Abstract
The reprogramming of adult cells into pluripotent cells or directly into alternative adult cell types represents a great potential technology for regenerative medicine. In the present study, the potential of key developmental adipogenic, neurogenic and hepatogenic regulators to reprogram human fibroblasts into adipocytes, neurocytes and hepatocytes was investigated. The results demonstrated that direct reprogramming of octamer-binding transcription factor 4 (Oct4) and CCAAT-enhancer-binding protein (C/EBP)β activated C/EBPα and peroxisome proliferator-activated receptor-γ expression, inducing the conversion of fibroblasts into adipocytes. Similarly, direct reprogramming of the transcription factors sex determining region-box 2, trans-acting T-cell specific transcription factor (GATA-3) and neurogenic differentiation 1 in fibroblasts may induce neurogenic differentiation through hemagglutinating virus of Japan envelope (HVJ-E) transfection. Moreover, hepatogenic differentiation was induced by combining the direct reprogramming of Oct4, GATA-3, hepatocyte nuclear factor 1 homeobox α and forkhead box protein A2 in fibroblasts. These results demonstrate that specific transcription factors and reprogramming factors are able to directly reprogram fibroblasts into adipogenic, neurogenic and hepatogenic differentiation lineages by HVJ-E transfection.
Keywords: reprogramming, fibroblast, transcription factor, adipocyte, neurocyte, hepatocyte
Introduction
It is believed that differentiation is predominantly a unidirectional and irreversible process that cells undertake during lineage commitment; however, this assumption has been challenged by the discovery of four transcription factors that are able to reprogram mouse and human cells into a pluripotent stage by transcription factor transduction (1). Overexpression of lineage-specific transcription factors has been widely utilized to alter cell fates for application in regenerative medicine or personalized disease modeling (2). The reprogramming of fibroblasts to induced pluripotent stem (iPS) cells raises the possibility that a somatic cell may be able to be reprogrammed to an alternative differentiated fate without first becoming a stem/progenitor cell (3–5). The ability to reprogram fibroblasts into iPS cells with four transcription factors may provide an alternative source of embryonic-like stem cells (6,7). A previous study has demonstrated that direct reprogramming of fibroblasts into cardiomyocyte-like cells in vitro is possible through the expression of three transcription factors, including Gata4, Mef2c and Tbx5 (3). Similarly, the effective cell-fate switching of fibroblasts into neuronal and hepatocyte lineages is generally achieved by the overexpression of lineage-instructive transcription factors (8,9). A recent study was able to reprogram human fibroblasts toward a cardiac fate, although these cells lacked mature cardiac functions (10). Furthermore, transdifferentiated cells are proliferatively arrested, which precludes them from expanding in large numbers for in vivo measurements and biomedical applications (11); however, generating sufficient iPS-derived adipogenic, neurogenic and hepatogenic cells that are pure, mature and that can be delivered safely remains challenging.
Previous findings suggest that forced expression of combinations of transcription factors, including octamer-binding transcription factor 4 (Oct4), sex determining region Y-box 2 (Sox2), Kruppel-like factor 4 (Klf4) and c-Myc, induces immortality and pluripotency in mammalian somatic cells (6). The combination of Oct4 with L-Myc induces the conversion of fibroblasts into functional osteoblasts (12). Moreover, Oct4 alone is sufficient for inducing neural conversion from human fibroblasts into osteoblasts for disease modeling, and co-expression of Oct4 and Sox2 enhances neural conversion from human fibroblasts (13). Intriguingly, transient induction of the four reprogramming factors (Oct4, Sox2, Klf4 and c-Myc) is able to efficiently transdifferentiate fibroblasts into functional neural stem/progenitor cells (NPCs) with appropriate signaling inputs (14); however, there are fewer reports on Sox2- or Oct4-induced adipogenic or hepatogenic conversion from human fibroblasts.
In the present study, key developmental adipogenic, neurogenic and hepatogenic regulators were examined for their ability to reprogram human fibroblasts into adipocytes, neurocytes and hepatocytes. It was demonstrated that human fibroblasts were successfully and directly reprogrammed into adipogenic, neurogenic and hepatogenic differentiation lineages by combining specific transcription factors with reprogramming factors.
