Abstract
Introduction
Non-invasive imaging plays a critical role in managing patients with cardiovascular disease. Although subjective visual interpretation remains the clinical mainstay, quantitative analysis facilitates objective, evidence-based management, and advances in clinical research. This has driven developments in computing and software tools aimed at achieving fully automated image processing and quantitative analysis. In parallel, machine learning techniques have been used to rapidly integrate large amounts of clinical and quantitative imaging data to provide highly personalized individual patient-based conclusions.
Areas covered
This review summarizes recent advances in automated quantitative imaging in cardiology and describes the latest techniques which incorporate machine learning principles. The review focuses on the cardiac imaging techniques which are in wide clinical use. It also discusses key issues and obstacles for these tools to become utilized in mainstream clinical practice.
Expert commentary
Fully-automated processing and high-level computer interpretation of cardiac imaging are becoming a reality. Application of machine learning to the vast amounts of quantitative data generated per scan and integration with clinical data also facilitates a move to more patient-specific interpretation. These developments are unlikely to replace interpreting physicians but will provide them with highly accurate tools to detect disease, risk-stratify, and optimize patient-specific treatment. However, with each technological advance, we move further from human dependence and closer to fully-automated machine interpretation.
Keywords: Artificial intelligence, machine learning, cardiac imaging, deep learning, image segmentation
1. Introduction
Noninvasive imaging plays a critical role in the diagnosis, outcome prediction and management of patients with cardiovascular disease. The quality and amount of imaging data acquired with each scan are continuously increasing in all modalities including computed tomography (CT), nuclear cardiology, echocardiography, and magnetic resonance imaging (MRI). In-depth reviews comparing various cardiac imaging modalities are available [1]. An overall, simplified comparison of primary cardiac imaging modalities is shown in Table 1. To date, the most fully automated approaches have been developed for nuclear cardiology, likely due to the lower image resolution and therefore simpler image analysis. However, advanced methods for other widely used cardiac modalities are being rapidly developed. This review focuses on the efforts in full automation of the widely used clinical imaging techniques – and on the efforts to derive the final diagnosis or prognosis by such automated techniques.
Table 1.
Overview of main clinical used cardiac imaging techniques.
| SPECT | PET | ECHO | MRI | CT | |
|---|---|---|---|---|---|
| Resolution (mm) | 7–15 | 4–10 | <1 | <1 2–3 (perfusion) | <1 |
| Signal Sensitivity (moles) | 10-12 - 10-9 | 10-12-10-9 | 10-5 -10-4 | 10-5 -10-4 | 10-5 -10-4 |
| Isotropic 3D imaging | Yes | Yes | No | No | Yes |
| Perfusion/ischemia | +++ | +++ | + | +++ | + |
| Function | + | + | ++ | +++ | + |
The importance of quantitative analysis – that is, deriving precise numerical physiological parameters from the imaging data – has become increasingly recognized for more sophisticated diagnosis, therapeutic monitoring, prognostication, and novel research purposes. Quantitative analysis has, therefore, become routine and even mandatory in many situations – this, in turn, has driven the development of advanced post-processing software tools which in some instances allow virtually fully automated image processing starting with the raw images. In recent reports, especially for nuclear cardiology, little if any physician supervision is required.
Currently, when automated processing methods are employed, a common workflow is that the physician performs a final quality control check and overview in their concluding report. However, with advancing machine intelligence, this final human check may also become surplus to requirements. Instead, software tools could provide this final quality check and in fact offer a more sophisticated, reproducible conclusion drawn from comparison with, not just one physician's career experience, but with massive ever-increasing training databases. Given the ever-abundant need for cost-effective diagnostic and treatment algorithms, supplanting the physician to achieve completely automated image processing, data analysis, quality control check, and final interpretation is not just an inspiring technical challenge but a valid option for reducing costs. It could be an imminent reality if this strategy is actively pursued by researchers and developed over the coming years.
Additionally, there is scope for machine learning to infiltrate and even outperform in skills traditionally reserved for expert readers. For example, could a disease diagnosis, or precise estimation of patient risk also be done by automated software tools? Can clinical information be rapidly integrated with imaging data to offer a more accurate final diagnosis and help select the most appropriate treatment on an individual patient basis? The possibilities for a sophisticated machine analysis are endless and not limited to a particular career experience or subjective biases, and not dependent on years of clinical training to reach their peak. This article provides insights into these probing questions, summarizes recent advances in automated quantitative imaging in cardiology and describes the latest techniques which incorporate machine learning principles. Finally, we offer an opinion into the direction of future development and the stand-off between machine and physician within cardiac imaging.
2. Automated image interpretation in cardiology
The multimodal images in cardiology require complex interpretation to arrive at patient diagnosis or assessment of patient risk. Often the first step is some form of image quantification either based on visual scoring or on computer analysis which will help the physician to diagnose the disease or assess patient risk. We will review some of the recent studies in this area, which describe automated methods for quantification of diagnostic cardiac images,
2.1. Nuclear cardiology
Myocardial perfusion SPECT and PET imaging (MPI) plays a crucial role in the diagnosis and management of coronary artery disease, providing key information concerning myocardial perfusion and ventricular function. At least 7 million patients undergo MPI in the USA annually [6]. As compared to SPECT, PET may provide benefits of lower radiation exposure and short image acquisition time. Quantitative PET techniques such as coronary blood flow may be superior for cardiovascular applications [7]. Nevertheless, by far most MPI worldwide are performed by SPECT. Consequently, the advanced methods allowing the full automation of the quantification are primarily optimized and validated for SPECT MPI. There are, however, equivalent automated tools available for PET image processing, including automated quantification of myocardial blood flow. Nuclear cardiology techniques are the most advanced with respect to image automation as compared to other imaging modalities in cardiology. It is, therefore, most likely that in this modality, the fully automated analysis and interpretation will become routine first. However, current reporting of nuclear cardiology scans remains primarily visual.
Despite the availability of automated tools, visual scoring of perfusion is typically currently performed by physicians with summed stress and summed rest scores based on the American Heart Association's (AHA) standard 17-segment polar map. This visual approach is time-consuming, subjective, and suffers from inter- [5], and intraobserver [4] variability, even when such scoring is performed by experienced physicians. Manual scoring of the stress and rest perfusion scans in nuclear cardiology is associated with significant variability. In Figure 1, we show the interobserver variability of two experienced physicians when interpreting MPI SPECT scans.
Figure 1.
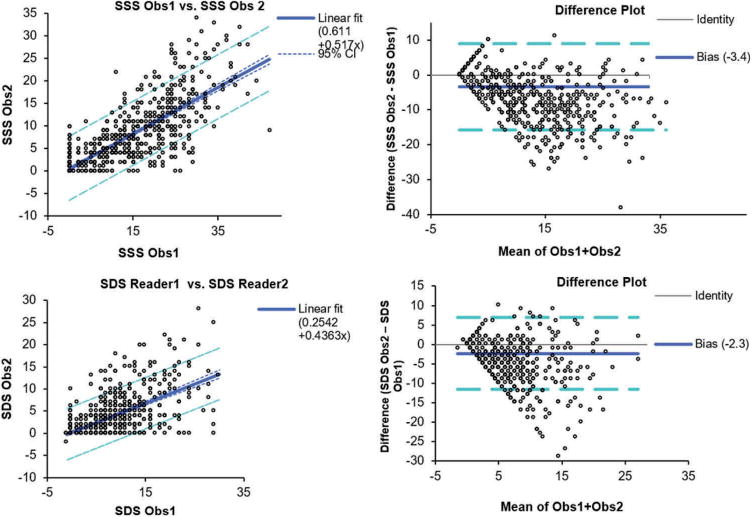
Inter-observer variability in nuclear cardiology.
An alternative approach to visual perfusion analysis is an automatic quantification of myocardial perfusion. This quantification approach is increasingly used in the clinical practice [3,8-10]. The initial step in the quantification of perfusion and function from MPI is the computer definition of the myocardium from gated and static perfusion image data and subsequent automatic reorientation of the images to standard coordinates (short axis/long axis of the heart) by the computer software. The accurate computer segmentation of the myocardium may be challenging due to potential perfusion defects, activity outside the myocardium, and image noise. Finally, the polar map representation of the myocardium is derived which is subsequently used for the quantification.
The quantification process is subsequently accomplished by statistical comparisons of pixels from test patients to other scans of normal patients internally stored by the quantification software. These comparisons allow identification of local areas of reduced myocardial perfusion, typically in polar map coordinates. The set of normal patients internally stored by the software is usually referred to as ‘normal database’ or ‘normal limits’ (when a collection of databases is considered, for example, stress and rest databases). This entire image analysis process can be almost entirely automated, with only a minority of image contours required some quick adjustments [11]. Incorrect definition of the myocardial contours in the minority of cases can result in false/positive defects, which can be mistaken for actual perfusion abnormalities. Therefore, currently, supervision by an operator is still required to verify myocardial segmentation. Typically, this is performed by the imaging technologist, before the scan interpretation by the physician.
Further automation of this process continues to be an active field of research, with several recent developments. For example, a new method that checks automatically derived myocardial contours for potential segmentation failures has been shown to reduce further the level of human supervision
Differences in summed stress scores (SSS) and summed difference scores (SDS) between 2 experienced observers reading 995 myocardial perfusion studies. The overall diagnostic agreement (SSS threshold 4) was 87%. required [11,12]. This approach has been compared to expert technologists and was found to be virtually equivalent (AUC = 0.96–1.00) to expert readers in the identification of failures.
Another recent development is group analysis where multiple studies are processed jointly to improve contour detection by avoiding interstudy inconsistencies. These new algorithms can lead to almost full automation of the image analysis steps with contour failure rate generally below 1% [2]. These recent software developments, combined with quality controls for automated left ventricle (LV) contours, would allow readers to target manual adjustment only to those few remaining studies flagged by the algorithm for potential errors. These tools, with further refinement, will ultimately allow entirely unsupervised automated perfusion scoring and quantification of myocardial function without compromising accuracy.
Relative quantification of MPI with normal databases is a powerful clinical tool which has been documented to rival expert physician's reading. For MPI SPECT, automated quantification of perfusion can be diagnostically equivalent to experienced readers [11], and has higher reproducibility [4]. In Figure 2, we show an overall diagnostic accuracy of the myocardial perfusion as compared to two experienced readers in a large population. The visual analysis in this study was performed in 4 steps. In the final step, the physicians had all the clinical information available. Despite that, we can see that the overall diagnostic accuracies of the physician reading and the computer analysis are similar. Most MPI scans in the USA are reported by provider-practice physicians outside of experienced academic settings; therefore, the accuracy and reproducibility of these readings may be even lower than that of the best experts. The automated systems are increasingly used clinically, but the final diagnosis and potential override of the results are possible by the clinician. To ensure standardization, we believe the fully automated results should be included in the final report.
Figure 2.
