Abstract
To ensure efficient genome duplication, cells have evolved numerous factors that promote unperturbed DNA replication, and protect, repair and restart damaged forks.
Here we identify DONSON as a novel fork protection factor, and report biallelic DONSON mutations in 29 individuals with microcephalic dwarfism. We demonstrate that DONSON is a replisome component that stabilises forks during genome replication. Loss of DONSON leads to severe replication-associated DNA damage arising from nucleolytic cleavage of stalled replication forks. Furthermore, ATR-dependent signalling in response to replication stress is impaired in DONSON-deficient cells, resulting in decreased checkpoint activity, and potentiating chromosomal instability. Hypomorphic mutations substantially reduce DONSON protein levels and impair fork stability in patient cells, consistent with defective DNA replication underlying the disease phenotype.
In summary, we identify mutations in DONSON as a common cause of microcephalic dwarfism, and establish DONSON as a critical replication fork protein required for mammalian DNA replication and genome stability.
Microcephalic primordial dwarfism (MPD) is the collective term for a group of human disorders characterised by intra-uterine and postnatal growth delay alongside marked microcephaly1, and includes disorders such as MOPD II, ATR/ATRIP-Seckel syndrome and Meier-Gorlin syndrome. Mutations in genes encoding either components of the DNA replication machinery (replisome) or genome stability proteins are a frequent cause of microcephalic dwarfism2–14.
During the course of normal DNA replication, a subset of replication forks may stall, causing ‘replication stress’15. This stalling can be caused by endogenous or exogenous sources, such as collision of the replisome with DNA lesions or the transcriptional machinery, or replication of difficult to replicate genomic regions. To facilitate efficient genome duplication, stalled replication forks must be stabilised and protected from collapse. Multiple factors safeguard replication fork stability, many of which function within the ATR-CHK1-dependent replication stress response16–18. This pathway ensures that fork stabilisation is tightly coordinated with a global reduction in DNA synthesis, allowing stalled or damaged forks to be repaired and restarted19,20.
Exome sequencing analysis of microcephalic dwarfism patients has identified several novel factors that regulate replication and/or the replication stress response. Using this strategy, we recently identified mutations in TRAIP in individuals with MPD5, and demonstrated that TRAIP is required for the response to replication-blocking DNA lesions. To identify similar disease-associated genes, we carried out whole exome sequencing of genetically uncharacterised patients with microcephaly. Here, we report the identification of DONSON as a new microcephalic dwarfism gene, and demonstrate that DONSON is a novel replisome component that maintains genome stability by protecting stalled/damaged replication forks.
Results
DONSON mutations identified in microcephalic dwarfism patients
Whole exome sequencing (WES) was undertaken on 26 patients with microcephaly and reduced stature. After aligning WES reads to the reference genome, variant calling, and filtering for rare variants (MAF <0.005), analysis under a recessive model of inheritance identified rare biallelic variants in the DONSON (‘Downstream neighbour of SON’) gene in nine affected individuals from seven families. Sanger capillary sequencing confirmed the presence of these mutations in these patients (P1-1 to P3, P6, and P9 to P11, Table 1). Subsequent re-sequencing of an additional 230 patients with primary microcephaly, microcephaly with reduced stature, or MPD, identified five additional families with compound heterozygous mutations in DONSON (P4, P5, P7, P8, P12; Table 1). All variants segregated amongst family members in a manner consistent with an autosomal recessive trait, and were present at a frequency of <0.5% in the ExAC database21.
Table 1.
Biallelic DONSON mutations identified in 29 individuals
| Patient | Country of Origin | Mutation 1 | Mutation 2 | Segregation |
|---|---|---|---|---|
| P1-1 | Italy | c.1251_1256delCTCTAA, p.Asn417_Ser418del | haplotype | Het, M |
| P1-2 | Italy | c.1251_1256delCTCTAA, p.Asn417_Ser418del | haplotype | Het, M |
| P2 | UK | c.877C>T, p.Arg293* | haplotype | Het, M, P |
| P3 | UK | c.1254dupT, p.Lys419* | haplotype | Het, M, P |
| P4 | UK | c.1686dupT, p.Asn563* | haplotype | Het, nps |
| P5 | Somalia | c.832T>C, p.Cys278Arg AND/OR c.845A>G, p.Tyr282Cys | haplotype | Het, M, P |
| P6 | USA | c.1282C>T, p.Gln428* | haplotype | Het, M, P |
| P7 | USA | c.1282C>T, p.Gln428* | haplotype | Het, nps |
| P8 | Italy | c.1474_1475delCA, p.Gln492Glufs*18 | c.786-7T>C | Het, M, P |
| P9 | Turkey | c.876C>G, p.Phe292Leu | c.1047-9A>G (SS) | Het, M |
| P10-1 | India | c.1628_1630dupAAA, p.Gln543_Ile544insLys | c.1032C>T, p.Ser344Ser (SS) | Het, M, P |
| P10-2 | India | c.1628_1630dupAAA, p.Gln543_Ile544insLys | c.1032C>T, p.Ser344Ser (SS) | Het, M, P |
| P11 | Saudi Arabia | c.786-22A>G (SS) | c.786-22A>G (SS) | Hom, M, P |
| P12 | Saudi Arabia | c.786-22A>G (SS) | c.786-22A>G (SS) | Hom, nps |
| P13-1 | Palestine | c.1337T>C, p.Met446Thr | c.1337T>C, p.Met446Thr | Hom, M, P |
| P13-2 | Palestine | c.1337T>C, p.Met446Thr | c.1337T>C, p.Met446Thr | Hom, M, P |
| P13-3 | Palestine | c.1337T>C, p.Met446Thr | c.1337T>C, p.Met446Thr | Hom, M, P |
| P14 | Saudi Arabia | c.786-22A>G (SS) | c.786-22A>G (SS) | Hom, M, P |
| P15 | Saudi Arabia | c.786-22A>G (SS) | c.786-22A>G (SS) | Hom, M, P |
| P16 | Saudi Arabia | c.786-22A>G (SS) | c.786-22A>G (SS) | Hom, M, P |
| P17 | Saudi Arabia | c.786-22A>G (SS) | c.786-22A>G (SS) | Hom, M, P |
| P18-1 | Saudi Arabia | c.786-22A>G (SS) | c.786-22A>G (SS) | Hom, M, P |
| P18-2 | Saudi Arabia | c.786-22A>G (SS) | c.786-22A>G (SS) | Hom, M, P |
| P18-3 | Saudi Arabia | c.786-22A>G (SS) | c.786-22A>G (SS) | Hom, M, P |
| P19 | Turkey | c.1297C>T, p.Pro433Ser | c.1297C>T, p.Pro433Ser | Hom, M, P |
| P20-1 | South Africa | c.1254dupT, p.Lys419* | c.1510G>A, p.Glu504Lys | Het, M, P |
| P20-2 | South Africa | c.1254dupT, p.Lys419* | c.1510G>A, p.Glu504Lys | Het, M, P |
| P21-1 | Saudi Arabia | c.1047-9A>G (SS) | c.1047-9A>G (SS) | Hom, M, P |
| P21-2 | Saudi Arabia | c.1047-9A>G (SS) | c.1047-9A>G (SS) | Hom, M, P |
‘Haplotype’ indicates the presence of three co-segregating variants: c.82A>C (p.Ser28Arg); c.786-33A>G; c.1466A>C (p.Lys489Thr). Hom, homozygous in affected individual; Het, compound heterozygous in affected individual; M, mutation identified in mother; P, mutation identified in father; nps, no parental samples available; SS, Splice site mutation. Reference sequence, NM_017613.3.
Two other concurrent molecular genetic studies provided further independent evidence to support the identification of DONSON as a novel human disease gene. Firstly, exome sequencing was carried out on a consanguineous Palestinian family previously reported to have a Fanconi Anaemia-like disorder22. These patients presented with microcephaly, short stature, slow growth and forearm and thumb dysplasia, although no individuals had haematological evidence of bone marrow failure. This WES analysis revealed a deleterious homozygous transition, c.1337T>C, resulting in substitution of a highly conserved residue (p.M446T) in all three affected individuals (P13-1, P13-2, P13-3; Table 1, Supplementary Fig. 1). Secondly, a study of five consanguineous families in Saudi Arabia with extreme microcephaly and short stature allowed a 1.6 Mb haplotype shared by all five families (combined multipoint LOD score Z= 8.0) to be mapped to a defined critical interval on chromosome 21 that contained DONSON. Whole exome and genome sequencing identified a single rare variant at this locus in DONSON, c.786-22A>G. Capillary sequencing confirmed this intronic variant to be homozygous in all seven affected individuals from this study (P14 to P18-3; Table 1), identical to that detected in two Saudi Arabian individuals present within the first study described above (P11, P12).
Subsequently, a further five individuals from three different families with DONSON mutations were identified in additional MPD patients recruited to two of the genetic studies described above (P19 to P21-2; Table 1).
DONSON mutations give rise to severe microcephaly with short stature
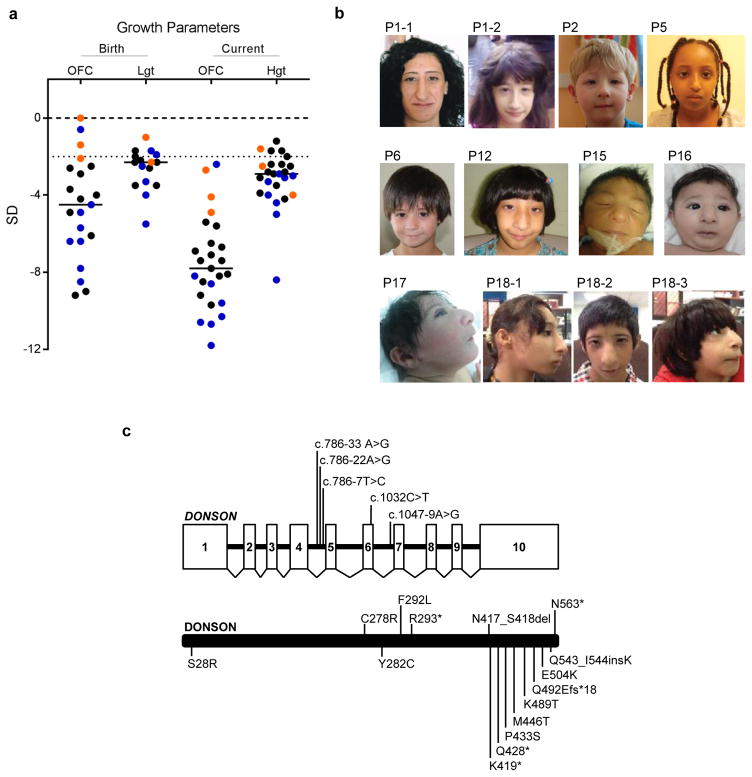
Despite their identification in separate studies, all patients with DONSON mutations had similar clinical phenotypes. Marked microcephaly was present (OFC −7.5 +/− 2.4 SD), with a substantial reduction in cerebral cortical size, along with decreased gyral folding evident on neuroimaging (Fig. 1a and Supplementary Fig. 2), similar to that previously reported for other primary microcephaly and microcephalic dwarfism patients5,23–25. Height was reduced (−3.2 +/− 1.4 SD), although much less so than head circumference (Fig. 1a), and to a lesser degree than observed in other microcephalic dwarfism-associated disorders (where height was typically ≤ −4 SD)2,3,5,8–10,24,26. Minor skeletal abnormalities were present in several patients, including fifth finger clinodactyly, syndactyly, brachydactyly, hypoplasia of carpal/metacarpal/phalangeal bones, or radial head dislocation (Supplementary Table 1). Absent/hypoplastic patellae were present in patients P12, P20-1 and P20-2. Notably, patient P19 had bilateral hypoplasia of the radius and thumb, which, together with the limb abnormalities displayed by P13-1 and 13-2, established radial ray defects as an uncommon but recurrent phenotype. In family P21, the most extreme phenotype was observed, with substantial limb shortening/reduction in association with foetal lethality (Supplementary Fig. 3). Aside from microcephaly, neither a recognisable facial phenotype (Fig. 1b) nor recurrent malformations affecting other organ systems were evident. Intellectual disability, if present, was typically mild.