Materials and methods
Generation of directly reprogramming cells
Human fibroblasts were seeded at 2×105 cells per well in Dulbecco's modified Eagle medium (DMEM; Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS; Thermo Fisher Scientific, Inc.). The following day, the media was changed to fresh media supplemented with hemagglutinating virus of Japan envelope (HVJ-E; catalogue no. ISK-GN-001-EX; Cosmo Bio Co., Ltd., Tokyo, Japan) transduction agent. Following overnight culture (37°C) in the protein transduction media, the media was changed to FBS-supplemented (10%) culture media and cells were cultured for an additional 48 h before repeating the same protein transduction cycle. Following two different rounds of protein transduction, cells were passaged and cultured in DMEM supplemented with 10% FBS. Media were changed every 3 to 4 days and cells were cultured for a further 20 days. The process for direct reprogramming of human fibroblasts is demonstrated in Fig. 1. In addition, neural differentiated fibroblasts (2×105/well) were plated onto poly-D-lysine/laminin coated plates and cultured in DMEM supplemented with 10% FBS, 1 mM β-mercaptoethanol (cat. no. 07604; Sigma-Aldrich; Merck kGaA, Darmstadt, Germany) and 10 ng/ml basic fibroblast growth factor (cat. no. F5392; Sigma-Aldrich; Merck kGaA) for 24 h at 37°C. Media was then changed to DMEM supplemented with 10% FBS, 5 mM KCl, 2 µM valproic acid, 10 µM forskolin, 1 µM hydrocortisone and 5 µg/ml insulin (all Shanghai Aladdin Bio-Chem Technology Co., Ltd, Shanghai, China) for 14 days at 37°C.
Figure 1.
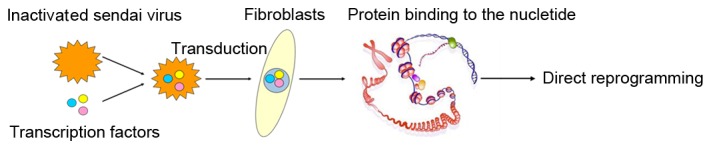
Schematic representation of the process used for the direct reprogramming of fibroblasts.
Localization of transfected reprogramming factors
In order to determine the intracellular localization of the transcription factors, protein-transduced cells were examined by fluorescent microscopy. Following 24 and 72 h of transduction, cells were harvested and washed twice (5 min/wash) with phosphate-buffered saline (PBS), then fixed in 4% (v/v) formaldehyde in PBS at room temperature for 1 min. Cells were washed twice (5 min/wash) with PBS and incubated with primary anti-His tag antibody (catalogue. no. 05-949; 1:1,000; Novagen; Merck KGaA), at room temperature for 1 h. Following washing three times with PBS, fluorescence conjugated secondary anti-mouse immunoglobulin G (H+L) Alexa Fluor 488 Conjugate (catalogue no. 4408S; 1:1,000; CST Biological Reagents Co., Ltd., Shanghai, China) was added and the cells were incubated in the dark for 1 h at room temperature. Cells were subsequently washed with PBS and nuclear DNA was labeled with Hoechst 33342 stain (1:3,000; Sigma-Aldrich; Merck KGaA). Images were captured using an Olympus DP70 microscope camera (Olympus Corp., Tokyo, Japan).
Immunofluorescence assay of differentiated iPS cells
Directly induced neurogenic cells were stained with anti-glial fibrillary acidic protein (catalogue no. 3655; 1:300), anti-nestin (catalogue no. 6994; 1:300; both CST Biological Reagents Co., Ltd.), anti-microtubule-associated protein 2 (MAP2; catalogue no. ab5392; 1:200) and anti-β-Tubulin III (catalogue no. ab18207; 1:200; both Abcam, Cambridge, UK) antibodies. Directly induced hepatocytes were stained with anti-Cytokeratin 19 antibody (cat. no. 4558; 1:400) and anti-Albumin antibody (cat. no. 4929; 1:400; both CST Biological Reagents Co., Ltd.), respectively.
Oil Red O staining
Cells were washed three times with PBS and fixed for 15 min with 3.7% formaldehyde at room temperature. Oil Red O (0.5% in isopropanol; catalogue no. O1391; Sigma Aldrich; Merck KGaA) was diluted with water (3:2) and incubated with the fixed cells at room temperature for 2 h. Cells were washed with water, then fat droplets in the stained cells were visualized by light microscopy and photographed using a Leica DM 2500 microscope (Leica Microsystems GmbH, Wetzlar, Germany).