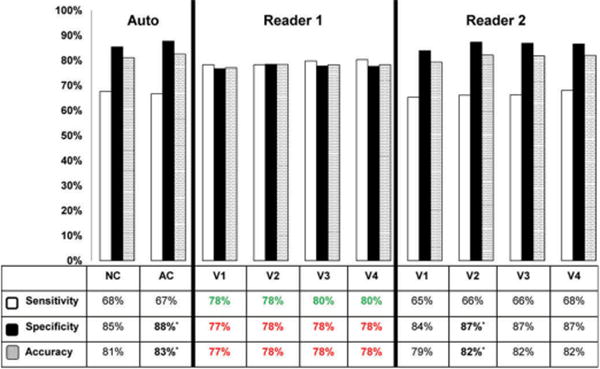
Diagnostic accuracy of automated versus visual analysis. A recent study confirmed that diagnostic accuracy for detecting CAD (≥70% stenosis) on a per-patient basis using automated methods is at least similar or marginally superior to that achieved by two expert visual readers. Comparisons were made for both attenuation-corrected (AC) and non-attenuation corrected (NC) data, and using variable amounts of imaging and clinical data available to the reader (V1-V4) with V4 representing full imaging and clinical information available. This research was originally published in JNM. Arsanjani R, Xu Y, Hayes SW, et al. Comparison of Fully Automated Computer Analysis and Visual Scoring for Detection of Coronary Artery Disease from Myocardial Perfusion SPECT in a Large Population. J Nucl Med. 2013;54:221–228. © by the Society of Nuclear Medicine and Molecular Imaging, Inc [11].
Although perfusion information is the primary information derived from MPI, several other imaging parameters can also be obtained in a fully automated fashion from the perfusion dataset. These include ejection fraction, diastolic/systolic volumes, motion thickening, transient ischemic dilation, myocardial mass, dyssynchrony parameters and myocardial blood flow. In total, several hundred quantitative image variables can be derived from a complete myocardial rest/stress MPI dataset. Currently, although these parameters are calculated by automated computer analysis, there are typically only subjectively incorporated in the final MPI diagnosis. This final human step introduces a layer of subjectivity into an otherwise fully quantitative dataset.
2.2. Cardiac CT
Coronary CT angiography (CTA) has recently emerged as a useful diagnostic test in selected stable but symptomatic patients needing noninvasive assessment of the coronary arteries [13–18]. It allows direct, noninvasive evaluation of the entire coronary tree. In 2014, ∼500,000 studies were performed in the USA and over 2 million worldwide, and these numbers are growing rapidly. The current clinical interpretation of coronary CTA uses subjective visual assessment of stenosis grade, with ≥50% considered significant. Coronary CTA is increasingly used for direct, noninvasive evaluation of the coronary arteries. Beyond stenosis, coronary CTA also permits assessment of atherosclerotic plaque (including total and non-calcified plaque burden) and coronary artery remodeling, previously only measurable through invasive techniques. However, standard visual interpretations have been shown to have a high rate of false-positive findings [14], which can lead to unnecessary additional testing and increased overall cost [19].
Over the last several years, several automated methods have been developed for standardized, semiautomated quantification of noncalcified and calcified plaques, and lumen measures from coronary CTA with [20–23], with research studies supporting this approach [23–25]. Such comprehensive quantification has been applied to large consecutive cohorts (800–1000 patients) [24]. An example of standardized software quantification in a plaque with high-risk adverse features is shown in Figure 3. Recent preliminary work has demonstrated that total plaque, non-calcified plaque burden measured by these tools – individualized per-patient measures of the coronary atherosclerotic burden – significantly improves prediction of lesion-specific ischemia by fractional flow reserve (FFR) over stenosis alone (area-under-receiver operator characteristics curve [AUC] 0.83 vs. 0.68) in lesions with intermediate stenosis [26]. Other automated approaches which can estimate FFR at any point of the vessel by computer processing from CTA have also been demonstrated [27].
Figure 3.
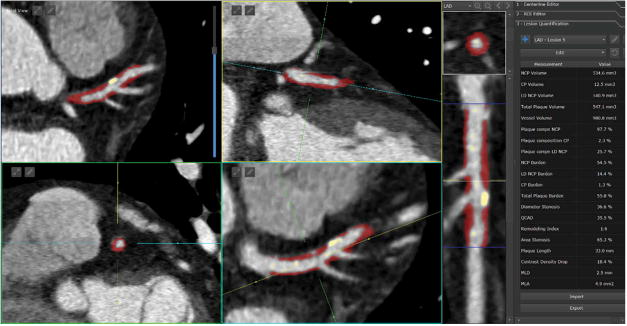
Standardized quantification of high-risk plaque lesion in the left anterior descending artery. Full color available online. The quantification allows standardized measurement of several parameters such as maximal stenosis, volumes of non-calcified (red) and calcified (yellow) plaques, total plaque volume, plaque composition, length of the lesion and drop of CT contrast.
Currently, although centerline extraction determination is mostly unsupervised, the accurate lumen segmentation for the determination of stenosis and detection and quantification of plaque may still require some manual subjective adjustments by the reporting physician. The CTA tools are not yet fully automated to the level achieved in nuclear cardiology, and still require significant time from a skilled operator for the contour adjustments. Nevertheless, it is probably a matter of time when these methods reach much higher level of automation, such as seen in nuclear cardiology studies. Unsupervised methods for the automated detection of subtle and significant coronary lesions from CTA have been recently demonstrated [28].
Apart from the primary application of cardiac CT in the coronary vessel analysis, CTA could be used for the assessment of perfusion and function and automated tools for these protocols that are being evaluated [29]. Furthermore, non-contrast cardiac CT can be utilized for detection of coronary calcium and thus the presence of atherosclerosis. These non-contrast images have lower resolution than CTA, and the lack of contrast can present difficulties in the automatic assignment of calcified lesions to correct coronary vessels. Several semiautomated and automated calcium scoring algorithms have been proposed using a publicly available standardized framework for the evaluation with 72 datasets [30]. Furthermore, methods for semi-automated measurements of epicardial coronary fat from non-contrast CT have also been developed and validated [31]; development of fully automated methods are currently in progress. These additional CT applications can in the future add to the comprehensive automated image assessment by this modality.
2.3. Echocardiography
Echocardiography is widely available, does not utilize any ionizing radiation and can be performed at the bedside. As a result, it is the most widely used noninvasive imaging technique in cardiology, with estimates as high as 7 million procedures annually in the USA, just under the Medicare program in 2011 [32]. In general, echocardiography is characterized by an excellent temporal resolution, allowing for real-time imaging and a relatively low cost. Echocardiography can be extended to the assessment of myocardial perfusion [33] in rest and stress conditions by administration of contrast material which changes reflective properties of the tissues, depending on the amount of contrast (which in turn depends on the blood flow). The high temporal sampling, the ability of realtime visualizations, and the absence of radiation dose make echocardiography an excellent imaging tool, not only for obtaining functional and structural insights about the myocardium, but also for the real-time image-guided cardiac interventions [34].
The acquisition of ultrasound images is still usually performed in 2D [35] using several standardized views from various parasternal, apical, and subcostal configurations of the transducer. The correct alignment of those views presents a challenge for sonographers. 3D-mode cardiac echo, which can be obtained with more complex transducers, solves this problem by the acquisition of volumes containing the myocardium. The 3D mode has the potential to derive more accurate and new quantitative parameters but suffers from reduced temporal resolution [36,37] and image quality compared to the 2D mode. The ultrasound imaging is constantly being improved by vendors who introduce new generation traducers (2D and 3D), faster electronics, and novel signal/ image processing methods. Over the years, this progress has led to radical improvements in image quality obtained in 2D and 3D echocardiography, and we expect this trend will continue in the future. The progress in technology also allows for the miniaturization of the ultrasound devices. It is currently possible to perform echocardiography studies using transducers which are directly connected to hand-held displays with no additional hardware needed. The signal from the transducer is processed by the hand-held device and displayed realtime to sonographer. This unprecedented mobility and flexibility in performing ultrasound examinations make the echo-cardiography one of the most important cardiac imaging modality with great prospects.
2.3.1. Segmentation
The most popular automated quantitative measurements obtained by echo are the ejection fraction and estimates of regional motion. These measurements can be obtained by tracking the endocardial wall in 2D and 3D movies. In a standard approach, automated image segmentation techniques are used to track the location of the endocardial wall and based on this, ejection fraction and myocardial wall motion are computed. The majority of analyses are done for 2D echo, where it is assumed that the main component of motion is visualized in the acquisition view. The process of tracking the endocardial wall is challenging and results in a relatively high interobserver variability [38,39]. Many approaches have been investigated in the past [40–45] in attempts to provide a robust solution. A thorough review of the segmentation algorithms for 2D ultrasound has been published [35].
For 3D transthoracic echocardiography, fully automated quantification software was developed that simultaneously detects LV and left atrium (LA) based on the detection of endocardial surfaces. In one approach, a model template describing the initial global shape and LV and LA chamber orientation is defined based on a large database of prior scans and followed by a patient-specific adaptation [46]. Once the model adopts to a current dataset, ventricle volumes, ejection fractions (EFs), 2D views and other parameters are derived from the 3D model and used for the cardiac function evaluation. The automatic model shows an excellent correlation with manually derived volumes from a single-beat 3D echocardiography in challenging atrial fibrillation patients [47].
2.3.2. Automated strain measurements
In recent years, quantitative values derived from echo sequences such as strain or strain rate have been shown to provide added diagnostic value. Strain is the fractional change in the length of myocardial tissue relative to its base length and is expressed as a percentage. Strain rate is the change in strain over time providing information on the speed of the deformation. Two general approaches can be used to measure strain which are based on tissue Doppler imaging (TDI) and speckle-tracking echocardiography (STE). In TDI, the strain rate can be directly calculated from the velocity calculated from Doppler measurements [48]. The strain can be obtained by integrating the strain rate over time. In STE, the noise structures (called speckles) on B-mode ultrasound images are identified and tracked by automated software frame-by-frame [49,50]. Based on those tracks and frame timings, the strain and the strain rate are computed. TDI measures the strain in a single direction whereas the STE measures the strain in 2D and 3D for a 2D ultrasound and 3D ultrasound, respectively. The ability to measure the strain and the strain rate in more than one dimension is a significant advantage of STE over TDI and contributed to substantial growth in the clinical utilization of this approach. One of the frequently used quantitative strain values is the global longitudinal strain (GLS) [51], which represents the average strain in longitudinal views. The GLS can be displayed either as a parametric map overlaid on original echo views or 17-segment AHA map. It has been shown that the GLS improves the diagnostic and prognostic accuracy of echocardiography in many LV dysfunctions. For example, the GLS and other strain-derived indicators are predictors of cardiac events [52], adverse events, and all-cause mortality [53].
For 2D ultrasound, the GLS can be measured on three standard apical echo views. The tracking points are first defined using semiquantitative approach. The strain is then automatically determined and can be displayed either as a parametric map or as a 17-segment AHA map. For the 3D ultrasound, the standard apical 2D views can be generated from the 3D data, and the strain can be obtained using 2D processing of those 2D-derived views.
2.3.3. Blood flow
Another potential application of quantitative imaging in echo-cardiography is the use of contrast echocardiography for estimation of the myocardial blood flow [54]. This approach may be useful for identification of patients with coronary stenosis [55]. According to a meta-analysis, the stress echocardiography with contrast has a similar diagnostic accuracy as SPECT MPI [56]. Although currently, the myocardial perfusion with contrast echocardiography is not a routine clinical examination, unlike other MPI techniques, it has the potential to be used at the bedside in the critical care setting [57]. The blood flow measurements with echocardiography are based on different principles as compared to other MPI modalities. The intravascular contrast agents used in myocardial perfusion contrast echocardiography consist of microbubbles (1–8 micro-millimiters in diameter) which have a high acoustic impedance created by water-gas interfaces. In a semiautomated quantification approach, the user defines the region of interest and the function of the microbubble concentration vs. time after the destruction of the microbubbles is determined. The slope of this curve is automatically analyzed to estimate the quantitative value of the blood flow [58]. Automated tools will be required for this method to reach broad clinical use.
