Abstract
Alterations in myelopoiesis are common across various tumor types, resulting in immature populations termed myeloid-derived suppressor cells (MDSCs). MDSC burden correlates with poorer clinical outcomes, credited to their ability to suppress antitumor immunity. MDSCs consist of two major subsets, monocytic and polymorphonuclear (PMN). Intriguingly, the latter subset predominates in many patients and tumor models, though the mechanisms favoring PMN-MDSC responses remain poorly understood. Ordinarily, lineage-restricted transcription factors regulate myelopoiesis that collectively dictate cell fate. One integral player is interferon regulatory factor-8 (IRF8), which promotes monocyte/dendritic cell differentiation while limiting granulocyte development. We recently showed that IRF8 inversely controls MDSC burden in tumor models, particularly the PMN-MDSC subset. However, where IRF8 acts in the pathway of myeloid differentiation to influence PMN-MDSC production has remained unknown. Here, we showed that: 1) tumor growth was associated with a selective expansion of newly defined IRF8lo granulocytic progenitors (GPs); 2) tumor-derived GPs had an increased ability to form PMN-MDSCs; 3) tumor-derived GPs shared gene expression patterns with IRF8−/− GPs, suggesting that IRF8 loss underlies GP expansion; and 4) enforced IRF8 overexpression in vivo selectively constrained tumor-induced GP expansion. These findings support the hypothesis that PMN-MDSCs result from selective expansion of IRF8lo GPs, and that strategies targeting IRF8 expression may limit their load to improve immunotherapy efficacy.
Keywords: Interferon regulatory factor-8, Myeloid-derived suppressor cell, Granulocyte-monocyte progenitor, Granulocyte progenitor, Tumor progression
Introduction
Perturbations in myelopoiesis are common in neoplastic disease, resulting in impaired myeloid responses evident at developmental or functional levels. At a developmental level, studies in animal models and patients reveal leukocytosis, characterized as either monocytosis or neutrophilia (1, 2). At a functional level, such myeloid populations appear morphologically immature and have a diminished capacity to stimulate effective immune responses (1). These cells have been broadly termed myeloid-derived suppressor cells (MDSCs) (3).
In mouse models, MDSCs are generally identified by co-expression of the canonical markers, CD11b and Gr-1 (4). More recently, MDSCs have been shown to comprise two major subsets, monocytic (M-MDSC) or polymorphonuclear (PMN-MDSC) based on differential expression of the Ly6C and Ly6G markers (4). In mouse tumor models, the elimination of MDSCs or their activities improves response to therapy, including chemotherapy (5–7), immunotherapy (8) and combination interventions (9, 10). In humans, MDSCs also stratify into M-MDSC and PMN-MDSC subsets, although the phenotypes used to distinguish them are more complex. Importantly, as in mouse models, MDSC presence in human cancer carries significant prognostic value (8, 11). Intriguingly, PMN-MDSCs predominate in both tumor models and cancer patients (12, 13); yet, the mechanisms behind this bias have remained incompletely understood (12). Thus, despite advances in our understanding of the importance of MDSCs in neoplasia, there is much less known regarding the precise origin of MDSCs, why PMN-MDSCs dominate the response and what transcriptional events govern this decision.
A current model for steady-state myelopoiesis posits that hematopoietic stem cells and multipotent progenitors differentiate along a series of lineage-restricted progenitor stages (14). Neoplasia disrupts this process. Wu, et al. (15) found that patients across a broad spectrum of tumor types have increased circulating hematopoietic progenitor cells, particularly granulocyte-monocyte progenitors (GMPs). Akin to earlier studies of MDSC biology, increased circulating GMPs were associated with poorer patient outcomes, defined by clinical stage and reduced time-to-progression. The production of and subsequent differentiation of GMPs are events regulated by lineage-instructive transcription factors. One such transcription factor, interferon regulatory factor-8 (IRF8), is specifically expressed at this stage in myeloid development and is hypothesized to dictate GMP differentiation into either granulocytes (IRF8lo) or monocytes (IRF8hi) (13, 16–18). This essential role for IRF8 was revealed using IRF8-deficient (IRF8−/−) mice. These mice develop a CML-like myeloproliferative disease characterized by the overproduction of both immature and mature granulocytes in the bone marrow and periphery (19). We recently found that the profound granulocytic phenotype observed in IRF8−/− mice is homologous to tumor-induced PMN-MDSCs at functional and gene expression levels (8). Moreover, we identified that IRF8 levels inversely controlled the size of the peripheral MDSC pool in mouse tumor models, particularly PMN-MDSCs. Importantly, we observed the inverse association between IRF8 expression and MDSC frequency in breast cancer patients, but not healthy donors (8). While these studies revealed IRF8 as an important negative regulator of MDSC production, particularly PMN-MDSCs, these observations were limited to cells in the periphery and did not examine events upstream in myeloid development.
Based on our work (8) and data from IRF8−/− models (19), we now hypothesized that IRF8 regulates tumor-induced PMN-MDSC production at the GMP stage or a downstream newly defined granulocyte progenitor (GP) stage (20). Altogether, our results show for the first time that PMN-MDSCs arise from a newly defined GP stage within the bone marrow and that IRF8 levels (and/or their downstream target genes) in those GPs guide their expansion or contraction.
Materials and Methods
Mice
All studies in mice were performed under protocols (1117M, 1108M) approved by the Institutional Animal Care and Use Committee of Roswell Park Cancer Institute. Homozygous IRF8-EGFP mice on a C57BL/6 (B6; H-2b) background were kindly provided by Dr. H. Morse (NIH, Bethesda, MD), and have been described (21). In this model, IRF8-EGFP levels can be determined under both homozygous and heterozygous breeding conditions (21). Accordingly, we bred IRF8-EGFP and BALB/c (H-2d) mice to generate F1 semi-syngeneic (H-2b/d) progeny for the 4T1 tumor studies. For the autochthonous mammary tumor model, we made use of transgenic mice expressing the polyomavirus middle T antigen under control of the MMTV promoter (also known as MTAG mice) on a B6 background kindly provided by Dr. S. Gendler (Mayo Clinic, Scottsdale, AZ) (22–24). As with the 4T1 model, we bred MTAG to IRF8-EGFP mice to generate progeny heterozygous for IRF8-EGFP expression. IRF8-deficient mice (IRF8−/−) were kindly provided by Dr. K. Ozato (NIH, Bethesda, MD). IRF8 transgenic (IRF8-Tg) mice, wherein IRF8 overexpression was controlled by the CMV promoter, were developed and propagated as described (8, 23, 25). We obtained wild-type female B6 and BALB/c mice were from Charles River (NCI Frederick, MD).
Cell Lines and Tumor Growth Experiments
The 4T1 mammary tumor cell line was obtained from ATCC (Manassas, VA) and maintained as described (22). AT-3 cells were originally derived from an MTAG mouse by our laboratory and maintained as described (22). 4T1 (5×104 cells) or AT-3 (5×105 cells) were implanted orthotopically into mammary gland #4 of mice at 8–12 weeks of age. 4T1 or AT-3 tumor growth was measured as described (8). Total tumor volume in female MTAG mice was quantified as described (22).
G-CSF Administration
Recombinant mouse G-CSF (Peprotech, Rocky Hill, NJ) was administered at 10μg/mouse subcutaneously for five consecutive days as described (24). Mice were euthanized on day six, and then bone marrow or spleen collected and processed for the indicated studies, as described below.
Bone Marrow Progenitor Analysis by Flow Cytometry
Bone marrow cells were collected by flushing femurs and tibias. Red blood cells were lysed using ACK lysis buffer. Cells were analyzed using an LSR II flow cytometer (BD) running Diva version 6.1.3 and data files were analyzed using FlowJo version 10. Bone marrow progenitors were defined as described (20, 26), and depicted in Fig. 1. Directly conjugated antibodies used for cell surface staining are listed in Supplemental Table 1.
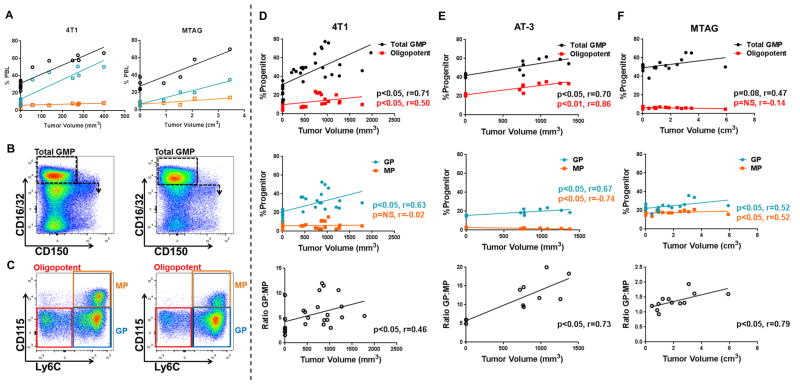
Figure 1. Myeloid progenitors in the bone marrow are altered during tumor growth.