Figure 1. DONSON mutations cause severe microcephaly and short stature.
(a) DONSON mutations result in severe prenatal-onset microcephaly, often associated with short stature. Length at birth (Lgt), current height (Hgt) and head circumference (OFC) plotted as z-scores (SD from population mean for age and sex). Black horizontal bars indicate mean values. Dashed line at −2 SD indicates cut-off for normal population distribution. Patients from the three independently identified DONSON patient cohorts are denoted by black (P1–P12 and P20), orange (P13), and blue (P14–19 and P21) circles. (b) Photographs of affected individuals with DONSON mutations demonstrating facial similarities. Written consent to publish photographs was obtained from the affected families. P, patient. (c) Schematics of the DONSON gene and protein indicating position of the identified mutations. DONSON mutations comprised a range of mutation classes (nonsense, frameshift, essential splice site, missense and intronic). The genomic structure is based on the longest ORF containing ten coding exons (white rectangles) (NM_017613.3). The positions of identified mutations affecting splicing are shown on the gene structure (top) and missense and truncating variants on the encoded protein (bottom).
In conclusion, the number of biallelic variants identified, combined with a common clinical presentation, provided strong evidence for DONSON being a novel human disease gene, associated with autosomal recessive inheritance. We therefore investigated the consequence of these mutations on DONSON protein function.
DONSON mutations markedly reduce protein levels
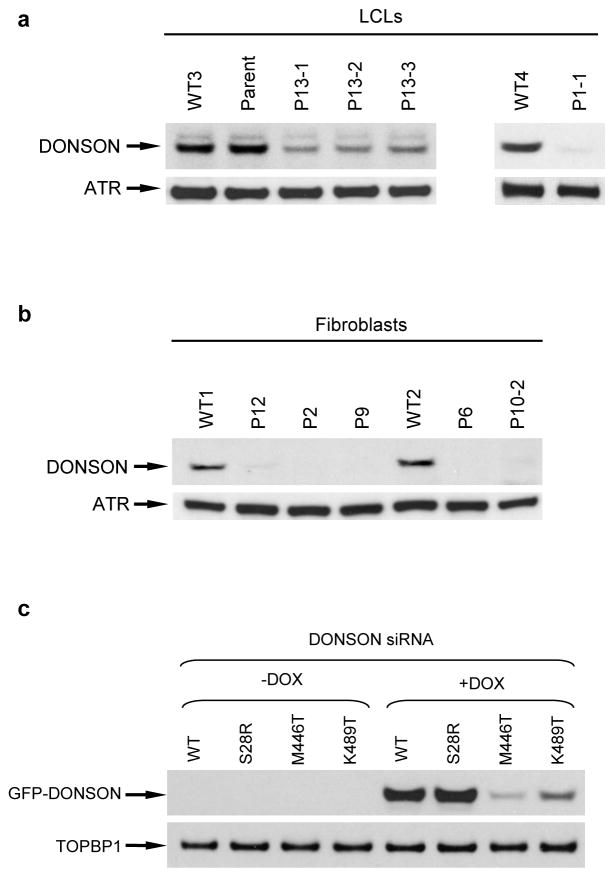
DONSON mutations were identified in 29 individuals, and comprised a range of mutation classes (Fig. 1c). Notably, no biallelic nonsense or frameshift mutations were observed, indicating that mutations likely resulted in partial loss of DONSON function. To investigate this, we established patient-derived primary fibroblast or lymphoblastoid cell lines representing a range of mutations. Immunoblotting demonstrated marked decreases in DONSON protein levels in all cell lines tested (Fig. 2a, b), establishing that the analysed mutations affected DONSON protein expression.
Figure 2. Mutations in DONSON affect protein levels.
(a–b) DONSON mutations result in severely reduced levels of DONSON protein. Immunoblotting of cell extracts from lymphoblastoid (a) and fibroblast (b) cell lines derived from patients with mutations in DONSON. ATR was used as a loading control. The two blots from (a) originate from two independent gels. (c) The K489T, but not the S28R variant, associated with the DONSON haplotype affects protein levels. Cells were treated with doxycycline 48 h post siRNA transfection, and harvested for Western blot analysis 24 h later (n=2). Exogenous DONSON were detected using an anti-GFP antibody respectively. TOPBP1 was used as a loading control. Depletion of endogenous DONSON in these cells was confirmed by immunoblotting (Supplementary Fig. 2).
Mutations in multiple families were associated with two different ancestral haplotypes (P1-1 to P7; P11, P12 and P14 to 18-3 respectively; Table 1) and were investigated in more detail. Firstly, as described above, nine individuals (P11, P12 and P14 to P18-3) were homozygous for the c.786-22A>G mutation, predicted to enhance a cryptic splice donor site within intron 4 (MaxEntScan)27. Consistent with a common ancestral founder, five consanguineous families of Saudi Arabian origin bearing this mutation (P14 to P18-3) shared a 1.6 Mb haplotype region of chromosome 21 (Supplementary Fig. 4). qRT-PCR analysis of RNA isolated from four patients with this variant demonstrated a significant decrease in full-length transcript, and increased skipping of exon 5 (Supplementary Fig. 5). This resulted in an out-of-frame mRNA predicted to undergo NMD, explaining the substantial reduction in protein levels seen in fibroblasts homozygous for this mutation (P12; Fig 2b).
Secondly, two missense variants in DONSON, p.S28R and p.K489T, and an intronic variant (c.786-33A>G) were present in seven individuals of European ancestry and one Somali (P1-1 to P7; Table 1). These were associated with a different ancestral haplotype, comprising a shared 127.7 kb genomic region (Supplementary Table 2). No other deleterious biallelic variants were present in the four other genes within this region. Despite the close proximity of the c.786-33A>G intronic variant to the Saudi Arabian c.786-22A>G mutation, the former change did not disrupt splicing between exons 4 and 5, as assessed by either mini gene splicing assays (Supplementary Fig. 6), or RT-PCR analyses of patient cell lines (data not shown), indicating this variant is unlikely to be pathogenic. Since DONSON protein levels were severely reduced in cells from patients inheriting this haplotype allele in combination with a truncating mutation (P2, P6), this suggested that either one or both of the missense variants associated with this haplotype (p.S28R, p.K489T) compromised protein levels (Fig. 2b).
To investigate these two variants, we established isogenic HeLa cell lines expressing doxycycline-inducible, siRNA-resistant wild type (WT) or mutant (p.S28R, p.K489T) GFP-tagged DONSON. Following siRNA depletion of endogenous DONSON (Supplementary Fig. 7), and induction of exogenous GFP-DONSON, immunoblotting revealed that the p.K489T mutation, but not p.S28R, reduced protein levels in a post-transcriptional manner (Fig. 2c and Supplementary Fig. 8). This suggested that the p.K489T substitution within the second haplotype causes the decreased DONSON protein levels observed in P2 and P6 (Fig. 2b). The K489T variant is present as a rare allele in the population, observed at a frequency of 0.00099 in the ExAC database21. In the patients reported here, it is always observed in trans with a frameshift or other protein-disrupting allele, suggesting that it is a functionally weak variant insufficient to cause disease alone, a conclusion supported by the presence of a single homozygous individual in ExAC.
Eight other point mutations were identified in patients: six missense substitutions, a two amino-acid deletion and an amino-acid insertion (Table 1 and Fig. 1c). Notably these were at highly conserved residues (Supplementary Fig. 1) and predicted to be deleterious (Alamut Visual). In agreement, exogenous expression of the p.M446T mutant resulted in significantly reduced protein levels (Fig. 2c), similar to cells derived from patients P13-1, P13-2 and P13-3. Furthermore, five of these mutations disrupted the subcellular localisation of GFP-DONSON (Supplementary Fig. 9), suggesting that these alterations also compromise DONSON protein function via protein mis-localisation.
Finally, an intronic mutation, c.1047-9A>G, was present in three individuals (P9, P21-1 and P21-2). qRT-PCR analysis of RNA isolated from the two patients homozygous for this variant (P21-1, P21-2) revealed a substantial reduction in DONSON transcript levels compared to normal controls (Supplementary Fig. 10). This variant was also observed in trans with the missense mutation F292L in patient P9. Since cells derived from this individual exhibited severely reduced levels of DONSON protein (Fig. 2b), it is likely that this intronic change also perturbs DONSON protein expression.
Taken together, the deleterious consequences of the identified mutations on splicing, transcript abundance, subcellular localisation and/or protein levels strengthened our conclusion that DONSON was a novel human disease gene. The fact that knockout of murine Donson leads to developmental lethality28, together with the presence of residual DONSON protein in patient-derived cell lines (Fig. 2a, b and Supplementary Fig. 11), supports the notion that the identified mutations are hypomorphic, retaining some residual function.
DONSON stabilises replication forks during normal DNA replication
While DONSON is highly conserved in metazoa and plants, its precise function(s) remained to be defined. Humpty-dumpty (hd), the Drosophila ortholog of DONSON, has been proposed to play a role in cell proliferation: Hd expression peaks during S-phase; hd mutants have an ‘egg shell’ phenotype; and clonal inactivation of hd impairs genome replication in larval tissues29.