2.4. Cardiac MRI
A standard clinical cardiac MRI (CMR) scan requires one hour of imaging time, followed by significant time for image postprocessing. Many automated approaches for CMR heart segmentation have been described [59–61]; however, large-scale clinical adoption of fully automated analysis methods for CMR analysis has not yet occurred. In many ways, CMR has the most to gain from automated segmentation because of the current lengthy and variable manual post-processing. Attempts at analysis automation are complicated by the variety of different CMR pulse sequences, scanning parameters and imaging protocols – each of which is tailored to the individual patient and the particular clinical question. Despite these limitations, there is a vast amount of ongoing work attempting to overcome these challenges and automate the key steps in CMR workflow.
2.4.1. Image acquisition
CMR requires highly skilled radiographers with sufficient knowledge of cardiac anatomy to prescribe appropriate imaging planes. Vendors have therefore invested in developing software tools capable of automated planning and acquisition. Some user input is still required for protocol selection, adjusting for patient parameters, identifying key anatomic reference points and providing final quality control. Nonetheless, semi-or near-automated image acquisition for CMR is a huge technological advance. Some of these tools have entered clinical mainstream providing highly reproducible acquisition, with increased time efficiency and minimal user input. Nevertheless, nonconventional anatomy such as congenital heart disease, complex scan protocols, and full user independence still pose significant challenges.
2.4.2. Segmentation & function
Heart chamber segmentation from CMR is an essential first step for the computation of functional indices such as ventricular volume, ejection fraction, mass, wall thickness, and wall motion. Manual delineation by experts is currently a standard practice – but it is a tedious, time-consuming process, prone to intra- and interobserver variability.
Although some relatively efficacious methods are commercially available for segmenting the LV on CMR [62], most still require a degree of user input for selecting key anatomic reference points, and confirmation that end-diastole and end-systole phases have been appropriately selected. Furthermore, quality control and fine adjustment remain user dependent. Moreover, as the geometry of the right ventricle (RV) is highly variable, automated algorithms for this chamber have proven more challenging to develop. Although several successful attempts have been described, none have proven robust enough to transition to mainstream, and RV segmentation remains a manual task requiring about 20 min per ventricle by a clinician.
The great need for automation has led to the development of a broad range of potential segmentation methods [63–66]. Shape prior information can also be used to guide the segmentation process, under the form of a statistical model, in a variational framework by using active shape and appearance models or via an atlas, using registration-based segmentation [67–73].
The challenges faced by all segmentation methods in CMR are: (i) fuzziness of cavity borders due to blood flow, acquisition artifacts, and partial volume effects, (ii) presence of papillary muscles in the LV and trabeculations in the RV, which have the same gray level as the surrounding myocardium but require exclusion during segmentation, and (iii) the complex patient-specific shape of the RV. Because isolated RV pathology is considered less common, most research effort has focused on the LV, leaving the problem of RV segmentation wide-open. Very recent techniques utilizing machine learning have been applied to the detection of motion patterns in RV to predict patient outcomes [74].
In summary, despite some success with semiautomated LV segmentation, fully automated chamber segmentation from CMR datasets is still a challenging problem due to several technical difficulties – especially for the RV. Current automated techniques suffer from poor robustness and accuracy and require large training datasets and a residual degree of user interaction.
2.4.3. Perfusion
Stress perfusion CMR, in particular, has become a first-line option in patients with suspected ischemic heart disease on the basis of data from several recent clinical studies and meta-analyses [75–78]. Studies such as CE-MARC (n = 752) have demonstrated higher diagnostic performance compared to SPECT. Higher diagnostic accuracy compared to nuclear MPI is considered to relate to the higher spatial resolution of perfusion CMR (2–3 mm in plane vs. 7–15 mm by SPECT vs. 4–10 mm by PET) which is particularly advantageous in females due to smaller hearts and in the setting of diffuse multivessel disease [79]. The scope for automated quantitative perfusion CMR in clinical practice and cardiovascular research is immense, but there remain several limitations that currently constrain this advance. The most important of these is a lack of standardization in image acquisition, contrast dosing protocols, post-processing and mathematical modeling. Efforts to standardize these aspects are currently under way [80,81].
In the interim, there are examples of successful in-house custom software solutions for small uniform datasets. For example, Tarroni et al. successfully demonstrated near-automated evaluation of stress/rest perfusion CMR using image noise density distribution for endocardial and epicardial border detection combined with non-rigid registration (n = 42) [82]. Noise characteristics of the blood pool and myocardium were used to facilitate automated endo-/ epicardial contouring. The only manual step was the placement of a seed point inside the LV cavity in a single frame and identification of the anterior RV insertion point. Contrast enhancement–time curves were automatically generated and used to calculate perfusion indices. Automated analysis of one sequence required <1 min and resulted in high-quality contrast enhancement curves both at rest and stress, showing expected patterns of the first-pass perfusion – compared to at least 10 min, and often 30 min for manual processing. The derived perfusion indices showed the same diagnostic accuracy as manual analysis (area under the receiver operating characteristic curve, up to 0.72 vs. 0.73).
2.4.4. Flow
The most common flow analysis performed in clinical CMR is that for the ascending and descending aorta. Automated quantitative flow analysis is dependent on the reliable identification of the vessel or region of interest, as well as its subsequent segmentation. To this end, Goel et al. demonstrated the feasibility of a robust fully automated tool to localize the ascending (AAo) and descending aorta (DAo) from acquired images using the Hough transform [83]. Hough transform assigns high values to circular objects and outputs these values as a spatial density map. This approach allowed to determine the location and size of the AAo and DAo in each phase of the cardiac cycle. The flow was then computed for the AAo and DAo for the whole cardiac cycle, using the Hough Transform for the approximate segmentation of the aorta. The final flow curve was generated by interpolating these flow values using a cubic spline. The algorithm was applied successfully to 1,884 participants. In the randomly selected 50-study validation set, linear regression showed an excellent correlation between the automated and manual methods for AAo flow (r = 0.99) and DAo flow (r = 0.99). Furthermore, Bland–Altman difference analysis demonstrated strong agreement with minimal bias for: AAo flow (mean difference (AM) = 0.47 ± 2.53 ml), and DAo flow (mean difference (AM) = 1.74 ± 2.47 ml).
2.4.5. Tissue characterization
Late gadolinium enhancement (LGE) images are typically used to detect regions of myocardial infarction or focal fibrosis with CMR. The distinction of normal from abnormal myocardium is usually clear -cut, and images can be rapidly interpreted visually with simple qualitative analysis. As the latter is not particularly burdensome, automated methods have not become particularly widespread or utilized for this purpose. Nonetheless, several studies have successfully shown the feasibility of automating segmentation of LGE images and quantifying scar burden as percentage myocardium or absolute volume [84–91]. Most methods employed simple standard deviation thresholding from a base healthy tissue intensity value. Other methods, such as full-width-at-half-maximum, maximum intensity projection and expectation-maximization fitting have also been proposed [85]. Any of these methods with appropriate refinement could be employed within a fully automated analysis workflow for CMR.
Currently, as with LGE imaging, interpretation of most additional forms of tissue characterization with CMR is based on qualitative analysis of visually detectable differences in the magnetic properties of tissues such as T1 and T2 relaxation times, rather than the quantitative values. Standardization of pulse sequences, as well as several quantitative parameters such as T1 and T2 mapping to assess diffuse tissue fibrosis and myocardial edema, are beginning to enter clinical routine and may ultimately lend themselves to more automated assessment.
3. Machine learning
Machine learning (ML) is a field of computer science that uses mathematical algorithms to identify patterns in data. These patterns can be used to predict or estimate various outcomes. ML algorithms typically build a model from available training data to make data-driven predictions or decisions. In recent years, ML techniques have emerged as highly effective methods for prediction and decision-making in a multitude of disciplines, including internet search engines, customized advertising, natural language processing, finance trending, and robotics [92,93]. These tools are increasingly becoming utilized in the medical field and in particular for analysis of images and data in cardiology.
Recently, novel approaches to image analysis and ML based on convolutional deep learning (CDL) gained enormous attention in recent years after the publication of CDL architecture [94,95] which performed considerably better than other types of state-of-the-art algorithms in image recognition task in ImageNet LSVRC-2010 contest for the recognition of natural images [96]. The algorithm classified images into 1000 categories and was trained based on 1.2 million training images. Since the publication of this work, the development of various implementations and applications skyrocketed. This growth is attributed to the availability of a large amount of data, innovations in CDL implementations (rectified linear units, dropout) [95,97] and the availability of relatively cheap computing resources provided by graphic processing units (GPUs), developed initially for the gaming industry.
The general idea of CDL is quite simple. In a standard machine learning, feature vectors (e.g. measurements) are derived from the images first. Subsequently, the machine learning (e.g. regression, support vector machines, random forests) is used to perform the classification. In CDL approach, a priori feature extraction is not usually performed, and raw images (or minimally processed images) are selected as the input to CDL network (Figure 4). The most important operations that CDL network performs are the series of convolution operations and image size reductions (frequently referred to as ‘pooling’). The number of layers (convolutional and pooling) varies depending on the task difficulty and the amount of available data and can vary from two layers for some small networks to 100s. The CDL is trained using annotated images (known ground truth). In this process, the parameters of convolutional kernels that define convolutional layers of the CDL are determined. Once trained, the network can be used for the prediction on previously unseen images.
Figure 4.
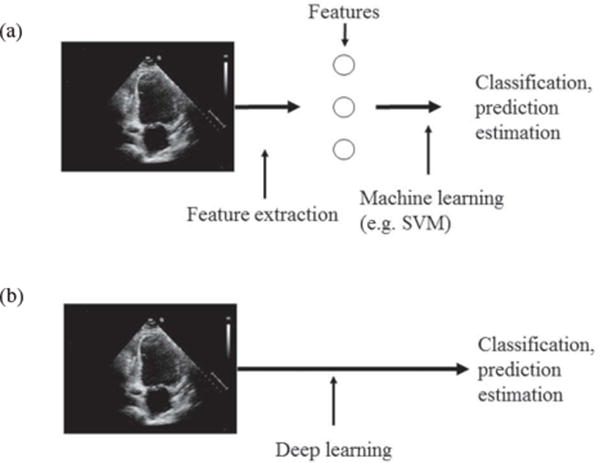
Traditional machine learning vs. deep learning. In typical machine learning (a) the pre-processing step extracts relevant features of the data (images) and then perform machine learning taking as an input extracted features. In deep learning (b) the deep network defines features as a part of learning process. The input to DL are raw data (images).
3.1. Application of machine learning to cardiac image analysis
Several applications of machine learning have been proposed for feature extraction and segmentation of cardiology images. Machine learning techniques can be utilized for automatic identification of lesions on coronary CTA images. For example, improvements for automatic lesion localization have been demonstrated by support vector machine method, which integrated several quantitative geometric and shape features (including stenosis, minimum luminal diameter, circularity, eccentricity), resulting in high sensitivity, specificity, and accuracy (93%, 95%, 94%) (Figure 5) [98]. Very recently, deep learning techniques have been applied for the identification of calcified plaques on CTA images, demonstrating improved accuracy over the existing methods [99].
Figure 5.
