Abstract
The highly polymorphic gene products of the classical MHC class I genes in humans (HLA-A, HLA-B, and HLA-C) play a critical role in the immune defense against intracellular infections. Because non-human primates are important models for AIDS vaccine research, rhesus monkeys from a thoroughly pedigreed and serotyped colony were subjected to full-length cDNA analysis of MHC class I gene transcripts. Rhesus macaques express multiple dominant Mamu-A and Mamu-B transcripts (majors) per chromosome, which are characterized by high expression levels. The presence of additional cDNAs with low levels of expression (minors) suggests evidence for transcriptional control of MHC class I genes. Moreover, phylogenetic analyses illustrate that most of the Mamu-A and Mamu-B loci/lineages identified display no or only limited levels of allelic polymorphism. Thus, MHC class I diversity in rhesus macaques is typified by the existence of an unmatched high number of Mamu-A and Mamu-B region configurations that exhibit polymorphism with regard to the number and combination of transcribed loci present per chromosome.
Keywords: AIDS, non-human primate
MHC class I glycoproteins are expressed on nucleated cells, and their biological function is to present foreign peptides of intracellular origin to cytotoxic T cells, which may result in the destruction of infected cells (1). In addition, inhibitory and stimulatory receptors on natural killer cells scan for the presence and absence of MHC class I molecules (2, 3). The polymorphism of the HLA-A, HLA-B, and HLA-C genes in the human population has been studied extensively, and hundreds of alleles have been identified (4). In contrast, the gene products of the nonclassical HLA-E, HLA-F, and HLA-G genes show low levels of polymorphism and a restricted tissue distribution and are thought to exert specialized functions (5, 6).
Considerable research has been conducted on the rhesus macaque (Macaca mulatta) MHC, because this species is widely used as a model for human diseases and organ transplantation. Simian immunodeficiency virus infection of macaques, for instance, is an important model for the study of AIDS (7–9). The rhesus macaque equivalents of the classical HLA-A and HLA-B genes (10–12) and the nonclassical HLA-E (13), HLA-F (14), and HLA-G (15) genes were identified and designated Mamu-A, Mamu-B, Mamu-E, Mamu-F, and Mamu-G, respectively. The latter appears to be a pseudogene, and its function may have been taken over by Mamu-AG, which is expressed on the rhesus monkey placenta and shares unique features with HLA-G (16, 17). Rhesus macaques possess an additional oligomorphic Mamu-B-like gene designated Mamu-I, which shares classical and nonclassical characteristics (18). The ortholoques of the HLA-C gene were found neither in rhesus macaques nor in any other species of Old World monkeys.
The organization of the rhesus macaque MHC class I region may be complex, because initial studies suggested that the Mamu-A and Mamu-B genes may have been duplicated (11, 19). The main question to be answered, however, revolves around the reported Mamu-A and Mamu-B sequences; the complex locus/allele relationships are not yet understood. The aim of this study was to shed light on the complexity of the class I region by providing a thorough inventory of the number of expressed Mamu-A and Mamu-B loci per chromosome by using a large panel of serotyped and pedigreed animals.
Materials and Methods
Animals and Cell Lines. The Biomedical Primate Research Centre houses a self-sustaining outbred colony of ≈1,000 rhesus macaques that have been pedigreed based on the segregation of serologically defined MHC haplotypes. Serotyping is performed by polyclonal sera raised by active immunizations. Serotypes are defined by a cluster of positive typing reactions. A blank serotype means that the typing reactions are not unambiguously interpretable. An inbreeding program resulted in a group of Mamu-A, Mamu-B, and Mamu-DR homozygous animals of consanguineous origin (20). The present Herpes papiotransformed B cell line cohort (≈100 individuals) consisted of samples originating mainly from Indian animals, as well as a few of Chinese and Burmese origin. Cell lines were selected in such a way that the panel covered all known Mamu-A and Mamu-B serotypes multiple times.
cDNA Cloning and Sequencing. RNA was isolated from B cells (RNeasy kit, Qiagen, Valencia, CA) and subjected to a One-Step RT-PCR kit, as recommended by the supplier. In these reactions, we used the primer sets 5′MAS/3′MAS and 5′MBS/3′MBS, which are specific for Mamu-A and Mamu-B transcripts, respectively (11). The final elongation step was extended to 30 min to generate a 3′dA overhang. The RT-PCR products were cloned by using the InsT/Aclone kit (Fermentas, St. Leon-Rot, Germany). After transformation colonies were picked for plasmid isolations (16–32 colonies for the Mamu-A transcript and 32–64 colonies for the Mamu-B transcript). Sequencing reactions were performed by using the BigDye terminator cycle sequencing kit, and samples were run on automated capillary sequencing systems (Applied Biosystems). All unreported Mamu-A and Mamu-B sequences and their corresponding accession numbers are depicted in Table 1. The sequences were named according to the proposal published in ref. 21.
