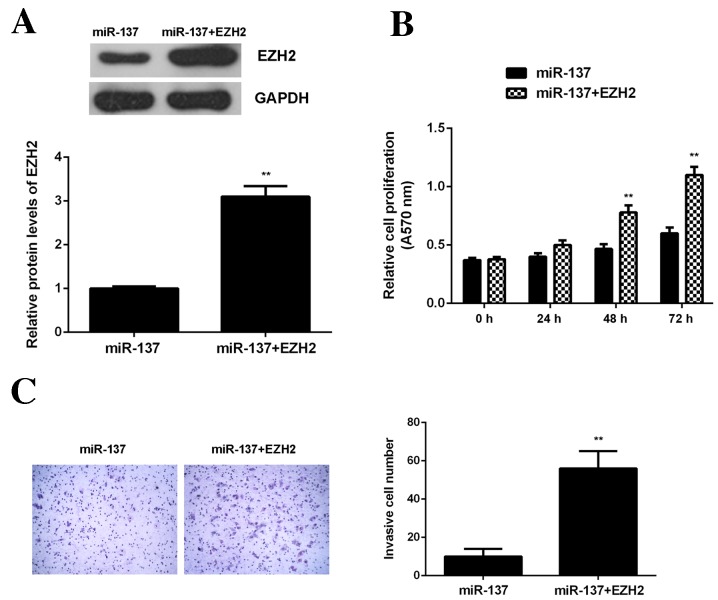
Overexpression of EZH2 attenuates the inhibitory effects of miR-137 on OS cell viability and invasion
It was then investigated whether EZH2 was involved in the miR-137-mediated viability and invasion of OS cells. miR-137-overexpressing U2OS cells were transfected with EZH2 expression plasmid. Following transfection, levels of EZH2 protein were significantly higher in the miR-137+EZH2 group compared with the miR-137 group (P<0.01; Fig. 7A). Data from the MTT and transwell assays further demonstrated that the viability and invasion of U2OS cells were significantly increased in the miR-137+EZH2 group compared with the miR-137 group (P<0.01; Fig. 7B and C). These findings suggest that miR-137 inhibits OS cell viability and invasion by directly targeting EZH2.
Figure 7.
U2OS cells were transfected with miR-137 mimic or co-transfected with miR-137 mimic and EZH2 expression plasmid. (A) Western blotting was performed to examine levels of EZH2 protein. (B) MTT assay was used to examine cell viability and (C) transwell assay was used to examine cell invasion. **P<0.01 vs. miR-137. miR, microRNA; EZH2, enhancer of zeste homologue 2.
Discussion
Until now, little has been known regarding the underlying regulatory mechanism of miR-137 in OS growth and metastasis. The present study demonstrated that miR-137 was significantly downregulated in OS tissues and cell lines, and it was determined that decreased miR-137 levels were significantly associated with lung metastasis and the advanced clinical stage of OS. Restoration of miR-137 expression significantly inhibited U2OS cell viability and invasion, while inhibiting miR-137 enhanced U2OS cell viability and invasion. Furthermore, EZH2 was found to be significantly upregulated in OS tissues and cell lines. EXH2 was identified as a direct target gene of miR-137 in U2OS cells and it was demonstrated that EZH2 expression was negatively regulated by miR-137 at the post-transcriptional level. Additionally, restoration of EZH2 expression significantly attenuated the suppressive effects of miR-137 on U2OS cell viability and invasion.
It has been reported that miR-137 generally serves a suppressive role in different types of cancer by targeting different oncogenes (15,16). miR-137 is downregulated in thyroid carcinoma and acts as a tumor suppressor by targeting Stromal cell-derived factor 1 and epidermal growth factor receptor (17,18). It can also improve the dexamethasone sensitivity in multiple myeloma cells via inhibition of microphthalmia-associated transcription factor as well as the Akt signaling pathway (6). Yang et al (7) have demonstrated that miR-137 inhibits the migration and invasion of non-small cell lung cancer cells by targeting bone morphogenetic protein 7. In the present study, it was demonstrated that miR-137 was significantly decreased in OS tissues compared with adjacent non-tumor tissues. Additionally, it was downregulated in four common human OS cell lines, including Saos-2, MG63, U2OS and SW1353, compared with the normal human osteoblast cell line hFOB. Similarly, Gao et al (8) reported that miR-137 was downregulated in OS tissues compared with paired normal larynx tissue, consistent with the findings of the current study. However, the clinical significance of miR-137 expression in OS has not previously been studied. The present study showed that decreased expression of miR-137 was significantly associated with lung metastasis and advanced clinical stage of OS, suggesting that the downregulation of miR-137 may contribute to the malignant progression of OS. Furthermore, Li et al (4) demonstrated that miR-137 had suppressive effects on the proliferation and migration of 143B and Saos-2 cells in vitro. In current study, it was demonstrated that overexpression of miR-137 significantly decreased U2OS cell viability and invasion, while knockdown of miR-137 promoted U2OS cell viability and invasion. Taken together, these results suggest that miR-137 acts as a tumor suppressor in OS.