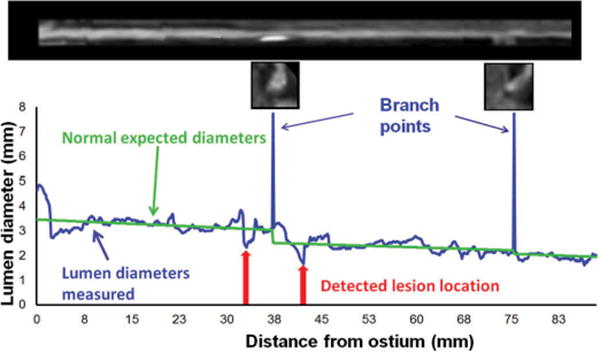
Automatic identification of coronary lesions from coronary CT Angiography by an algorithm based on machine learning. An example of lumen segmentation with lesion detection. This research was originally published in Journal of Medical Imaging. Kang D, Dey D, Slomka PJ, et al. Structured Learning Algorithm for Detection of Nonobstructive and Obstructive Coronary Plaque Lesions from Coronary CT Angiography. 2(1), 014003 (Mar 06 2015) [98].
Machine learning techniques have also been also applied to echocardiography. One of the most frequent tasks in echocardiography is to determine the exact position of the cardiac wall on frames of echo movies. This determination is needed for accurate estimation of quantitative parameters such as EF or LV mass. Because of high correlated noise and possible poor image quality, this task may be quite difficult. Artificial algorithms based on deep learning architectures can be used for automatic segmentation of the ventricle walls [94,100–102] or the detection of the bounding box containing heart valves in 2D echocardiography [103]. Recently published work shows promising results of utilizing speckle-tracking data and ML for automated discrimination of hypertrophic cardiomyopathy from physiological hypertrophy seen in athletes [104]. These encouraging results are steps toward a general artificial intelligence model of 2D echocardiographic data interpretation.
The control of image quality and identification of correction positioning of echocardiography views are also interesting applications of machine learning. The quality of echo images varies depending on the experience of the sonographer, patient cooperation, or ability to position the probe in an optimal location. Performing echocardiography imaging on obese or overweight patients may also be difficult due to problems with close positioning of the probe to the myocardium. Deep learning can be used to determine the quality of ultrasound [104,105] which may be a useful tool if available in real-time.
In CMR, the lack of standard units (compared, for example, to the Hounsfield scale in CT) makes it more challenging to directly apply simple intensity-based segmentation techniques, and images can also suffer from magnetic field in homogeneities and respiration artifacts. Accordingly, a machine learning solution to facilitate automated segmentation to obtain consistent measurements, and to save clinicians' time, is highly desirable. Initial approaches incorporating deep learning strategies have been demonstrated the segmentation of CMR images [106,107]. Nevertheless, for the reasons described above – predominantly the heterogeneity of imaging pulse sequences in MRI – this has been a difficult challenge, but it remains an area of active research, especially since societal guidelines push toward more standardized acquisition sequences [108].
Machine learning methods could also be used on a higher level to combine multiple automatically extracted image (quantitative parameters), into a derived overall metric with higher diagnostic power. For example, functional cardiac parameters and perfusion parameters both carry some diagnostic information and physicians try to combine this information in their minds when arriving at the final diagnosis. Studies have been reported, in which the overall diagnostic accuracy of conventional MPI was demonstrated to be improved by combining perfusion and functional parameters, utilizing a support vector machines (SVM) machine learning algorithm [109]. Perfusion deficits, ischemic changes, and ejection fraction changes between stress and rest were derived by the quantitative software. The polynomial SVM model demonstrated overall per-patient improvement in diagnostic accuracy of MPI as compared to individual quantitative parameters (86% vs. 81%; p < 0.01). The overall AUCs for detection of the obstructive disease were also significantly improved.
3.2. Combining clinical variables and risk factors with images for higher level interpretation
The final clinical diagnosis performed by the physician usually includes more information than what is available only from the images. When physicians review the patient scan, typically they consider information such as patient age, history, and other clinical findings, which together form a pre-test risk profile. Although this task is currently routinely performed by physicians during reporting, the precise probability of disease based on multiple factors cannot be reliably estimated by subjective human interpretation – particularly as the number of clinical and quantitative variables available for clinical consideration grows.
Several recent studies demonstrated that it is feasible to combine such clinical information with quantitative image information and subsequently derive enhanced probability or percentile diagnostic or prognostic scores to predict disease or patient outcome. Such tools could be valuable for the physicians since they would allow them to assign precise patient-specific risk scores for a particular outcome. This development could consequently lead to the optimized selection of various therapies, based on the quantitative rather than subjective risk assessment.
3.3. Studies combining clinical and image information
3.3.1. Enhanced diagnosis
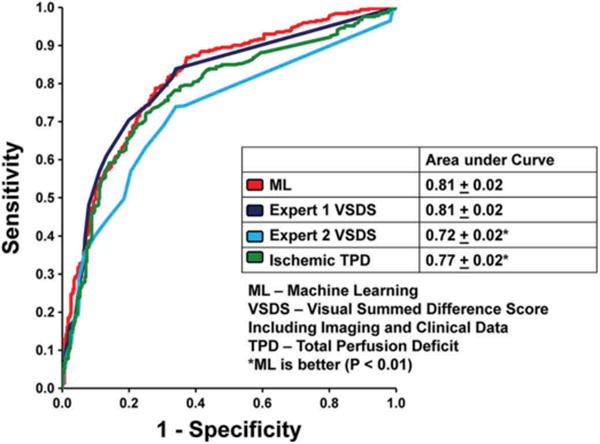
Several studies have been recently reported where such high-level interpretation is attempted. In one study, a boosting machine learning method (LogitBoost) [110] was utilized. This machine learning algorithm allows ensemble learning (a group of weak learners combined) by sequentially applying a classification algorithm to repeatedly reweighted versions of the training data and then taking a weighted majority vote of the sequence of classifiers. Such boosting algorithms are widely used in machine learning. The algorithm was trained in a 10-fold cross-validation experiment and compared it to quantitative perfusion assessment from SPECT MPI images (Total perfusion deficit-TPD), and visual scores by the experienced physicians in a large patient cohort with MPI scans (n = 1181), where expanded clinical information was recorded. When clinical information was provided to LogitBoost in addition to the imaging features LogitBoost achieved higher accuracy (87%) than one of the expert readers (82%) or automated TPD (83%; p < 0.01), and higher AUC (0.94 ± 0.01) than TPD (0.88 ± 0.01) or 2 visual readers (0.89,0.85; p < 0.001), for the detection of disease (Figure 6) [111].
Figure 6.

Application of machine learning to automated quantitation. When clinical and imaging information was provided to the LogitBoost machine learning technique in a large study (n = 1181), it achieved a significantly higher diagnostic accuracy for detection of significant CAD (87%) than one of the expert readers (82%) or TPD (83%; P < 0.01); and a higher AUC (0.94 ± 0.01) than TPD (0.88 ± 0.01) or 2 visual readers (0.89,0.85; P < 0.001). With kind permission from Springer Science+Business Media: Journal of Nuclear Cardiology, Improved accuracy of myocardial perfusion SPECT for detection of coronary artery disease by machine learning in a large population, Volume 20, 2013, Page 558, Arsanjani R, Xu Y, Dey D, et al., Figure 3, © by the American Society of Nuclear Cardiology [111].
3.3.2. Prediction of revascularization and ischemia
One issue facing clinicians is the choice of the possible treatments. Machine learning was used to integrate clinical data and quantitative image features for prediction of early revascularization in patients with suspected coronary artery disease. This approach was evaluated in 713 MPI studies with correlating invasive angiography within 90 days after the initial MPI scan. Several automatically derived image features along with clinical parameters including patient gender, history of hypertension and diabetes mellitus, ST depression on baseline ECG, ECG and clinical response during stress, and post-ECG probability were integrated by ML algorithm to predict revascularization events. The AUC for revascularization prediction by machine learning was similar to that of one reader and superior to another reader (Figure 7). Thus, machine learning approach was found to be comparable or better than the experienced reader in the prediction of the early revascularization after MPI.
Figure 7.
Nuclear cardiology prediction of revascularization after the scan. Automated measures have similar area under the ROC curve as one experienced reader and much higher than second reader in N = 713 patients. With kind permission from Springer Science+Business Media: Journal of Nuclear Cardiology, Prediction of revascularization after myocardial perfusion SPECT by machine learning in a large population, Volume 22, 2015, Page 882, Arsanjani R, Dey D, Khachatryan T, et al., Figure 2, © by the American Society of Nuclear Cardiology [112].
Machine learning has also been applied to integrate coronary CTA imaging to integrate biomarkers from quantitative CTA – to improve diagnostic accuracy. It has been demonstrated that a machine learning score, combining quantitative stenosis and plaque measures with machine learning, significantly improves the prediction of impaired regional myocardial flow reserve over stenosis [AUC 0.83 vs. 0.68] [114]. This preliminary data provides evidence that objective risk scores combining quantitative stenosis and plaque can improve prediction of ischemia from standard CTA, without the need for additional testing.
3.3.3. Machine learning for outcome prediction
Ultimately machine learning models are probably most useful in the quantitative assessment of risk. A recent study in the European Heart Journal demonstrated that outcomes such as all-cause mortality could be accurately predicted by machine learning algorithms which combine image information obtained from coronary CTA and clinical information [113]. A large, prospective multinational registry of patients undergoing CTA was utilized to assess the feasibility and accuracy of ML to predict 5-year all-cause mortality, and then compare the performance to existing clinical or imaging metrics.
The analysis included 10,030 patients with suspected coronary artery disease and 5-year follow-up from the CONFIRM (Coronary CT Angiography EvaluatioN For Clinical Outcomes: An InteRnational Multicenter) registry. All patients underwent coronary CTA as their standard of care. 25 clinical and 44 CTA parameters were evaluated, including segment stenosis score (SSS), segment involvement score (SIS), modified Duke index (DI), the number of segments with noncalcified, mixed or calcified plaques, age, sex, gender, standard cardiovascular risk factors and Framingham risk score (FRS). ML involved automated feature selection by information gain ranking, model building with a boosted ensemble algorithm, and ten-fold stratified cross-validation. ML exhibited a higher area under receiver operator characteristics curve compared to the FRS or coronary CTA severity scores alone (SSS, SIS, DI) for predicting all-cause mortality (ML: 0.79 vs. FRS: 0.61, SSS: 0.64, SIS: 0.64, DI: 0.62; p < 0.001) (Figure 8). By net reclassification improvement metric, it could be shown that significant percentage of patients could be more appropriately assigned a risk category when such new ML score was used (Table 2). The 5-year risk of mortality was significantly better than existing clinical or coronary CTA metrics alone. The observed efficacy suggests ML has an important clinical role in evaluating prognostic risk in individual patients with suspected coronary artery disease.
Figure 8.
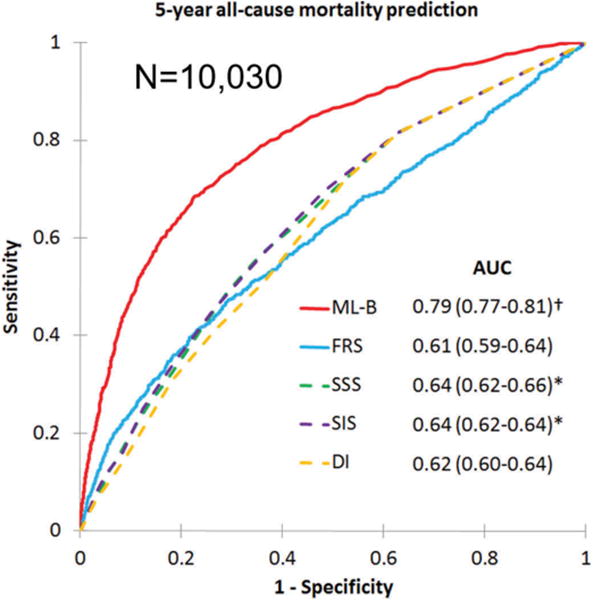
5-year all-cause prediction of mortality. Machine learning boosting model (ML-B) was able to predict mortality much better than the existing imaging scores derived by visual expert analysis (Summed segmental scores-SSS, Segment Involvement scores-SIS) or clinical scores such Duke index (DI) or Framingham Risk Score (FRS). Reproduced, with permission, from Manish Motwani et al. Machine Learning For Prediction Of All-Cause Mortality In Patients With Suspected Coronary Artery Disease: A 5-Year Multicentre Prospective Registry Analysis. European Heart Journal (2016) DOI: http://dx.doi.org/10.1093/eurheartj/ehw188. Published by Oxford University Press on behalf of the European Society of Cardiology [113].