Table 1. Unreported Mamu-A and Mamu-B sequences.
| Alleles | Accession nos. | Reference animals |
|---|---|---|
| Mamu-A | ||
| *0602 | AJ542567 | KM, 8653 |
| *0505 | AJ551315 | 1VJ, 1IH |
| *0506 | AJ551316 | BB58, BB10 |
| *0507 | AJ551317 | 8745 |
| *0509 | AJ551318 | 2B, 2G |
| *0510 | AJ551319 | Ri260 |
| *0511 | AJ551320 | 1KM, 1VV |
| *0703 | AJ542568 | C77, 9222 |
| *1305 | AJ551321 | 3238, 8813 |
| *1306 | AJ542570 | 98049 |
| *1602 | AJ542572 | 98049 |
| *19 | AJ542573 | 1VJ, 1IH |
| *21 | AJ542574 | 9133, 8813 |
| *23 | AJ542575 | 1JT, 9222 |
| *24 | AJ542576 | 9133, 8813 |
| *25 | AJ542577 | 2B, 2G |
| *26 | AJ542578 | 8827, 8769 |
| *27 | AJ542579 | 9151, 1ZA |
| *28 | AJ542580 | C77 |
| Mamu-B | ||
| *0602 | AJ844596 | 9178, KP |
| *0702 | AJ556875 | 1GG, 8884 |
| *0703 | AJ556876 | BB10, BB113 |
| *19 | AJ556877 | 1RK, 1JT |
| *20 | AJ556878 | 1VJ, B21 |
| *21 | AJ556879 | 1VJ, B21 |
| *22 | AJ556880 | 1VJ, 9151 |
| *24 | AJ556881 | 1RK, 1JT |
| *26 | AJ844602 | MR |
| *27 | AJ556882 | MR, 3019 |
| *28 | AJ556883 | 1VJ, B21 |
| *29012 | AJ556884 | 2AK, 1IH |
| *3002 | AJ556885 | 1GG, 8884 |
| *3601 | AJ556886 | 2BZ, 1QA |
| *3602 | AJ556887 | BB36, BB78 |
| *37 | AJ556888 | 2BZ, 2QA |
| *38 | AJ556889 | 8827, 1GG |
| *39 | AJ556890 | BB10, BB58 |
| *40 | AJ556891 | 2B, 2G, 2V |
| *41 | AJ556892 | 1OX, 2CA |
| *43 | AJ556893 | MR, 3019 |
| *44 | AJ556894 | 2B, 2G, 2V |
| *4501 | AJ556895 | 2BZ, 1QA |
| *4502 | AJ556896 | BB36, BB78 |
| *46 | AJ556897 | 1RK, 8822 |
| *4701 | AJ556898 | 8822, 8827 |
| *4702 | AJ556899 | B65 |
| *48 | AJ556900 | 1OX, 2CA |
| *49 | AJ844603 | 3C, 96084 |
| *5002 | AJ620415 | 3238, EKK |
| *5301 | AJ556901 | M14 |
| *5302 | AJ844604 | 3C, MR |
| *54 | AJ556902 | 1PV |
| *55 | AJ556903 | 1PV, 1GX |
| *57 | AJ844605 | MR, 96084 |
| *5802 | AJ556904 | 1PV, 1GX |
| *6002 | AJ556905 | 8745 |
| *61 | AJ556906 | 8745 |
| *63 | AJ556907 | 1PV, 3019 |
| *64 | AJ556908 | 1OX, 9202 |
| *65 | AJ620416 | 3238, EKK |
| *66 | AJ844597 | 95044, 8704 |
| *67 | AJ844598 | 95044, 8704 |
| *68 | AJ844599 | 95044, 8704 |
| *69 | AJ844601 | 1PH, 9208 |
| *70 | AJ844600 | 3C, MR |
| *71 | AJ849330 | 9178 |
Names, EMBL accession numbers, and reference animals are provided.
Locus-Specific PCR and Phylogenetic Analysis. To establish the presence of Mamu-A2(A*05) or Mamu-A4(A*14) alleles within cDNA samples, the following primer sets were used: (5′A*05) GCCCCCAGGCTCGCACTCCTTGAGA and (3′A*05) CTSGCCCTCCAGGTAGGCTCTCCA; (5′A*14) GGGACCCGACGGGCGCCTCCAA and (3′A*14) GGCCCTCCAGGTAGACTCTGTC. The 5′A*05 primer annealed in exon 2 and the 3′-primer in exon 3, respectively. The A*14 primers both have annealing sites in exon 3. Amplifications were carried out starting with 2 min at 94°C, followed by 25 cycles of 94°C, 65°C, and 72°C, for 1 min each. The PCR products were subjected to direct sequencing, as described above. Neighbor-joining trees were constructed as published in refs. 20 and 22. Bootstrap values based on 1,000 replications are indicated.
