Abstract
Background
Since 2012, WHO guidelines for the prevention of mother-to-child transmission (PMTCT) of HIV-1 in resource-limited settings recommend the initiation of lifelong antiretroviral combination therapy (cART) for all pregnant HIV-1 positive women independent of CD4 count and WHO clinical stage (Option B+). However, long-term outcomes regarding development of drug resistance are lacking until now. Therefore, we analysed the emergence of drug resistance mutations (DRMs) in women initiating Option B+ in Fort Portal, Uganda, at 12 and 18 months postpartum (ppm).
Methods and findings
124 HIV-1 positive pregnant women were enrolled within antenatal care services in Fort Portal, Uganda. Blood samples were collected at the first visit prior starting Option B+ and postpartum at week six, month six, 12 and 18. Viral load was determined by real-time RT-PCR. An RT-PCR covering resistance associated positions in the protease and reverse transcriptase HIV-1 genomic region was performed. PCR-positive samples at 12/18 ppm and respective baseline samples were analysed by next generation sequencing regarding HIV-1 drug resistant variants including low-frequency variants. Furthermore, vertical transmission of HIV-1 was analysed. 49/124 (39.5%) women were included into the DRM analysis. Virological failure, defined as >1000 copies HIV-1 RNA/ml, was observed in three and seven women at 12 and 18 ppm, respectively. Sequences were obtained for three and six of these. In total, DRMs were detected in 3/49 (6.1%) women. Two women displayed dual-class resistance against all recommended first-line regimen drugs. Of 49 mother-infant-pairs no infant was HIV-1 positive at 12 or 18 ppm.
Conclusion
Our findings suggest that the WHO-recommended Option B+ for PMTCT is effective in a cohort of Ugandan HIV-1 positive pregnant women with regard to the low selection rate of DRMs and vertical transmission. Therefore, these results are encouraging for other countries considering the implementation of lifelong cART for all pregnant HIV-1 positive women.
Introduction
Mother-to-child transmission (MTCT) is the most common way of HIV-1 infection among children under the age of 15[1, 2]. Vertical transmission of the virus can occur during pregnancy, labour and delivery or through breastfeeding. Without any intervention, the risk of transmission ranges between 15% and 45%. Since the end of the last century, the use of antiretroviral (ARV) drugs for the prevention of mother-to-child transmission (PMTCT) has unfolded a potential to reduce the rate of MTCT to <5% [1, 3–5]. However, the frequent emergence of drug resistance mutations (DRMs) linked with temporary single- or combination- ARV regimens has to be viewed as a major threat for future treatment success in mothers and their children by reducing the susceptibility to ARV drugs[6–11]. Even in complex PMTCT regimens with longer intake duration, the level of acquired DRMs remains high as long as they are applied only temporarily, as discontinuing ARV intake after delivery and possible restart during subsequent pregnancies or for later antiretroviral therapy (ART) promotes the emergence of DRMs[12, 13].
Since 2012, the WHO PMTCT guidelines recommend “Option B+”, which stands for the initiation of lifelong combination ART (cART) for all pregnant and breastfeeding women living with HIV-1 regardless of CD4 count or WHO clinical stage[14]. The recommended regimen consists of two nucleoside reverse transcriptase inhibitors (NRTIs) combined with a non-nucleoside reverse transcriptase inhibitor (NNRTI) (first-line regimen), or a protease inhibitor (PI) (second-line regimen). Infants born to HIV-1-infected mothers also receive nevirapine (NVP, an NNRTI) or zidovudine from birth until six weeks of age. While the early start and lifelong intake of cART increases the effectiveness regarding PMTCT by lastingly suppressing viral loads[15], the approach also focuses on improving maternal health and reducing mortality and morbidity of HIV-1 positive women. The continuous intake of cART is reported to decrease or avoid resistance development[16], but suboptimal adherence and loss to follow-up due to individual, structural or community factors still pose a risk for the selection of drug resistance even under Option B+[17, 18]. Besides the need to overcome this huge barrier, surveillance of DRMs is a key factor to optimize treatment outcomes and support national, regional and global decision-making regarding the choice of first-line regimens[6].