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) of differentiated iPS cells
Total RNA was extracted from directly induced neurogenic and directly induced hepatocyte cells using TRIzol Reagent (Thermo Fisher Scientific, Inc., Waltham, MA, USA), according to the manufacturer's instructions. RNA was treated with RNase-free DNase I (Promega Corporation, Madison, WI, USA) to prevent contamination of trace genomic DNA. cDNA was then synthesized from RNA in a reaction mixture (20 µl) containing 2 µg total RNA, using a M-MuLV Reverse Transcriptase and Oligo d(T)23 VN primers (New England Biolabs, Inc., Ipswich, MA, USA). qPCR was performed using an iTaq Universal SYBR Green Supermix (Bio-Rad Laboratories, Inc., Hercules, CA, USA), according to the manufacturer's instructions. Following initial denaturation at 95°C for 3 min, 40 cycles of denaturation were performed at 95°C for 15 sec, followed by annealing at 56°C for 20 sec and extension at 72°C for 20 sec. The Cq (quantification cycle fluorescence value) was calculated using SDS 2.1 software (Applied Biosystems; Thermo Fisher Scientific, Inc.), and relative expression levels of mRNA were calculated using the ΔΔCq method (15) and normalized to the internal control GAPDH. To assess cell programming of iPS cells, the following primer sequences were used: For CCAAT-enhancer-binding protein-α (C/EBPα), forward, 5′-TCCCTAGTGTTGGCTGGAAG-3′ and reverse, 5′-CAGTAGGATGGTGCCTGCTG-3′, for PPARγ, forward, 5′-AAAGAAGCGGTGAACCACTGATA-3′ and reverse, 5′-AATGGCATCTCTGTGTCAACCA-3′, for nestin, forward, 5′-AACCACAGGAGTGGGAACTG-3′ and reverse, 5′-TCTGGCATTGACTGAGCAAC-3′, for MAP2, forward, 5′-GCAGTTCTCAAAGGCTAGAC-3′ and reverse, 5′-ATCGTGACCTGAACC-3′, for E-cadherin, forward, 5′-GGCACAGATGGTGTGATTAC-3′ and reverse, 5′-GAGCACCTrCCATGACAGA-3′, for Albumin, forward, 5′-AGAAAGTACCCCAAGTGTCAA-3′ and reverse, 5′-AGCTGCGAAATCATCCATAAC-3′, for tryptophan 2,3-dioxygenase (TDO), forward, 5′-TGCTCAAGGTGATAGCTCGGA-3′ and reverse, 5′-AGGAGCTTGAAGATGACCACCA-3′ and for GAPDH, forward, 5′-ACAGGGGAGGTGATAGCATT-3′ and reverse, 5′-GACCAAAACTTCATACATCTC-3′. Three replicates were completed for each gene.
Statistical analysis
Statistical analyses were performed using PRISM v.5.0 (GraphPad Software, Inc., La Jolla, CA, USA). Data are presented as the mean ± standard deviation for each group. Student's unpaired t-test and one-way analysis of variance were used for statistical evaluation. P<0.05 was considered to indicate a statistically significant difference.
Results
Transfected transcription factors localize to the nucleus
To determine the intracellular localization of transduced transcription factors by HVJ-E, anti-His tag antibody was utilized and proteins were observed using microscopy. One day subsequent to viral transduction, 90% living cells were positive for His-tag transcription factors, and all anti-His-tagged proteins localized to the nucleus. Fluorescence intensity decreased markedly 72 h after transfection, with only ~15% of the fibroblasts exhibiting fluorescence at 72 h, suggesting a gradual degradation of the anti-His-tagged transcription factors. These results demonstrated that HVJ-E is an efficient vehicle for protein transduction into fibroblasts, and transducted transcription factors remain in the nucleus for at least 72 h (Fig. 2).
Figure 2.
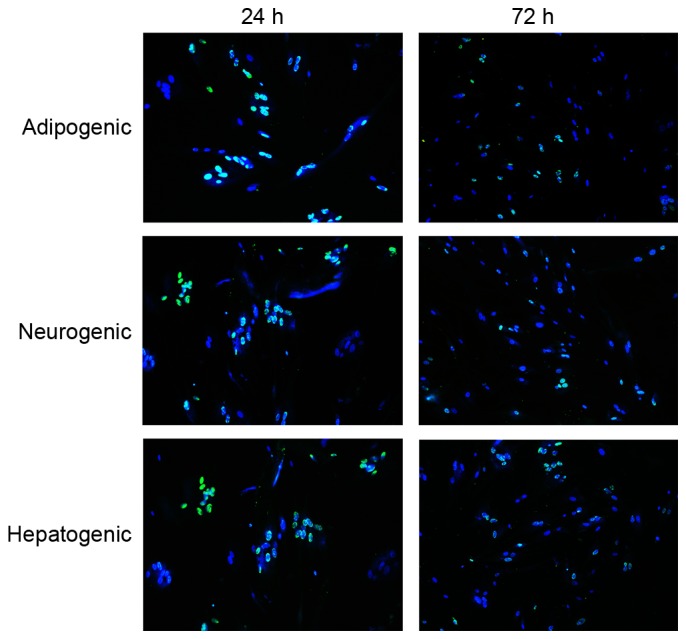
Protein transduction and visualization in human fibroblasts 24 and 72 h after hemagglutinating virus of Japan envelope transduction of fibroblasts. Cells were stained with Hochest 33342 (blue) for nuclei and immunostained with anti-His antibody (green) for transcription factors (magnification, ×100).