(A) Percentages of total myeloid cells (CD11b+ cells; black lines) or the granulocytic (Gr-1hiCD115−; blue lines) or monocytic fractions (Gr-1intCD115+; orange lines) from the peripheral blood of 4T1 tumor-bearing (left) or MTAG (right) mice plotted in relation to tumor volume (each point is a measurement from an individual mouse). (B) Gating strategy used for total GMPs (NTB: 35.6% vs. 4T1: 65.4%) and newly defined downstream progenitor subsets, oligopotent GMPs (NTB: 22.8% vs. 4T1: 14.2%), GPs (NTB: 53.2% vs. 4T1: 73.2%), MPs (NTB: 22.5% vs. 4T1: 11.8%) (C) from bone marrow of NTB (left) and 4T1 (right) tumor-bearing mice. Progenitor populations were calculated as a percentage of the Lin−cKit+Sca-1− gate and plotted by tumor volume with total and oligopotent GMPs (top), GPs and MPs (middle) and GP:MP ratio (bottom) for the 4T1, AT-3 and MTAG models (D – F, respectively). 4T1, AT-3 and MTAG data compiled from multiple experiments. For D, n= 8 for NTB or n=24 for 4T1; E, n=3 for NTB or n=8 for AT-3; F, n= 7 for WT or n=12 for MTAG.
In vitro Differentiation of Bone Marrow Progenitors
Lineage depletion of bone marrow cells was performed as described (27) using anti-mouse Abs reactive against Ter-119, Gr-1 and B220 (eBiosience). Lineage-negative (Lin−) bone marrow cells were stained with rat anti-mouse mAbs to c-kit, Sca-1, CD16/32, CD150, CD115 and Ly6C to label GMP subpopulations. Cells were sorted using the FACSAria cell sorter running Diva acquisition. GPs were sorted to >90% purity and incubated with SCF (10ng/ml) plus IL-3 (50ng/ml), G-CSF or M-CSF (50ng/ml) (all from PeproTech) for 4 days. Cells were then washed and immunostained with antibodies to CD11b, F4/80, Ly6C and Ly6G and analyzed.
Suppression Assays
Sorted GPs from pooled bone marrow cells were used either immediately or cultured with G-CSF, as described above. GPs were then co-incubated with splenocytes, which had been stained with CellTrace Violet (Thermo Fisher Scientific, C34557) per manufacturer’s instructions, at a 1:2 ratio for 72 hours in 96-well-round bottomed plates. Dilution of CellTrace Violet in CD4+ or CD8+ T cells was analyzed by flow cytometry. Percent suppression was calculated by (ΔMFI anti-CD3 stimulated T cells minus ΔMFI GP co-cultured T cells) divided by ΔMFI anti-CD3 stimulated T cells. ΔMFI calculated by subtraction from unstimulated controls.
RNA-seq studies
Total RNA from frozen cell pellets were prepared using the miRNeasy micro kits (Qiagen), following manufacturer’s instructions, with a DNAse digest performed on the column to prevent carryover of genomic DNA before RNA elution. Eluted RNA samples were quantified using a Qubit fluorometer (Thermo Fisher Scientific) and evaluated for degradation using a 4200 Tapestation (Agilent Technologies). RNA sequencing libraries were prepared with the TruSeq Stranded Total RNA kit (Illumina) from 100ng total RNA following manufacturer’s instructions. After ribosomal RNA depletion, remaining RNA was purified, fragmented and reverse transcribed into first strand cDNA using random primers. The RNA template was removed and a replacement strand, incorporating dUTP in place of dTTP was synthesized to generate ds cDNA. AMPure XP beads were used to separate the ds cDNA from the second strand reaction mix resulting in blunt-ended cDNA. A single ‘A’ nucleotide was then added to the 3′ ends of the blunt fragments. Multiple indexing adapters, containing a single ‘T’ nucleotide on the 3′ end of the adapter, were ligated to the ends of the ds cDNA, preparing them for hybridization onto a flow cell. Adapter ligated libraries were amplified by PCR, purified using Ampure XP beads, and validated for appropriate size on a 4200 TapeStation D1000 Screentape (Agilent Technologies). The libraries were quantified using KAPA Biosystems qPCR kit and pooled together in an equimolar fashion. The pool was denatured and diluted to 16pM for On-Board Cluster Generation and sequencing on a HiSeq2500 sequencer using the appropriate paired-end cluster kit and rapid mode SBS reagents following the manufacturer’s recommended protocol (Illumina).
Raw reads passed quality filter from Illumina RTA were first pre-processed using FASTQC(v0.10.1) for sequencing base quality control to be mapped to the latest mouse reference genome (mm10) and RefSeq annotation database using Tophat(v2.0.13) (28). Second round of QC using RSeQC (29) was applied to mapped bam files to identify potential RNA-seq library preparation problems. From the mapping results, read counts for genes were obtained by HTSeq (30) using intersection-strict option. Differentially expressed genes were identified using DESeq2 (31), a variance-analysis package developed to infer the statically significant difference in RNA-seq data. Gene fold changes for sample without replicates were calculated using regularized-log2 transformation in DESeq2 R package. RNA-seq data are deposited under accession number GSE87800, and can be found via the following link: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE87800
Real-time RT-PCR analysis was performed as described (8) to validate the RNA-seq results. The following were the primer sequences used in this study: GAPDH: forward primer (FP), 5′-CATCACCATCTTCCAGGAGCG-3′; reverse primer (RP), 5′-ACGGACACATTGGGGGTAGG -3′; IRF8: FP, 5′-CGTGGAAGACGAGGTTACGCTG-3′; RP, 5′-GCTGAATGGTGTGTGTCATAGGC-3′; CXCR2: FP, 5′-GGTCGTACTGCGTATCCTGCCTCA-3′; RP, 5′-TAGCCATGATCTTGAGAAGTCCAT-3′; MMP9: FP, 5′-CCACCACAACTGAACCACAG-3′; RP, 5′-AGTAAGGAAGGGGCCCTGTA-3′; KLF4: FP, 5′-GTGCCCCGACTAACCGTTG-3′; RP, 5′-GTCGTTGAACTCCTCGGTCT-3′; G-CSFR: FP, 5′-TCATGGCCACCAGTCGGGC-3′; RP, 5′-CACGCTGGAGTCCCAGAAG -3′.
Statistical Analysis
Data was analyzed using GraphPad (Version 6.07) and SAS/STAT (Version 9.4) software. Data recorded as the mean ± SEM of the indicated number of mice, or biologic or experimental replicates. Relationships between myeloid frequencies and tumor volume in any single genotype were analyzed using Spearman correlation and linear regression. IRF8 expression levels within populations and/or population frequencies obtained at experimental endpoints were compared using nonparametric t-tests where indicated. Additional statistical analysis was performed for Figure 7 to compare the relationships between progenitor frequencies and tumor volume between WT and IRF8-Tg mice. The difference in slopes was estimated using this genotype-marker interaction. Whether the slopes were different was determined by the 95% confidence interval for the difference, or its p-value. Separate statistical models were fit for each marker. Models were fit using Ordinary Least Squares. In all cases, p-values < 0.05 were considered statistically significant.
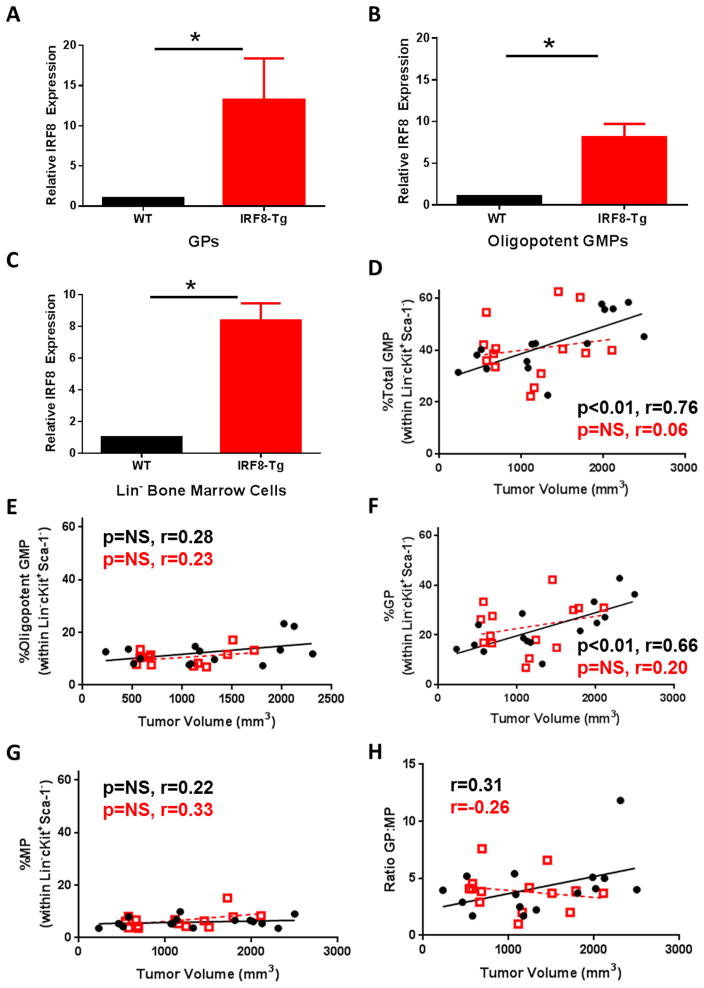
Figure 7. Enhanced IRF8 expression restrains GP production.