Until now, population-based Sanger sequencing has been the gold standard for the screening of HIV-1 genotypic resistance. However, population-based sequencing is not able to detect low-frequency HIV-1 drug resistant variants at levels below 20%[12]. Nevertheless, studies show that also low-frequency HIV-1 drug resistant variants can have an influence on the success of cART[8, 19]. With its high analytical sensitivity, next generation sequencing (NGS) has overcome the limitations of traditional Sanger sequencing and therefore gained increasing importance in the last years[20–22].
Uganda is classified as a HIV-1 high burden country with a prevalence around 7.4% in the general population in 2013[23]. In 2012 Uganda started implementing Option B+ which was scaled-up countrywide since then, increasing the percentage of pregnant women under ART from 86% in 2011 to 94% in 2014[23]. Until now, long-term data on resistance development during intake of Option B+ in a real-life scenario are scarce. In fact, there is only one study from Malawi, reporting DRMs in 8.5% of women at 12 months postpartum (ppm)[24].
The aim of this study was to assess the emergence of HIV-1 DRMs against three major drug classes, NRTIs, NNRTIs and PIs, leading to resistance against the recommended first- and second-line drugs for PMTCT in Fort Portal, Uganda. Using a combination of conventional and real-time RT-PCR assays as well as the NGS platform MiSeq (Illumina) the development and variation in time of DRMs could be assessed.
Materials and methods
Ethic statement
Ethical approval was obtained from the Committee of Higher Degrees, Research and Ethics, College of Health Sciences, Makerere University Kampala, by the Ugandan National Council for Science and Technology and by the Ethical Committee of Charité - Universitätsmedizin Berlin. We obtained informed written consent from all participants involved in the study and treated data strictly confidential. Participants could withdraw their study participation at all times without any negative consequences for their treatment in hospital.
Clinical samples and study design
Between January 2013 and December 2013, women attending antenatal care (ANC) at Virika Hospital and the Regional Referral Hospital in Fort Portal, Uganda, were recruited for a longitudinal prospective cohort study as a part of a larger study on Option B+ outcomes. According to the WHO and Ugandan PMTCT guidelines from 2012, HIV-1 positive pregnant women were enrolled on Option B+, composed of a single-pill based on a NRTI/NNRTI, fixed-dose combination of efavirenz/lamivudine/tenofovir (EFV/3TC/TDF) as first-line regimen; newborns received NVP from birth up to six weeks of life, regardless of infant feeding method[14]. Women were included in the study due to following eligibility criteria: Informed written consent, age ≥18 years, confirmed pregnancy and positive HIV-1 status without prior ART enrolment.
Women were followed-up at subsequent ANC and postpartal care visits until 18 ppm. Plasma samples from women were collected at the first ANC visit before initiating Option B+ (baseline sample), at subsequent ANC visits, at six weeks postpartum (ppw) as well as six, 12 and 18 ppm. From infants, plasma samples were obtained or blood from heel pricks was spotted onto filter papers (Dried Blood Spots, DBS) at the same time intervals. In the present study, which aims to analyse the emergence of DRMs at least 1 year after initiation of Option B+ in ANC only women with a sample at 12 and/or 18 ppm and a respective baseline sample available were included.
Extraction of viral nucleic acids
Nucleic acids were extracted using the NucliSens easyMAG system (Biomerieux) according to the manufacturer`s instructions. Briefly, plasma samples were centrifuged for 10 min at 4°C with 3000 g to remove cryoprecipitate. Subsequently, 500μl of plasma were transferred into 2 ml of lysis buffer, mixed and incubated at room temperature for 10 min. After adding 100 μl of magnetic silica solution, total nucleic acids were extracted by automated magnetic separation (Generic 2.0.1) and finally eluted in 60 μl of elution buffer. Extracted nucleic acids were aliquoted in portions of 10 μl and immediately stored at -80°C until further use.
For the extraction of total nucleic acids from DBS, 2 blood spots (50 μl blood each) were cut out from the filter paper and transferred to 2 ml of lysis buffer. After shaking for one hour at room temperature with 300 rpm, 1.8 ml of lysated eluate was used for automated nucleic acid extraction performed as described above.