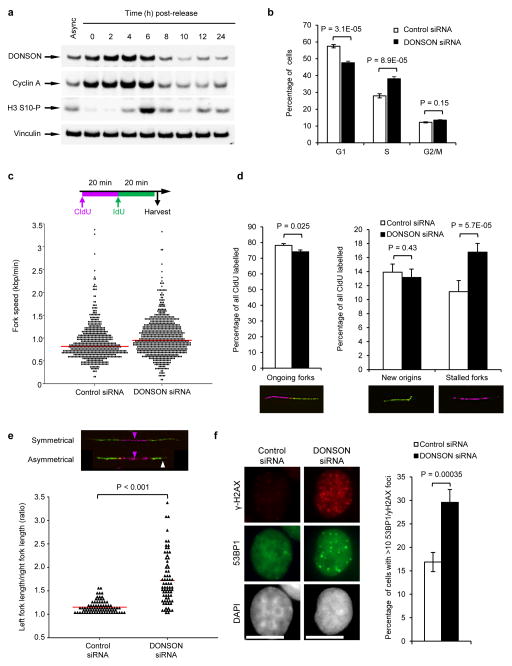
In light of this, we investigated whether human DONSON might play a similar role. After synchronising cells with a double thymidine block, we observed that human DONSON, like Hd, was also maximally expressed during S-phase, mirroring Cyclin A expression (Fig. 3a). Furthermore, depletion of DONSON resulted in a significant increase in BrdU-positive cells observed by FACS, consistent with a role in promoting efficient S-phase progression (Fig. 3b). Given these data, we next used DNA fibre analysis to assess whether DONSON depletion led to decreased DNA replication fork progression. Although fork progression rates did not decrease in cells lacking DONSON (Fig. 3c), compromising DONSON expression increased spontaneous replication fork stalling, with a concomitant decrease in the number of ongoing forks (Fig. 3d). Moreover, we also observed increased replication fork asymmetry in cells depleted of DONSON, indicating replication fork instability (Fig. 3e). Together, this suggests that the increase in BrdU-positive DONSON-depleted cells may reflect prolonged S-phase due to stalled replication forks, rather than a global reduction in DNA synthesis.
Figure 3. Loss of DONSON results in spontaneous replication fork stalling and increased genome instability.
(a) DONSON protein levels are increased during S-phase. HeLa cells were synchronised in S-phase using a double thymidine block, released, harvested at the indicated time points, and immunoblotting was performed (n=2). Cyclin A and phospho-histone H3 Ser-10 are markers of S/G2 and M phase respectively. Vinculin represents a loading control. (b) S-phase is prolonged upon DONSON depletion. HeLa cells transfected with the indicated siRNAs were pulsed with BrdU, fixed and analysed by FACS (n=4; error bars indicate SD). (c–e) Replication fork analysis of HeLa cells transfected with control or DONSON siRNA and pulsed with CldU and IdU. (c) Top: Schematic of DNA fibre analysis. Bottom: loss of DONSON does not decrease replication fork velocity. Replication fork speed (kb/min) was determined (n=5). (d) DONSON depletion results in spontaneous fork stalling. Percentages of ongoing replication forks, new origins and stalled replication forks in cells from (c) were quantified (n=3). (e) DONSON depletion leads to replication fork asymmetry. Top: example images; magenta arrows indicate origins of replication; white arrow denotes fork asymmetry. Bottom: plot indicates the ratio of left/right fork track lengths of bidirectional replication forks in cells from (c). Red lines denote median ratios (n=3). (f) Loss of DONSON increases spontaneous γH2AX/53BP1 foci formation. HeLa cells transfected with the indicated siRNAs were immunostained with antibodies to 53BP1 and γH2AX (left panel), and the percentage of cells with >10 53BP1 and γH2AX foci were quantified using fluorescence microscopy (right panel; n=5; >300 cells per sample per independent experiment). Scale bar; 10 μm.
Since DNA replication is closely linked with genome stability15,30,31, we reasoned that loss of DONSON would lead to a failure to complete timely replication and increased S-phase DNA damage. To test this hypothesis, we combined immunofluorescence of γ-H2AX and 53BP1 (markers of DNA damage and DNA double strand breaks respectively) with EdU labelling to identify S-phase cells. We observed that a significant proportion of DONSON-depleted cells exhibited spontaneous γ-H2AX and 53BP1 foci (Fig. 3f), of which the majority occurred in S-phase cells (Supplementary Fig. 12a–d), consistent with the identification of DONSON as a potential genome stability regulator by high-throughput siRNA screening32. Using pulsed-field gel electrophoresis, we also observed elevated levels of DNA double strand breaks in the absence of DONSON (Supplementary Fig. 12e). Taken together, these data support a role for DONSON in maintaining replication fork stability during unperturbed DNA replication, and demonstrate that spontaneous DNA damage arises in replicating cells in the absence of DONSON.
DONSON is a component of the replisome
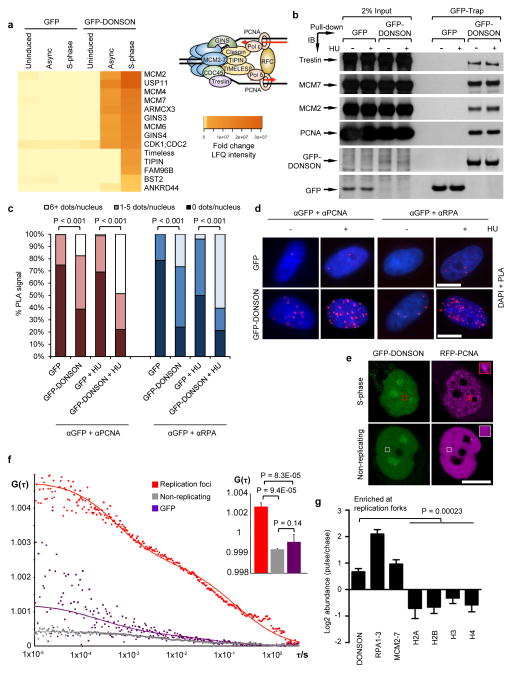
To shed further light on the role of DONSON in regulating replication fork stability, we carried out mass spectrometry screening to identify interaction partners of GFP-tagged DONSON. Amongst the interactors, we detected multiple replication proteins including subunits of the MCM helicase and the GINS complex (Fig. 4a and Supplementary Table 3). To confirm these findings, we carried out pull-down analyses coupled with immunoblotting to identify GFP-DONSON binding proteins. Consistent with our mass spectrometry data, we detected interactions between GFP-DONSON and the replisome components MCM2, MCM7, Treslin and PCNA (Fig. 4b), suggesting that DONSON associated with the replisome.
Figure 4. DONSON localizes to the replication fork.
(a–d) DONSON interacts with multiple components of the replication machinery. (a) GFP or GFP-DONSON was precipitated by GFP-Trap, from asynchronous cells or cells accumulated in S-phase with 2 mM HU treatment for 24 h. Heatmap denotes significant interactions identified by mass spectrometry (n=3). Inset: Schematic of the mammalian replisome with selected replication factors. (b) 293FT cells were transfected with the indicated expression vectors in the presence/absence of HU. GFP or GFP-DONSON were isolated by GFP-Trap and co-precipitating proteins visualised by immunoblotting (n=2). Benzonase Nuclease was included to exclude DNA-mediated interactions. The bottom two panels are scanned images of Ponceau S-stained nitrocellulose membrane. (c–d) DONSON localises in close proximity to replication forks. (c–d) PLA was carried out on cells from (a) using the indicated antibodies in the presence/absence of HU (n=2). (c) Quantification of PLA signals. (d) Representative PLA images. (e–f) DONSON interacts with PCNA at replication foci in live cells. (e) Representative confocal images of live cells expressing GFP-DONSON and RFP-PCNA. Boxes indicate representative regions used for FCCS analysis. (f) FCCS measurements of GFP-DONSON and RFP-PCNA reveal significant cross-correlation at replication foci at similar concentrations. Average cross-correlation curves are shown from cells expressing GFP-DONSON in replication foci (red) or non-replicating (grey) cells, or GFP-expressing S-phase nuclei (purple). Inset: Mean cross-correlation amplitude values from multiple cells (error bars indicate SD; n=4, 3 and 5). Increased G(τ) values indicate higher degree of cross-correlation between GFP-DONSON and RFP-PCNA in replication foci. See also Supplementary Fig. 11. (g) iPOND was performed on 293T (n=3), HeLa (n=2) and HCT116 (n=2) cells, and EdU-coprecipitates analysed by mass spectrometry. Data represents the combination of all seven experiments. Log2 abundance denotes the ratio of proteins at nascent DNA compared to mature chromatin. Values >0 represent proteins enriched at forks, whilst values ≤ 0 denote chromatin-bound factors. Scale bars; 10 μm.
We next used three complementary techniques to assess whether DONSON localised to sites of DNA replication. Firstly, we carried out proximity ligation assays (PLA) of GFP-DONSON with the replication proteins PCNA and RPA. In line with DONSON being closely associated with the replication machinery, we observed robust PLA signals between GFP-DONSON and both PCNA and RPA (Fig. 4c, d). We next performed Fluorescence Cross-Correlation Spectroscopy (FCCS)33,34 in live HeLa cells stably co-expressing RFP-PCNA and GFP-DONSON, to measure the degree of co-diffusion of these molecules. Significantly increased co-diffusion of PCNA and DONSON was observed in S-phase PCNA foci, but not in nuclei of non-replicating cells (Fig. 4e–f and Supplementary Fig. 13), indicating that these proteins interacted during DNA replication. Finally, we utilised iPOND (isolation of proteins on nascent DNA)35 combined with mass spectrometry to ascertain whether DONSON is present on newly replicated DNA. Crucially, this approach demonstrated that DONSON, like MCMs and RPA, was significantly enriched at replication forks compared to mature chromatin (Fig. 4g).
Collectively, these data strongly support the conclusion that DONSON is a novel replisome component that plays a role in promoting fork stability.
DONSON depletion impairs cell-cycle checkpoint activation
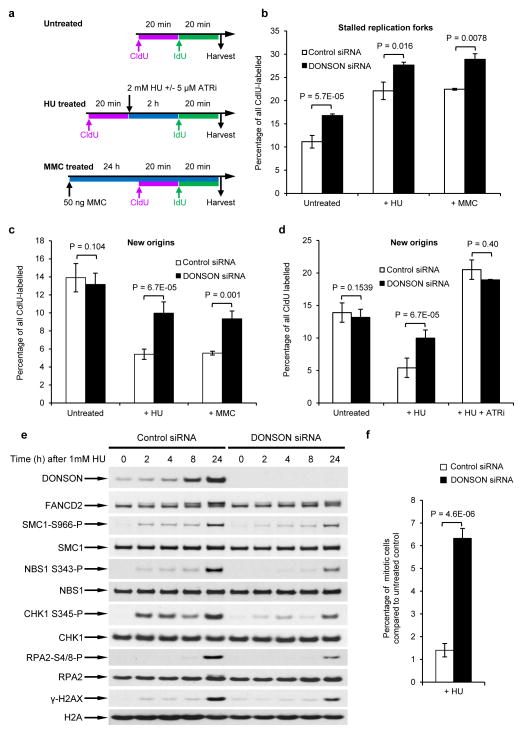
Since our data suggested that DONSON functions to protect replication forks during unperturbed DNA replication, we extended our findings to evaluate the role of DONSON in preventing replication fork stalling following exogenous replication stress. Exposure to the replication stress-inducing agents hydroxyurea (HU) and mitomycin C (MMC) induced significantly more fork stalling in DONSON-depleted cells than in control cells (Fig. 5a–b). DONSON depletion also resulted in a failure to suppress new origin firing upon exogenous replication stress (Fig. 5c). Since suppression of new origin firing reflects checkpoint activity, this suggests that DONSON is required for efficient activation of the intra-S phase checkpoint. To further investigate this, we measured activation of this checkpoint after inhibition of ATR (VE821; ATRi), the apical kinase which governs the replication stress response18. Upon HU exposure and ATR inhibition, we observed no difference in the number of new origins fired between control or DONSON-depleted cells, indicating that DONSON and ATR may function within the same pathway to activate the intra-S phase checkpoint (Fig. 5d).