Results and Discussion
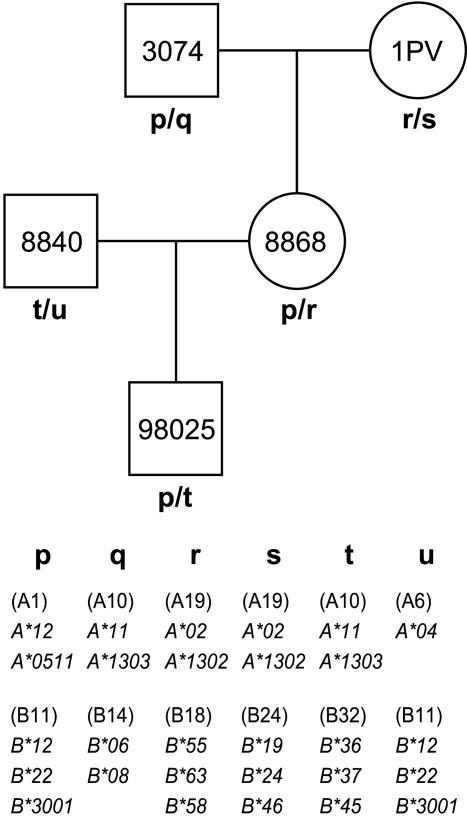
Definition of Mamu-A and Mamu-B Region Configurations and Identification of Mamu-A1. The cDNAs and the serologically defined Mamu-A and Mamu-B specificities segregate in a Mendelian manner (Fig. 1). In analogy to Mamu-DR, the Mamu-A and Mamu-B region configurations display variation with regard to the number and combination of Mhc class I loci present per haplotype. Serotype names are used for the identification of different region configurations.
Fig. 1.
Segregation of serotypes and Mamu-A and Mamu-B cDNAs in a Mendelian fashion in a rhesus macaque family. The maternal and paternal chromosomes are designated p–u. Serotypes are shown in brackets, and cDNAs are depicted in italics.
In contrast to the single HLA-A locus in humans, rhesus macaques express multiple Mamu-A-like cDNAs, illustrating the presence of compound loci segregating on a single chromosome (Fig. 1). Because of the extent and quality of the B cell line panel, it was possible to unravel the complex relationships that exist between serotypes and cDNAs. As can be seen, for all A serotypes, an allele of a polymorphic locus can be defined (Table 2). One-dimensional isoelectric focusing analyses illustrated that the corresponding gene products are characterized by relatively high expression levels (data not shown). For most of these structures, simian immunodeficiency virus epitopes have been defined (23–29), indicating that they act as bona fide MHC class I molecules.
Table 2. Mamu-A serotypes and major cDNA sequences detected.
| cDNAs
|
|||||
|---|---|---|---|---|---|
| Serotype | Mamu-A1 | Mamu-A2 | Mamu-A3 | Mamu-A4 | Configuration |
| A1 | A*12 | A*0511 | — | A*14 | I |
| A3 | A*19 | A*0505 | — | A*14 | I |
| A5 | A*27 | A*05 | — | A*14 | I |
| A9 | A*01 | A*0504 | — | A*14 | I |
| A22 | A*0701/02/03 | A*0507 | — | A*14 | I |
| A23 | A*0601/02 | A*0501 | — | A*14 | I |
| Blank | A*25 | A*0509 | — | A*14 | I |
| A28 | A*21 | A*24 | A*1305 | A*14 | II |
| A10 | A*11 | A*0503 | A*1303 | A*14 | II |
| A21 | A*1602 | — | A*1306 | A*14 | III |
| A19 | A*02 | — | A*1302 | A*14 | III |
| A6 | A*04 | — | — | A*14 | IV |
| A33 | A*23 | — | — | A*14 | IV |
| A27 | A*26 | — | — | A*14 | IV |
| Blank | A*28 | — | — | A*14 | IV |
The Mamu-A*05 allele depicted by only two digits has not yet been assigned unambiguously. Proposed locus names and configuration definitions are indicated. —, Indicates the absence of cDNAs.
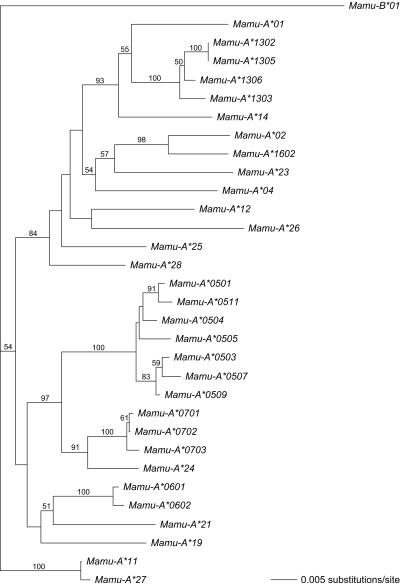
In the large outbred colony studied, most serotypes are represented by one allele of the polymorphic A locus (Table 2). A few exceptions to this rule were observed. For instance, the A22 and A23 serotypes are represented by three different Mamu-A*07 and two Mamu-A*06 alleles, respectively, whereas an earlier reported allele (11) differs slightly from Mamu-A*1602. It is concluded that, in the colonies analyzed thus far (mostly Indian animals), the lineages encoded by the locus controlling the serotype (proposed name Mamu-A1) display low degrees of allelic variation. Similar to the human system, however, phylogenetic comparisons illustrated that the various lineages controlled by this locus are characterized by large genetic distances (Fig. 2).