Amplification of nucleic acids
For amplification of viral RNA three different PCRs were performed; two conventional RT-PCR assays to identify DMRs in the protease (PR) and reverse transcriptase (RT) genomic region of the HIV-1 polymerase (pol)-region and a real-time TaqMan RT-PCR to determine the HIV-1 viral load. With the priority of obtaining as many sequences as possible for DRM analysis and due to the limited material available, first conventional RT-PCR assays were performed, followed by HIV-1 viral load measurement.
For conventional RT-PCR two overlapping fragments of 576 bp length (fragment 1) containing the codons 1 to 99 of the PR and codons 1 to 93 of the RT and of 718 bp length (fragment 2) containing the codons 22 to 260 of the RT [25] were amplified using the Qiagen One Step RT PCR kit and an Eppendorf Mastercycler pro. The resulting PR-RT sequence (fragment 1+2, position 2277–3302 on HXB2 genome, Acc. No. K03455.1) covers all resistance-associated mutations of PR and RT. In the following text viral sequences named “PR”, “RT” and “PR-RT” refer to the corresponding PCR-product of “fragment 1, 2 or 1+2”.
Conventional RT-PCR assays were performed on all maternal 12 and 18 ppm samples as well as respective baseline samples. Additional samples from earlier time points (6 ppw or 6 ppm) were subjected to the PCRs if maternal HIV-1 sequences displayed DRMs. Samples resulting in RT-PCR fragments were applied to NGS. Infant 12/18 ppm samples were analysed by conventional RT-PCR. In case of a positive result infants were considered to be HIV-1 infected.
The HIV-1 viral load was determined using a quantitative real-time TaqMan RT-PCR (MX3005P qPCR system from Agilent) with primers located in the long terminal repeat (LTR) of the HIV-1 genome. Primers and probe for the amplification of a 118 base pair (bp) fragment were used as described by Cleland et al., 2001[26]. An external standard curve generated from known concentrations of the laboratory strain HTLVIIIB (Acc. No. K03455.1) was carried out on each plate to enable absolute quantification of HIV-1 genome equivalents in every sample. Viral load was determined for all maternal baseline samples and post partum samples resulting in RT-PCR fragments. In this study, samples with HIV-1 viral loads >1000 copies/ml were defined being a result of “virological failure” (VF).
Next generation sequencing (NGS)
Amplicons were purified using Agencourt AMPure® XP Beads (Beckman Coulter) and quantified using Quant-iT™ PicoGreen® dsDNA Reagent (Invitrogen). The Nextera XT® kit (Illumina) was used to prepare the NGS libraries. Sequencing was performed on the Illumina MiSeq® employing the 2 x 200 or 2 x 300 bp paired end modus. The NGS data were processed through an in-house bioinformatics pipeline making use of the tools Trimmomatic (version 0.33)[27], FLASH (version 1.2.11)[28], and BWA (version 0.7.10)[29], all wrapped in a Python script. After adapter-clipping, trimming and merging, five iterations of mapping against a reference sequence (HXB2, Acc. No. K03455.1) were performed. Potential insertions and deletions were considered during the mapping cycles. The final output was subject to further Python scripts, which enabled codon-based variant detection and generation of a consensus sequence containing ambiguities according to an adjustable threshold. A Sanger-like threshold of 20% was applied in general. Moreover, the sensitivity and specificity for the detection of low-frequency drug resistant variants was determined using mixtures of recombinant HIV-1 from the wild-type pNL4.3 clone (Acc. No. M19921) and a mutant pNL4.3 derived clone that harbours twelve DRMs within pol. Cloning and generation of the recombinant virus has been described previously[6, 30]. Twenty-eight mixtures ranging from 1% to 50% mutant in wild type were prepared in viral loads ranging from 103 to 106 copies/ml. In mixtures with viral loads <105 copies/ml, the sensitivity of detection varied and the noise was higher than in mixtures with viral loads ≥105 copies/ml. Consequently, NGS thresholds for low-frequency variants of 3% and 10% for samples with low and high viral load, respectively, were determined.