Table 2.
Risk Reclassification table for prediction of mortality by machine learning (ML) combining coronary CT angiography versus existing clinical score – Framingham risk score (FRS).
| FRS risk category | Machine learning risk category | Total | ||
|---|---|---|---|---|
| Low | Intermediate | High | ||
| Death, n | ||||
| Low | 38 | 89 | 108 | 235 |
| Intermediate, | 22 | 124 | 60 | 206 |
| High | 11 | 108 | 185 | 304 |
| Total | 71 | 321 | 353 | 745 |
| No Death, n | ||||
| Low | 2193 | 1578 | 256 | 4027 |
| Intermediate | 1209 | 1700 | 209 | 3118 |
| High | 487 | 1169 | 484 | 2140 |
| Total | 3889 | 4447 | 949 | 9285 |
Overall NRI index (95% CI) 0.24 (0.19–0.30) p < 0.0001
4. Issues and potential pitfalls
The technological development described above are encouraging. There are however several foreseeable obstacles and possible pitfalls when these technologies are translated into clinical use.
4.1. Availability of the data
Although computer modeling and image processing techniques have advanced tremendously in the recent years, one key issue is the availability of the data for training and validation of the new methods. There are several legal barriers in the medical community when it comes to medical data sharing. Even fully de-identified data usually require investigational research board approvals before any sharing or analysis, and typically these datasets are not available publicly. There are often legal barriers, and the data ownership is not always clear. These obstacles, unfortunately, will hinder the optimization of the image processing and analysis methods, especially if these methods are based on machine learning. In particular, deep learning models require large datasets during the training phase to obtain sufficient accuracy [115]. If large repositories of well-characterized image sets and associated clinical outcomes were publicly available, it would facilitate further development and optimization of these tools for the physicians. Such data sharing should be justified especially when the collection of these datasets are funded by public research funds.
Encouragingly, the recent biobanks and international consortia managing medical imaging databases, such as the UK Biobank [116], the Cardiac Atlas Project [117], or the VISCERAL Project [118], have solved many of these difficult problems in ethics of data sharing, medical image organization, and data distribution – in particular, when aggregating the data from multiple sources.
For the image segmentation tasks, valuable open datasets have been provided online under the Grand Challenge initiative [119]. This site contains large, extensively validated and annotated training datasets for the segmentation of LV by MRI, CT, and echocardiography. Especially noteworthy, is perhaps the Kaggle (universal platform for testing various machine learning algorithms) second annual Data Science Bowl challenge with over 1000 2D cardiac MRI datasets [120]. This dataset is larger than any other dataset previously compiled for this modality and will facilitate new automated algorithm developments. An online competition system with cash prize has been set up for the researchers where their algorithms can be evaluated on this dataset. There are also smaller (<100 patients) validated datasets available, for example for the segmentation of coronary arteries from CT angiography [121]. Such worthwhile initiatives will facilitate further development of automated tools for cardiac modalities, including novel machine-learning approaches.
4.2. Data standardization
The picture archiving and communication system (PACS) and the Digital Imaging and Communications in Medicine (DICOM) standards have been invaluable in providing a consistency platform for data. However, studies coming from multiple centers often use specific nomenclature, follow different guidelines or utilize different acquisition protocols. In these cases, even these standards are not sufficient. For example, there is no standard way to store some of the key image-related information (e.g. cardiac acquisition plane) as the naming often depends on the custom set up of the scanning terminal and the language practiced at the imaging center. Even standard DICOM tags often contain vendor specific nomenclature. Such differences reduce our ability to effectively query and explore the databases for relevant images.
4.3. Physicians resistance
The emergence of high-level image analysis and image interpretation tools may perhaps evoke some fear in the medical profession about their work being somehow replaced by such automated analysis and interpretation. However, these developments should be viewed by the physicians in a positive light, since they will allow to achieve a more optimal, answer personalized and a superior tool for the quantitative risk or diagnostic assessment for their patients. Indeed, these tools will enable meaningful implementation of precision-medicine principles by the physicians. Thus, the overall ability to improve patients' well-being by the medical care will be significantly enhanced. That should be a compelling argument in overcoming any resistance to the clinical translation of these new tools.
4.4. Integration of the electronic patient record with imaging data
When making a diagnosis, physicians integrate clinical history with imaging data. As described above, it has been demonstrated that computer algorithms can derive risk scores for patients utilizing such integrated information. However, in clinical practice sometimes the electronic patient record is not interfaced directly with the imaging software and workstations. It would not be feasible to manually enter and transfer multiple clinical variables for each patient to obtain integrated analysis or risk score. There are initiatives in the cardiology community which can help in this respect. For example, in nuclear cardiology community, the American Society of Nuclear Cardiology has led efforts to gather all relevant clinical data via the ImageGuide registry [122]. Reporting software vendors are cooperating and will be providing appropriate interfaces to hospital systems to collect and transmit such data linking these with the images. This infrastructure could be potentially used to feed the machine-learning models. Consequently, this would allow the risk score to be automatically computed and be available during reporting.
4.5. External validation
Computer algorithms trained by image and clinical data from one institution or on a particular type of equipment may not perform optimally if patient or image characteristics changes. Therefore, machine learning software should be developed and validated with multicenter registries covering a broad spectrum of patient demographics. Formal validations on data external to the training population may need to be performed. However, this may be difficult due to the potential data sharing barriers described above. In addition, one key aspect of machine learning in a medical environment is that incoming patient characteristics and disease patterns may change in time even in the same center and therefore machine learning models may need to be adapted incrementally to adjust for this. The exact mechanisms for such continuing evolution of medical decision support systems have not yet been worked out in clinical practice.
4.6. Legal issues
One potential obstacle is the issue of the clinical clearance for the fully automated by the appropriate agencies (US FDA, CE Mark). Typically, most current diagnostic software tools considered as Class 2 (medium-risk) devices, which are involved in the diagnosis but are not providing a complete and final diagnosis. Fully automated systems would be likely classified as Class 3 (high-risk) devices with much higher bar with respect to performance and validation. This issue is similar to that facing the developers of self-driving cars where even if the statistical performance may better than that of the best driver, an individual error leading to an accident is still a liability. In the medical field, it is probably also not enough to demonstrate the higher overall performance as compared to the expert physicians, as there may be individual cases (patients) where the automated system would still provide an incorrect answer, despite overall superiority. The other potential legal problem is the validation and certification of a system which is designed to continuously learn from new patient data. Some new guidelines will have to be established for the metrics of performance of such devices, as current validation regiments assume static system performance. It is likely that a physician override and check will still be required for such systems in the foreseeable future.
5. Expert commentary
Computer analysis and high-level interpretation of cardiac imaging are becoming feasible. In cardiology, the field of nuclear cardiology is most advanced in this respect and may be viewed as a harbinger of things to come. Recent advances in machine learning will facilitate further rapid development. It is highly beneficial to combine image information with clinical data to provide optimal diagnosis and prognosis. Although quantitative tools are used routinely in the clinical practice, especially in nuclear cardiology, the higher-level tools combining multiple features and clinical data are not yet prevalent. However, in several recent research studies, they have been demonstrated to show a very high potential. These upcoming developments will not replace the role of physicians but will provide them with highly accurate tools to detect disease, stratify risk in an easy to understand manner and optimize patient-specific treatment and further tests.
6. Five-year view
It is likely that in a 5-year time the level of automation for analysis and interpretation will be significantly raised, compared to what is possible today. It is likely, that entirely unsupervised extraction of all image parameters will be possible for nuclear cardiology, and minimal supervision will be required for other modalities. Greater standardization of acquisition protocols will be needed to maximize the potential gains from automation and machine learning. This goal will require significant support from the vendors but also from the medical centers - to facilitate data sharing. Fully quantitative diagnostic and risk stratification scores will be developed for clinicians and these will become integrated with the imaging software. Risk stratification will transition from oversimplified population-based risk scores to machine-learning-based metrics incorporating a large number of clinical and imaging variables in real-time beyond the limits of human cognition - this will deliver highly accurate and individual personalized risk assessments and facilitate tailored management plans. However, the clinical translation of these exciting techniques will depend on many factors outside of technological progress, such as aspects related to logistical, legal, standardization, and reimbursement.
Key issues.
Data availability for algorithm training
Potential resistance from the medical community
The level of automation for the image processing methods
Integration of multiple imaging features
The need to validate algorithms in large populations
Seamless connection of clinical and imaging data
Acknowledgments
Funding: This work was supported in part by grant R01HL089765 (PI: Piotr Slomka) and grant R01HL133616-01 (PI: Damini Dey) from the National Heart, Lung, and Blood Institute/National Institute of Health (NHLBI/NIH).
Cedars-Sinai receives royalties for licensing of quantitative perfusion software, a portion of which is shared with the inventors of which PJ. Slomka, DS. Berman and G. Germano are among. Arkadiusz Sitek is an employee of Philips Research North America.
Footnotes
Declaration of interest: The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
References
Papers of special note have been highlighted as either of interest (•) or of considerable interest (••) to readers.