Fig. 2.
Phylogenetic tree of full-length Mamu-A sequences. The tree was constructed according to the neighbor-joining method, and bootstrap values based on 1,000 replications are indicated.
The Mamu-A2(*05/*24) Locus. In addition to the polymorphic Mamu-A transcript, some region configurations express a Mamu-A*05 cDNA (Table 2). In our panel, at least eight highly related alleles are observed, each of which seems to segregate with a particular serotype. For example, Mamu-A*0501 appears to be unique for the A23 serotype, whereas the Mamu-A*0504 allele is found only in A9-positive animals (Table 2).
Urvater et al. (29) reported that the Mamu-A*05 and Mamu-A*07 genes are characterized by a 162-bp insertion in intron 2, which was confirmed for Mamu-A*05, but the Mamu-A*07 genes in our panel seem to lack the insert. The Mamu-A*24 genes, however, possess this intron modification. Based on the presence of the shared insert and the supporting phylogenetic tree (Fig. 2), Mamu-A*05 and Mamu-A*24 are considered to represent two lineages controlled by one locus (Table 2). It is highly probable that the Mamu-A*05/*24 locus arose from a duplication/recombination in which the Mamu-A*07 gene was involved, subsequently followed by the intron 2 insertion. This alteration is also observed in Mafa-A*05 genes of cynomolgus macaques (Macaca fascicularis), indicating that this locus is at least 2.5 million years old. The Mamu-A*05/*24 locus (proposed name Mamu-A2), characterized by differential distribution, might encode a nonclassical class I protein with a specialized function in the immune response. A similar situation may exist for the Patr-AL locus in chimpanzees (30).
The Mamu-A3(*13) and Mamu-A4(*14) Loci. Four Mamu-A*13 alleles were reported in ref. 31. In our panel, two alleles designated Mamu-A*1305 and Mamu-A*1306, were detected, and they segregate with the A28 and A21 serotypes, respectively (Table 2). Based on the presence of two other Mamu-A loci in, for instance, A10-positive animals, Mamu-A*13 must represent a third locus (proposed name Mamu-A3) present in only two region configurations (Table 2).
Mamu-A*14 sequences, which are characterized by a unique two-triplet deletion in the first exon, are present in all animals (Table 2). Apparently this locus displays no polymorphism in our population of Indian animals. The situation for this locus is very similar to the Mamu-I locus that arose from a duplication event and, as a consequence, shares unique features with Mamu-B (18). A proposed name for this locus is Mamu-A4.
Mamu-A Region Configurations. The four Mamu-A region configurations identified express not only the invariant Mamu-A4 gene but also one Mamu-A1 gene controlling the serotype and, in most cases, one or two additional genes belonging to either the Mamu-A2 or Mamu-A3 loci (Table 2). In contrast to humans, who may express maximally two distinct HLA-A molecules, a heterozygous rhesus macaque may express maximally up to seven different Mamu-A-like molecules. In general, however, the different Mamu-A loci and/or lineages display low levels of allelic polymorphism. Recently, a rhesus macaque Mhc class I region was sequenced (32), and the data are fully compatible with the Mamu-A region configuration IV as is present in A6 serotyped animals (Table 2).
Mamu-B Serotypes Are Defined by Combinations of cDNAs with High Expression Levels. For each B serotype, at least one unique combination of cDNAs was detected (Table 3). As can be seen, most B serotypes are typified by the presence of two, three, and in one case even four cDNAs. For the corresponding gene products of these cDNAs, simian immunodeficiency virus epitopes recognized by cytotoxic T lymphocyte have been defined (8, 12, 27, 29, 33, 34). In some samples, cDNAs with low expression levels were detected next to the major cDNAs with high expression levels. These “minors” are not listed in Table 3.