Oct4 and C/EBPβ directly reprogram fibroblasts into adipocytes
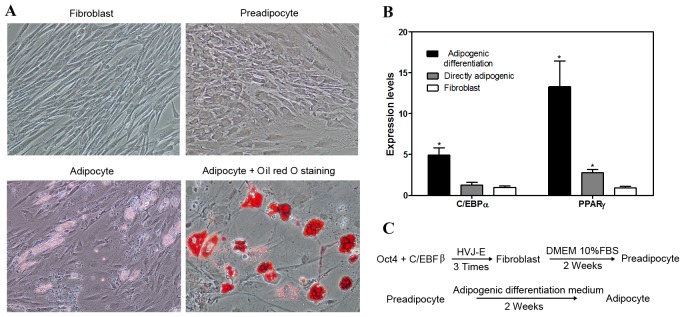
The C/EBP family of transcriptional regulators is critically important for the activation of adipogenic genes during differentiation. C/EBPβ is rapidly induced upon adipocyte differentiation and is responsible for activating the adipogenic regulators C/EBPα and PPAR, which together activate the majority of genes expressed in differentiating adipocytes (16). Oct4 attenuates the elevated expression levels of a group of epithelial cell transforming sequence genes (17); therefore, Oct4 and C/EBPα were selected for the present study to induce fibroblasts into pre-adipocytes by HVJ-E transfection. Two weeks after direct adipogenic reprogramming, morphological observations indicated that the fibroblasts had changed to round pre-adipocytes and, after an additional 2 weeks, adipogenic differentiation medium lipid droplets were observed by Oil Red O staining in the cells (Fig. 3A). RT-qPCR analysis confirmed that C/EBPα and PPARγ gene expression levels increased in the direct adipogenic reprogramming and adipogenic differentiation groups (Fig. 3B). This increase was significant (P<0.05) in the adipogenic differentiation C/EBPα and PPARγ groups and significant (P<0.05) in the PPARγ directly adipogenic group, compared with the fibroblast control group. The protocol for direct reprogramming of fibroblasts into adipocytes is presented in Fig. 3C.
Figure 3.
Direct adipogenic reprogramming. (A) Morphology and Oil Red O staining. Fibroblasts were directly induced into preadipocytes, followed by differentiation into adipocytes in DMEM with supplements. Lipid droplets were stained by Oil Red O (magnification, ×100). (B) Reverse transcription-quantitative polymerase chain reaction analysis of expression levels of C/EBPα and PPARγ marker genes in the adipogenic group. (C) Procedure of direct adipocyte reprogramming. Data are expressed as mean ± standard deviation (n=3 per group). *P<0.05 vs. fibroblast control group. C/EBPα, CCAAT-enhancer-binding protein α; PPARγ, peroxisome proliferator-activated receptor γ; HVJ-E, hemagglutinating virus of Japan envelope; Oct4, octamer-binding transcription factor 4; DMEM, Dulbecco's modified Eagle medium; FBS, fetal bovine serum.
Sox2, GATA-3 and neurogenic differentiation 1 (NEUROD1) induce direct neurogenic reprogramming
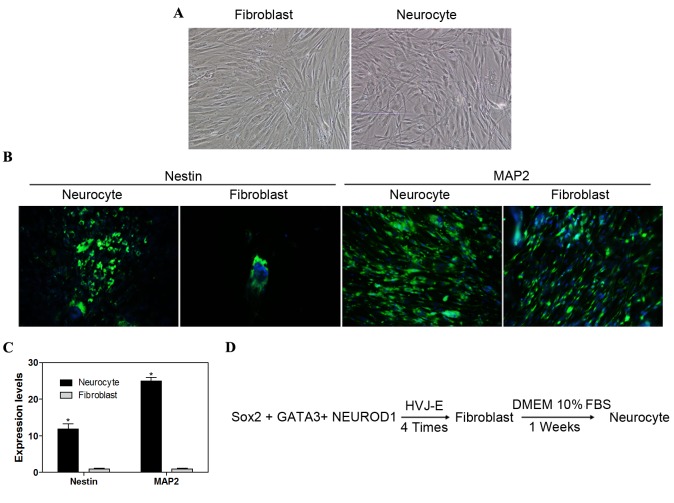
Nestin is an intermediate filament protein expressed in dividing cells during the early stages of development in the nervous system (18). MAP2 is involved in microtubule assembly, which is an essential step in neurogenesis (19). Seven days after induction, the cells began to develop neuronal morphology, including the development of soma, axons and dendrites (Fig. 4A). The directly reprogrammed cells were positive for nestin and MAP2 protein, which are expressed by numerous cell types of the central nervous system, and the levels of nestin and MAP2 were markedly increased in neurocytes as compared to fibroblasts (Fig. 4B). Furthermore, RT-qPCR analysis demonstrated that nestin and MAP2 expression levels at the early stage of NPCs, and the mRNA levels of nestin and MAP2 were significantly higher (P<0.05) in neurocytes compared with fibroblasts (Fig. 4C). The protocol for direct reprogramming of fibroblasts into neurocytes is presented in Fig. 4D. These observations demonstrated that fibroblasts are able to generate NPCs and differentiate further into mature neurons.