Detection of viral drug resistance mutations and determination of HIV-1 subtype
Only samples with successfully generated sequence were included in the final DRM-analysis. DRMs were determined according to the Stanford University’s HIV-1 drug resistance database[31] and by the International Antiviral Society USA (IAS-USA)[32].
The prevalence of pre-existing transmitted drug resistance mutations (TDRMs) in baseline samples was assessed using the WHO surveillance drug resistance mutations list (SDRM)[33]. Therefore, resistance mutations in the PR and RT were considered.
In contrast, the prevalence of DRMs selected by the implemented NRTI/NNRTI regimen was only analysed in the viral RT sequence of post partum samples, as no selection of DRMs in PR was expected. Based on sequence availability, comparison of viral RT sequences from baseline samples to respective sequences from follow-up (FUP) samples after delivery was performed to identify emerging DRMs acquired during the PMTCT regimen. However, the extent of DRM-selection in FUP samples without the respective baseline sequence could be assessed by monitoring their proportion compared to the respective 6 ppw and 6 ppm samples. Drug resistant low-frequency variants were defined as DRMs detected by NGS below the Sanger-like cut-off of 20%.
HIV-1 subtypes were determined on basis of the Sanger-like NGS sequences with an ambiguity threshold of 20%. HIV-1 subtypes were determined using the REGA HIV-1 subtyping tool v3.0[34].
Results
Study population
124 women attending ANC between January and December 2013 were tested HIV-1 positive. All women were drug naïve at enrolment and received the recommended first-line regimen consisting of EFV/3TC/TDF. Socio-demographic and clinical baseline data, as well as information on the antenatal adherence of the study group were published by Schnack et al., 2016[35].
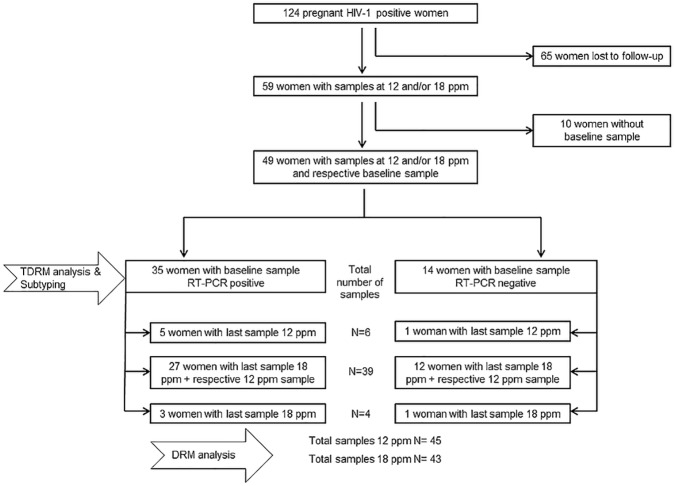
Only 59/124 (47.6%) women completed the study-period at 18 or at least 12 ppm. However, ten of them were excluded due to missing baseline samples. These 49 women finally constituted to the present study cohort for genotypic resistance testing (Fig 1). Both, the 12 and 18 ppm sample were available for 39 women, while either the 12 or 18 ppm sample was available for six and four women, respectively, resulting in overall 49 women with 45 samples from 12 ppm and 43 samples from 18 ppm. Baseline, 6 ppw and 6 ppm samples were available for 49/49, 43/49 and 44/49 women, respectively (6 ppw and 6 ppm not shown in Fig 1).
Fig 1. Sample availability.
Analysing the last available sample of involved infants, in 43 infants at 18 ppm and six infants at 12 ppm, no HIV-1 infection was detected.
Genotypic resistance testing and virological failure
Assessment of TDRMs was carried out for 35/49 RT-PCR positive baseline samples, resulting in 27 PR-RT sequences, six PR sequences, and two RT sequences (Table 1).