Table 3. Mamu-B serotypes and corresponding major cDNA sequences.
| Serotypes | cDNAs | |||
|---|---|---|---|---|
| B2a | B*24 | B*38 | B*4701/02 | — |
| B2b | B*38 | B*46 | B*4701 | — |
| B11a | B*38 | B*3001 | B*12 | B*22 |
| B11b | — | B*3001 | B*12 | B*22 |
| B13 | B*48 | B*64 | B*41 | — |
| B14 | B*08 | — | B*06 | — |
| B17 | B*17 | — | B*2901 | — |
| B18 | B*55 | B*63 | B*5802 | — |
| B20 | B*21 | B*28 | B*20 | — |
| B24 | B*24 | B*46 | B*19 | — |
| B25 | B*5002 | B*65 | B*69 | — |
| B26a | B*01 | B*3002 | B*0701/02 | — |
| B26b | B*01 | B*6002 | B*61 | — |
| B26c | B*01 | B*46 | B*0702 | — |
| B29 | B*44 | — | B*40 | — |
| B31 | B*66 | B*67 | B*68 | — |
| B32 | B*3601/02 | B*37 | B*4501/02 | — |
| B34 | B*27 | B*43 | — | — |
| B35 | B*0602 | B*71 | — | — |
| blanka | B*01 | B*39 | B*0703 | — |
| blankb | B*01 | B*53 | B*0702 | — |
Sequences are listed in a random order and are represented by four digits when polymorphism for the respective lineage has been documented. —, Indicates the absence of cDNAs.
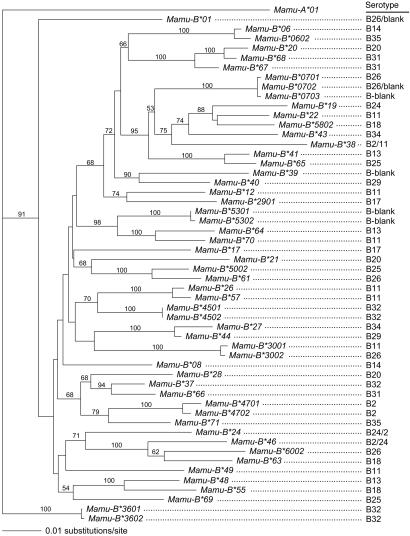
For some serotypes, at least two different combinations of cDNAs were observed. For example, the Mamu-B*01, Mamu-B*3002, and Mamu-B*0701 sequences were found in Indian animals with the B26 serotype, whereas, in Chinese animals, the Mamu-B*01, Mamu-B*6002, and Mamu-B*61 sequences were observed (Table 3). A few highly related sequences are situated on different Mamu-B region configurations. For instance, the Mamu-B*3001 and Mamu-B*3002 alleles, differing for three base pairs, were detected in the case of the B11 and B26 serotypes, respectively (Table 3). Furthermore, different Mamu-B region configurations may share identical sequences, as was the case for the Mamu-B*46 (Table 3). In general, the different Mamu-B loci/lineages display no or limited levels of allelic variation and are characterized by large genetic distances (Fig. 3). In total, 21 unique cDNA combinations were identified (Table 3) that appear to segregate in families with the relevant serotype (Fig. 1).
Fig. 3.
Phylogenetic tree of full-length Mamu-B cDNAs. The tree was constructed according to the neighbor-joining method, and bootstrap values based on 1,000 replications are indicated. Serotypes have been provided.
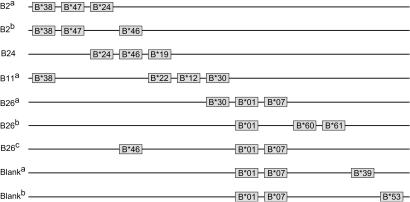
Mamu-B Region Diversity Is About Loci. In contrast to Mamu-A, the definition of loci appears to be difficult for the Mamu-B region. A schematic representation of nine selected Mamu-B region configurations (differing in number and combination of genes) was made to illustrate the complex locus/allele relationships (Fig. 4). For animals with the B2 serotypes, two distinct Mamu-B region configurations were observed sharing the Mamu-B*38 and Mamu-B*47 genes that, by definition, represent different loci (Fig. 4). The B2a serotype possesses an additional Mamu-B*24 sequence, whereas the other harbors Mamu-B*46. These genes represent a third and fourth Mamu-B locus, as is supported by B24-seropositive animals, where those loci are found on the same configuration together with Mamu-B*19. Hence, only these three Mamu-B region configurations define five Mamu-B loci, which apparently display no polymorphism (Table 3).
Fig. 4.
Schematic representation of nine Mamu-B region configurations. The physical order and distance of the genes responsible for the major cDNA transcripts have not yet been defined. Vertical alignment of cDNAs reflects identical locus designation at the genomic level.
For the B11a serotype, the Mamu-B*38, Mamu-B*3001, Mamu-B*12, and Mamu-B*22 sequences were found to segregate on one chromosome and, thus, must represent four distinct loci. The Mamu-B*19, Mamu-B*22, and Mamu-B*58 cDNAs share a unique one-codon deletion in exon 5 and cluster tightly together in the phylogenetic tree (Fig. 3). For that reason, the Mamu-B*19 sequence (B24) and Mamu-B*22 cDNA (B11a) are considered to represent one locus (Fig. 4). The Mamu-B*38 locus is shared with the B2 serotyped cells, whereas the Mamu-B*12 and Mamu-B*30 sequences are considered to represent two additional Mamu-B loci.