Quantitative RT-PCR analysis of IRF8 levels in sorted GPs (A), oligopotent GMPs (B) or lineage-depleted bone marrow cells (C) of naïve WT or IRF8-Tg mice. (D–H) WT (n=15) (black circles) or IRF8-Tg (n=14) (red squares) mice were implanted with AT-3 tumor cells, as in Fig. 1. Bone marrow progenitor cell analysis was performed, as described earlier. Frequency of total GMPs (D), oligopotent GMPs (E), GPs (F) and MPs (G) plotted in relation to tumor volume and trend lines generated by linear regression analysis. (H) Ratio of GPs to MPs was also calculated. (D–H) P-values based on Spearman correlation of the individual lines. Data compiled from three experiments whereby each data point is from an individual mouse.
Results
Tumor growth significantly elevates GP frequencies in the bone marrow
While MDSCs result from a defect in myeloid differentiation, definitive evidence tying the origin of MDSCs to a precise myeloid progenitor in the bone marrow has been lacking. To investigate where and how tumor growth impairs myelopoiesis in the bone marrow, we focused on well-established mouse models of mammary cancer that are proficient at generating MDSCs. We utilized two orthotopic implantable models and one autochthonous model. For the former model, we made use of: 1) 4T1, a spontaneously derived mammary tumor syngeneic to BALB/c mice (8, 24, 32, 33); and 2) AT-3, a spontaneously derived mammary carcinoma established from the MMTV-PyMT autochthonous tumor model in the C57BL/6 background (also known at the ‘MTAG’ model for middle T Ag) (22). For the autochthonous model, we made use of the MTAG mouse itself, which spontaneously develops multifocal mammary tumors beginning at ~14-weeks of age (reflecting tissue-specific expression of the PyMT oncogene driven by the MMTV promoter) (22, 23, 34). These models have been studied extensively both in MDSC biology and mammary tumor progression and, consistent with previous studies, we observed a significant expansion of CD11b+ myeloid cells in the peripheral blood (Fig. 1A for 4T1 and MTAG models). These cells were primarily Gr-1hiCD115− granulocytes, with no appreciable increase in CD115+ monocyte frequency. We previously showed that such granulocytes are bona fide MDSCs based on their ability to inhibit T cell proliferation (8, 23, 24).
To quantify the impact of tumor burden on progenitor populations in the bone marrow, we performed comprehensive flow analysis on cells isolated from each of the models over the course of tumor growth (Fig. 1). Our analysis initially focused on GMPs, the bifurcation point between monocytic and granulocytic differentiation. As classically defined (26), we assessed total GMPs based on the following immunophenotype: Lineage (Lin) −Sca-1−ckit+CD16/32+CD150− (Fig. 1B). First, with regard to the 4T1 model and consistent with previous studies (15, 35), 4T1 tumor growth was accompanied by a significant rise in the total GMP population (Fig. 1B, right). We then refined our analysis based on a recent study that demonstrated classically defined GMPs can be subdivided into three distinct populations based on additional markers of monocytic or granulocytic progenitors and their differentiation potential (20) (Fig. 1C). Oligopotent GMPs, which retain equal capacity for differentiation into granulocytes or monocytes are now defined as Ly6C−CD115lo/−. Ly6C+CD115lo/− cells are enriched for granulocyte progenitors (GPs), while Ly6C+CD115+ cells are enriched for monocyte progenitors (MPs).
Analysis of these novel populations in 4T1 tumor-bearing mice revealed a small but significant increase in oligopotent GMPs with increasing tumor burden (Fig. 1D top). In contrast, GPs significantly increased during tumor growth while MP frequency was largely unaffected (Fig. 1D middle). Thus, the increase in GPs appeared to account for the majority of the increase observed for classically defined total GMPs. To reflect further on how tumor growth skews GMP responses towards the granulocytic lineage, we calculated the ratio of GPs to MPs, which positively correlated with 4T1 tumor volume (Fig. 1D bottom). As with the 4T1 model, total GMPs in the AT-3 model were significantly increased (Fig. 1E top). Similarly, oligopotent GMPs (Fig. 1E top) as well as GPs were elevated, while MPs seemed to decrease (Fig. 1E middle). Here again, the ratio of GPs to MPs was significantly correlated with tumor volume (Fig. 1E bottom). Lastly, in the autochthonous model (Fig. 1F), total GMPs also modestly expanded, and the expansion largely correlated with a significant rise in GPs, which was reinforced by the elevated GP:MP ratio, a finding which was consistent with both orthotopic implantable models (Fig. 1D & E). Collectively, these data indicated that the rise in total GMPs was mainly due to the expansion of GPs, revealing an early myeloid bias toward the granulocytic lineage in tumor-bearing mice not previously recognized using the classical GMP phenotype. This is the first description of these progenitor populations in tumor-bearing mice, and the first observation that their frequencies during tumor growth are differentially altered.
Tumor-induced GPs differentiate into PMN-MDSCs
Although we found that GPs significantly expanded in tumor-bearing mice, it was unclear whether these cells were indeed precursors to MDSCs. To determine whether tumor-induced GPs are developmentally distinct from their control counterparts, we sorted them from the bone marrow of 4T1 tumor-bearing mice (4T1-GPs) or non-tumor-bearing mice (NTB-GPs) (Supplemental Fig. 1A), followed by culture with the indicated myelopoietic cytokine(s) and analysis for differential expression of Ly6G and Ly6C, markers used to distinguish PMN-MDSCs from M-MDSCs (4). Under all treatment conditions, GPs preferentially differentiated into CD11b+Ly6CloLy6G+ cells (Fig. 2A & B; Supplemental Fig. 1B), a phenotype shared with PMN-MDSCs (4). The ability of GPs to differentiate largely into CD11b+Ly6CloLy6G+ cells was determined by calculating cell percentages and absolute numbers (Fig. 2A & B) relative to the total bone marrow counts (Supplemental Fig. 1C). In contrast, much less monocytic differentiation was observed (Fig. 2C & D), in agreement with the characterization of Ly6C+CD115lo/− cells as GPs (20). Importantly, 4T1-GPs resulted in greater granulocyte differentiation compared to those from NTB mice, except after treatment with G-CSF, a prototypic granulocyte driver (Fig. 2A & B). Morphological assessment of these cultures identified various states of differentiation, with stem cell factor (SCF)+IL-3 cultures appearing least differentiated, followed by untreated cells appearing mainly as early granulocytes, and G-CSF-treated cells largely appearing as differentiated, banded granulocytes (Supplemental Fig. 1D). Based on cell percentages, a modest population of CD11b+Ly6ChiLy6G− monocytic cells appeared after culture with M-CSF (Fig. 2C; Supplemental Fig. 1B). These cells also expressed the mature macrophage marker, F4/80, and displayed ‘foamy’ morphology consistent with a heavily vacuolated cytoplasm (Supplemental Fig. 1D & E). However, after correcting for absolute cell number, the overall change in the monocytic/macrophage population was minimal compared to what we observed with the granulocytic population (Fig. 2B & D; Supplemental Fig. 1E). Despite culture with M-CSF, 4T1-GPs produced fewer monocytic cells, expressed less F4/80, and were less differentiated (based on morphology).
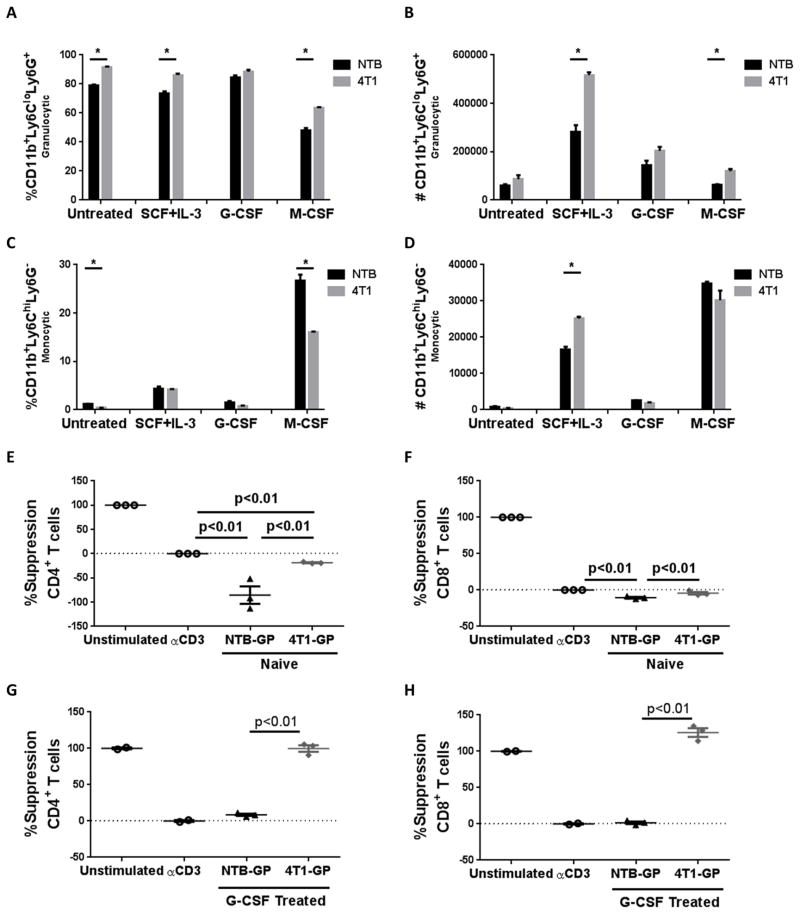
Figure 2. Tumor-induced progenitors are skewed toward granulocytic differentiation.