EZH2 is a histone methyltransferase and is the catalytic subunit of polycomb repressive complex 2, a highly conserved protein complex that regulates gene expression by methylating lysine 27 on histone H3 (9). It has been demonstrated that EZH2 serves a promoting role in different types of human cancer, meaning that it may be a promising therapeutic target to treat cancer (10,11). Chen et al (10) have reported that EZH2 may promote the proliferation of colorectal cancer stem cells by upregulating p21cip1-Wnt/β-catenin signaling. Jang et al (11) indicated that EZH2 expression was significantly upregulated in luminal A breast cancer and this upregulation of expression was associated with poor overall survival. Recently, EZH2 was found to be significantly upregulated in OS and high expression of EZH2 was significantly associated with more aggressive tumor behavior and poorer outcomes in patients with OS (12). Furthermore, knockdown of EZH2 by small interfering RNA or its specific inhibitor 3-deazaneplanocin A significantly inhibited OS cell growth, migration, proliferation and invasion, and inhibited the action of cancer stem cells (12) Therefore, EZH2 may be a novel therapeutic target to treat OS.
In the present study, EZH2 was found to be significantly upregulated in OS tissues and cell lines, and was identified as a direct target gene of miR-137. In addition, the expression of EXH2 protein was negatively regulated by miR-137 in U2OS cells. Therefore, the down regulation of miR-137 may contribute towards the upregulation of EZH2 in OS, further promoting its malignant progression. The present study further indicated that EZH2 overexpression significantly attenuated the suppressive effects of miR-137 on U2OS cell viability and invasion, suggesting that miR-137 inhibits the viability of OS cells by targeting EZH2. Apart from miR-137, a number of other miRs, including miR-138, miR-101 and miR-26, were found to serve a suppressive role in OS by directly targeting EZH2 (19–21). Furthermore, the targeting relationship between miR-137 and EZH2 was found in glioblastoma, neuroblastoma and melanoma (22–24). Therefore, the results of the current study increase the understanding regarding the miR-137/EZH2 axis in human cancer.
In conclusion, the present study demonstrates that miR-137, which is downregulated in OS, acts as a tumor suppressor in OS by targeting EZH2. Therefore, the miR-137/EZH2 axis may be a useful therapeutic target in developing treatments for OS.
References
- 1.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Bassampour SA, Abdi R, Bahador R, Shakeri M, Torkaman A, Yahaghi E, Taheriazam A. RETRACTED ARTICLE: Downregulation of miR-133b/miR-503 acts as efficient prognostic and diagnostic factors in patients with osteosarcoma and these predictor biomarkers are correlated with overall survival. Tumour Biol. 2015 Aug 16; doi: 10.1007/s13277-015-3918-9. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 3.Chen G, Fang T, Huang Z, Qi Y, Du S, Di T, Lei Z, Zhang X, Yan W. MicroRNA-133a Inhibits Osteosarcoma Cells Proliferation and Invasion via Targeting IGF-1R. Cell Physiol Biochem. 2016;38:598–608. doi: 10.1159/000438653. [DOI] [PubMed] [Google Scholar]
- 4.Li ZM, Zhang HY, Wang YX, Wang WB. MicroRNA-137 is downregulated in human osteosarcoma and regulates cell proliferation and migration through targeting FXYD6. J Drug Target. 2016;24:102–110. doi: 10.3109/1061186X.2015.1057149. [DOI] [PubMed] [Google Scholar]
- 5.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 6.Zhang B, Ma L, Wei J, Hu J, Zhao Z, Wang Y, Chen Y, Zhao F. miR-137 suppresses the phosphorylation of AKT and improves the dexamethasone sensitivity in multiple myeloma cells via targeting MITF. Curr Cancer Drug Targets. 2016;16:807–817. doi: 10.2174/1568009616666160203114140. [DOI] [PubMed] [Google Scholar]
- 7.Yang YR, Li YX, Gao XY, Zhao SS, Zang SZ, Zhang ZQ. MicroRNA-137 inhibits cell migration and invasion by targeting bone morphogenetic protein-7 (BMP7) in non-small cell lung cancer cells. Int J Clin Exp Pathol. 2015;8:10847–10853. [PMC free article] [PubMed] [Google Scholar]
- 8.Gao S, Wang J, Xie J, Zhang T, Dong P. Role of miR-138 in the regulation of larynx carcinoma cell metastases. Tumour Biol. 2015 Oct 24; doi: 10.1007/s13277-015-4244-y. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 9.Italiano A. Role of the EZH2 histone methyltransferase as a therapeutic target in cancer. Pharmacol Ther. 2016;165:26–31. doi: 10.1016/j.pharmthera.2016.05.003. [DOI] [PubMed] [Google Scholar]