Figure 4.
Direct neurogenic reprogramming. (A) Induced neurocytes exhibited typical neuron morphology with soma, dendrites and axons (magnification, ×100). (B) Nestin and MAP2 markers were identified by immunofluorescence staining (magnification ×100). (C) Reverse transcription-quantitative polymerase chain reaction analysis of MAP2 and nestin expression levels in differentiated iPS cells. (D) Procedure of direct neurogenic reprogramming. Data are expressed as mean ± standard deviation (n=3 in each group). *P<0.05 vs. fibroblast control group. MAP2, microtubule-associated protein 2; iPS, induced pluripotent stem; Sox2, sex determining region Y-box 2; NEUROD1, neurogenic differentiation 1; HVJ-E, hemagglutinating virus of Japan envelope; DMEM, Dulbecco's modified Eagle medium; FBS, fetal bovine serum.
Oct4 and GATA-3 induce direct hepatogenic reprogramming
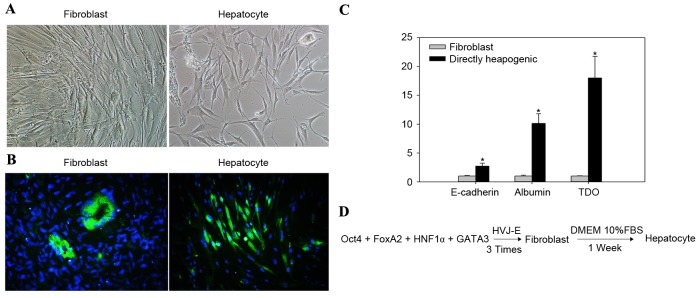
Hepatocytes are derived from the endoderm. Oct4 attenuates the expression of ectodermal genes (20). Trans-acting T-cell specific transcription factor (GATA3) is required for the development of the endoderm (21). One week subsequent to three repetitions of direct hepatogenic reprogramming, the morphology of fibroblasts altered from spindle shaped to specific hepatogenic cells (Fig. 5A). TDO, a liver-specific cytosolic hemoprotein, is a rate-limiting enzyme in L-tryptophan catabolism and a key serotonergic determinant (22). Glucocorticoids transcriptionally activate the TDO gene, resulting in the induction of enzyme expression (22). In the present study, upregulated TDO expression levels in the hepatocyte group were confirmed by immunofluorescence staining (Fig. 5B). RT-qPCR results demonstrated that TDO expression levels were significantly higher (P<0.05) in the directly reprogrammed hepatocyte group compared with the TDO expression levels in the fibroblast group. Serum albumin is produced in the liver, dissolved in the blood and is the most abundant blood protein in mammals. Albumin is essential for maintaining the oncotic pressure required for the distribution of body fluids between blood vessels and body tissues (23). In the absence of albumin, the high pressure in the blood vessels would force more fluids out into the tissues. In the present study, albumin expression confirmed the differentiation from fibroblasts to hepatocytes. Albumin expression levels were significantly higher (P<0.05) in the directly reprogrammed hepatocyte group compared with those in the fibroblast group. Furthermore, levels of E-cadherin expression were significantly increased in the directly reprogrammed hepatocyte group, relative to those in the fibroblast group (P<0.05; Fig. 5C). Fig. 5D presents the schematic protocol for direct reprogramming of fibroblasts into hepatocytes.
Figure 5.
Direct hepatogenic reprogramming. (A) One week after direct hepatogenic reprogramming, cells exhibited typical endodermal cell morphology (magnification, ×100). (B) TDO marker was identified by immunofluorescence staining (magnification, ×100). (C) Gene expression levels of E-cadherin, albumin and TDO were significantly increased in hepatocytes compared with the fibroblast control groups. (D) Procedure of direct hepatogenic reprogramming. Data are expressed as mean ± standard deviation (n=3 in each group). *P<0.05 vs. fibroblast control group. TDO, tryptophan 2,3-dioxygenase; Oct4, octamer-binding transcription factor 4; FoxA2, forkhead box protein A2; HNF1α, hepatocyte nuclear factor 1 homeobox α; GATA3, trans-acting T-cell specific transcription factor; HVJ-E, hemagglutinating virus of Japan envelope; DMEM, Dulbecco's modified Eagle medium; FBS, fetal bovine serum.