Figure 5. Depletion of DONSON compromises activation of cell cycle checkpoints.
(a–c) Loss of DONSON results in replication fork instability that is exacerbated by replication stress. (a) HeLa cells transfected with either control or DONSON siRNA were pulsed with CldU, exposed to 2 mM HU for 2 h, and then pulsed with IdU. Alternatively, cells were exposed to 50 ng/ml MMC for 24 h, and pulsed with sequential pulses of CldU and IdU (see schematic). DNA fibres were quantified, and the percentage of (b) stalled forks and (c) new origins are displayed (in all cases n=3). (d) Loss of DONSON is epistatic with ATR inhibition. Replication fork analysis of HeLa cells transfected with either control or DONSON siRNA. Cells were pulsed with CldU, exposed to 2 mM HU +/− 5μM ATR inhibitor for 2 h, and then pulsed with IdU (n=3). New origins (2nd label origin) were counted as an indicator of intra-S phase checkpoint activation. (e) Cells lacking DONSON exhibit defective or delayed ATR activation in response to replication stress. Whole cell extracts of HeLa cells transfected with either control or DONSON siRNA were subjected to immunoblot analysis using the indicated antibodies following treatment with 1 mM HU (n=2). (f) The percentage of mitotic cells following exposure to 1 mM HU for 24 h (from (e)) was determined by flow cytometry, using antibodies to phosphorylated histone H3-Ser10 (a marker of mitosis) (n=5).
We next examined whether the ATR-dependent replication stress response was functional in the absence of DONSON. We first monitored ATR pathway activation in DONSON-depleted cells treated with HU or MMC by immunoblotting, using phospho-specific antibodies to known ATR substrates. This analysis revealed that cells lacking DONSON failed to efficiently phosphorylate a number of ATR substrates, such as CHK1 and NBS1, in response to HU or MMC (Fig. 5e and Supplementary Fig. 14a). Moreover, ATR autophosphorylation on T1989, another marker of ATR activation36, was reduced (Supplementary Fig. 14b). Loss of DONSON also significantly increased mitotic indices following exposure to HU or MMC as measured by FACS, demonstrating that DONSON-depleted cells fail to efficiently activate the G2/M checkpoint in response to replication stress (Fig. 5f and Supplementary Fig. 14c). We next determined whether the reduced ATR signalling observed was due to decreased levels of RPA-coated ssDNA, which is the stimulus for ATR activation. Surprisingly, DONSON-depleted cells exhibited elevated levels of RPA-coated ssDNA following HU treatment (Supplementary Fig. 15), consistent with defective activation of the ATR-dependent replication stress response.
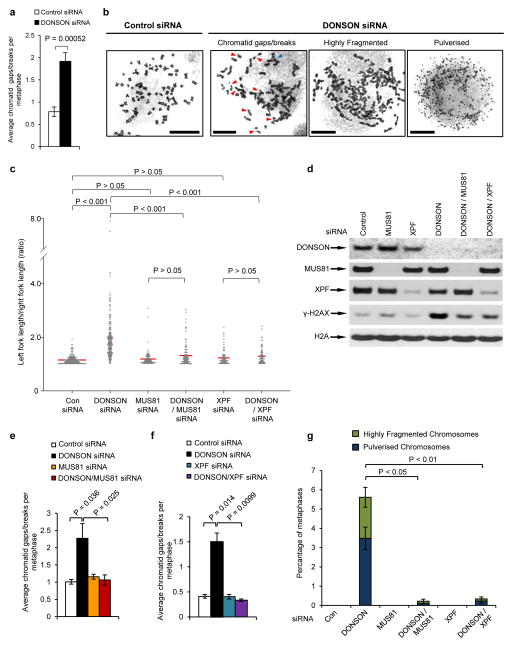
Dysregulated DNA replication combined with impaired intra-S phase checkpoint signalling, such as in ATR-deficient cells37–41, gives rise to extensive chromosome breakage and genome instability. Consistent with this, we observed significantly elevated levels of spontaneous micronuclei and chromatid gaps/breaks in cells lacking DONSON (Fig. 6a and Supplementary Fig. 16a), which was exacerbated by exposure to HU or MMC (Supplementary Fig. 16a–c). We also observed spontaneously-arising highly-fragmented or completely pulverised metaphase chromosomes in cells lacking DONSON, which increased upon exogenous replication stress (Fig. 6b and Supplementary Fig. 16d).
Figure 6. Increased spontaneous chromosome breakage and fragmentation of mitotic chromosomes in DONSON-depleted cells.
(a,b) Metaphases chromosomes from DONSON or control siRNA transfected HeLa cells were visualised by Giemsa staining and light microscopy. (a) Quantification of average numbers of chromatid gaps/breaks per metaphase (n=6; >50 metaphases per sample per experiment). (b) Representative images of normal chromosomes, chromosomes containing gaps/breaks, highly fragmented and pulverized chromosomes. Red arrows denote chromatid gaps/breaks; blue arrows indicate chromosomal exchanges. Scale bar; 10 μm. (c–g) Loss of the structure-specific nucleases MUS81 or XPF significantly reduces the spontaneous replication fork asymmetry and genome instability in DONSON-depleted cells. (c) Cells transfected with the indicated siRNAs were pulsed with CldU and IdU. Replication fork asymmetry was measured as in (Fig. 3e). The red lines denotes median ratios (n=3). (d) Co-depletion of MUS81 or XPF with DONSON reduces levels of spontaneous DNA damage. Extracts from cells transfected with the indicated siRNAs were subjected to SDS-PAGE and immunoblotting using the antibodies indicated. (e–f) Co-depletion of MUS81 (e) or XPF (f) reduces chromosomal aberrations in cells lacking DONSON. Quantification of the average number of chromatid gaps/breaks per metaphase in cells transfected with control, DONSON, MUS81 and/or XPF siRNA. At least 50 metaphases per experiment were counted (n=3). (g) Quantification of the average percentage of metaphases containing highly fragmented chromosomes or pulverized chromosomes in cells transfected with the indicated siRNAs. At least 50 metaphases per experiment were counted (n=3).
Together, these data confirm that upon exogenous replication stress, DONSON is required to stabilise stalled replication forks, efficiently activate the intra-S and G/2M cell-cycle checkpoints, and maintain genome stability.
Cleavage of stalled replication forks in DONSON-deficient cells
It has been proposed that the spontaneous DNA damage arising in ATR-deficient cells is due to processing of stalled/damaged forks by SLX4-associated structure-specific nucleases, such as MUS81, SLX1 and XPF42–46. We therefore postulated that the replication abnormalities and chromosomal aberrations of DONSON-deficient cells might arise via similar mechanisms. Indeed, the spontaneous replication fork asymmetry and H2AX phosphorylation exhibited by DONSON-depleted cells were partially reduced by co-depletion of either MUS81 or XPF (Fig. 6c, d). Moreover, co-depletion of MUS81 or XPF also reduced chromosome breakage and genomic pulverisation in these cells (Fig. 6e–g). From this, we conclude that the severe genome instability apparent in the absence of DONSON is due to nucleolytic processing of damaged replication forks by structure-specific nucleases.
Replication stress-induced genomic damage in DONSON patient cells
To link the role of DONSON in regulating replication fork stability and the phenotype of patients with DONSON mutations, we characterised replication dynamics and genomic stability of patient-derived fibroblasts. All DONSON patient-derived cell lines examined (P2, P6, P9, P10-2, P12) showed higher levels of spontaneous fork asymmetry and fork stalling than cells from unaffected individuals (Supplementary Figs. 17a, 18). Furthermore, patient-derived cells also exhibited elevated fork asymmetry and fork stalling following HU exposure, combined with defective intra-S phase checkpoint activation (Supplementary Fig. 17a, 18). Finally, levels of S-phase DNA damage and chromosome breakage were also elevated in these cell lines (Supplementary Fig. 17b–c). Together, these observations provide a potential pathological explanation for the clinical phenotype.
In addition, using isogenic cell lines inducibly expressing GFP-DONSON (Fig. 2c), we observed that expression of the haplotype-associated S28R mutant, but not the K489T variant, complemented loss of endogenous DONSON by rescuing the spontaneous fork stalling observed upon DONSON depletion (Supplementary Fig. 19). This is consistent with K489T being the pathogenic variant within the haplotype region (Fig. 2c).
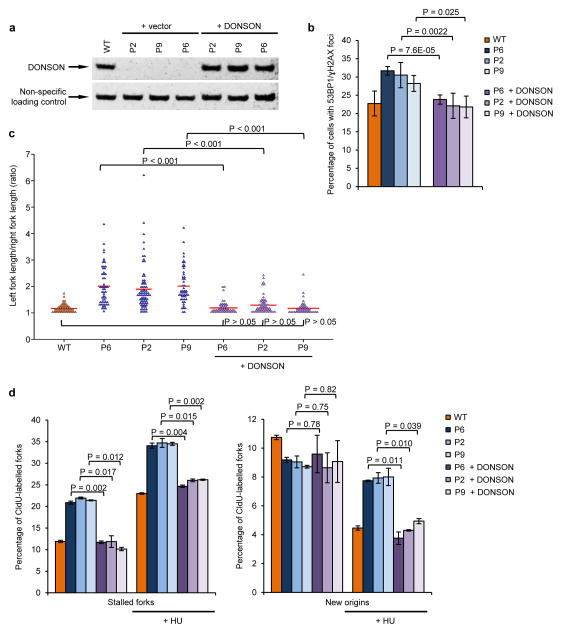
Finally, we set out to demonstrate that the patient-associated cellular phenotypes were directly due to mutation of DONSON. We first established three paired isogenic cell lines via transduction of patient-derived fibroblasts with retroviral expression vectors encoding WT DONSON or an empty vector (Fig. 7a). Importantly, the spontaneous DNA damage, replication fork stalling, replication fork asymmetry and intra-S phase checkpoint defect were all corrected by expression of WT DONSON (Fig. 7b–d and Supplementary Fig. 20), confirming that these phenotypes were directly due to DONSON deficiency. Lastly, using one of these cell lines, we also observed that inhibition of ATR and mutation of DONSON are epistatic with regard to the observed replication abnormalities (Supplementary Fig. 21).