- 1.Heo R, Nakazato R, Kalra D, et al. Noninvasive imaging in coronary artery disease. Semin Nucl Med. 2014;44(5):398–409. doi: 10.1053/j.semnuclmed.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Germano G, Kavanagh PB, Ruddy TD, et al. “Same-patient processing” for multiple cardiac SPECT studies. 2. Improving quantification repeatability. J Nucl Cardiol. 2016;23(6):1442–1453. doi: 10.1007/s12350-016-0674-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu YH. Quantification of nuclear cardiac images: the Yale approach. J Nucl Cardiol. 2007;14(4):483–491. doi: 10.1016/j.nuclcard.2007.06.005. [DOI] [PubMed] [Google Scholar]
- 4.Xu Y, Hayes S, Ali I, et al. Automatic and visual reproducibility of perfusion and function measures for myocardial perfusion SPECT. J Nucl Cardiol. 2010;17(6):1050–1057. doi: 10.1007/s12350-010-9297-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berman DS, Kang X, Gransar H, et al. Quantitative assessment of myocardial perfusion abnormality on SPECT myocardial perfusion imaging is more reproducible than expert visual analysis. J Nucl Cardiol. 2009;16(1):45–53. doi: 10.1007/s12350-008-9018-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Einstein AJ. Effects of radiation exposure from cardiac imaging: how good are the data? J Am Coll Cardiol. 2012;59(6):553–565. doi: 10.1016/j.jacc.2011.08.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sciagra R, Passeri A, Bucerius J, et al. Clinical use of quantitative cardiac perfusion PET: rationale, modalities and possible indications. Position paper of the cardiovascular committee of the European Association of Nuclear Medicine (EANM) Eur J Nucl Med Mol Imaging. 2016;43(8):1530–1545. doi: 10.1007/s00259-016-3317-5. [DOI] [PubMed] [Google Scholar]
- 8.Germano G, Kavanagh PB, Slomka PJ, et al. Quantitation in gated perfusion SPECT imaging: the Cedars-Sinai approach. J Nucl Cardiol. 2007;14(4):433–454. doi: 10.1016/j.nuclcard.2007.06.008. [DOI] [PubMed] [Google Scholar]
- 9.Garcia EV, Faber TL, Cooke CD, et al. The increasing role of quantification in clinical nuclear cardiology: the emory approach. J Nucl Cardiol. 2007;14(4):420–432. doi: 10.1016/j.nuclcard.2007.06.009. [DOI] [PubMed] [Google Scholar]
- 10.Ficaro EP, Lee BC, Kritzman JN, et al. Corridor4DM: the Michigan method for quantitative nuclear cardiology. J Nucl Cardiol. 2007;14(4):455–465. doi: 10.1016/j.nuclcard.2007.06.006. [DOI] [PubMed] [Google Scholar]
- 11••.Arsanjani R, Xu Y, Hayes SW, et al. Comparison of fully automated computer analysis and visual scoring for detection of coronary artery disease from myocardial perfusion SPECT in a large population. J Nucl Med. 2013;54(2):221–228. doi: 10.2967/jnumed.112.108969. A large study comparing the visual and automatic analysis of nuclear cardiology studies. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu Y, Kavanagh P, Fish M, et al. Automated quality control for segmentation of myocardial perfusion SPECT. J Nucl Med. 2009;50(9):1418–1426. doi: 10.2967/jnumed.108.061333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Taylor AJ, Cerqueira M, Hodgson JM, et al. ACCF/SCCT/ACR/AHA/ ASE/ASNC/NASCI/SCAI/SCMR 2010 appropriate use criteria for cardiac computed tomography: a report of the american college of cardiology foundation appropriate use criteria task force, the society of cardiovascular computed tomography, the american college of radiology, the american heart association, the american society of echocardiography, the american society of nuclear cardiology, the north american society for cardiovascular imaging, the society for cardiovascular angiography and interventions, and the society for cardiovascular magnetic resonance. J Am Coll Cardiol. 2010;56(22):1864–1894. doi: 10.1016/j.jacc.2010.07.005. [DOI] [PubMed] [Google Scholar]
- 14.Budoff MJ, Dowe D, Jollis JG, et al. Diagnostic performance of 64-multidetector row coronary computed tomographic angiography for evaluation of coronary artery stenosis in individuals without known coronary artery disease: results from the prospective multi-center ACCURACY (Assessment by Coronary Computed Tomographic Angiography of Individuals Undergoing Invasive Coronary Angiography) trial. J Am Coll Cardiol. 2008;52(21):1724–1732. doi: 10.1016/j.jacc.2008.07.031. [DOI] [PubMed] [Google Scholar]
- 15.Hausleiter J, Meyer T, Hadamitzky M, et al. Non-invasive coronary computed tomographic angiography for patients with suspected coronary artery disease: the Coronary Angiography by Computed Tomography with the Use of a Submillimeter resolution (CACTUS) trial. Eur Heart J. 2007;28(24):3034–3041. doi: 10.1093/eurheartj/ehm150. [DOI] [PubMed] [Google Scholar]
- 16.Achenbach S, Ropers U, Kuettner A, et al. Randomized comparison of 64-slice single- and dual-source computed tomography coronary angiography for the detection of coronary artery disease. J Am Coll Cardiol Cardiovascular Imaging. 2008;1(2):177–186. doi: 10.1016/j.jcmg.2007.11.006. [DOI] [PubMed] [Google Scholar]
- 17.Miller JM, Rochitte CE, Dewey M, et al. Diagnostic performance of coronary angiography by 64-row CT. N Engl J Med. 2008;359(22):2324–2336. doi: 10.1056/NEJMoa0806576. [DOI] [PubMed] [Google Scholar]
- 18.Meijboom WB, Van Mieghem CAG, Mollet NR, et al. 64-slice computed tomography coronary angiography in patients with high, intermediate, or low pretest probability of significant coronary artery disease. J Am Coll Cardiol. 2007;50:1469–1475. doi: 10.1016/j.jacc.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 19.Min JK, Shaw LJ, Berman DS. The present state of coronary computed tomography angiography: a process in evolution. J Am Coll Cardiol. 2010;55(10):957–965. doi: 10.1016/j.jacc.2009.08.087. [DOI] [PubMed] [Google Scholar]
- 20.Dey D, Schepis T, Marwan M, et al. Automated three-dimensional quantification of non-calcified coronary plaque from coronary CT angiography: comparison with intravascular ultrasound. Radiology. 2010;257(2):516–522. doi: 10.1148/radiol.10100681. [DOI] [PubMed] [Google Scholar]
- 21.Dey D, Cheng VY, Slomka PJ, et al. Automated 3-dimensional quantification of noncalcified and calcified coronary plaque from coronary CT angiography. J Cardiovasc Comput Tomogr. 2009;3(6):372–382. doi: 10.1016/j.jcct.2009.09.004. [DOI] [PubMed] [Google Scholar]
- 22.Schuhbaeck A, Dey D, Otaki Y, et al. Interscan reproducibility of quantitative coronary plaque volume and composition from CT coronary angiography using an automated method. Eur Radiol. 2014;24(9):2300–2308. doi: 10.1007/s00330-014-3253-3. [DOI] [PubMed] [Google Scholar]
- 23.Dey D, Achenbach S, Schuhbaeck A, et al. Comparison of quantitative atherosclerotic plaque burden from coronary CT angiogra-phy in patients with first acute coronary syndrome and stable coronary artery disease. J Cardiovasc Comput Tomogr. 2014;8(5):368–374. doi: 10.1016/j.jcct.2014.07.007. [DOI] [PubMed] [Google Scholar]
- 24.Kral BG, Becker LC, Vaidya D, et al. Noncalcified coronary plaque volumes in healthy people with a family history of early onset coronary artery disease. Circ Cardiovas Imag. 2014;7(3):446–453. doi: 10.1161/CIRCIMAGING.113.000980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hell MM, Dey D, Marwan M, et al. Non-invasive prediction of hemodynamically significant coronary artery stenoses by contrast density difference in coronary CT angiography. Eur J Radiol. 2015;84(8):1502–1508. doi: 10.1016/j.ejrad.2015.04.024. [DOI] [PubMed] [Google Scholar]
- 26.Diaz-Zamudio M, Dey D, Schuhbaeck A, et al. Automated quantitative plaque burden from coronary CT angiography noninva-sively predicts hemodynamic significance by using fractional flow reserve in intermediate coronary lesions. Radiology. 2015;276(2):408–415. doi: 10.1148/radiol.2015141648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gaur S, Ovrehus KA, Dey D, et al. Coronary plaque quantification and fractional flow reserve by coronary computed tomography angiography identify ischaemia-causing lesions. Eur Heart J. 2016;37(15):1220–1227. doi: 10.1093/eurheartj/ehv690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kang D, Slomka PJ, Nakazato R, et al. Automated knowledge-based detection of nonobstructive and obstructive arterial lesions from coronary CT angiography. Med Phys. 2013;40(4):041912. doi: 10.1118/1.4794480. [DOI] [PubMed] [Google Scholar]
- 29.Mor-Avi V, Kachenoura N, Maffessanti F, et al. Three-dimensional quantification of myocardial perfusion during regadenoson stress computed tomography. Eur J Radiol. 2016;85(5):885–892. doi: 10.1016/j.ejrad.2016.02.028. [DOI] [PubMed] [Google Scholar]
- 30.Wolterink JM, Leiner T, de Vos BD, et al. An evaluation of automatic coronary artery calcium scoring methods with cardiac CT using the orCaScore framework. Med Phys. 2016;43(5):2361–2373. doi: 10.1118/1.4945696. [DOI] [PubMed] [Google Scholar]
- 31.Dey D, Wong ND, Tamarappoo B, et al. Computer-aided non-contrast CT-based quantification of pericardial and thoracic fat and their associations with coronary calcium and metabolic syndrome. Atherosclerosis. 2010;209(1):136–141. doi: 10.1016/j.atherosclerosis.2009.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.https://www.ncbi.nlm.nih.gov/books/NBK208663/. 2016.