Discussion
Several studies have reported the direct conversion of fibroblasts into iPS cells, where the resulting cells are non-proliferating and terminally differentiated (24,25). The present study demonstrated that human fibroblasts are able to be directly reprogrammed into functional and proliferating adipocytes, neurocytes and hepatocytes through cell activation by specific transcription factors and reprogramming factors, including Oct4, Sox2, GATA3, C/EBF, NEUROD1, forkhead box protein A2 and hepatocyte nuclear factor 1 homeobox α.
There is mounting evidence to suggest that the differentiation of preadipocytes is regulated, in part, by a cascade of transcriptional events involving activation of the C/EBPs and PPARγ (16,26,27). Moreover, inhibition of C/EBPβ activity not only blocks C/EBPα and PPARγ expression, but it also renders the preadipocytes dependent on an exogenous PPARγ ligand for differentiation into adipocytes (28). In the present study, it was demonstrated that direct reprogramming to overexpress C/EBPβ activated C/EBPα and PPARγ expression, resulting in the conversion of fibroblasts into adipocytes. Additionally, Oct4, as well as C/EBPβ, was identified to be involved in the complex program of adipogenic gene expression during terminal preadipocyte differentiation.
GATA-3 belongs to the GATA family and is a zinc finger transcription factor that is expressed in T-cell lineages, as well as in the nervous system, during development (29). GATA-3 is able to regulate luminal epithelial cell differentiation in the mammary gland (30). In addition, the GATA-3 transcription factor is required for the survival of embryonic and adult sympathetic neurons. The GATA-3 transcription factor exhibits a specific and restricted expression pattern in the developing and adult mouse brain (31). Emerging evidence demonstrates that GATA-3 is involved in the development of serotonergic neurons in the caudal raphe nuclei (31). NEUROD1 is predominantly expressed in the nervous system late in development; therefore, it is more likely to be involved in terminal differentiation, neuronal maturation and survival (32,33). Sox2 curtails a group of mutual exclusion genes during reprogramming (34); therefore, Sox2, GATA-3 and NEUROD1 were selected in the present study to induce the conversion of fibroblasts into neurocytes by HVJ-E transfection. In the present study, it was demonstrated that direct reprogramming of Sox2, GATA3 and NEUROD1 in fibroblasts induced neurogenic differentiation by HVJ-E transfection. Similarly, hepatogenic differentiation was induced by combining Oct4 and GATA-3 in the direct reprogramming of fibroblasts. GATA-3 expression in hepatocytes increases during human development and erythropoietin mRNA accumulation is unaltered in mutant mice lacking GATA-3 (35). Considering this, the results of the present study are consistent with other emerging evidence to suggest a vital role for specific transcription and reprogramming factors in the direct reprogramming of fibroblasts into iPS cells.
In conclusion, the present study demonstrated that specific transcription and reprogramming factors are able to directly reprogram fibroblasts into adipogenic, neurogenic and hepatogenic differentiation by HVJ-E transfection. Future investigation in human cells and advances in safe delivery of defined factors will be necessary to advance this technology for use in potential regenerative therapies.
References
- 1.Batta K, Florkowska M, Kouskoff V, Lacaud G. Direct reprogramming of murine fibroblasts to hematopoietic progenitor cells. Cell Rep. 2014;9:1871–1884. doi: 10.1016/j.celrep.2014.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vierbuchen T, Wernig M. Direct lineage conversions: Unnatural but useful? Nat Biotechnol. 2011;29:892–907. doi: 10.1038/nbt.1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ieda M, Fu JD, Delgado-Olguin P, Vedantham V, Hayashi Y, Bruneau BG, Srivastava D. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell. 2010;142:375–386. doi: 10.1016/j.cell.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xue Y, Ouyang K, Huang J, Zhou Y, Ouyang H, Li H, Wang G, Wu Q, Wei C, Bi Y, et al. Direct conversion of fibroblasts to neurons by reprogramming PTB-regulated microRNA circuits. Cell. 2013;152:82–96. doi: 10.1016/j.cell.2012.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang P, Zhang L, Gao Y, He Z, Yao D, Wu Z, Cen J, Chen X, Liu C, Hu Y, et al. Direct reprogramming of human fibroblasts to functional and expandable hepatocytes. Cell stem cell. 2014;14:370–384. doi: 10.1016/j.stem.2014.01.003. [DOI] [PubMed] [Google Scholar]