GPs were sorted from the bone marrow of NTB or 4T1 tumor-bearing mice (≥90% purity) and cultured for 4 days in vitro as described in the Methods. Frequencies (A) or absolute numbers (B) of granulocytic (CD11b+Ly6CloLy6G+) cells or frequencies (C) or absolute numbers (D) of monocytic (CD11b+Ly6ChiLy6G−) cells were determined after the indicated cytokine treatment. GPs (from NTB or 4T1 tumor-bearing mice) were sorted in a similar manner as in (A) and co-cultured with CellTrace Violet-stained syngeneic splenocytes (1:2 ratio for 72 hours) in the presence of anti-CD3 mAb. GPs were tested either directly after sorting (E, F) or after 4 days of in vitro incubation with G-CSF (G, H). Additional controls included CellTrace Violet-stained splenocytes cultured without GPs in the absence or presence of anti-CD3 mAb. Percent suppression for CD4+ or CD8+ T cell proliferation was calculated, as described in the Methods. Each point is a technical replicate of at least three determinations. Results shown are representative of two independent experiments.
To determine whether 4T1-GPs were functionally distinct from NTB-GPs, we tested their ability to suppress T cell proliferation. GPs from NTB or 4T1 tumor-bearing mice either used directly after sorting or after culture with G-CSF to facilitate differentiation, were co-incubated with anti-CD3-stimulated splenocytes (Fig. 2E–H). T cell subset-specific proliferation was measured by a modified CFSE-based flow cytometry assay (i.e., CellTrace Violet). Neither freshly isolated NTB-GPs nor 4T1-GPs expressed suppressive activity; rather, they stimulated CD4+ or CD8+ T cell proliferation, though 4T1-GPs appeared to be less ‘immune activating’ (Fig. 2E & F). In contrast to freshly isolated GPs, we observed highly significant CD4+ or CD8+ T cell suppression by G-CSF-cultured 4T1-GPs, but little-to-no suppression by NTB-GPs (Fig. 2G & H). Together, these data indicated that tumor growth skewed the differentiation of GPs toward PMN-MDSCs.
IRF8 expression is inversely linked to tumor-induced GP expansion
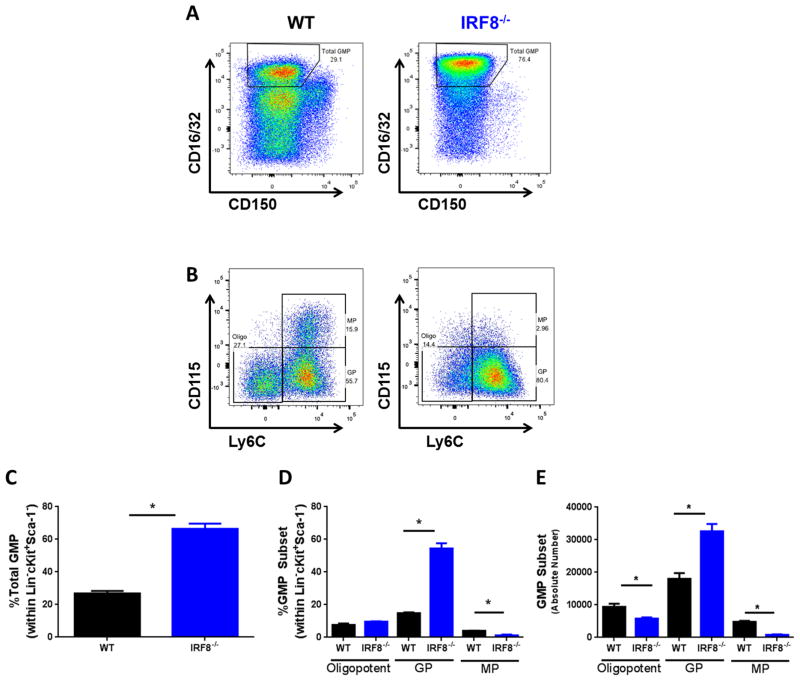
We previously demonstrated that IRF8 expression by peripheral myeloid cells inversely related to the quantity of MDSCs in tumor-bearing mice (8). However, whether changes in progenitor IRF8 expression levels contribute to such MDSC responses remained unclear. To investigate this question, we first examined myeloid progenitors in WT and IRF8−/− mice. Similar to previous studies (36, 37), IRF8−/− mice displayed a significant expansion of total GMPs compared to WT controls (Fig. 3A & C). Importantly, as in tumor-bearing mice, this expansion was largely attributable to GPs (Fig. 3B, D & E). These findings are consistent with the well-recognized role of IRF8 in constraining granulocytic differentiation.
Figure 3. IRF8 loss is associated with expansion of GPs.
(A) Gating strategy and calculated frequencies (C) of total GMPs from the bone marrow of littermate-matched WT and IRF8−/− mice. (B) Gating strategy for the indicated myeloid progenitor subsets of WT or IRF8−/− mice shown in A, along with their calculated frequency (D) and absolute number (E). n=3 mice each of the indicated genotype. *p < 0.01
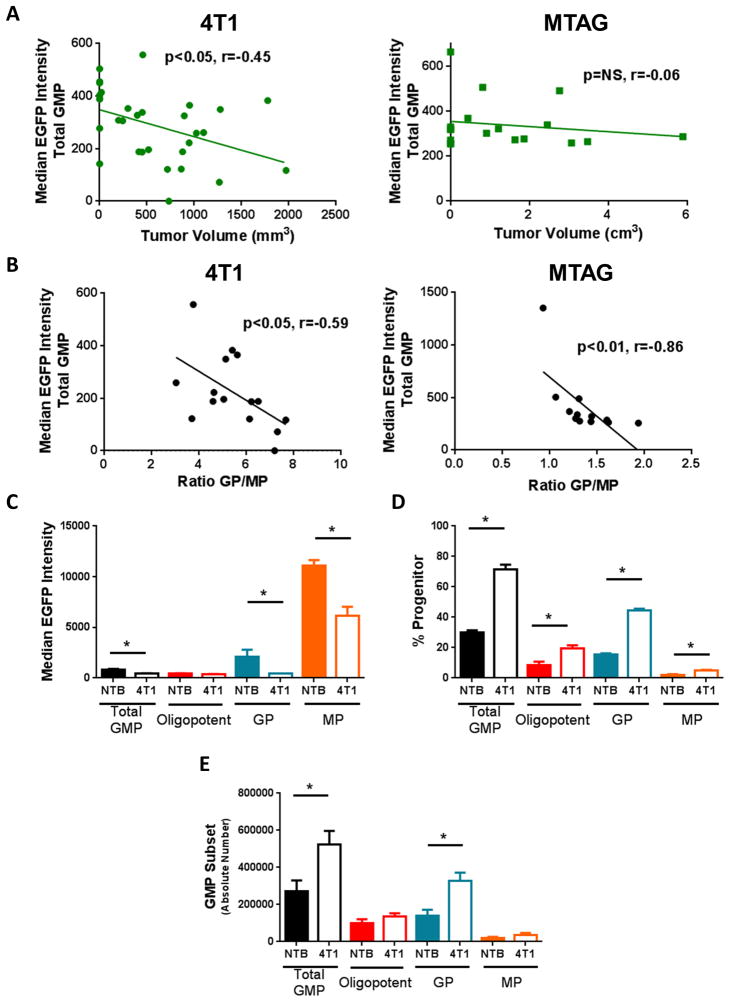
To explore tumor-induced changes in IRF8 expression during myelopoiesis, we used a recently generated transgenic knock-in mouse expressing an IRF8-EGFP fusion protein at the endogenous IRF8 locus (21, 38). Here, the stop codon of exon 9 of the mouse IRF8 locus was replaced with an EGFP sequence that results in the production of an IRF8-EGFP fusion protein under the control of endogenous IRF8 regulatory elements. Importantly, EGFP is an authentic surrogate marker of IRF8 expression, as IRF8 mRNA is found exclusively in the EGFP+, but not the EGFP− fraction and the knock-in does not alter IRF8 biology, as myeloid differentiation or function remains intact (21, 38). Moreover, IRF8 levels as measured by EGFP expression in various mature and immature myeloid populations paralleled IRF8 levels as measured by conventional mRNA analysis (21, 38). We first observed a significant decline in IRF8 expression by total GMPs over the course of 4T1 tumor growth, with a more modest effect in MTAG mice (Fig. 4A). These findings were reinforced by noting significant negative correlations between IRF8 levels in the total GMP population vs. the GP to MP ratio for both 4T1 and MTAG models (Fig. 4B). In agreement with our previous work (8), IRF8 expression was significantly reduced in peripheral CD11b+Gr-1+ cells during tumor growth (Supplemental Fig. 2A & B). Upon further examination, total GMPs had a bimodal expression of IRF8, which corresponded to unique IRF8 expression levels in each progenitor subset. Oligopotent GMPs and GPs were primarily IRF8lo/−, while MPs were primarily IRF8hi (Fig. 4C). The frequency and absolute number of IRF8lo/− GPs was greatly elevated in tumor-bearing mice, contributing to the IRF8 decrease in total GMPs (Fig. 4D & E). The decrease in IRF8 in total GMPs from tumor-bearing mice is likely due to expansion of the IRF8lo/− GPs.