Table 1. Outcome sample analysis.
| N (%) | No. PCR | Viral load tested for | Succesful NGS for | |
|---|---|---|---|---|
| positive of | No. of samples and | No. of samples | ||
| No. analysed | median viral load (IQR) | analysed and | ||
| in copies/ml | sequence results | |||
| No. of women included in | 49 | |||
| DRM analysis | ||||
| Baseline samples for No. of women | 49 (100) | 35/49 | 42/49 | 35/35 |
| 4.9x103 | 27 PR-RT | |||
| (1.2x104-1.1x103) | 6 PR | |||
| 2 RT | ||||
| No. of samples taken at time point | ||||
| 6 ppw | 43 (87.8) | 2/3 | 2/2 | 2/2 |
| 4.1x103 | 2 PR-RT | |||
| (5.1x103-3.2x103) | ||||
| 6 ppm | 44 (89.8) | 3/3 | 3/3 | 3/3 |
| 1.5x104 | 3 PR-RT | |||
| (1.8x104-9.3x103) | ||||
| 12 ppm | 45 (91.8) | 5/45 | 5/5 | 5/5 |
| 1.1x104 | 5 PR-RT | |||
| (1.3x104-0) | ||||
| 18 ppm | 43 (87.8) | 7/43 | 7/7 | 6/7 |
| 1.1x104 | 5 PR-RT | |||
| (5.2x104-4.3x103) | 1 RT |
No.: number.
Furthermore, PR-RT sequences could be obtained for 2/3 samples at 6 ppw, 3/3 samples at 6 ppm, 5/5 samples at 12 ppm and 5/7 samples at 18 ppm. One 18 ppm sequence covered the RT fragment only and one sequence generated by NGS failed the inclusion criteria due to a low number of reads. In total 52 samples were analysed by NGS. The mean read length ranged from minimal 137 to maximal 219 bp. After pre-processing, ~18.000 to ~537.000 valid reads led to mean coverages of ~1.100 to ~126.000.
Viral loads could be detected in 42/49 (85.7%) baseline samples (median 4.9x103; IQR 1.2x104-1.1x103), in 3/5 12 ppm samples and 7/7 18 ppm samples. For all three samples with VF, defined as viral loads >1000 copies/ml, at 12 ppm the respective 18 ppm sample likewise displayed VF. In total, 3/45 and 7/43 women at 12 and 18 ppm (corresponding to 7/49 women at 12/18 ppm) experienced VF corresponding to 93.3% and 85.7% of women at 12 and 18 ppm reaching viral suppression.
Drug resistance mutations at baseline and subtyping
According to the sequence availability of baseline samples TDRMs in the PR and RT of HIV-1 could be analysed for 33 and 29 of 35 women, respectively (Table 1). In none of the 29 RT-sequences NRTI- or NNRTI-selected mutations according to the WHO-SDRM list[33] could be detected. Likewise, no PI-selected TDRMs were found in the 33 PR-sequences. However, considering the IAS-USA mutation list[32] the Q58E mutation in the PR was detected in one sample at baseline. Furthermore, a large number of minor PI resistance mutations were found in the baseline samples[32]. The most prevalent minor PI resistance mutation was M36I (31/33), followed by H69K (24/33) and L89M (20/33). Further detected minor PI resistance mutations were L10I and L10V (each 1/33), G16E (4/33), K20I and K20R (4/33 and 10/33, respectively), 33 (1/33), D60E (4/33), I62V (5/33), L63P (11/33), I64V and I64M (10/33and 1/33, respectively), V82I (3/33) and I93L (1/33).
HIV-1 subtypes were determined from PR-RT baseline sequences of 35 women. Subtype A1 was predominant in the study cohort (19/35; 54.3%), followed by subtype D (7/35; 20.0%). 14.3% (5/35) of women were infected with recombinant HIV-1 strains, while 8.6% (3/35) and 2.9% (1/35), were infected with subtype G and C HIV-1, respectively.
Drug resistance mutations at 12 and 18 ppm
HIV-1 RT genomic region could be analysed for 5/5 12 ppm and 6/7 18 ppm samples, corresponding to seven women (Table 1). DRMs were detected in 3/5 12 ppm and 3/6 18 ppm HIV-1 samples (Table 2), corresponding to three women. Hence, HIV-1 DRMs were detected in 3/49 (6.1%) women at 12 and/or 18 ppm.