- 6.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 7.Qian L, Huang Y, Spencer CI, Foley A, Vedantham V, Liu L, Conway SJ, Fu JD, Srivastava D. In vivo reprogramming of murine cardiac fibroblasts into induced cardiomyocytes. Nature. 2012;485:593–598. doi: 10.1038/nature11044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Du Y, Wang J, Jia J, Song N, Xiang C, Xu J, Hou Z, Su X, Liu B, Jiang T, et al. Human hepatocytes with drug metabolic function induced from fibroblasts by lineage reprogramming. Cell stem cell. 2014;14:394–403. doi: 10.1016/j.stem.2014.01.008. [DOI] [PubMed] [Google Scholar]
- 9.Kim HS, Kim J, Jo Y, Jeon D, Cho YS. Direct lineage reprogramming of mouse fibroblasts to functional midbrain dopaminergic neuronal progenitors. Stem cell Res. 2014;12:60–68. doi: 10.1016/j.scr.2013.09.007. [DOI] [PubMed] [Google Scholar]
- 10.Nam YJ, Song K, Luo X, Daniel E, Lambeth K, West K, Hill JA, DiMaio JM, Baker LA, Bassel-Duby R, Olson EN. Reprogramming of human fibroblasts toward a cardiac fate; Proc Natl Acad Sci USA; 2013; pp. 5588–5593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Graf T, Enver T. Forcing cells to change lineages. Nature. 2009;462:587–594. doi: 10.1038/nature08533. [DOI] [PubMed] [Google Scholar]
- 12.Yamamoto K, Kishida T, Sato Y, Nishioka K, Ejima A, Fujiwara H, Kubo T, Yamamoto T, Kanamura N, Mazda O. Direct conversion of human fibroblasts into functional osteoblasts by defined factors; Proc Natl Acad Sci USA; 2015; pp. 6152–6157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mitchell RR, Szabo E, Benoit YD, Case DT, Mechael R, Alamilla J, Lee JH, Fiebig-Comyn A, Gillespie DC, Bhatia M. Activation of neural cell fate programs toward direct conversion of adult human fibroblasts into tri-potent neural progenitors using OCT-4. Stem Cells Dev. 2014;23:1937–1946. doi: 10.1089/scd.2014.0023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim J, Efe JA, Zhu S, Talantova M, Yuan X, Wang S, Lipton SA, Zhang K, Ding S. Direct reprogramming of mouse fibroblasts to neural progenitors; Proc Natl Acad Sci USA; 2011; pp. 7838–7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 16.Zuo Y, Qiang L, Farmer SR. Activation of CCAAT/enhancer-binding protein (C/EBP) alpha expression by C/EBP beta during adipogenesis requires a peroxisome proliferator-activated receptor-gamma-associated repression of HDAC1 at the C/ebp alpha gene promoter. J Biol Chem. 2006;281:7960–7967. doi: 10.1074/jbc.M510682200. [DOI] [PubMed] [Google Scholar]
- 17.Chuang CY, Lin KI, Hsiao M, Stone L, Chen HF, Huang YH, Lin SP, Ho HN, Kuo HC. Meiotic competent human germ cell-like cells derived from human embryonic stem cells induced by BMP4/WNT3A signaling and OCT4/EpCAM (epithelial cell adhesion molecule) selection. J Biol Chem. 2012;287:14389–14401. doi: 10.1074/jbc.M111.338434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jin Z, Liu L, Bian W, Chen Y, Xu G, Cheng L, Jing N. Different transcription factors regulate nestin gene expression during P19 cell neural differentiation and central nervous system development. J Biol Chem. 2009;284:8160–8173. doi: 10.1074/jbc.M805632200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mortazavi Y, Sheikhsaran F, Khamisipour GK, Soleimani M, Teimuri A, Shokri S. The evaluation of nerve growth factor over expression on neural lineage specific genes in human mesenchymal stem cells. Cell J. 2016;18:189–196. doi: 10.22074/cellj.2016.4313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kotkamp K, Mössner R, Allen A, Onichtchouk D, Driever W. A Pou5f1/Oct4 dependent Klf2a, Klf2b, and Klf17 regulatory sub-network contributes to EVL and ectoderm development during zebrafish embryogenesis. Dev Biol. 2014;385:433–447. doi: 10.1016/j.ydbio.2013.10.025. [DOI] [PubMed] [Google Scholar]