Figure 4. IRF8 levels in Total GMPs predict GP:MP ratio.
(A) Median IRF8-EGFP expression by total GMPs in the bone marrow was plotted by tumor volume for 4T1 tumor-bearing mice (left) or total tumor volume for MTAG mice (right). (B) The intensity of IRF8 expression in total GMPs was plotted against the GP:MP ratio, revealing a negative correlation for 4T1 tumor-bearing (left) and MTAG mice (right). Each data point in panels A & B is from an individual mouse. (C) Differential IRF8 expression levels in GMP subsets at endpoint, accounting for the bimodal distribution of IRF8 in total GMPs. Frequency (D) and absolute number (E) (i.e., calculated by multiplying the total number of bone marrow cells collected by the percentage of each subset determined by flow cytometry) of GMP subsets at endpoint in NTB and 4T1 tumor-bearing mice. (n=5 mice each in panels C–E).
IRF8 loss is associated with altered gene expression in tumor-induced GPs
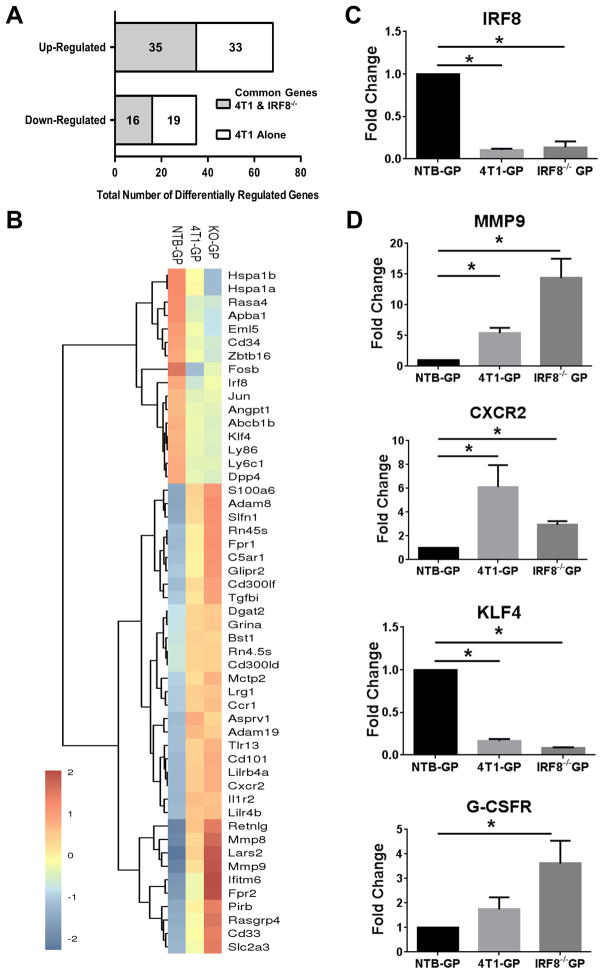
To understand better at a molecular level why 4T1-GPs were distinct from NTB-GPs, and to determine whether such differences were IRF8-regulated, we performed RNA-seq analyses. To that end, we performed three comparisons: first, we identified differentially expressed genes (> 2-fold) between NTB-GPs vs. 4T1-GPs. Secondly, we identified differentially expressed genes between NTB-GPs vs. IRF8−/− GPs. Third, we identified differentially expressed genes in common between 4T1-GPs and IRF8−/− GPs. Comparison of NTB- and 4T1-GP identified 103 up- or down-regulated genes (Fig. 5A). Based on cluster analysis, we identified 51 genes (35 upregulated and 16 downregulated) that were common to IRF8−/− GPs (Fig. 5A & B, Supplemental Table 2). Thus, nearly 50% of the tumor-induced changes in GP gene expression were potentially associated with IRF8 loss (Fig. 5A). Importantly, IRF8 represents one of the downregulated genes (Fig. 5C). We selected three additional genes (MMP9, CXCR2, and KLF4) for validation by qRT-PCR analysis, based on their relationship with hematopoiesis/myelopoiesis and/or MDSC biology (Fig. 5D). We also evaluated GP expression of the G-CSF receptor by qRT-PCR analysis. Both tumor-bearing (IRF8lo) GPs and IRF8−/− GPs expressed higher levels of G-CSF receptor compared to the controls (Fig. 5D), consistent with a greater propensity for G-CSF responsiveness (Fig. 2). Together, these data are consistent with the hypothesis that tumor growth fundamentally alters GPs at a molecular level and that changes in IRF8 expression contribute to this altered GP phenotype.
Figure 5. Tumor-induced GPs share a gene expression profile with IRF8−/− GPs.
(A) Data showing the number of differentially regulated genes (> 2-fold up or down) common to 4T1-GPs and IRF8−/−-GPs (gray area) or limited to 4T1-GPs (white area) compared to GPs of NTB mice. (B) Heat-map of genes in A, along with cluster analysis for NTB-, 4T1- and IRF8−/−-GPs. (C) qRT-PCR validation of IRF8 levels in the indicated GP populations. (D) qRT-PCR analysis of the indicated genes associated with MDSC biology from NTB-, 4T1- or IRF8−/− sorted GPs. All values normalized to GPs of NTB mice. *p<0.01 for 3 or more technical replicates using RNA from the RNA-seq analysis.
G-CSF recapitulates the myeloid progenitor imbalance seen in tumor-bearing hosts
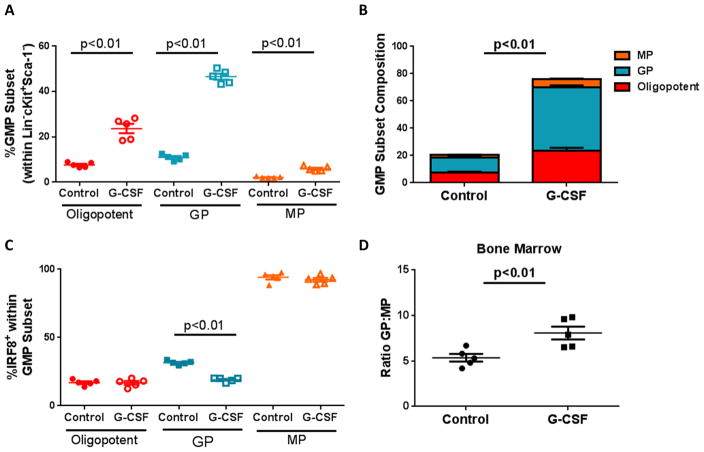
Our previous work supported a network by which tumor-derived G-CSF represented a major mechanism for the production of PMN-MDSCs (8). To examine whether G-CSF also mediates the observed myeloid progenitor imbalance, we utilized recombinant G-CSF protein (rG-CSF) known to induce PMN-MDSCs in the periphery (24). We injected naïve IRF8-EGFP mice with rG-CSF and, following treatment, observed splenomegaly as previously reported (24) (Supplemental Fig. 2C). This splenomegaly strongly reflected the expansion of granulocytic over monocytic cells (Supplemental Fig. 2D–E), accompanied by a reduction in peripheral expression of IRF8 (Supplemental Fig. 2F). In the bone marrow, we observed an expansion of oligopotent GMPs as well as GPs compared to the vehicle-treated controls (Fig. 6A; Supplemental Fig. 2H & I). MPs were expanded, albeit, to a lesser extent. Together, total GMPs expanded four-fold, with the greatest contribution by GPs (Fig. 6B). Comparable to high 4T1 tumor burden, G-CSF administration reduced the percentage of IRF8+ GPs, and altered the GP to MP ratio (Fig. 6C & D). Collectively, these data support the hypothesis that G-CSF is an integral tumor-derived factor facilitating the myeloid progenitor imbalance and subsequent production of PMN-MDSCs.
Figure 6. Impact of G-CSF treatment on IRF8 expression and myeloid progenitor cell frequencies.
Recombinant G-CSF was administered (10μg/mouse) for five consecutive days in IRF8-EGFP mice. Mice were rested for 24 hours, after which bone marrow was collected and analyzed for the indicated myeloid progenitor subset, as in Fig 1. (A–B) Total GMP frequencies are shown in control- or G-CSF-treated mice, along with a breakdown of the different myeloid progenitor subsets. IRF8 expression (C) and GP:MP ratio (D) were also analyzed. Data representative of two experiments (n=5 for all groups).
Enforced IRF8 overexpression limits aberrant myelopoiesis during tumor growth
Our data indicate that tumor growth expands IRF8lo/− GPs. To determine mechanistically whether this expansion is IRF8-regulated, we utilized a transgenic mouse model (IRF8-Tg), wherein IRF8 overexpression is controlled using the CMV promoter (23, 25) which would ensure transgene expression within bone marrow populations. We reasoned that transgene overexpression of IRF8 in the myeloid compartment would render these cells more refractory to tumor-induced alterations. Previously, such enforced IRF8 overexpression reduced MDSC accumulation in the periphery, as well as their pro-tumorigenic behavior (23). Under steady state conditions, however, all hematologic parameters measured within the leukocyte compartment (i.e., CBC and spleen) of both WT and IRF8-Tg groups of mice were within normal range (25). First, we verified significant enhancement of IRF8 expression in flow-purified GPs of non-tumor-bearing IRF8-Tg vs. WT mice (Fig. 7A). We extended this analysis to oligopotent GMPs, as well as lineage-negative bone marrow cells as source of all progenitors and found a significant enhancement of IRF8 compared to the controls (Fig. 7B & C).