- 33.Bierig SM, Mikolajczak P, Herrmann SC, et al. Comparison of myo-cardial contrast echocardiography derived myocardial perfusion reserve with invasive determination of coronary flow reserve. Eur J Echocardiogr. 2009;10(2):250–255. doi: 10.1093/ejechocard/jen217. [DOI] [PubMed] [Google Scholar]
- 34.Silvestry FE, Kerber RE, Brook MM, et al. Echocardiography-guided interventions. J Am Soc Echocardiogr. 2009;223:213–231. doi: 10.1016/j.echo.2008.12.013. quiz 316-7. [DOI] [PubMed] [Google Scholar]
- 35.Noble JA, Boukerroui D. Ultrasound image segmentation: a survey. IEEE Trans Med Imaging. 2006;25(8):987–1010. doi: 10.1109/tmi.2006.877092. [DOI] [PubMed] [Google Scholar]
- 36.Bhan A, Kapetanakis S, Monaghan MJ. Three-dimensional echocar-diography. Heart. 2010;96(2):153–163. doi: 10.1136/hrt.2009.176347. [DOI] [PubMed] [Google Scholar]
- 37.Weyman AE. The Year in Echocardiography. J Am Coll Cardiol. 2010;53(17):1558–1567. doi: 10.1016/j.jacc.2009.01.042. [DOI] [PubMed] [Google Scholar]
- 38.Hoffmann R, Von Bardeleben S, Kasprzak JD, et al. Analysis of regional left ventricular function by cineventriculography, cardiac magnetic resonance imaging, and unenhanced and contrast-enhanced echocardiography: a multicenter comparison of methods. J Am Coll Cardiol. 2006;47:121–128. doi: 10.1016/j.jacc.2005.10.012. [DOI] [PubMed] [Google Scholar]
- 39.Johri AM, Picard MH, Newell J, et al. Can a teaching intervention reduce interobserver variability in lvef assessmenta quality control exercise in the echocardiography lab. JACC Cardiovascular Imaging. 2011;4:821–829. doi: 10.1016/j.jcmg.2011.06.004. [DOI] [PubMed] [Google Scholar]
- 40.Haak A, Vegas-Sanchez-Ferrero G, Mulder HW, et al. Segmentation of multiple heart cavities in 3-D transesophageal ultrasound images. IEEE Trans Ultrason Ferroelectr Freq Control. 2015;62(6):1179–1189. doi: 10.1109/TUFFC.2013.006228. [DOI] [PubMed] [Google Scholar]
- 41.Stebbing RV, Namburete AI, Upton R, et al. Data-driven shape parameterization for segmentation of the right ventricle from 3D +t echocardiography. Med Image Anal. 2015;21(1):29–39. doi: 10.1016/j.media.2014.12.002. [DOI] [PubMed] [Google Scholar]
- 42.Chalana V, Linker DT, Haynor DR, et al. A multiple active contour model for cardiac boundary detection on echocardiographic sequences. IEEE Trans Med Imaging. 1996;15(3):290–298. doi: 10.1109/42.500138. [DOI] [PubMed] [Google Scholar]
- 43.Friedland N, Adam D. Automatic ventricular cavity boundary detection from sequential ultrasound images using simulated annealing. IEEE Trans Med Imaging. 1989;8(4):344–353. doi: 10.1109/42.41487. [DOI] [PubMed] [Google Scholar]
- 44.Hozumi T, Yoshida K, Yoshioka H, et al. Echocardiographic estimation of left ventricular cavity area with a newly developed automated contour tracking method. J Am Soc Echocardiogr. 1997;10(8):822–829. doi: 10.1016/s0894-7317(97)70042-7. [DOI] [PubMed] [Google Scholar]
- 45.Mignotte M, Meunier J. A multiscale optimization approach for the dynamic contour-based boundary detection issue. Comput Med Imaging Graph. 2001;25(3):265–275. doi: 10.1016/s0895-6111(00)00075-6. [DOI] [PubMed] [Google Scholar]
- 46.Tsang W, Salgo IS, Medvedofsky D, et al. Transthoracic 3D echo-cardiographic left heart chamber quantification using an automated adaptive analytics algorithm. JACC Cardiovasc Imaging. 2016;9(7):769–782. doi: 10.1016/j.jcmg.2015.12.020. [DOI] [PubMed] [Google Scholar]
- 47.Otani K, Nakazono A, Salgo IS, et al. Three-dimensional echocardio-graphic assessment of left heart chamber size and function with fully automated quantification software in patients with atrial fibrillation. J Am Soc Echocardiogr. 2016;29(10):955–965. doi: 10.1016/j.echo.2016.06.010. [DOI] [PubMed] [Google Scholar]
- 48.Urheim S, Edvardsen T, Torp H, et al. Myocardial strain by doppler echocardiography. Validation of a new method to quantify regional myocardial function. Circulation. 2000;102(10):1158–1164. doi: 10.1161/01.cir.102.10.1158. [DOI] [PubMed] [Google Scholar]
- 49.Leitman M, Lysyansky P, Sidenko S, et al. Two-dimensional strain-a novel software for real-time quantitative echocardiographic assessment of myocardial function. J Am Soc Echocardiogr. 2004;17(10):1021–1029. doi: 10.1016/j.echo.2004.06.019. [DOI] [PubMed] [Google Scholar]
- 50.Toyoda T, Baba H, Akasaka T, et al. Assessment of regional myo-cardial strain by a novel automated tracking system from digital image files. J Am Soc Echocardiogr. 2004;17(12):1234–1238. doi: 10.1016/j.echo.2004.07.010. [DOI] [PubMed] [Google Scholar]
- 51.Stanton T, Leano R, Marwick TH. Prediction of all-cause mortality from global longitudinal speckle strain: comparison with ejection fraction and wall motion scoring. Circ Cardiovasc Imaging. 2009;2(5):356–364. doi: 10.1161/CIRCIMAGING.109.862334. [DOI] [PubMed] [Google Scholar]
- 52.Russo C, Jin Z, Elkind MS, et al. Prevalence and prognostic value of subclinical left ventricular systolic dysfunction by global longitudinal strain in a community-based cohort. Eur J Heart Fail. 2014;16(12):1301–1309. doi: 10.1002/ejhf.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ersboll M, Valeur N, Mogensen UM, et al. Prediction of all-cause mortality and heart failure admissions from global left ventricular longitudinal strain in patients with acute myocardial infarction and preserved left ventricular ejection fraction. J Am Coll Cardiol. 2013;61(23):2365–2373. doi: 10.1016/j.jacc.2013.02.061. [DOI] [PubMed] [Google Scholar]
- 54.Lepper W, Belcik T, Wei K, et al. Myocardial contrast echocardiography. Circulation. 2004;109(25):3132–3135. doi: 10.1161/01.CIR.0000132613.53542.E9. [DOI] [PubMed] [Google Scholar]
- 55.Arnold JR, Karamitsos TD, Pegg TJ, et al. Adenosine stress myocardial contrast echocardiography for the detection of coronary artery disease: a comparison with coronary angiography and cardiac magnetic resonance. JACC Cardiovasc Imaging. 2010;3(9):934–943. doi: 10.1016/j.jcmg.2010.06.011. [DOI] [PubMed] [Google Scholar]
- 56.Stress echocardiography with contrast for the diagnosis of coronary artery disease: an evidence-based analysis. Ont Health Technol Assess Ser. 2010;10(10):1–59. [PMC free article] [PubMed] [Google Scholar]
- 57.Orde S, McLean A. Bedside myocardial perfusion assessment with contrast echocardiography. Critical Care (London, England) 2016;20:58. doi: 10.1186/s13054-016-1215-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wei K, Jayaweera AR, Firoozan S, et al. Quantification of myocardial blood flow with ultrasound-induced destruction of microbubbles administered as a constant venous infusion. Circulation. 1998;97(5):473–483. doi: 10.1161/01.cir.97.5.473. [DOI] [PubMed] [Google Scholar]
- 59.Peng P, Lekadir K, Gooya A, et al. A review of heart chamber segmentation for structural and functional analysis using cardiac magnetic resonance imaging. Magma. 2016;29(2):155–195. doi: 10.1007/s10334-015-0521-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Karim R, Housden RJ, Balasubramaniam M, et al. Evaluation of current algorithms for segmentation of scar tissue from late gadolinium enhancement cardiovascular magnetic resonance of the left atrium: an open-access grand challenge. J Cardiovasc Magn Reson. 2013;15:105. doi: 10.1186/1532-429X-15-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tobon-Gomez C, Geers A, Peters J, et al. Benchmark for algorithms segmenting the left atrium from 3D CT and MRI datasets. IEEE Trans Med Imaging. 2015;34(7):1460–1473. doi: 10.1109/TMI.2015.2398818. [DOI] [PubMed] [Google Scholar]
- 62.O'Donnell T, Funka-Lea G, Tek H, et al. Comprehensive cardiovascular image analysis using MR and CT at siemens corporate research. Int J Comput Vis. 2006;70(2):165–178. [Google Scholar]
- 63.Frangi AF, Niessen WJ, Viergever MA. Three-dimensional modeling for functional analysis of cardiac images, a review. IEEE Trans Med Imaging. 2001;20(1):2–5. doi: 10.1109/42.906421. [DOI] [PubMed] [Google Scholar]
- 64.Pednekar A, Kurkure U, Muthupillai R, et al. Automated left ventricular segmentation in cardiac MRI. IEEE Trans Biomed Eng. 2006;53(7):1425–1428. doi: 10.1109/TBME.2006.873684. [DOI] [PubMed] [Google Scholar]
- 65.Lynch M, Ghita O, Whelan PF. Automatic segmentation of the left ventricle cavity and myocardium in MRI data. Comput Biol Med. 2006;36(4):389–407. doi: 10.1016/j.compbiomed.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 66.Kurkure U, Pednekar A, Muthupillai R, et al. Localization and segmentation of left ventricle in cardiac cine-MR images. IEEE Trans Biomed Eng. 2009;56(5):1360–1370. doi: 10.1109/TBME.2008.2005957. [DOI] [PubMed] [Google Scholar]
- 67.Grosgeorge D, Petitjean C, Caudron J, et al. Automatic cardiac ventricle segmentation in MR images: a validation study. Int J Comput Assist Radiol Surg. 2011;6(5):573–581. doi: 10.1007/s11548-010-0532-6. [DOI] [PubMed] [Google Scholar]
- 68.Kaus MR, Von Berg J, Weese J, et al. Automated segmentation of the left ventricle in cardiac MRI. Med Image Anal. 2004;8(3):245–254. doi: 10.1016/j.media.2004.06.015. [DOI] [PubMed] [Google Scholar]
- 69.Mitchell SC, Lelieveldt BP, van der Geest RJ, et al. Multistage hybrid active appearance model matching: segmentation of left and right ventricles in cardiac MR images. IEEE Trans Med Imaging. 2001;20(5):415–423. doi: 10.1109/42.925294. [DOI] [PubMed] [Google Scholar]
- 70.van Assen HC, Danilouchkine MG, Frangi AF, et al. SPASM: a 3D-ASM for segmentation of sparse and arbitrarily oriented cardiac MRI data. Med Image Anal. 2006;10(2):286–303. doi: 10.1016/j.media.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 71.Abi-Nahed J, Jolly MP, Yang GZ. Robust active shape models: A robust, generic and simple automatic segmentation tool. International conference on medical image computing and computer-assisted intervention; 2006 Springer. 2006;9(Pt 2):1–8. doi: 10.1007/11866763_1. [DOI] [PubMed] [Google Scholar]
- 72.Andreopoulos A, Tsotsos JK. Efficient and generalizable statistical models of shape and appearance for analysis of cardiac MRI. Med Image Anal. 2008;12(3):335–357. doi: 10.1016/j.media.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 73.Lorenzo-Valdés M, Sanchez-Ortiz GI, Elkington AG, et al. Segmentation of 4D cardiac MR images using a probabilistic atlas and the EM algorithm. Med Image Anal. 2004;8(3):255–265. doi: 10.1016/j.media.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 74.Dawes TJ, De Marvao A, Shi W, et al. Machine learning of three-dimensional right ventricular motion enables outcome prediction in pulmonary hypertension: a cardiac MR imaging study. Radiology. 2017:161315. doi: 10.1148/radiol.2016161315. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Greenwood JP, Maredia N, Younger JF, et al. Cardiovascular magnetic resonance and single-photon emission computed tomography for diagnosis of coronary heart disease (CE-MARC): a prospective trial. Lancet. 2012;379(9814):453–460. doi: 10.1016/S0140-6736(11)61335-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Schwitter J, Wacker CM, Wilke N, et al. MR-IMPACT II: magnetic resonance imaging for myocardial perfusion assessment in coronary artery disease trial: perfusion-cardiac magnetic resonance vs. single-photon emission computed tomography for the detection of coronary artery disease: a comparative multicentre, multivendor trial. Eur Heart J. 2013;34(10):775–781. doi: 10.1093/eurheartj/ehs022. [DOI] [PubMed] [Google Scholar]
- 77.Hamon M, Fau G, Nee G, et al. Meta-analysis of the diagnostic performance of stress perfusion cardiovascular magnetic resonance for detection of coronary artery disease. J Cardiovasc Magn Reson. 2010;12:29. doi: 10.1186/1532-429X-12-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jaarsma C, Leiner T, Bekkers SC, et al. Diagnostic performance of noninvasive myocardial perfusion imaging using single-photon emission computed tomography, cardiac magnetic resonance, and positron emission tomography imaging for the detection of obstructive coronary artery disease: a meta-analysis. J Am Coll Cardiol. 2012;59(19):1719–1728. doi: 10.1016/j.jacc.2011.12.040. [DOI] [PubMed] [Google Scholar]