Table 2. Outcome of detection of DRMs in three women.
| Sample | Baseline | 6 ppw | 6 ppm | 12 ppm | 18 ppm | |
|---|---|---|---|---|---|---|
| ID | ||||||
| 1 | VL | 1.2x104 | 6.0x103 | 1.5x104 | 1.3x104 | 4.8x104 |
| (copies/ml) | ||||||
| RT-PCR | pos | pos | pos | pos | pos | |
| DRMs (%) | wt | wt | *K103N (16.8) | **K103N (6.32) | K103N (82.2) | |
| 2 | VL | 5.5x104 | - | 3.5x103 | 1.1x104 | 5.8x103 |
| (copies/ml) | ||||||
| RT-PCR | pos | neg | pos | pos | pos | |
| DRMs (%) | wt | ns | K70E (99.6) | K70E (99.7) | K70E (99.7) | |
| M184V (99.4) | M184V (99.5) | M184V (96.8) | ||||
| K103N (99.2) | K103N (99.1) | K103N (99.4) | ||||
| E138Q (99.3) | E138Q (99.5) | V108I (29.6) | ||||
| *P225H (13.6) | E138Q (99.6) | |||||
| P225H (76.2) | ||||||
| 3 | VL | 2.0x104 | 2.2x103 | 2.1x104 | 1.7x104 | 1.3x105 |
| (copies/ml) | ||||||
| RT-PCR | neg | pos | pos | pos | pos | |
| DRMs (%) | ns | M184V (85.4) | M184V (94.7) | M184V (97.2) | M41L (41.7) | |
| K103N (84.2) | T215Y (21.5) | **L210W (4.3) | L74V (35.0) | |||
| V179T (32.2) | K103N (89.4) | T215Y (90.4) | M184V (99.4) | |||
| *V179T (13.0) | K103N (99.4) | L210W (71.2) | ||||
| T215Y (99.5) | ||||||
| *A98G (4.4) | ||||||
| L100I (93.8) | ||||||
| K103N (98.9) | ||||||
| *V108I (6.0) | ||||||
| *V179T (3.7) | ||||||
| P225H (20.2) |
VL: viral load; wt: wild type; nd: not detected; ns: no sequence from sample; -: exclusion according to sample inclusion criterion
* Low-frequency drug resistant variant
** Low-frequency drug resistant variant below the NGS cut-off, defined dependent on viral load.
In one woman (Sample ID 1) HIV-1 resistance against EFV and NVP caused by the K103N mutation in the RT was selected. This woman had reported full adherence until 12 ppm and stated at 18 ppm not having taken pills for >seven days. The two other women (Sample ID 2 and 3) showed dual-class drug resistance against NRTIs and NNRTIs, including all drugs recommended for first-line ART. In both women, mutations K103N, P225H and M184V were detected, although they reported full adherence until 18 ppm. Outcome of the DRM analysis for the remaining 46 women is listed in S1 Table.
Low-frequency drug resistant viral variants were found in only one of the three women at 18 ppm (Sample ID 3: V108I 6.0% and A98G 4.4%). At 12 ppm, low-frequency drug resistant variants were detected in another woman (Sample ID 2: P225H 13.6%). Furthermore, low-frequency drug resistant variants–but below the defined cut-off dependent on the viral load of the sample—were also found in two women at 12 ppm (Sample ID 1: K103N 6.3% and sample ID 3: L210W 4.3%). Nevertheless, all low-frequency drug resistant variants found at 12 ppm, independent if below or above the defined NGS cut-off, were detected in the respective 18 ppm sample selected as main viral variant (Table 2). All women with DRMs at 12/18 ppm were of subtype D.
Discussion
Our study is among the first to provide data on the selection of DRMs until at least 1 year after initiation of Option B+ for PMTCT.