All cells with the B26 and blank serotypes share the Mamu-B*01 cDNA, and its large phylogenetic distance from known Mamu-B sequences (Fig. 3) is an indication that it represents yet another Mamu-B locus (Fig. 4). The Mamu-B*30 locus in B11-serotyped cells is shared with the B26a region configuration, whereas the B26c, B2b and B24 serotypes share the Mamu-B*46 locus. Mamu-B*07, which lacks the characteristic one-codon deletion present in Mamu-B*19 and Mamu-B*22 (Fig. 3), is considered to define a separate locus. Neither the sequences nor the phylogenetic analyses provide any argument for grouping the Mamu-B*39, Mamu-B*53, Mamu-B*60, or Mamu-B*61 sequences together with earlier defined loci. Hence, this small sample provides evidence that at least as many as 13 different Mamu-B-like loci can be defined in the case of only five serotypes. In total, 21 different Mamu-B region configurations are defined (Table 3), indicating that the number of Mamu-B loci may be much higher.
Recently, it was documented that the rhesus MHC class I region contains at least 18 apparently functional Mamu-B-like genes (32). The reported Mamu-B region is in agreement with our B11-like configurations; however, the genomic data documented the presence of additional functional genes such as Mamu-B*26, Mamu-B*49, Mamu-B*53, Mamu-B*57, and Mamu-B*70. RT-PCR studies indeed demonstrated that for some B11 region configurations additional minor class I cDNAs can be detected in small numbers (Table 4). This finding indicates that the various Mamu-B genes exhibit differential transcription levels. The present report defines for a considerable number of Mamu-A and Mamu-B configurations the dominant cDNAs characterized by the highest expression levels. Knowledge of the exact genetic order and gene content of a substantial number of Mamu-B region configurations is an absolute prerequisite to define a sensible nomenclature system.
Table 4. MHC class I transcription level analysis of B11 serotyped cells.
| Animal | Serotype | B*12 | B*3001 | B*38 | B*22 | B*53 | B*49 | B*57 | B*70 | B*26 |
|---|---|---|---|---|---|---|---|---|---|---|
| 3C | 11, 11 | ++ | ++ | — | + | +/- | +/- | — | +/- | — |
| MR | 34, 11 | ++ | ++ | — | + | +/- | +/- | +/- | +/- | |
| 96084 | 11, 11 | ++ | ++ | ++ | + | +/- | +/- | +/- | +/- | — |
The plus and minus signs are an indication of the level of expression. Major cDNAs are indicated by ++ (more expressed) or + (less expressed), and the minors are indicated by +/-. —, Indicates the absence of cDNA.
Duplications and Crossing-Over Events. The present results strongly suggest that diversity within the Mamu-B region is maintained by duplication and reshuffling of Mamu-B loci. This model is in sharp contrast to the situation in humans, where only one HLA-B gene is present per haplotype, and this locus displays a high degree of allelic polymorphism. Allelic polymorphism is virtually absent for the Mamu-B region. The differential gene number observed for the Mamu-A and Mamu-B regions is probably maintained by unequal crossing-over events. In this respect, the MHC class I region may resemble the human KIR loci (2, 3) and the HLA-DR and Mamu-DR regions (35). Before the unequal crossing over, several rounds of duplications must have expanded the number of class I genes in macaques. Such an expanded MHC class I gene repertoire is not unique and has been reported for some rodents (36), cattle (37), birds (38), and macaques (5, 11, 39). This study, however, illustrates that within a given population, individuals may display huge differences in the number of expressed class I genes and in the combinations of those genes as they segregate on a chromosome.
Differential Strategies to Cope with Parasites: Polymorphism Versus Diversity. The HLA-A, HLA-B, and HLA-C genes display a high degree of polymorphism in the human population. Because of this allelic polymorphism, individual variation is thought to reduce the chance that one pathogen can sweep through the entire population. Rhesus macaque populations seem to have banked on an alternative strategy, because allelic polymorphism appears to be virtually absent for the MHC class I genes. In contrast to humans, who normally have a fixed set of classical HLA genes, rhesus macaque populations are characterized by the presence of an abundance of Mamu-A and Mamu-B region configurations displaying polymorphism with regard to the number and combination of expressed loci present per chromosome. Diversity in gene number, but also gene combinations, is a bona fide strategy to ensure that different individuals mount distinct responses against the same pathogen. Moreover, the Mamu-A and Mamu-B region configurations themselves seem to be subject to frequent crossing-over processes, even further maximizing and maintaining diversity in the population. Additional layers of polymorphism may be added by differential expression levels.
Acknowledgments
We thank Henk van Westbroek for preparation of the figures and Ms. Donna Devine for editing the manuscript. This work was supported in part by National Institutes of Health Grant 1R24 RR16038-01.