- 21.Zhang C, Ye X, Zhang H, Ding M, Deng H. GATA factors induce mouse embryonic stem cell differentiation toward extraembryonic endoderm. Stem Cells Dev. 2007;16:605–613. doi: 10.1089/scd.2006.0077. [DOI] [PubMed] [Google Scholar]
- 22.Liao M, Pabarcus MK, Wang Y, Hefner C, Maltby DA, Medzihradszky KF, Salas-Castillo SP, Yan J, Maher JJ, Correia MA. Impaired dexamethasone-mediated induction of tryptophan 2,3-dioxygenase in heme-deficient rat hepatocytes: Translational control by a hepatic eIF2alpha kinase, the heme-regulated inhibitor. J Pharmacol Exp Ther. 2007;323:979–989. doi: 10.1124/jpet.107.124602. [DOI] [PubMed] [Google Scholar]
- 23.Sverdlov M, Shajahan AN, Minshall RD. Tyrosine phosphorylation-dependence of caveolae-mediated endocytosis. J Cell Mol Med. 2007;11:1239–1250. doi: 10.1111/j.1582-4934.2007.00127.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Caiazzo M, Dell'Anno MT, Dvoretskova E, Lazarevic D, Taverna S, Leo D, Sotnikova TD, Menegon A, Roncaglia P, Colciago G, et al. Direct generation of functional dopaminergic neurons from mouse and human fibroblasts. Nature. 2011;476:224–227. doi: 10.1038/nature10284. [DOI] [PubMed] [Google Scholar]
- 25.Liu X, Li F, Stubblefield EA, Blanchard B, Richards TL, Larson GA, He Y, Huang Q, Tan AC, Zhang D, et al. Direct reprogramming of human fibroblasts into dopaminergic neuron-like cells. Cell Res. 2012;22:321–332. doi: 10.1038/cr.2011.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jimenez MA, Akerblad P, Sigvardsson M, Rosen ED. Critical role for Ebf1 and Ebf2 in the adipogenic transcriptional cascade. Mol Cell Biol. 2007;27:743–757. doi: 10.1128/MCB.01557-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mori T, Sakaue H, Iguchi H, Gomi H, Okada Y, Takashima Y, Nakamura K, Nakamura T, Yamauchi T, Kubota N, et al. Role of Kruppel-like factor 15 (KLF15) in transcriptional regulation of adipogenesis. J Biol Chem. 2005;280:12867–12875. doi: 10.1074/jbc.M410515200. [DOI] [PubMed] [Google Scholar]
- 28.Hamm JK, Park BH, Farmer SR. A role for C/EBPbeta in regulating peroxisome proliferator-activated receptor gamma activity during adipogenesis in 3T3-L1 preadipocytes. J Biol Chem. 2001;276:18464–18471. doi: 10.1074/jbc.M100797200. [DOI] [PubMed] [Google Scholar]
- 29.Hong SJ, Choi HJ, Hong S, Huh Y, Chae H, Kim KS. Transcription factor GATA-3 regulates the transcriptional activity of dopamine beta-hydroxylase by interacting with Sp1 and AP4. Neurochem Res. 2008;33:1821–1831. doi: 10.1007/s11064-008-9639-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Asselin-Labat ML, Sutherland KD, Barker H, Thomas R, Shackleton M, Forrest NC, Hartley L, Robb L, Grosveld FG, van der Wees J, et al. Gata-3 is an essential regulator of mammary-gland morphogenesis and luminal-cell differentiation. Nat Cell Biol. 2007;9:201–209. doi: 10.1038/ncb1530. [DOI] [PubMed] [Google Scholar]
- 31.van Doorninck JH, van der Wees J, Karis A, Goedknegt E, Engel JD, Coesmans M, Rutteman M, Grosveld F, De Zeeuw CI. GATA-3 is involved in the development of serotonergic neurons in the caudal raphe nuclei. J Neurosci. 1999;19:RC12. doi: 10.1523/JNEUROSCI.19-12-j0002.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cho JH, Tsai MJ. The role of BETA2/NeuroD1 in the development of the nervous system. Mol Neurobiol. 2004;30:35–47. doi: 10.1385/MN:30:1:035. [DOI] [PubMed] [Google Scholar]
- 33.Breslin MB, Zhu M, Lan MS. NeuroD1/E47 regulates the E-box element of a novel zinc finger transcription factor, IA-1, in developing nervous system. J Biol Chem. 2003;278:38991–38997. doi: 10.1074/jbc.M306795200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu Q, Zhang L, Su P, Lei X, Liu X, Wang H, Lu L, Bai Y, Xiong T, Li D, et al. MSX2 mediates entry of human pluripotent stem cells into mesendoderm by simultaneously suppressing SOX2 and activating NODAL signaling. Cell Res. 2015;25:1314–1332. doi: 10.1038/cr.2015.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dame C, Sola MC, Lim KC, Leach KM, Fandrey J, Ma Y, Knöpfle G, Engel JD, Bungert J. Hepatic erythropoietin gene regulation by GATA-4. J Biol Chem. 2004;279:2955–2961. doi: 10.1074/jbc.M310404200. [DOI] [PubMed] [Google Scholar]