Next, to test our hypothesis that tumor induced myeloid progenitor bias was controlled by IRF8 expression levels, we performed a new series of experiments and orthotopically implanted IRF8-Tg and WT littermates with AT-3 cells, followed by analysis of the different progenitors during tumor growth (Fig. 7D–H). The rationale for choosing AT-3 cells for these experiments reflects the fact that these cells are fully syngeneic to this mouse model. Similar to the 4T1 model, and as with our earlier findings with AT-3 cells (Fig. 1), WT mice showed a significant association between tumor volume and frequency of total GMPs (Fig. 7D), attributable mainly to a rise in GPs (Fig. 7F). Consistent with this interpretation, we observed no significant differences in oligopotent GMP and MP frequencies (Fig. 7E & G, respectively). In contrast, total GMPs from IRF8-Tg mice did not significantly correlate with tumor burden (Fig. 7D), and none of the three subsets changed in association with tumor volume (Fig. 7D–H). Although there seemed to be more variability in progenitor cell frequencies, such as the GPs and total GMPs in IRF8-Tg mice compared to the WT controls, statistical analysis revealed no significant differences in such frequencies between the two groups of mice. Importantly, the GP to MP ratio for AT-3-bearing WT mice positively associated with tumor burden, whereas IRF8-Tg mice showed either a negative or non-association with tumor burden (Fig. 7H). Altogether, these results show that enforced IRF8 expression selectively mitigates GP expansion in tumor-bearing hosts, thus accounting for the reduction of PMN-MDSCs previously observed in the periphery.
Discussion
MDSCs comprise two major subsets, monocytic and PMN; yet, the precise origin of each has remained unclear. Here, we provide the first evidence of a precise myeloid progenitor population that gives rise to PMN-MDSCs, and an underlying basis for their production. First, we showed that tumor growth induces the expansion of a newly defined and previously unrecognized GP population. Secondly, GP expansion is IRF8-dependent, either due to the preferential expansion of an IRF8lo/− population or repression of IRF8 expression in oligopotent GMPs or GPs. Thirdly, increasing expression of IRF8 rescues this tumor-induced myeloid imbalance, suggesting therapies that modulate IRF8 levels can reduce MDSC burden and enhance immunotherapy. Thus, our work has refined our current understanding of how tumor growth drives MDSC development, particularly PMN-MDSCs, at a progenitor level.
We are the first to identify IRF8 expression within novel progenitors in the bone marrow of mice bearing orthotopic and spontaneous mammary tumors. Our work refines previous reports of expanded myeloid progenitors during tumor growth (15, 35, 39) to specify expansion of a recently defined GP population (20). When these GPs were first defined, antibody-based flow cytometric analysis suggested higher IRF8 expression in these cells than was observed here. In contrast, we used a recently developed IRF8-EGFP mouse model with validated IRF8 expression levels and bioactivity (21, 40). In our study, RNA analysis additionally verified low IRF8 mRNA levels in GPs, which further decreased in GPs from tumor-bearing hosts. The apparent reduction in IRF8 expression within total GMPs was largely attributable to the expansion of IRF8lo/− GPs.
While we found that IRF8 levels are low in GPs and even lower in tumor GPs (Fig. 5), IRF8 may play an important role in cell survival, as suggested in the study by Yanez et al. (20). Consequently, it is possible that reduced IRF8 levels in tumor GPs compared to control GPs, may render tumor GPs less sensitive to apoptosis. IRF8 is a known regulator of anti-apoptotic molecules, such as Bcl-2 (41), and IRF8-deficiency in hematopoietic cells has been associated with increased resistance to apoptosis (42). Interestingly, downregulation of IRF8 expression by MDSCs reduces their susceptibility to immune-mediated apoptosis, enabling these cells to persist in cancer models (43). Thus, the larger accumulation of GPs in tumor-bearing mice compared to the controls may reflect multiple mechanisms including a reduced ability to die and turnover, which require further investigation. Interestingly, MPs did not appreciably expand during tumor growth in these models and appeared to maintain high IRF8 expression levels. In IRF8−/− mice, we found reduced MP frequencies compared to the wild-type controls, lower than that reported elsewhere (20). While the reasons underlying the differences in MP frequency remain unclear, both their study and ours highlight the importance of IRF8 in regulating the balance and fate of GPs vs. MPs. Future studies are necessary, however, to investigate the role of IRF8 in the fate and function of tumor-induced MPs.
Clearly, the biology of GPs vs. MPs is distinct, and it is likely that the mechanisms regulating their expression of IRF8 are different as well. The notion of developmental or functional plasticity is common in lymphoid and myeloid biology (44), and likely due to the engagement of other transcription factors. For example, recent studies showed that IRF8 drives a pro-monocytic program through interactions with, and sequestration of, the transcription factor C/EBPα, thereby preventing C/EBPα-stimulated granulocytic differentiation (45). Interestingly, we found that Kruppel-like factor KLF4, which is critical for monocyte differentiation (46), was reduced in 4T1- and IRF8−/−-GPs compared to NTB-GPs, which may in part explain the greater granulocyte bias in these GPs. IRF8-mediated monocyte differentiation also requires PU.1 as a binding partner (47, 48). Therefore, expression of these and other relevant IRF8 binding partners in different myeloid progenitors deserves further exploration to clarify how IRF8 is regulated during transition between these progenitor stages over the course of tumor growth.
A major question facing the MDSC field is how do immature myeloid cells become immune suppressive MDSCs? A two signal model has been proposed, in which 1) immature myeloid cells expand due to inhibition of their terminal differentiation, followed by 2) an activation phase, converting ‘normal’ immature cells to MDSCs (12). In vivo, we have seen that IRF8-deficiency expands myeloid progenitors. In vitro, freshly isolated tumor-induced GPs were not suppressive, consistent with earlier work reporting that CD11b+Gr-1+ cells from the bone marrow of tumor-bearing mice do not suppress compared to similar cells isolated from the periphery (49). However, after four days in culture with G-CSF, we found that tumor-induced GPs produced significantly more suppressive MDSCs than those of NTB hosts. These data show that tumor-induced GPs, despite sharing the same comprehensive flow marker phenotype as NTB-GPs, are precursors to suppressive PMN-MDSCs. In our prior studies (8), we found that IRF8 expression levels did not necessarily correlate with immunosuppressive activity. Specifically, we found that while MDSCs from IRF8−/− mice were suppressive, MDSCs from tumor-bearing IRF8-Tg mice were equally suppressive despite the fact that IRF8 levels remained high. IRF8 levels, however, inversely controlled MDSC frequencies in the spleen and tumor microenvironment. These findings indicated that IRF8 expression levels influenced MDSC numbers, but not necessarily the immune suppressive function of the remaining or residual MDSC population, consistent with the conclusion that IRF8 acts developmentally.
RNA-seq studies further support the idea that tumor-induced GPs are distinct from NTB-GPs, although the precise genes that confer immune suppressive behavior require detailed investigation. However, we did identify other MDSC-related genes differentially expressed by 4T1-GPs and IRF8−/− GPs, compared to those of NTB-GPs. For example, we verified upregulation of MMP9 and IL-8 receptor (also known as CXCR2) in 4T1- and IRF8−/−-GPs compared to NTB-GPs. MMP9 has been associated with MDSC-mediated remodeling of the extracellular matrix and promotion of tumor angiogenesis (50), while CXCR2 is associated with MDSC trafficking and resistance to immunotherapy (51). These data support the hypothesis that interfering with IRF8 expression by either an active mechanism of repression or a passive failure to activate underlies both the first and second signals in this model.
We and other laboratories have previously implicated G-CSF as a primary driver of dysregulated myelopoiesis in these tumor models (8, 24, 52). We previously showed that tumor-derived G-CSF induces PMN-MDSCs, based on loss-of-function (G-CSF silencing) or gain-of-function (ectopic G-CSF overexpression) experiments consistent with the notion that MDSC production is G-CSF dependent. Moreover, we showed that recombinant G-CSF administration alone recapitulates this MDSC outcome based on phenotypic, immune suppressive and gene expression profiling studies. Thus, the production of G-CSF by cancer cells provides at least one molecular basis for PMN-MDSC generation. Administration of recombinant G-CSF largely recapitulated the bone marrow cellular phenotype seen during mammary tumor growth. Previous work from our laboratory also identified G-CSF acting through STAT3 to downregulate IRF8 expression in peripheral myeloid cells (8). Whether STAT3 mediates repression of IRF8 during transition to the GP stage requires further study. Enforced IRF8 overexpression was able to redirect these expanded progenitors toward a more normalized GP:MP ratio, as well as a decrease in MDSCs in the periphery and an increased antitumor response when combined with a second therapy (8, 23). It is important to recognize that the IRF8-Tg model is not GMP or GP specific and, thus, may affect IRF8 levels broadly. However, our analyses involved a detailed inspection of changes in specific myeloid progenitors, and the ones most profoundly altered during tumor growth were the GPs. Importantly, while there is a positive relationship between tumor growth and total GMP or GP frequency in WT mice, this relationship does not exist in IRF8-Tg mice. The apparent, albeit not statistically significant, variability in total GMP or GP proportions in IRF8-Tg mice likely reflects additional complex biologic mechanisms, which requires further study. These may include the nature and/or quantity of tumor-derived factors that influence progenitor cell expansion, which may vary from mouse to mouse; potential variation in the absolute levels of IRF8 overexpression; IRF8-independent mechanisms; and the interrelationship among these complex biologic factors.