Data deposition: The sequences reported in this paper have been deposited in the EMBL database (accession nos. AJ542567–AJ542580, AJ551315–AJ551321, AJ556875–AJ556908, AJ620415, AJ620416, AJ844596–AJ844602, and AJ849330).
References
- 1.Parham, P. & Ohta, T. (1996) Science 272, 67–74. [DOI] [PubMed] [Google Scholar]
- 2.Vilches, C. & Parham, P. (2002) Annu. Rev. Immunol. 20, 217–251. [DOI] [PubMed] [Google Scholar]
- 3.Martin, M. P., Bashirova, A., Traherne, J., Trowsdale, J. & Carrington, M. (2003) J. Immunol. 172, 2192–2195. [DOI] [PubMed] [Google Scholar]
- 4.Robinson, J., Waller, M. J., Parham, P., de Groot, N., Bontrop, R., Kennedy, L. J., Stoehr, P. & Marsh, S. G. (2003) Nucleic Acids Res. 31, 311–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ishitani, A., Sageshima, N., Lee, N., Dorofeeva, N., Hatake, K., Marquardt, H. & Geraghty, D. E. (2003) J. Immunol. 171, 1376–1384. [DOI] [PubMed] [Google Scholar]
- 6.Shiroishi, M., Tsumoto, K., Amano, K., Shirakihara, Y., Colonna, M., Braud, V. M., Allan, D. S., Makadzange, A., Rowland-Jones, S., Willcox, B., et al. (2003) Proc. Natl. Acad. Sci. USA 100, 8856–8860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barouch, D. H., Santra, J. E., Schmnitz, J. E., Kuroda, M. J., Fu, T. M., Wagner, W., Bilska, M., Craiu, A., Zheng, X. X., Krivulka, G. R., et al. (2000) Science 290, 486–492. [DOI] [PubMed] [Google Scholar]
- 8.Evans, D. T., O'Connor, D. H., Jing, P., Dzuris, J. L., Sidney, J., da Silva, J., Allen, T. M., Horton, H., Venham, J. E., Rudersdorf, R. A., et al. (1999) Nat. Med. (London) 5, 1270–1276. [DOI] [PubMed] [Google Scholar]
- 9.Schmitz, J. E., Kuroda, M. J., Santra, S., Sasseville, V. G., Simon, M. A., Lifton, M. A., Racz, P., Tenner-Racz, K., Dalesandro, M., Scallon, B. J., et al. (1999) Science 283, 857–860. [DOI] [PubMed] [Google Scholar]
- 10.Miller, M. D., Yamamoto, H., Hughes, A. L., Watkins, D. I. & Letvin, N. L. (1991) J. Immunol. 147, 320–329. [PubMed] [Google Scholar]
- 11.Boyson, J. E., Shufflebotham, C., Cadavid, L. F., Urvater, A. J., Knapp, L.A., Hughes, A. L. & Watkins, D. I. (1996) J. Immunol. 156, 4656–4665. [PubMed] [Google Scholar]
- 12.Vogel, T. U., Evans, D. T., Urvater, J. A., O'Connor, D. H., Hughes, A. L. & Watkins, D. I. (1999) Immunol. Rev. 167, 327–337. [DOI] [PubMed] [Google Scholar]
- 13.Boyson, J. E., McAdam, S. N., Gallimore, A., Golos, T. G., Liu, X., Gotch, F. M., Hughes, A. L. & Watkins, D. I. (1995) Immunogenetics 41, 59–68. [DOI] [PubMed] [Google Scholar]
- 14.Otting, N. & Bontrop, R. E. (1993) Immunogenetics 38, 141–145. [DOI] [PubMed] [Google Scholar]
- 15.Boyson, J. E., Iwanaga, K. K., Golos, T. G. & Watkins, D. I. (1996) J. Immunol. 157, 5428–5437. [PubMed] [Google Scholar]
- 16.Boyson, J. E., Iwanaga, K. K., Golos, T. G. & Watkins, D. I. (1997) J. Immunol. 159, 3311–3321. [PubMed] [Google Scholar]
- 17.Ryan, A. F., Grendell, R. L., Geraghty, D. E. & Golos, T. G. (2002) J. Immunol. 169, 673–683. [DOI] [PubMed] [Google Scholar]
- 18.Urvater, J. A., Otting, N., Loehrke, J. H., Rudersdorf, R., Piekarczyk, M. S., Golos, T. G., Hughes, A. L., Bontrop, R. E. & Watkins, D. I. (2000) J. Immunol. 164, 1386–1398. [DOI] [PubMed] [Google Scholar]
- 19.Geraghty, D. E., Daza, R., Williams, L. M., Vu, Q. & Ishitani, A. (2002) Immunol. Rev. 190, 69–85. [DOI] [PubMed] [Google Scholar]
- 20.Bontrop, R. E., Otting, N., de Groot, N. G. & Doxiadis, G. G. (1999) Immunol. Rev. 167, 339–350. [DOI] [PubMed] [Google Scholar]