- 10.Chen JF, Luo X, Xiang LS, Li HT, Zha L, Li N, He JM, Xie GF, Xie X, Liang HJ. EZH2 promotes colorectal cancer stem-like cell expansion by activating p21cip1-Wnt/β-catenin signaling. Oncotarget. 2016;7:41540–41558. doi: 10.18632/oncotarget.9236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jang SH, Lee JE, Oh MH, Lee JH, Cho HD, Kim KJ, Kim SY, Han SW, Kim HJ, Bae SB, Lee HJ. High EZH2 protein expression is associated with poor overall survival in patients with luminal a breast cancer. J Breast Cancer. 2016;19:53–60. doi: 10.4048/jbc.2016.19.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun R, Shen J, Gao Y, Zhou Y, Yu Z, Hornicek F, Kan Q, Duan Z. Overexpression of EZH2 is associated with the poor prognosis in osteosarcoma and function analysis indicates a therapeutic potential. Oncotarget. 2016;7:38333–38346. doi: 10.18632/oncotarget.9518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chong Y, Zhang J, Guo X, Li G, Zhang S, Li C, Jiao Z, Shao M. MicroRNA-503 acts as a tumor suppressor in osteosarcoma by targeting L1CAM. PLoS One. 2014;9:e114585. doi: 10.1371/journal.pone.0114585. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 14.Duan Z, Choy E, Harmon D, Liu X, Susa M, Mankin H, Hornicek F. MicroRNA-199a-3p is downregulated in human osteosarcoma and regulates cell proliferation and migration. Mol Cancer Ther. 2011;10:1337–1345. doi: 10.1158/1535-7163.MCT-11-0096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Silber J, Lim DA, Petritsch C, Persson AI, Maunakea AK, Yu M, Vandenberg SR, Ginzinger DG, James CD, Costello JF, et al. miR-124 and miR-137 inhibit proliferation of glioblastoma multiforme cells and induce differentiation of brain tumor stem cells. BMC Med. 2008;6:14. doi: 10.1186/1741-7015-6-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shen H, Wang L, Ge X, Jiang CF, Shi ZM, Li DM, Liu WT, Yu X, Shu YQ. MicroRNA-137 inhibits tumor growth and sensitizes chemosensitivity to paclitaxel and cisplatin in lung cancer. Oncotarget. 2016;7:20728–20742. doi: 10.18632/oncotarget.8011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dong S, Jin M, Li Y, Ren P, Liu J. miR-137 acts as a tumor suppressor in papillary thyroid carcinoma by targeting CXCL12. Oncol Rep. 2016;35:2151–2158. doi: 10.3892/or.2016.4604. [DOI] [PubMed] [Google Scholar]
- 18.Luo Y, Li X, Dong J, Sun W. microRNA-137 is downregulated in thyroid cancer and inhibits proliferation and invasion by targeting EGFR. Tumour Biol. 2016;37:7749–7755. doi: 10.1007/s13277-015-4611-8. [DOI] [PubMed] [Google Scholar]
- 19.Zhu Z, Tang J, Wang J, Duan G, Zhou L, Zhou X. MiR-138 acts as a tumor suppressor by targeting EZH2 and enhances cisplatin-induced apoptosis in osteosarcoma cells. PLoS One. 2016;11:e0150026. doi: 10.1371/journal.pone.0150026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang H, Meng Y, Cui Q, Qin F, Yang H, Chen Y, Cheng Y, Shi J, Guo Y. MiR-101 Targets the EZH2/Wnt/β-catenin the pathway to promote the osteogenic differentiation of human bone marrow-derived mesenchymal stem cells. Sci Rep. 2016;6:36988. doi: 10.1038/srep36988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lu J, He ML, Wang L, Chen Y, Liu X, Dong Q, Chen YC, Peng Y, Yao KT, Kung HF, Li XP. MiR-26a inhibits cell growth and tumorigenesis of nasopharyngeal carcinoma through repression of EZH2. Cancer Res. 2011;71:225–233. doi: 10.1158/0008-5472.CAN-10-1850. [DOI] [PubMed] [Google Scholar]
- 22.Sun J, Zheng G, Gu Z, Guo Z. MiR-137 inhibits proliferation and angiogenesis of human glioblastoma cells by targeting EZH2. J Neurooncol. 2015;122:481–489. doi: 10.1007/s11060-015-1753-x. [DOI] [PubMed] [Google Scholar]
- 23.Ren X, Bai X, Zhang X, Li Z, Tang L, Zhao X, Li Z, Ren Y, Wei S, Wang Q, et al. Quantitative nuclear proteomics identifies that miR-137-mediated EZH2 reduction regulates resveratrol-induced apoptosis of neuroblastoma cells. Mol Cell Proteomics. 2015;14:316–328. doi: 10.1074/mcp.M114.041905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luo C, Tetteh PW, Merz PR, Dickes E, Abukiwan A, Hotz-Wagenblatt A, Holland-Cunz S, Sinnberg T, Schittek B, Schadendorf D, et al. miR-137 inhibits the invasion of melanoma cells through downregulation of multiple oncogenic target genes. J Invest Dermatol. 2013;133:768–775. doi: 10.1038/jid.2012.357. [DOI] [PubMed] [Google Scholar]