Figure 7. DONSON patient cells have spontaneous defects in replication fork progression that result in DNA damage.
(a) Complementation of patient-derived fibroblasts with WT DONSON. Fibroblasts derived from DONSON patients P2, P6 and P9 were infected with retroviruses encoding either WT DONSON or an empty vector. DONSON expression was determined by immunoblotting. A non-specific cross-reactive protein represents a loading control. (b) Expression of WT DONSON in patient fibroblasts rescues elevated levels of spontaneous DNA damage. The percentage of cells from (a) with 53BP1/γH2AX foci was quantified by immunostaining (n=3). (c) DNA fibre analysis of complemented DONSON patient fibroblasts pulsed with CldU and IdU. Fork asymmetry was quantified. Plot indicates ratios of left/right fork track lengths of bidirectional replication forks. The red lines denote median ratios. (n=3). (d) The percentage of stalled forks and new origins from cells in (c) was quantified (n=3). Ongoing forks are shown in (Supplementary Fig. 19).
Discussion
Here we identify DONSON as a novel human disease gene, and describe 29 individuals with a range of mutations in DONSON, establishing such alterations as a frequent cause of microcephalic dwarfism. Since normal embryonic development requires rapid cellular proliferation47,48 it is exquisitely sensitive to genetic perturbations that impact DNA replication1–3,6. A failure to complete timely genome duplication will profoundly affect the number of cells generated during embryonic development. For example, hypomorphic mutations in ATR result in severe microcephaly and growth retardation, both in humans and in a murine model2,3,49,50, due to the role that ATR plays in preventing replication stress during development49–50. We propose that mutation of DONSON similarly reduces the number of cells generated during development via a failure to maintain replication fork stability in the presence of endogenous replication stress, thus explaining the decreased organism size observed. Furthermore, since brain development requires rapid proliferation of neural progenitor cells within a limited timeframe47, it is particularly sensitive to disruptive genetic perturbations. This may explain why brain development is disproportionately affected in these individuals compared to growth.
DONSON has no predicted domain structure or paralogs, and previous characterisation has been limited to two previous studies: an siRNA screen proposing that DONSON regulates genome stability, and a study of its Drosophila ortholog Humpty-dumpty suggesting a role in cell proliferation29,32. Consistent with this, we establish that DONSON is a replisome component that ensures replication fork stability, and promotes efficient activation of both intra-S and G2/M cell-cycle checkpoints upon exogenous replication stress. Loss of DONSON leads to increased spontaneous stalling of replication forks, which are subsequently cleaved into replication-associated DNA double strand breaks by structure-specific nucleases. Defective cell-cycle checkpoint activation in DONSON-deficient cells then allows transmission of these breaks into mitosis, accounting for the elevated chromosomal damage and genome fragmentation observed (Supplementary Fig. 22). Additional studies will be important to confirm this model, and to investigate whether DONSON is a constitutive component of the replisome, or whether it is recruited to a subset of replication forks. Furthermore, establishing which replisome components DONSON directly interacts with, and the functional importance of these associations, will also inform understanding of its biological function.
The mechanism by which DONSON ensures replication fork stability and promotes checkpoint activation remains to be defined. We propose that in addition to being a replisome component, DONSON is also involved in promoting the ATR-CHK1 replication stress response, since we observed that DONSON-depleted cells exhibit defective activation of cell cycle checkpoints and reduced ATR-dependent signalling in response to exogenous replication stress. This hypothesis is supported by the observation that impaired replication alone, such as that arising from a hypomorphic mutation in MCM4 (MCM4Chaos3/Chaos3), does not give rise to decreased CHK1 phosphorylation or increased new origin firing upon replication stress51. However, it is unclear whether DONSON functions directly or indirectly to regulate the ATR-CHK1 pathway. Our demonstration that cells lacking DONSON do not exhibit a global reduction in replication, or decreased levels of RPA-coated ssDNA, indicates that loss of DONSON does not affect the cells ability to generate the primary stimulus for ATR pathway activation. Based on this, we propose that either DONSON directly activates ATR, in a manner similar to TOPBP152 or ETAA153,54, or functions indirectly to regulate other factors necessary for efficient ATR-CHK1 signalling, such as the MRE11/RAD50/NBS1 (MRN)55 complex or TIPIN/Timeless19,20. Since DONSON does not contain a canonical ATR activation domain, which is found in both TOPBP1 and ETAA1, we favour the latter possibility. However, how DONSON acts to promote ATR signalling is not yet clear, and future work will be critical in establishing whether this is direct or indirect.
It is also evident that the cellular phenotype of cells lacking DONSON cannot be explained solely by abnormal DNA replication or defective ATR-dependent signalling. In particular, exposure of cells lacking ATR to exogenous replication stress results in highly elevated levels of H2AX phosphorylation, a situation not observed upon DONSON loss (Fig. 5e), despite the presence of substantial amounts of DNA damage. Therefore, whilst our observations are consistent with a role for DONSON in promoting ATR-CHK1 signalling, DONSON may also impact on other pathways that promote H2AX phosphorylation at the replication fork, for example those governed by ATM or the MRN complex.
In summary, we have identified DONSON as a novel disease gene that plays a key role in regulating cellular replication and cell cycle checkpoints. Further studies examining how DONSON functions will provide fundamental insight into how cells maintain replication fork integrity, and how these pathways prevent human disease.
Online Methods
Research subjects
Genomic DNA from the affected children and family members was extracted from peripheral blood using standard methods or saliva samples using Oragene collection kits according to the manufacturer’s instructions. Informed consent was obtained from all participating families. Ethics for the studies were approved by the Scottish Multicentre Research Ethics Committee (04:MRE00/19), by an IRB-approved research protocol (KFSHRC RAC# 2080006), and via the ’National Gene Mapping’ protocol by Guy’s and St. Thomas’ National Health Service (NHS) Foundation Trust local research ethics committee (ref.: 08/H0802/84, “Systematic Characterization of Genes in Inherited Disorders”). In addition, ethical approval for linkage studies on the genetics of Fanconi anaemia in 1989 were obtained from the Guy’s Hospital Research Ethics Committee (ref. EC89/10/27)22, with further approval for mutation analysis on existing samples in 1996 (ref. 96/3/9). Parents provided written consent for the publication of photographs of the affected individuals.
Exome sequencing and haplotype analysis
Exome sequencing of genomic DNA and variant filtering was performed as described previously12. Cohort resequencing was performed by Sanger sequencing of PCR products representing all coding exons of DONSON (primer sequences are detailed in Supplementary Table 4), with variant calling using MutationSurveyor (SoftGenetics Inc.). Haplotype analysis was undertaken by SNP genotyping both patients using Affymetrix CytoScan 750K arrays. Genotypes were generated using Affymetrix Genotyping Console software and examined manually. The pathogenic impact of DONSON mutations was predicted using Alamut Visual Software (Interactive Biosoftware Inc).
Cell culture and generation of cell lines
Lymphoblastoid cell lines (LCLs) were maintained in RPMI 1640 supplemented with 15% FBS, L-glutamine and penicillin/streptomycin antibiotics. LCLs were generated in house from peripheral blood samples by EBV transformation using standard methods. Dermal primary fibroblasts were grown from skin-punch biopsies in AmnioMax medium (Life Technologies) and then maintained in DMEM supplemented with 10% FBS, 5% L-glutamine and 5% penicillin/streptomycin antibiotics. Patient cell lines were validated using Sanger sequencing and immunoblotting. 293FT (Invitrogen) and HeLa (ATCC) cells were maintained in DMEM supplemented with 10% FBS, 5% L-glutamine and 5% penicillin/streptomycin antibiotics.
Stable cell lines were generated by Flp recombinase–mediated integration using HeLa-Flp-In T-REx host cells (gift from S. Taylor, University of Manchester) transfected with pcDNA5/FRT/TO-EGFP (vector only or EGFP-TRAIP) and pCAGGS-Flp.e (gift from D.-J. Kleinjan, University of Edinburgh). Transfected cells were selected using 5 μg/ml blasticidin and 400 μg/ml hygromycin, and the resulting colonies were then expanded for testing. Protein expression was induced with 1 μg/ml doxycycline (Sigma-Aldrich) treatment.
Primary fibroblasts derived from patients 2, 6 and 9 were immortalized with TERT retroviral supernatant with 4 μg/ml polybrene and infected with pMSCV-vector only or pMSCV-DONSON. Selection was performed using 750 ng/ml puromycin (Clontech) and 500 μg/ml neomycin (Invitrogen). Expression of the protein was verified by immunoblotting (Fig. 7a). All cell lines were routinely tested for mycoplasma.
Cell treatments
Plasmids and siRNA oligos were transfected in Opti-MEM reduced serum medium using Lipofectamine 2000 and Oligofectamine (Life Technologies) respectively according to the manufacturer’s guidelines. A custom siRNA targeting lacZ has previously been described56, and was used as a control siRNA. A DONSON siRNA SMARTpool (Dharmacon) was used for all siRNA transfections except when transfecting the HeLa Flp-In/T-Rex cells expressing an exogenous, siRNA-resistant, GFP-tagged DONSON construct. In this case a custom DONSON siRNA sequence (CCTGTGGACTGGAGTATTAdTdT) was used (Dharmacon). MUS81 siRNA SMARTpool and XPF siRNA SMARTpool (Dharmacon) were used where indicated. Transfected cells in both cases were analysed at 48–72 h post transfection. Where indicated, cells were treated with 1–2 mM hydroxyurea (Sigma-Aldrich), 50 ng/ml mitomycin C (Sigma-Aldrich) or 2 mM thymidine (Sigma-Aldrich). The ATR inhibitor (VE-821; Selleck Chemicals) was used at 5 μM. dNTP analogues EdU, CldU and IdU were purchased from Sigma Aldrich, and were used as indicated.
RT-PCR
Total RNA was extracted from cell lines using the RNeasy kit (Qiagen) according to the manufacturer’s instructions. DNA was removed by treatment with DNase I (Qiagen), and cDNA was generated using random oligomer primers and AMV RT (Roche). The RT-PCR primer pairs used are detailed in Supplementary Table 4.
DNA expression constructs
pEGFP-DONSON expression construct was created by cloning the human DONSON ORF into the pDONR221 Gateway shuttle vector (Invitrogen). WT DONSON was amplified from cDNA and recombined into the pDEST-EGFP vector to generated a GFP-tagged DONSON expression construct. The DONSON ORF was made siRNA resistant using site-directed mutagenesis (Agilent Technologies) by altering the following nucleotides: CCTGTGGACTGGAGTATTA was changed to CCCGTAGATTGGTCTATCA. Patient-associated mutations were engineered into the pEGFP-DONSON expression plasmid using site-directed mutagenesis according to the manufacturer’s instructions. (All primers are detailed in Supplementary Table 4).
The retroviral expression construct expressing DONSON was created by recombination between the pDONR221-DONSON vector and a Gateway-compatible pMSCVneo retroviral expression construct (Clontech).