In summary, our study pinpoints a novel downstream GMP subset expanded in mammary tumor models and is likely an initial developmental source of MDSCs, particularly the PMN-MDSCs. We have extended the translational significance of these findings by showing that restoring IRF8 expression can correct the balance of myeloid progenitors. While further studies are warranted to examine events up or downstream of IRF8 expression that positively or negatively alter MDSC production, this work identifies a previously undescribed role of IRF8 in mitigating tumor-induced aberrant myelopoiesis. The conceptual value of enhancing IRF8 expression to curtail MDSC escalation has significant therapeutic implications in cases where a competent myeloid compartment is required for efficacy.
Supplementary Material
Acknowledgments
Grant Support: This work was supported by R01CA140622 and R01CA172105 from the National Cancer Institute/NIH (to SIA), an Alliance Developmental Award from the Roswell Park Alliance Foundation (to SIA, as well as a separate one to MJN), an NIH training grant T32CA085183 (to CSN) and a pre-doctoral fellowship award F30CA200133 (to LBM). All RPCI core resources were supported through an NCI Cancer Center Support Grant CA016056.
We thank Mary Lynn Hensen for excellent technical support, particularly for maintaining all mouse colonies, Dr. Vishala Neppalli (RPCI) for assistance with morphologic assessment of the various progenitor populations; and Drs. K. Ozato (NIH), H. Morse (NIH) and S. Gendler (Mayo Clinic) for providing important mouse models.
Footnotes
Conflicts of Interest: All authors declare no conflicts of interest related to this work
References
- 1.Messmer MN, Netherby CS, Banik D, Abrams SI. Tumor-induced myeloid dysfunction and its implications for cancer immunotherapy. Cancer Immunol Immunother. 2015;64:1–13. doi: 10.1007/s00262-014-1639-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shoenfeld Y, Tal A, Berliner S, Pinkhas J. Leukocytosis in non hematological malignancies--a possible tumor-associated marker. J Cancer Res Clin Oncol. 1986;111:54–58. doi: 10.1007/BF00402777. [DOI] [PubMed] [Google Scholar]
- 3.Gabrilovich DI, Bronte V, Chen SH, Colombo MP, Ochoa A, Ostrand-Rosenberg S, Schreiber H. The terminology issue for myeloid-derived suppressor cells. Cancer Res. 2007;67:425. doi: 10.1158/0008-5472.CAN-06-3037. author reply 426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Youn JI, Nagaraj S, Collazo M, Gabrilovich DI. Subsets of myeloid-derived suppressor cells in tumor-bearing mice. J Immunol. 2008;181:5791–5802. doi: 10.4049/jimmunol.181.8.5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Suzuki E, Kapoor V, Jassar AS, Kaiser LR, Albelda SM. Gemcitabine selectively eliminates splenic Gr-1+/CD11b+ myeloid suppressor cells in tumor-bearing animals and enhances antitumor immune activity. Clin Cancer Res. 2005;11:6713–6721. doi: 10.1158/1078-0432.CCR-05-0883. [DOI] [PubMed] [Google Scholar]
- 6.Vincent J, Mignot G, Chalmin F, Ladoire S, Bruchard M, Chevriaux A, Martin F, Apetoh L, Rebe C, Ghiringhelli F. 5-Fluorouracil selectively kills tumor-associated myeloid-derived suppressor cells resulting in enhanced T cell-dependent antitumor immunity. Cancer Res. 2010;70:3052–3061. doi: 10.1158/0008-5472.CAN-09-3690. [DOI] [PubMed] [Google Scholar]
- 7.Sevko A, Michels T, Vrohlings M, Umansky L, Beckhove P, Kato M, Shurin GV, Shurin MR, Umansky V. Antitumor effect of paclitaxel is mediated by inhibition of myeloid-derived suppressor cells and chronic inflammation in the spontaneous melanoma model. J Immunol. 2013;190:2464–2471. doi: 10.4049/jimmunol.1202781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Waight JD, Netherby C, Hensen ML, Miller A, Hu Q, Liu S, Bogner PN, Farren MR, Lee KP, Liu K, Abrams SI. Myeloid-derived suppressor cell development is regulated by a STAT/IRF-8 axis. J Clin Invest. 2013;123:4464–4478. doi: 10.1172/JCI68189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mahoney KM, Rennert PD, Freeman GJ. Combination cancer immunotherapy and new immunomodulatory targets. Nat Rev Drug Discov. 2015;14:561–584. doi: 10.1038/nrd4591. [DOI] [PubMed] [Google Scholar]
- 10.Curran MA, Montalvo W, Yagita H, Allison JP. PD-1 and CTLA-4 combination blockade expands infiltrating T cells and reduces regulatory T and myeloid cells within B16 melanoma tumors. Proc Natl Acad Sci U S A. 2010;107:4275–4280. doi: 10.1073/pnas.0915174107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Solito S, Marigo I, Pinton L, Damuzzo V, Mandruzzato S, Bronte V. Myeloid-derived suppressor cell heterogeneity in human cancers. Ann N Y Acad Sci. 2014;1319:47–65. doi: 10.1111/nyas.12469. [DOI] [PubMed] [Google Scholar]
- 12.Condamine T, Mastio J, Gabrilovich DI. Transcriptional regulation of myeloid-derived suppressor cells. J Leukoc Biol. 2015;98:913–922. doi: 10.1189/jlb.4RI0515-204R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Abrams SI, Netherby CS, Twum DY, Messmer MN. Relevance of Interferon Regulatory Factor-8 Expression in Myeloid-Tumor Interactions. J Interferon Cytokine Res. 2016;36:442–453. doi: 10.1089/jir.2015.0174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yamamoto R, Morita Y, Ooehara J, Hamanaka S, Onodera M, Rudolph KL, Ema H, Nakauchi H. Clonal analysis unveils self-renewing lineage-restricted progenitors generated directly from hematopoietic stem cells. Cell. 2013;154:1112–1126. doi: 10.1016/j.cell.2013.08.007. [DOI] [PubMed] [Google Scholar]
- 15.Wu WC, Sun HW, Chen HT, Liang J, Yu XJ, Wu C, Wang Z, Zheng L. Circulating hematopoietic stem and progenitor cells are myeloid-biased in cancer patients. Proc Natl Acad Sci U S A. 2014;111:4221–4226. doi: 10.1073/pnas.1320753111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tamura T, Nagamura-Inoue T, Shmeltzer Z, Kuwata T, Ozato K. ICSBP directs bipotential myeloid progenitor cells to differentiate into mature macrophages. Immunity. 2000;13:155–165. doi: 10.1016/s1074-7613(00)00016-9. [DOI] [PubMed] [Google Scholar]
- 17.Tsujimura H, Nagamura-Inoue T, Tamura T, Ozato K. IFN consensus sequence binding protein/IFN regulatory factor-8 guides bone marrow progenitor cells toward the macrophage lineage. J Immunol. 2002;169:1261–1269. doi: 10.4049/jimmunol.169.3.1261. [DOI] [PubMed] [Google Scholar]
- 18.Tamura T, Ozato K. ICSBP/IRF-8: its regulatory roles in the development of myeloid cells. J Interferon Cytokine Res. 2002;22:145–152. doi: 10.1089/107999002753452755. [DOI] [PubMed] [Google Scholar]
- 19.Holtschke T, Lohler J, Kanno Y, Fehr T, Giese N, Rosenbauer F, Lou J, Knobeloch KP, Gabriele L, Waring JF, Bachmann MF, Zinkernagel RM, Morse HC, 3rd, Ozato K, Horak I. Immunodeficiency and chronic myelogenous leukemia-like syndrome in mice with a targeted mutation of the ICSBP gene. Cell. 1996;87:307–317. doi: 10.1016/s0092-8674(00)81348-3. [DOI] [PubMed] [Google Scholar]
- 20.Yanez A, Ng MY, Hassanzadeh-Kiabi N, Goodridge HS. IRF8 acts in lineage-committed rather than oligopotent progenitors to control neutrophil vs monocyte production. Blood. 2015;125:1452–1459. doi: 10.1182/blood-2014-09-600833. [DOI] [PubMed] [Google Scholar]
- 21.Wang H, Yan M, Sun J, Jain S, Yoshimi R, Abolfath SM, Ozato K, Coleman WG, Jr, Ng AP, Metcalf D, DiRago L, Nutt SL, Morse HC., 3rd A reporter mouse reveals lineage-specific and heterogeneous expression of IRF8 during lymphoid and myeloid cell differentiation. J Immunol. 2014;193:1766–1777. doi: 10.4049/jimmunol.1301939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stewart TJ, Abrams SI. Altered immune function during long-term host-tumor interactions can be modulated to retard autochthonous neoplastic growth. J Immunol. 2007;179:2851–2859. doi: 10.4049/jimmunol.179.5.2851. [DOI] [PubMed] [Google Scholar]