- 21.Klein, J., Bontrop, R. E., Dawkins, R. L., Erlich, H. A., Gyllensten, U. B., Heise, E. R., Jones, P. P., Parham, P., Wakeland, E. K. & Watkins, D. I. (1990) Immunogenetics 31, 217–219. [DOI] [PubMed] [Google Scholar]
- 22.de Groot, N. G., Otting, N., Doxiadis, G. G., Balla-Jhagjhoorsingh, S. S., Heeney, J. L., van Rood, J. J., Gagneux, P. & Bontrop, R. E. (2002) Proc. Natl. Acad. Sci. USA 99, 11748–11753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yasutomi, Y., McAdam, S. N., Boyson, J. E., Piekarczyk, M. S., Watkins, D. I. & Letvin, N. L. (1995) J. Immunol. 154, 2516–2522. [PubMed] [Google Scholar]
- 24.Voss, G. & Letvin, N. L. (1996) J. Virol. 70, 7335–7340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Allen, T. M., Sidney, J., del Guercio, M. F., Glickman, R. L., Lensmeyer, G. L., Wiebe, D. A., DeMars, R., Pauza, C. D., Johnson, R. P., Sette, A., et al. (1998) J. Immunol. 160, 6062–6071. [PubMed] [Google Scholar]
- 26.O'Connor, D. H., Mothe, B. R., Weinfurter, J. T., Fuenger, S., Rehrauer, W. M., Jing, P., Rudersdorf, R. R., Liebl, M. E., Krebs, K., Vasquez, P., et al. (2003) J. Virol. 77, 9029–9040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Newberg, M. H., Kuroda, M. J., Charini, W. A., Miura, A., Lord, C. I., Schmitz, J. E., Gorgone, D. A., Lifton, M. A., Kuus-Reichel, K. & Letvin, N. L. (2002) Virology 301, 365–373. [DOI] [PubMed] [Google Scholar]
- 28.Vogel, T. U., Friedrich, T. C., O'Connor, D. H., Rehrauer, W., Dodds, E. J., Hickman, H., Hildebrand, W., Sidney, J., Sette, A., Hughes, A. L., et al. (2002) J. Virol. 76, 11623–11636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Urvater, J. A., Steffen, S. R., Rehrauer, W. & Watkins, D. I. (2000) Tissue Antigens 55, 153–156. [DOI] [PubMed] [Google Scholar]
- 30.Adams, E. J., Cooper, S. & Parham, P. (2001) J. Immunol. 167, 3858–3869. [DOI] [PubMed] [Google Scholar]
- 31.Urvater, J. A., McAdam, S. N., Loehrke, J. H., Allen, T. M., Moran, J. L., Rowell, T. J., Rojo, S., Lopez de Castro, J. A., Taurog, J. D. & Watkins, D. I. (2000) Immunogenetics 51, 314–325. [DOI] [PubMed] [Google Scholar]
- 32.Daza-Vamenta, R., Glusman, G., Rowen, L., Guthrie, B. & Geraghthy, D. E. (2004) Genome Res. 14, 1501–1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miller, M. D., Lord, C. I., Stallard, V., Mazzara, G. P. & Letvin, N. L. (1990) J. Immunol. 144, 122–128. [PubMed] [Google Scholar]
- 34.Evans, D. T., Jing, P., Allen, T. M., O'Connor, D. H., Horton, H., Venham, J. E., Piekarczyk, M., Dzuris, J., Dykhuzen, M., Mitchen, J., et al. (2000) J. Virol. 74, 7400–7410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Doxiadis, G. G., Otting, N., de Groot, N. G., Noort, R. & Bontrop, R. E. (2001) J. Immunol. 164, 3193–3199. [DOI] [PubMed] [Google Scholar]
- 36.Delarbre, C., Jaulin, C., Kourilski, P. & Gachelin, G. (1992) Immunogenetics 37, 29–38. [DOI] [PubMed] [Google Scholar]
- 37.Holmes, E. C., Roberts, A. F. C., Staines, K. A. & Ellis, S. A. (2003) Immunogenetics 55, 193–202. [DOI] [PubMed] [Google Scholar]
- 38.Shiina, T., Shimizu, S., Hosomichi, K., Kohara, S., Watanabe, S., Hanzawa, K., Bech, S., Kulski, J. Z. & Inoko, H. (2004) J. Immunol. 172, 6751–6763. [DOI] [PubMed] [Google Scholar]
- 39.Uda, A., Tanabayashi, K., Yamada, Y. K., Akari, H., Lee, Y. Mukai, R., Terao, K. & Yamada, A. (2004) Immunogenetics 56, 155–163. [DOI] [PubMed] [Google Scholar]