The human telomerase reverse transcriptase (hTERT) expressing retroviral construct used to immortalise patient-derived human fibroblasts was a kind gift from Bob Weinberg (Addgene plasmid: #1771).
Minigene splicing reporter assay
A 1.58 kb stretch of the DONSON gene encompassing the 3′ end of intron 3, exon 4, intron 4, exon 5 and the 5′ end of intron 5 was amplified using DNA from a healthy individual and DONSON patients (carrying mutation c.786-22A>G or c.786-33A>G) using the DONSON-int3-SalI-F and DONSON-int5-SpeI-R primers, and cloned into the RHCglo vector57 using the SalI and SpeI restriction sites. Site-directed mutagenesis was used to introduce the DONSON intron 4 splice acceptor mutation (c.786-1G>A) into the splicing reporter construct. HeLa cells were transfected with each individual splicing mutation reporter construct using Lipofectamine 2000 according the manufacturer’s instructions. 24 h post-transfection, cells were harvested, total cellular RNA was extracted and cDNA generated using Superscript III reverse transcriptase first-stand synthesis system (Invitrogen). PCR was carried out using primers (RSV_minigene_F and RSV_minigene_R) to the 5′ and 3′ ends of the artificial exons present in the RHCglo vector. DONSON WT and mutant cDNA amplicons were resolved on a 2% agarose gel to visualise differences in splicing. Individual PCR products were subsequently cloned into the pGEM-T Easy Vector (Promega) and sequenced to verify the exon content of each transcript. All relevant primers are detailed in Supplementary Table 4.
iPOND
iPOND was performed as previously described35,58. Briefly, exponentially growing cells were incubated with 10 μM EdU for 10 min, cross-linked with 1% formaldehyde, harvested and permeabilised. For pulse-chase controls, cells were incubated in 10 μM EdU for 10 min, washed in media containing 10 μM thymidine, then incubated with media containing 10 μM thymidine for 1 h, before being cross-linked. Biotin azide was covalently attached to EdU within newly replicated DNA using a Click reaction, and EdU containing DNA was precipitated using Streptavidin agarose beads. Edu co-precipitates were then analysed by mass spectrometry. Log2 abundance values represent the ratio of proteins found in EdU-pulsed samples compared to those pulse-chased with EdU-thymidine.
Immunoblot analysis and antibodies
Whole cell extracts were obtained by sonication in UTB buffer (8 M Urea, 50 mM Tris, 150 mM β-mercaptoethanol) and analysed by SDS-PAGE following standard procedures. Protein samples were run on 6–12% acrylamide SDS-PAGE or 4–12% NuPage mini-gels (Life Technologies) and transferred onto nitrocellulose membrane. Immunoblotting was performed using antibodies to: Cyclin A (Santa Cruz, sc-751; 1:1,000), CHK1 (Santa Cruz, sc-8408; 1:1,000), CHK2 (Santa Cruz, sc-5278; 1:1000), FANCD2 (Santa Cruz, sc-20022; 1:1000), MCM2 (BD Transduction Laboratories, 610700; 1:10000), MCM7 (Santa Cruz, sc-56324; 1:1000), MUS81 (Santa Cruz, sc-53382; 1:2000); XPF (Santa Cruz, sc-136153; 1:1000); H2A (Millipore, 07-146; 1:3000), γ-H2AX (Millipore, 05-636; 1:3000), RPA2 (Millipore, NA18; 1:1000), phospho-histone H3 Ser-10-P (Millipore); pS343-NBS1 (Abcam, 47272; 1:500); NBS1 (Genetex, GTX70224; 1:10000); ATR (Bethyl Laboratories, A300-137A; 1:1000), pS345-CHK1 (Cell Signaling Technology, 2341; 1:100), pS4/S8-RPA2 (Bethyl Laboratories, A300-245A; 1:1,000), pS966-SMC1 (Bethyl Laboratories, A300-050A; 1:1,000), SMC1 (Bethyl Laboratories, A300-055A; 1:1,000), Treslin (Bethyl Laboratories, A303-472A; 1:1,000); TOPBP1 (Bethyl Laboratories; A300-111A; 1:1000); Vinculin (Sigma-Aldrich, V9264; 1:1,000); α-Tubulin (Sigma-Aldrich, T5168; 1:4000); GFP (Roche, 11814460001; 1:500). The polyclonal anti-DONSON antibody was generated by immunising rabbits with a GST-fusion protein encoding aa 1-125 of human DONSON. Antibody was affinity-purified from rabbit sera (Eurogentec) and specificity established using lysates from patient cells and DONSON siRNA-transfected cells.
Loading controls for all blots derive from reprobing the same membrane, except for phospho-antibody blots, where paired gels were run simultaneously, and blotted in parallel for phosphorylated and total proteins.
Immunofluorescence and fluorescent microscopy
siRNA transfected HeLa cells or passage-matched TERT-immortalized fibroblasts were seeded on coverslips 24 h before extraction/fixation. To visualise cells undergoing DNA replication, cells incubated in medium containing 10 μM EdU for 10–30 min prior to harvesting. To remove soluble proteins before immunofluorescence, cells were pre-extracted for 10 min on ice with ice-cold buffer (25 mM HEPES, pH 7.4, 50 mM NaCl, 1 mM EDTA, 3 mM MgCl2, 300 mM sucrose and 0.5% Triton X-100) and then fixed with 4% paraformaldehyde for 15 min. For analysis of cells transfected with GFP-tagged protein, cells were fixed and permeabilised by incubation with ice-cold methanol for 20 minutes. EdU immunolabeling was performed using Click-iT EdU Imaging Kit (Invitrogen, C10337) according to the manufacturer’s protocol. Cells were stained for 53BP1 (Novus Biologicals, NB100-304; 1:1,000) and/or γH2AX (Millipore, 05-636; 1:1000) and stained with secondary antibodies conjugated to Alexa Fluor-568 (Life Technologies) and DAPI.
For quantification of signal-integrated densities of γH2AX staining, cells were stained with an antibody specific to γH2AX (Millipore, 05-636; 1:1000), images were visualized using a Zeiss Axioplan 2 microscope with iVision software (BioVision Technologies) and captured using a 40× oil-immersion objective. For quantification of signal-integrated densities of RPA staining, cells were stained with RPA2 antibody (Millipore, NA18; 1:200), images were visualized using a Nikon Eclipse Ni microscope with NIS-Elements software (Nikon Instruments) and captured using a 100× oil-immersion objective. Nuclei were segmented on the basis of DAPI staining and then signal-integrated density of γH2AX or RPA staining quantified for each nuclear region using ImageJ software (US National Institutes of Health). For quantification of γH2AX staining, more than 100 EdU positive cells and 50 EdU negative cells were analyzed per experiment per condition, and for quantification of RPA staining, more than 200 cells were analyzed per experiment per condition. Exposure time, binning, microscope settings and light source intensity were kept constant for all the samples in all cases.
For quantification of native BrdU foci cells were incubated in medium containing 10 μM BrdU for 24 h prior to harvesting. Six hours prior to harvesting, 2 mM HU was added to the media. To visualise ssDNA foci, cells were extracted for 10 min on ice with ice-cold buffer (25 mM HEPES, pH 7.4, 50 mM NaCl, 1 mM EDTA, 3 mM MgCl2, 300 mM sucrose and 0.5% Triton X-100) and then fixed with 4% paraformaldehyde for 15 min. After fixation, cells were washed with PBS and blocked in 3% FCS in PBS for 30 min at room temperature. ssDNA was visualised using a BrdU antibody (Abcam, ab6326; 1:500). To denature DNA cells were incubated in 2 M HCl in PBS for 30 min prior to addition of the BrdU antibody. Images were acquired as for γH2AX quantification and foci were quantified using ImageJ-based script. Nuclei were defined on the basis of DAPI staining and native BrdU foci were detected using “Find maxima” function of ImageJ within each nuclear region. Exposure time, binning, microscope settings, light source intensity and the noise level in “Find maxima” function were kept constant for all the samples within each individual experiment. More than 100 cells were analyzed per experiment per condition.
Metaphase spreads
Chromosomal aberrations were scored in Giemsa-stained metaphase spreads as previously described56. Briefly, demecolcine (Sigma Aldrich) was added at a final concentration of 0.2 μg/ml 4 h prior to harvesting. Cells were harvested by trypsinisation, subjected to hypotonic shock for 1 hour at 37°C in 0.3 M sodium citrate and fixed in 3:1 methanol:acetic acid solution. Cells were dropped onto acetic acid humidified slides, stained for 15 minutes in Giemsa-modified solution (Sigma; 5% v/v in H2O) and washed in water for 5 minutes.
DNA fibre spreading assay
Passage-matched primary, TERT-immortalized fibroblasts or siRNA transfected HeLa cells were pulse labeled with CldU for 20 min, washed with media and damaged with 2 mM hydroxyurea for 2 h before being pulse labeled with IdU for 40 min. Alternatively, 50 ng/ml mitomycin C was added to the cells 24 h before CldU pulse labeling and left on during 20 min CldU and 20 min ldU pulse labeling. Cells were harvested by trypsinization, and cell pellets were washed in PBS. 5 × 105 cells were lysed directly onto glass slides using spreading buffer (200 mM Tris-HCl, pH 7.5, 50 mM EDTA, 0.5% SDS) and fixed in methanol/acetic acid (3:1 ratio). Following 2.5 M HCl denaturation, CldU was detected using rat anti-BrdU (clone BU1/75, ICR1; Abcam, ab6326; 1:750), and IdU was detected using mouse anti-BrdU (clone B44; BD Biosciences, 347583; 1:750). Slides were then fixed in 4% paraformaldehyde before immunostaining with secondary antibodies conjugated to Alexa Fluor-594 or Alexa Fluor-488 (Life Technologies). Labeled DNA fibers were visualized using a Nikon Eclipse Ni microscope with NIS-Elements software (Nikon Instruments). Images were captured using 40× oil-immersion objectives and were recoloured and analyzed using ImageJ software (US National Institutes of Health). For quantification of replication structures, at least 400 structures were counted per experiment. Tract lengths were measured using ImageJ (National Institutes of Health; http://rsbweb.nih.gov/ij/). To calculate fork velocity, arbitrary length values were converted into micrometers using the scale bars created by the microscope, with 1 μm equivalent to 2.59 kb59. Replication fork speed (kb/min) was then determined by dividing the length of CldU and IdU tracks (in kb) from ongoing forks by the pulse time.
FACS analysis
For BrdU analysis, HeLa cells were pulse labeled with 10 μM BrdU for 30 min before fixation with 70% ethanol at −20 °C for 16 h. Cells were then digested with 1 mg/ml pepsin and denatured with 2 M HCl before washing with PBS and blocking in 0.5 % BSA, 0.5 % Tween-20. BrdU labeling was detected using anti-BrdU antibody (Abcam, ab6326; 1:75) and FITC-conjugated anti-rat secondary antibody. DNA content was assessed by staining with 50 μg/ml propidium iodide. Cells were sorted on a BD Biosciences FACS Aria II and data were analyzed using FlowJo software (v7.6.1, Tree Star).