- 23.Stewart TJ, Liewehr DJ, Steinberg SM, Greeneltch KM, Abrams SI. Modulating the expression of IFN regulatory factor 8 alters the protumorigenic behavior of CD11b+Gr-1+ myeloid cells. J Immunol. 2009;183:117–128. doi: 10.4049/jimmunol.0804132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Waight JD, Hu Q, Miller A, Liu S, Abrams SI. Tumor-derived G-CSF facilitates neoplastic growth through a granulocytic myeloid-derived suppressor cell-dependent mechanism. PLoS One. 2011;6:e27690. doi: 10.1371/journal.pone.0027690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stewart TJ, Greeneltch KM, Reid JE, Liewehr DJ, Steinberg SM, Liu K, Abrams SI. Interferon regulatory factor-8 modulates the development of tumour-induced CD11b+Gr-1+ myeloid cells. J Cell Mol Med. 2009;13:3939–3950. doi: 10.1111/j.1582-4934.2009.00685.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pronk CJ, Rossi DJ, Mansson R, Attema JL, Norddahl GL, Chan CK, Sigvardsson M, Weissman IL, Bryder D. Elucidation of the phenotypic, functional, and molecular topography of a myeloerythroid progenitor cell hierarchy. Cell Stem Cell. 2007;1:428–442. doi: 10.1016/j.stem.2007.07.005. [DOI] [PubMed] [Google Scholar]
- 27.Nemeth MJ, Curtis DJ, Kirby MR, Garrett-Beal LJ, Seidel NE, Cline AP, Bodine DM. Hmgb3: an HMG-box family member expressed in primitive hematopoietic cells that inhibits myeloid and B-cell differentiation. Blood. 2003;102:1298–1306. doi: 10.1182/blood-2002-11-3541. [DOI] [PubMed] [Google Scholar]
- 28.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang L, Wang S, Li W. RSeQC: quality control of RNA-seq experiments. Bioinformatics. 2012;28:2184–2185. doi: 10.1093/bioinformatics/bts356. [DOI] [PubMed] [Google Scholar]
- 30.Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pulaski BA, Ostrand-Rosenberg S. Mouse 4T1 breast tumor model. Curr Protoc Immunol. 2001;Chapter 20(Unit 20):22. doi: 10.1002/0471142735.im2002s39. [DOI] [PubMed] [Google Scholar]
- 33.Aslakson CJ, Miller FR. Selective events in the metastatic process defined by analysis of the sequential dissemination of subpopulations of a mouse mammary tumor. Cancer Res. 1992;52:1399–1405. [PubMed] [Google Scholar]
- 34.Guy CT, Cardiff RD, Muller WJ. Induction of mammary tumors by expression of polyomavirus middle T oncogene: a transgenic mouse model for metastatic disease. Mol Cell Biol. 1992;12:954–961. doi: 10.1128/mcb.12.3.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Casbon AJ, Reynaud D, Park C, Khuc E, Gan DD, Schepers K, Passegue E, Werb Z. Invasive breast cancer reprograms early myeloid differentiation in the bone marrow to generate immunosuppressive neutrophils. Proc Natl Acad Sci U S A. 2015;112:E566–575. doi: 10.1073/pnas.1424927112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koenigsmann J, Rudolph C, Sander S, Kershaw O, Gruber AD, Bullinger L, Schlegelberger B, Carstanjen D. Nf1 haploinsufficiency and Icsbp deficiency synergize in the development of leukemias. Blood. 2009;113:4690–4701. doi: 10.1182/blood-2008-05-158485. [DOI] [PubMed] [Google Scholar]
- 37.Scheller M, Foerster J, Heyworth CM, Waring JF, Lohler J, Gilmore GL, Shadduck RK, Dexter TM, Horak I. Altered development and cytokine responses of myeloid progenitors in the absence of transcription factor, interferon consensus sequence binding protein. Blood. 1999;94:3764–3771. [PubMed] [Google Scholar]
- 38.Newman DM, Leung PS, Putoczki TL, Nutt SL, Cretney E. Th17 cell differentiation proceeds independently of IRF8. Immunol Cell Biol. 2016;94:796–801. doi: 10.1038/icb.2016.33. [DOI] [PubMed] [Google Scholar]
- 39.Pu S, Qin B, He H, Zhan J, Wu Q, Zhang X, Yang L, Qu C, Zhou Z. Identification of early myeloid progenitors as immunosuppressive cells. Sci Rep. 2016;6:23115. doi: 10.1038/srep23115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Newman DM, Leung PS, Putoczki TL, Nutt SL, Cretney E. Th17 cell differentiation proceeds independently of IRF8. Immunol Cell Biol. 2016 doi: 10.1038/icb.2016.33. [DOI] [PubMed] [Google Scholar]
- 41.Burchert A, Cai D, Hofbauer LC, Samuelsson MK, Slater EP, Duyster J, Ritter M, Hochhaus A, Muller R, Eilers M, Schmidt M, Neubauer A. Interferon consensus sequence binding protein (ICSBP; IRF-8) antagonizes BCR/ABL and down-regulates bcl-2. Blood. 2004;103:3480–3489. doi: 10.1182/blood-2003-08-2970. [DOI] [PubMed] [Google Scholar]
- 42.Gabriele L, Phung J, Fukumoto J, Segal D, Wang IM, Giannakakou P, Giese NA, Ozato K, Morse HC., 3rd Regulation of apoptosis in myeloid cells by interferon consensus sequence-binding protein. J Exp Med. 1999;190:411–421. doi: 10.1084/jem.190.3.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hu X, Bardhan K, Paschall AV, Yang D, Waller JL, Park MA, Nayak-Kapoor A, Samuel TA, Abrams SI, Liu K. Deregulation of apoptotic factors Bcl-xL and Bax confers apoptotic resistance to myeloid-derived suppressor cells and contributes to their persistence in cancer. J Biol Chem. 2013;288:19103–19115. doi: 10.1074/jbc.M112.434530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Orkin SH, Zon LI. Hematopoiesis and stem cells: plasticity versus developmental heterogeneity. Nat Immunol. 2002;3:323–328. doi: 10.1038/ni0402-323. [DOI] [PubMed] [Google Scholar]
- 45.Kurotaki D, Yamamoto M, Nishiyama A, Uno K, Ban T, Ichino M, Sasaki H, Matsunaga S, Yoshinari M, Ryo A, Nakazawa M, Ozato K, Tamura T. IRF8 inhibits C/EBPalpha activity to restrain mononuclear phagocyte progenitors from differentiating into neutrophils. Nat Commun. 2014;5:4978. doi: 10.1038/ncomms5978. [DOI] [PubMed] [Google Scholar]
- 46.Feinberg MW, Wara AK, Cao Z, Lebedeva MA, Rosenbauer F, Iwasaki H, Hirai H, Katz JP, Haspel RL, Gray S, Akashi K, Segre J, Kaestner KH, Tenen DG, Jain MK. The Kruppel-like factor KLF4 is a critical regulator of monocyte differentiation. EMBO J. 2007;26:4138–4148. doi: 10.1038/sj.emboj.7601824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Carotta S, Willis SN, Hasbold J, Inouye M, Pang SH, Emslie D, Light A, Chopin M, Shi W, Wang H, Morse HC, 3rd, Tarlinton DM, Corcoran LM, Hodgkin PD, Nutt SL. The transcription factors IRF8 and PU.1 negatively regulate plasma cell differentiation. J Exp Med. 2014;211:2169–2181. doi: 10.1084/jem.20140425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tamura T, Kurotaki D, Koizumi S. Regulation of myelopoiesis by the transcription factor IRF8. Int J Hematol. 2015;101:342–351. doi: 10.1007/s12185-015-1761-9. [DOI] [PubMed] [Google Scholar]
- 49.Ugel S, De Sanctis F, Mandruzzato S, Bronte V. Tumor-induced myeloid deviation: when myeloid-derived suppressor cells meet tumor-associated macrophages. J Clin Invest. 2015;125:3365–3376. doi: 10.1172/JCI80006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang L, DeBusk LM, Fukuda K, Fingleton B, Green-Jarvis B, Shyr Y, Matrisian LM, Carbone DP, Lin PC. Expansion of myeloid immune suppressor Gr+CD11b+ cells in tumor-bearing host directly promotes tumor angiogenesis. Cancer Cell. 2004;6:409–421. doi: 10.1016/j.ccr.2004.08.031. [DOI] [PubMed] [Google Scholar]
- 51.Highfill SL, Cui Y, Giles AJ, Smith JP, Zhang H, Morse E, Kaplan RN, Mackall CL. Disruption of CXCR2-mediated MDSC tumor trafficking enhances anti-PD1 efficacy. Sci Transl Med. 2014;6:237ra267. doi: 10.1126/scitranslmed.3007974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kowanetz M, Wu X, Lee J, Tan M, Hagenbeek T, Qu X, Yu L, Ross J, Korsisaari N, Cao T, Bou-Reslan H, Kallop D, Weimer R, Ludlam MJ, Kaminker JS, Modrusan Z, van Bruggen N, Peale FV, Carano R, Meng YG, Ferrara N. Granulocyte-colony stimulating factor promotes lung metastasis through mobilization of Ly6G+Ly6C+ granulocytes. Proc Natl Acad Sci U S A. 2010;107:21248–21255. doi: 10.1073/pnas.1015855107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.