- 79.Greenwood JP, Motwani M, Maredia N, et al. Comparison of cardiovascular magnetic resonance and single-photon emission computed tomography in women with suspected coronary artery disease from the Clinical Evaluation of Magnetic Resonance Imaging in Coronary Heart Disease (CE-MARC) Trial. Circulation. 2014;129(10):1129–1138. doi: 10.1161/CIRCULATIONAHA.112.000071. [DOI] [PubMed] [Google Scholar]
- 80.Schulz-Menger J, Bluemke DA, Bremerich J, et al. Standardized image interpretation and post processing in cardiovascular magnetic resonance: society for Cardiovascular Magnetic Resonance (SCMR) board of trustees task force on standardized post processing. J Cardiovasc Magn Reson. 2013;15:35. doi: 10.1186/1532-429X-15-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81••.Kramer CM, Barkhausen J, Flamm SD, et al. Standardized cardiovascular magnetic resonance (CMR) protocols 2013 update. J Cardiovasc Magn Reson. 2013;15:91. doi: 10.1186/1532-429X-15-91. Describing efforts to standardize the field of CMR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tarroni G, Corsi C, Antkowiak PF, et al. Myocardial perfusion: near-automated evaluation from contrast-enhanced MR images obtained at rest and during vasodilator stress. Radiology. 2012;265(2):576–583. doi: 10.1148/radiol.12112475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Goel A, McColl R, Ks K, et al. Fully automated tool to identify the aorta and compute flow using phase-contrast MRI: validation and application in a large population based study. J Magn Reson Imaging. 2014;40(1):221–228. doi: 10.1002/jmri.24338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Karim R, Bhagirath P, Claus P, et al. Evaluation of state-of-the-art segmentation algorithms for left ventricle infarct from late Gadolinium enhancement MR images. Med Image Anal. 2016;30:95–107. doi: 10.1016/j.media.2016.01.004. [DOI] [PubMed] [Google Scholar]
- 85.Karim R, Housden RJ, Balasubramaniam M, et al. Evaluation of current algorithms for segmentation of scar tissue from late gadolinium enhancement cardiovascular magnetic resonance of the left atrium: an open-access grand challenge. J Cardiovasc Magn Reson. 2013;15(1):105. doi: 10.1186/1532-429X-15-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kolipaka A, Chatzimavroudis GP, White RD, et al. Segmentation of non-viable myocardium in delayed enhancement magnetic resonance images. Int J Cardiovasc Imaging (Formerly Card Imaging) 2005;21(2):303–311. doi: 10.1007/s10554-004-5806-z. [DOI] [PubMed] [Google Scholar]
- 87.Yan AT, Shayne AJ, Brown KA, et al. Characterization of the peri-infarct zone by contrast-enhanced cardiac magnetic resonance imaging is a powerful predictor of post-myocardial infarction mortality. Circulation. 2006;114(1):32–39. doi: 10.1161/CIRCULATIONAHA.106.613414. [DOI] [PubMed] [Google Scholar]
- 88.Positano V, Pingitore A, Giorgetti A, et al. A fast and effective method to assess myocardial necrosis by means of contrast magnetic resonance imaging. J Cardiovasc Magn Reson. 2005;7(2):487–494. doi: 10.1081/jcmr-200053630. [DOI] [PubMed] [Google Scholar]
- 89.Detsky JS, Paul G, Dick AJ, et al. Reproducible classification of infarct heterogeneity using fuzzy clustering on multicontrast delayed enhancement magnetic resonance images. IEEE Trans Med Imaging. 2009;28(10):1606–1614. doi: 10.1109/TMI.2009.2023515. [DOI] [PubMed] [Google Scholar]
- 90.Tao Q, Milles J, Zeppenfeld K, et al. Automated segmentation of myocardial scar in late enhancement MRI using combined intensity and spatial information. Magn Reson Med. 2010;64(2):586–594. doi: 10.1002/mrm.22422. [DOI] [PubMed] [Google Scholar]
- 91.Lu Y, Yang Y, Connelly KA, et al. Automated quantification of myocardial infarction using graph cuts on contrast delayed enhanced magnetic resonance images. Quant Imaging Med Surg. 2012;2(2):81–86. doi: 10.3978/j.issn.2223-4292.2012.05.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Waljee AK, Higgins PD. Machine learning in medicine: a primer for physicians. Am J Gastroenterol. 2010;105(6):1224–1226. doi: 10.1038/ajg.2010.173. [DOI] [PubMed] [Google Scholar]
- 93.Deo RC. Machine Learning in Medicine. Circulation. 2015;132(20):1920–1930. doi: 10.1161/CIRCULATIONAHA.115.001593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sengupta PP, Huang YM, Bansal M, et al. Cognitive machine-learning algorithm for cardiac imaging a pilot study for differentiating constrictive pericarditis from restrictive cardiomyopathy. Circ Cardiovas Imag. 2016;9(6):e004330. doi: 10.1161/CIRCIMAGING.115.004330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Krizhevsky A, Sutskever I, Hinton GE. Imagenet classification with deep convolutional neural networks. Advances in neural information processing systems. 2012:1097–1105. [Google Scholar]
- 96.Russakovsky O, Deng J, Su H, et al. ImageNet Large Scale Visual Recognition Challenge. Arxiv. 2014:14090575. Cs. [Google Scholar]
- 97.Srivastava N, Hinton GE, Krizhevsky A, et al. a simple way to prevent neural networks from overfitting. J Machine Learn Res. 2014;15:1929–1958. [Google Scholar]
- 98.Kang D, Dey D, Slomka PJ, et al. Kuo CCJ structured learning algorithm for detection of nonobstructive and obstructive coronary plaque lesions from coronary CT angiography. J Med Imaging. 2015;2(1):014003. doi: 10.1117/1.JMI.2.1.014003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99••.Wolterink JM, Leiner T, Bd DV, et al. Automatic coronary artery calcium scoring in cardiac CT angiography using paired convolutional neural networks. Med Image Anal. 2016;34:123–136. doi: 10.1016/j.media.2016.04.004. First application of deep learning to coronary plaque detection. [DOI] [PubMed] [Google Scholar]
- 100.Carneiro G, Nascimento JC. Combining multiple dynamic models and deep learning architectures for tracking the left ventricle endocardium in ultrasound data. IEEE Trans Pattern Anal Mach Intell. 2013;35(11):2592–2607. doi: 10.1109/TPAMI.2013.96. [DOI] [PubMed] [Google Scholar]
- 101.Knackstedt C, Bekkers SC, Schummers G, et al. Fully automated versus standard tracking of left ventricular ejection fraction and longitudinal strain: the FAST-EFs Multicenter Study. J Am Coll Cardiol. 2015;66(13):1456–1466. doi: 10.1016/j.jacc.2015.07.052. [DOI] [PubMed] [Google Scholar]
- 102.Belous G, Busch A, Rowlands D. Segmentation of the left ventricle from ultrasound using random forest with active shape model. Artificial Intelligence, Modelling and Simulation (AIMS), 2013 1st International Conference on; 2013: IEEE; 2013; pp. 315–319. [Google Scholar]
- 103.Ghesu FC, Krubasik E, Georgescu B, et al. Marginal space deep learning: efficient architecture for volumetric image parsing. IEEE Trans Med Imaging. 2016;35(5):1217–1228. doi: 10.1109/TMI.2016.2538802. [DOI] [PubMed] [Google Scholar]
- 104.Narula S, Shameer K, Salem Omar AM, et al. Machine-learning algorithms to automate morphological and functional assessments in 2D echocardiography. J Am Coll Cardiol. 2016;68(21):2287–2295. doi: 10.1016/j.jacc.2016.08.062. [DOI] [PubMed] [Google Scholar]
- 105.Luong C, Abdi A, Jue J, et al. Automatic quality assessment of echo apical 4-chamber images using computer deep learning. Am Heart Assoc. 2015;2015:2015. [Google Scholar]
- 106.Avendi MR, Kheradvar A, Jafarkhani H. A combined deep-learning and deformable-model approach to fully automatic segmentation of the left ventricle in cardiac MRI. Med Image Anal. 2016;30:108–119. doi: 10.1016/j.media.2016.01.005. [DOI] [PubMed] [Google Scholar]
- 107.Emad O, Yassine IA, Fahmy AS. Automatic localization of the left ventricle in cardiac MRI images using deep learning. Conf Proc IEEE Eng Med Biol Soc. 2015;2015:683–686. doi: 10.1109/EMBC.2015.7318454. [DOI] [PubMed] [Google Scholar]
- 108.Suinesiaputra A, Cowan BR, Al-Agamy AO, et al. A collaborative resource to build consensus for automated left ventricular segmentation of cardiac MR images. Med Image Anal. 2014;18(1):50–62. doi: 10.1016/j.media.2013.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Arsanjani R, Xu Y, Dey D, et al. Improved accuracy of myocardial perfusion SPECT for the detection of coronary artery disease using a support vector machine algorithm. J Nucl Med. 2013;54(4):549–555. doi: 10.2967/jnumed.112.111542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Friedman J, Hastie T, Tibshirani R. Additive logistic regression: a statistical view of boosting (with discussion and a rejoinder by the authors) The Annals of Statistics. 2000;28(2):337–407. [Google Scholar]
- 111.Arsanjani R, Xu Y, Dey D, et al. Improved accuracy of myocardial perfusion SPECT for detection of coronary artery disease by machine learning in a large population. J Nucl Cardiol. 2013;20(4):553–562. doi: 10.1007/s12350-013-9706-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Arsanjani R, Dey D, Khachatryan T, et al. Prediction of revascularization after myocardial perfusion SPECT by machine learning in a large population. J Nucl Cardiol. 2015;22(5):877–884. doi: 10.1007/s12350-014-0027-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113••.Motwani M, Dey D, Berman DS, et al. Machine learning for prediction of all-cause mortality in patients with suspected coronary artery disease: a 5-year multicentre prospective registry analysis. Eur Heart J. 2016:pii. doi: 10.1093/eurheartj/ehw188. ehw188. &lsqbEpub ahead of print]. Large and first-in-kind study of predicting mortality by machine learning by combining clinical and imaging data. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Dey D, Diaz Zamudio M, Schuhbaeck A, et al. Relationship between quantitative adverse plaque features from coronary computed tomography angiography and downstream impaired myocardial flow reserve by 13N-ammonia positron emission tomography: a pilot study. Circ Cardiovasc Imaging. 2015;8(10):e003255. doi: 10.1161/CIRCIMAGING.115.003255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115••.Greenspan H, Van Ginneken B, Summers RM. Guest Editorial Deep Learning in Medical Imaging: overview and Future Promise of an Exciting New Technique. IEEE Trans Med Imaging. 2016;35(5):1153–1159. In-depth editorial on prospects of deep learning in medical imaging. [Google Scholar]
- 116.Petersen SE, Matthews PM, Bamberg F, et al. Imaging in population science: cardiovascular magnetic resonance in 100,000 participants of UK Biobank-rationale, challenges and approaches. J Cardiovasc Magn Reson. 2013;15(1):1. doi: 10.1186/1532-429X-15-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Fonseca CG, Backhaus M, Bluemke DA, et al. The Cardiac Atlas Project —an imaging database for computational modeling and statistical atlases of the heart. Bioinformatics. 2011;27(16):2288–2295. doi: 10.1093/bioinformatics/btr360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Jimenez del Toro OA, Goksel O, Menze B, et al. VISCERAL–VISual Concept Extraction challenge in RAdioLogy: ISBI 2014 challenge organization. Proceedings VISCERAL Challenge at ISBI. 2014;1194:6–15. [Google Scholar]
- 119.https://grand-challenge.org/All_Challenges/. 2016.
- 120.https://www.kaggle.com/c/second-annual-data-science-bowl. 2016
- 121.Kirişli HA, Schaap M, Metz CT, et al. Standardized evaluation framework for evaluating coronary artery stenosis detection, stenosis quantification and lumen segmentation algorithms in computed tomography angiography. Med Image Anal. 2013;17(8):859–876. doi: 10.1016/j.media.2013.05.007. [DOI] [PubMed] [Google Scholar]
- 122.Tilkemeier PL, Mahmarian JJ, Wolinsky DG, et al. ImageGuide Update. J Nucl Cardiol. 2015;22(5):994–997. doi: 10.1007/s12350-015-0217-1. [DOI] [PubMed] [Google Scholar]