For mitotic analysis and immuno-detection of phospho-histone H3 (Ser10), HeLa cells were harvested, fixed, permeabilised 24 h post exposure to HU or MMC, as previously described56. Cells were analysed using an Accuri flow cytometer (BDBiosciences) in conjunction with CFlowplus software. Data represents that obtained from at least 30,000 cells.
Immunoprecipitation and GFP-Trap
293FT cells transfected with plasmids encoding GFP-DONSON or GFP were untreated, or exposed to 2 mM HU for 16 h and harvested. Cells were then incubated in lysis buffer (150 mM NaCl, 50 mM Tris HCl pH7.5, 2 mM MgCl2, 1 % NP40, 90 U/ml Benzonase (Novagen) and Protease Inhibitor Cocktail EDTA free (Roche)) for 30 min with rotation at 4 °C. The resultant cell lysates were pre-cleared at 44,000 rpm at 4 °C for 30 min.
For immunoprecipitations, 3 mg of lysate was immunoprecipitated with 5 μg of antibody, immune complexes collected with Protein A-Sepharose (Sigma-Aldrich). Complexes were washed with wash buffer (150 mM NaCl, 50 mM Tris HCl pH 7.5, 0.5 % NP40, and Complete Protease Inhibitor Cocktail (Roche)) and analysed by SDS-PAGE.
For GFP-Trap, 3 mg lysates were incubated with GFP-Trap agarose beads (ChromoTek) at 4 °C for 5 h. The resulting GFP-Trap complexes were washed with wash buffer as above and analysed by SDS-PAGE. Experiments were carried out in the presence of Benzonase Nuclease to exclude the possibility of interactions being mediated by DNA.
For mass spectrometry analysis, GFP or GFP-DONSON were isolated from tetracycline induced, or uninduced, Flp-In T-REx HeLa cell extracts by incubation with GFP-trap magnetic agarose beads (Chromotek) for 2 hours on a Kingfisher Duo robotic handling station (Thermo). Asynchronous cells and S-phase accumulated cells, using a 24 h treatment with 2 mM HU, were analysed. On-bead digest and mass spectrometry were performed as described60. Data represents three independent experiments for each condition, analysed by back-to-back MS and quantified by Label free quantification (LFQ). Proteins were identified and quantified with the MaxQuant 1.5 software suite by searching against the Uniprot human database. M(ox) and protein N-terminal acetylation were set as variable, and carbamylation as a fixed modification, with a 1 % FDR. Contaminants and reverse data base hits were deleted. Protein significantly enriched by GFP-DONSON were selected on the basis of p-value <0.05, and >2 fold change from asynchronous to S-phase, as identified by Student t-test and ratio cut-off against the respective negative control LFQ data as determined by MaxQuant (p<0.05; 2-fold).
Proximity ligation assay (PLA)
PLA was carried out as described in5,56. Briefly, cells from GFP or GFP-DONSON Flp-In T-REx HeLa cell lines were treated with 1ug/ml doxycycline and fixed/extracted after 24 h. For PCNA visualisation, cells were fixed with methanol at −20 °C for 10 min followed by a 5 min extraction in 0.3% Triton-X100 in PBS. For RPA visualisation, cells were pre-extracted in nuclear extraction buffer for 5 minutes on ice, and fixed in 3.6% paraformaldehyde for 10 minutes at room temperature. Cells were then incubated in anti-PCNA (PC10, 1:500, Santa Cruz) or anti-RPA (NA18; 1:500; Merck-Millipore) antibodies along with anti-GFP antibody (ab6556, 1:500, Abcam), and in situ proximity ligation was performed using a Duolink Detection Kit (Sigma Aldrich). Nuclear foci were imaged using a Nikon Eclipse Ni-U microscope equipped with a 100X oil lens in conjunction with a Zyla camera, and images were acquired using Elements software (Nikon). More than 200 cells were analysed per experiment per condition.
Fluorescence Cross-Correlation Spectroscopy (FCCS)
HeLa cells stably expressing GFP-DONSON and mCherry-PCNA (construct kindly provided by C. Lukas, Copenhagen; referred to as RFP-PCNA) were used for FCCS. For all details on Fluorescence Microscopy Imaging and FCS/FCCS, refer to Supplementary Note.
Statistical Analyses
Statistical differences were analyzed by: two-tailed Student T-Test (Fig. 3b, 3d, 3f, 4f, 4g, 5b–d, 5f, 6a, 6e, 6f, 7b, 7d and Supplementary Fig. 5c, 8b, 9a, 12c, 13i, 14c, 16a–c, 17a–c, 19, 20, 21); Mann-Whitney rank sum test (Fig. 3e, 6c, 7c and Supplementary Fig. 12b, 15a, 15c); and Chi-Squared Test (Fig 4c, 6g and Supplementary Fig. 16d). n refers to number of independent experiments unless indicated. Error bars represent standard error of the mean (s.e.m.) unless specified.
Data Availability
The NGS data used in the manuscript can be obtained from the European Genome-phenome Archive (EGA) under accession EGAS00001002224. NGS datasets on patients P14-P18, p21 are not available due to institutional IRB restrictions. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD005690.
Supplementary Material
Acknowledgments
We would like to thank the families and clinicians for their involvement and participation. We are grateful to R. S. Taylor (University of Manchester) and D.-J. Kleinjan (University of Edinburgh), J. Bartek and C. Bartek (Copenhagen) for their kind gift of reagents. We thank E. Freyer, J. Wills, J. Ding, A. Fluteau, C. Keith, D. Longman and the IGMM FACS, core sequencing and mass-spectrometry facilities for technical assistance and advice. The Walking With Giants Foundation and Potentials Foundation supported the Primordial Dwarfism Registry (M.B.B.). This work is supported by funding from Cancer Research UK (C17183/A13030) (G.S.S., M.R.H. and A.V.), the Medical Research Council (MR/M009882/1) (J.J.R.), Worldwide Cancer Research (13-1012) (A.Z.), Birmingham Children’s Hospital Research Foundation (BCHRF400) (R.M.A.M.), University of Birmingham (J.J.R., R.M.A.M., A.B.), Newlife Foundation for Disabled Children (P.C., L.S.B.), Medical Research Scotland (L.S.B.) and the National Institute for Health Research (NIHR) Biomedical Research Centre at Guy’s and St Thomas’ NHS Foundation Trust and King’s College London (H.B., A. Amar, N.J.P, M.A.S., C.G.M.), German Federal Ministry of Education and Research (BMBF) (1GM1404; E-RARE network EuroMicro) (G. Yigit), KSCDR funding and KACST grant 09-MED941-20 (F.S.A.), Swiss National Science Foundation (P2ZHP3_158709) (O.M.), Medical Research Council, the Lister Institute for Preventative Medicine and the European Research Council (ERC, 281847) (A.P.J.).
Footnotes
Author Contributions
J.J.R., M.R.H., P.C., O.M., A.Z., A.L., R.M.A.M., A.B. and G.S.S. designed and performed the cell biology experiments. J.E.M., L.S.B., R.S, C.V.L., F.S.A, M.A.S., C.G.M., Y.L., S.M. and G. Yigit performed NGS sequencing and analysis. L.S.B., P.C., R.C.C., R.S., A.V., J.E.M., M.A.S., C.V.L., Z.T., M.A.M.R., H.B., A. Amar., S.M., A. Almoisheer, H.S.A. and N.J.P. performed sequencing, genotyping, linkage analysis, analysis of splicing and other molecular genetics experiments. D.C. and S.R.W. performed iPOND experiments. P.T., D.K.P. and K.S. performed FCCS analysis. A.V.K. performed mass spectrometry analysis. E.F., M.Z.S., S.A.T., A. Alswaid, S.A., J.Y.A., M.A.B., A.F.B., L.C., H.C., A.D., R.F., E.H., E.F.P., A.P., L.S., S.T., G. Yoon., J.A., P.N., A.J.Q., B.D.H., M.A. and R.H. contributed clinical cases and clinical data and analysis for the study. M.B.B., C.A.W., J.E.M., L.S.B, A.M.R.T., F.S.A., C.G.M. and A.P.J. recruited study cohorts, and performed review of phenotypes and sample collection. J.J.R., M.R.H., L.S.B., A.P.J. and G.S.S. wrote the manuscript. The study was planned and supervised by G.S.S., C.G.M., F.S.A and A.P.J.
Competing Financial Interests Statement
The authors declare no competing financial interests.
References
- 1.Klingseisen A, Jackson AP. Mechanisms and pathways of growth failure in primordial dwarfism. Genes Dev. 2011;25:2011–24. doi: 10.1101/gad.169037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.O’Driscoll M, Ruiz-Perez VL, Woods CG, Jeggo PA, Goodship JA. A splicing mutation affecting expression of ataxia-telangiectasia and Rad3-related protein (ATR) results in Seckel syndrome. Nat Genet. 2003;33:497–501. doi: 10.1038/ng1129. [DOI] [PubMed] [Google Scholar]
- 3.Ogi T, et al. Identification of the first ATRIP-deficient patient and novel mutations in ATR define a clinical spectrum for ATR-ATRIP Seckel Syndrome. PLoS Genet. 2012;8:e1002945. doi: 10.1371/journal.pgen.1002945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.German J. Bloom’s syndrome. I. Genetical and clinical observations in the first twenty-seven patients. Am J Hum Genet. 1969;21:196–227. [PMC free article] [PubMed] [Google Scholar]
- 5.Harley ME, et al. TRAIP promotes DNA damage response during genome replication and is mutated in primordial dwarfism. Nat Genet. 2016;48:36–43. doi: 10.1038/ng.3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qvist P, et al. CtIP Mutations Cause Seckel and Jawad Syndromes. PLoS Genet. 2011;7:e1002310. doi: 10.1371/journal.pgen.1002310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rosin N, et al. Mutations in XRCC4 cause primary microcephaly, short stature and increased genomic instability. Hum Mol Genet. 2015;24:3708–17. doi: 10.1093/hmg/ddv115. [DOI] [PubMed] [Google Scholar]
- 8.Bicknell LS, et al. Mutations in the pre-replication complex cause Meier-Gorlin syndrome. Nat Genet. 2011;43:356–9. doi: 10.1038/ng.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bicknell LS, et al. Mutations in ORC1, encoding the largest subunit of the origin recognition complex, cause microcephalic primordial dwarfism resembling Meier-Gorlin syndrome. Nat Genet. 2011;43:350–5. doi: 10.1038/ng.776. [DOI] [PubMed] [Google Scholar]
- 10.Guernsey DL, et al. Mutations in origin recognition complex gene ORC4 cause Meier-Gorlin syndrome. Nat Genet. 2011;43:360–4. doi: 10.1038/ng.777. [DOI] [PubMed] [Google Scholar]
- 11.Fenwick AL, et al. Mutations in CDC45, Encoding an Essential Component of the Pre-initiation Complex, Cause Meier-Gorlin Syndrome and Craniosynostosis. Am J Hum Genet. 2016;99:125–38. doi: 10.1016/j.ajhg.2016.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Murray JE, et al. Extreme growth failure is a common presentation of ligase IV deficiency. Hum Mutat. 2014;35:76–85. doi: 10.1002/humu.22461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Murray JE, et al. Mutations in the NHEJ component XRCC4 cause primordial dwarfism. Am J Hum Genet. 2015;96:412–24. doi: 10.1016/j.ajhg.2015.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shaheen R, et al. Genomic analysis of primordial dwarfism reveals novel disease genes. Genome Res. 2014;24:291–9. doi: 10.1101/gr.160572.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zeman MK, Cimprich KA. Causes and consequences of replication stress. Nat Cell Biol. 2014;16:2–9. doi: 10.1038/ncb2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300:1542–8. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
- 17.MacDougall CA, Byun TS, Van C, Yee MC, Cimprich KA. The structural determinants of checkpoint activation. Genes Dev. 2007;21:898–903. doi: 10.1101/gad.1522607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nam EA, Cortez D. ATR signalling: more than meeting at the fork. Biochem J. 2011;436:527–36. doi: 10.1042/BJ20102162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chou DM, Elledge SJ. Tipin and Timeless form a mutually protective complex required for genotoxic stress resistance and checkpoint function. Proc Natl Acad Sci U S A. 2006;103:18143–7. doi: 10.1073/pnas.0609251103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kemp MG, et al. Tipin-replication protein A interaction mediates Chk1 phosphorylation by ATR in response to genotoxic stress. J Biol Chem. 2010;285:16562–71. doi: 10.1074/jbc.M110.110304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lek M, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–91. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Milner RD, Khallouf KA, Gibson R, Hajianpour A, Mathew CG. A new autosomal recessive anomaly mimicking Fanconi’s anaemia phenotype. Arch Dis Child. 1993;68:101–3. doi: 10.1136/adc.68.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Germanaud D, et al. Simplified gyral pattern in severe developmental microcephalies? New insights from allometric modeling for spatial and spectral analysis of gyrification. Neuroimage. 2014;102(Pt 2):317–31. doi: 10.1016/j.neuroimage.2014.07.057. [DOI] [PubMed] [Google Scholar]
- 24.Martin CA, et al. Mutations in PLK4, encoding a master regulator of centriole biogenesis, cause microcephaly, growth failure and retinopathy. Nat Genet. 2014;46:1283–92. doi: 10.1038/ng.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Trimborn M, et al. Mutations in microcephalin cause aberrant regulation of chromosome condensation. Am J Hum Genet. 2004;75:261–6. doi: 10.1086/422855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Griffith E, et al. Mutations in pericentrin cause Seckel syndrome with defective ATR-dependent DNA damage signaling. Nat Genet. 2008;40:232–6. doi: 10.1038/ng.2007.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yeo G, Burge CB. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J Comput Biol. 2004;11:377–94. doi: 10.1089/1066527041410418. [DOI] [PubMed] [Google Scholar]
- 28.Mouse Genome Database (MGD) at the Mouse Genome Informatics website. Vol. 2016 (ed. Laboratory, T.J.) (Bar Harbor, Maine).
- 29.Bandura JL, et al. humpty dumpty is required for developmental DNA amplification and cell proliferation in Drosophila. Curr Biol. 2005;15:755–9. doi: 10.1016/j.cub.2005.02.063. [DOI] [PubMed] [Google Scholar]
- 30.Cortez D. Preventing replication fork collapse to maintain genome integrity. DNA Repair (Amst) 2015;32:149–57. doi: 10.1016/j.dnarep.2015.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Errico A, Costanzo V. Mechanisms of replication fork protection: a safeguard for genome stability. Crit Rev Biochem Mol Biol. 2012;47:222–35. doi: 10.3109/10409238.2012.655374. [DOI] [PubMed] [Google Scholar]
- 32.Fuchs F, et al. Clustering phenotype populations by genome-wide RNAi and multiparametric imaging. Mol Syst Biol. 2010;6:370. doi: 10.1038/msb.2010.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Papadopoulos DK, et al. Probing the kinetic landscape of Hox transcription factor-DNA binding in live cells by massively parallel Fluorescence Correlation Spectroscopy. Mech Dev. 2015;138(Pt 2):218–25. doi: 10.1016/j.mod.2015.09.004. [DOI] [PubMed] [Google Scholar]
- 34.Bacia K, Schwille P. Practical guidelines for dual-color fluorescence cross-correlation spectroscopy. Nat Protoc. 2007;2:2842–56. doi: 10.1038/nprot.2007.410. [DOI] [PubMed] [Google Scholar]
- 35.Sirbu BM, et al. Analysis of protein dynamics at active, stalled, and collapsed replication forks. Genes Dev. 2011;25:1320–7. doi: 10.1101/gad.2053211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu S, et al. ATR autophosphorylation as a molecular switch for checkpoint activation. Mol Cell. 2011;43:192–202. doi: 10.1016/j.molcel.2011.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Durkin SG, Arlt MF, Howlett NG, Glover TW. Depletion of CHK1, but not CHK2, induces chromosomal instability and breaks at common fragile sites. Oncogene. 2006;25:4381–8. doi: 10.1038/sj.onc.1209466. [DOI] [PubMed] [Google Scholar]
- 38.Ozeri-Galai E, Schwartz M, Rahat A, Kerem B. Interplay between ATM and ATR in the regulation of common fragile site stability. Oncogene. 2008;27:2109–17. doi: 10.1038/sj.onc.1210849. [DOI] [PubMed] [Google Scholar]
- 39.Toledo LI, et al. ATR prohibits replication catastrophe by preventing global exhaustion of RPA. Cell. 2013;155:1088–103. doi: 10.1016/j.cell.2013.10.043. [DOI] [PubMed] [Google Scholar]
- 40.Brown EJ, Baltimore D. ATR disruption leads to chromosomal fragmentation and early embryonic lethality. Genes Dev. 2000;14:397–402. [PMC free article] [PubMed] [Google Scholar]
- 41.Brown EJ, Baltimore D. Essential and dispensable roles of ATR in cell cycle arrest and genome maintenance. Genes Dev. 2003;17:615–28. doi: 10.1101/gad.1067403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Forment JV, Blasius M, Guerini I, Jackson SP. Structure-specific DNA endonuclease Mus81/Eme1 generates DNA damage caused by Chk1 inactivation. PLoS One. 2011;6:e23517. doi: 10.1371/journal.pone.0023517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Couch FB, et al. ATR phosphorylates SMARCAL1 to prevent replication fork collapse. Genes Dev. 2013;27:1610–23. doi: 10.1101/gad.214080.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ragland RL, et al. RNF4 and PLK1 are required for replication fork collapse in ATR-deficient cells. Genes Dev. 2013;27:2259–73. doi: 10.1101/gad.223180.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hodskinson MR, et al. Mouse SLX4 is a tumor suppressor that stimulates the activity of the nuclease XPF-ERCC1 in DNA crosslink repair. Mol Cell. 2014;54:472–84. doi: 10.1016/j.molcel.2014.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Svendsen JM, et al. Mammalian BTBD12/SLX4 assembles a Holliday junction resolvase and is required for DNA repair. Cell. 2009;138:63–77. doi: 10.1016/j.cell.2009.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Takahashi T, Nowakowski RS, Caviness VS., Jr The cell cycle of the pseudostratified ventricular epithelium of the embryonic murine cerebral wall. J Neurosci. 1995;15:6046–57. doi: 10.1523/JNEUROSCI.15-09-06046.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Snow MHL. Gastrulation in the mouse: growth and regionalization of epiblast. J Embryol Exp Morphol. 1977;42:293–303. [Google Scholar]
- 49.Murga M, et al. A mouse model of ATR-Seckel shows embryonic replicative stress and accelerated aging. Nat Genet. 2009;41:891–8. doi: 10.1038/ng.420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Despras E, Daboussi F, Hyrien O, Marheineke K, Kannouche PL. ATR/Chk1 pathway is essential for resumption of DNA synthesis and cell survival in UV-irradiated XP variant cells. Hum Mol Genet. 2010;19:1690–701. doi: 10.1093/hmg/ddq046. [DOI] [PubMed] [Google Scholar]
- 51.Kawabata T, et al. Stalled fork rescue via dormant replication origins in unchallenged S phase promotes proper chromosome segregation and tumor suppression. Mol Cell. 2011;41:543–53. doi: 10.1016/j.molcel.2011.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kumagai A, Lee J, Yoo HY, Dunphy WG. TopBP1 activates the ATR-ATRIP complex. Cell. 2006;124:943–55. doi: 10.1016/j.cell.2005.12.041. [DOI] [PubMed] [Google Scholar]
- 53.Bass TE, et al. ETAA1 acts at stalled replication forks to maintain genome integrity. Nat Cell Biol. 2016;18:1185–1195. doi: 10.1038/ncb3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Haahr P, et al. Activation of the ATR kinase by the RPA-binding protein ETAA1. Nat Cell Biol. 2016;18:1196–1207. doi: 10.1038/ncb3422. [DOI] [PubMed] [Google Scholar]
- 55.Duursma AM, Driscoll R, Elias JE, Cimprich KA. A role for the MRN complex in ATR activation via TOPBP1 recruitment. Mol Cell. 2013;50:116–22. doi: 10.1016/j.molcel.2013.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Higgs MR, et al. BOD1L Is Required to Suppress Deleterious Resection of Stressed Replication Forks. Mol Cell. 2015;59:462–77. doi: 10.1016/j.molcel.2015.06.007. [DOI] [PubMed] [Google Scholar]
- 57.Singh G, Cooper TA. Minigene reporter for identification and analysis of cis elements and trans factors affecting pre-mRNA splicing. Biotechniques. 2006;41:177–81. doi: 10.2144/000112208. [DOI] [PubMed] [Google Scholar]
- 58.Sirbu BM, Couch FB, Cortez D. Monitoring the spatiotemporal dynamics of proteins at replication forks and in assembled chromatin using isolation of proteins on nascent DNA. Nat Protoc. 2012;7:594–605. doi: 10.1038/nprot.2012.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Conti C, et al. Replication fork velocities at adjacent replication origins are coordinately modified during DNA replication in human cells. Mol Biol Cell. 2007;18:3059–67. doi: 10.1091/mbc.E06-08-0689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Turriziani B, et al. On-beads digestion in conjunction with data-dependent mass spectrometry: a shortcut to quantitative and dynamic interaction proteomics. Biology (Basel) 2014;3:320–32. doi: 10.3390/biology3020320. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The NGS data used in the manuscript can be obtained from the European Genome-phenome Archive (EGA) under accession EGAS00001002224. NGS datasets on patients P14-P18, p21 are not available due to institutional IRB restrictions. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD005690.