According to WHO recommendations the success of HIV-1 drug treatment is defined by a viral load suppression in >85% of individuals after 12 months of ART [36]. This criterion was met in our study with 93.3% (42/45) of women at 12 and 85.7% (36/43) at 18 ppm reaching viral suppression. First results of women enrolled in Option B+ in Malawi (recommended first-line regimen: EFV/3TC/TDF) report viral suppression in 91.5% of women at 12 ppm, which is comparable to our results[24]. Data from other countries implementing Option B+ are not available until now. However, in another study from Malawi, where pregnant women initiating cART between 2008 and 2011 were followed up until 24 ppm, viral suppression was achieved in 87.1% of women at 12 and 89.9% at 18 ppm, respectively[37]. In a cohort of pregnant women in Tanzania, women initiating cART between 2004 and 2006 had a viral suppression rate of only 47% at 12 ppm[38]. However, in both studies, women initiated cART for their own health due to a low CD4 cell count at baseline. Furthermore, in both settings an NVP-based drug regimen was the first-line recommendation at the time of study. Therefore, these data are not directly comparable to the data from our study, where drug naïve women initiated cART independent of CD4 cell count. The high virological suppression rates in our study as in Malawi support the latest WHO guidelines moving away from different “options” for PMTCT to the early, lifelong cART initiation for all pregnant HIV-1 positive women regardless of clinical or CD4 cell count or stage of disease[39]. Nevertheless, a known problem assessing the potential success of PMTCT interventions in low-income settings is the high number of women lost to follow-up[40]. Taking into account the loss to follow-up rate of 52.4% in this study, the true viral suppression rate is probably much lower and fails to meet the criteria for successful HIV-1 drug treatment. Furthermore, using the reversed approach of testing in our study–conventional RT-PCR followed by viral load measurement–we might have missed women with detectable viral load (due to the sensitivity of the conventional RT-PCR of ≥1000 copies/ml). However, the application of both assays (RT-PCR and viral load measurement) to all baseline samples revealed only 7/49 RT-PCR negative samples with viral load (which was below 1000 copies/ml for all seven cases) indicating that all women experiencing VF were identified by our testing scheme.
It has been shown that stopping and restarting an NNRTI-based regimen poses a risk for emergence of DRMs and may influence suppression rates when restarting a similar regimen[13, 41]. In our study, DRMs were detected in 6.1% (3/49) of women at 12 and/or 18 ppm. The absence of DRMs in the sequence of baseline samples of two women (Sample ID 1+2) enabled to identify the DRMs at 12/18 ppm as newly acquired (Table 2). For the third woman (Sample ID 3) the corresponding baseline viral sequence was missing due to a negative result in the two conventional RT-PCR assays. However, increasing levels of DRMs from 6 ppw up to highest levels at 18 ppm points to an ongoing selection of DRMs. Among women displaying VF almost half of them (42.9%; 3/7) selected for HIV-1 DRMs. Two of them harboured multiple DRMs, resulting in dual-class resistance against NRTIs and NNRTIs, including those recommended as first-line therapy for Option B+ (EFV/3TC/TDF). Although all three women reported full or almost full adherence, the selection of drug resistance mutations as well as the nearly constant detection of viral loads above 1000 copies/ml in the follow-up samples indicate suboptimal drug concentrations. However, besides drug adherence, other factors like pharmacogenomic issues or drug interactions with additional therapeutic agents against co-morbidities might contribute to changes in drug levels or drug half life, resulting in the development of DRMs[42]. Furthermore Kyeyune et al., observed a higher frequency of treatment failure and drug resistance in subtype D versus subtype A infected individuals[43]. All three women experiencing treatment failure in this study were of subtype D indicating a possible influence of the subtype, maybe in combination with other adverse factors, on treatment success. However, to confirm this hypothesis a closer examination of more subtype D infected individuals with and without treatment failure would be required. The NNRTI-associated mutation K103N was observed in all these three women remarkably present as the main viral variant. In Malawi, where low-frequency HIV-1 drug resistant variants were not analysed, DRMs were present in 8.5% of women at 12 ppm, corresponding to a comparable percentage of 40% for women with detectable viral load. In line with our results, they also detected the K103N in all women with DRMs. In the above mentioned studies from Tanzania and Malawi with women initiating life-long cART due to low CD4 counts, DRMs were observed in 34% and 6% of women at 12 ppm, respectively[37, 38]. However, in the Malawian cohort only 15/27 samples with detectable HIV-1 RNA were sequenced, which possibly leads to underestimation of the DRM. Data from the pre-Option B+ era report DRMs (including low-frequency variants) in 40% of infected Tanzanian women receiving a drug regimen resembling Option A between delivery and 16 weeks postpartum[6]. Compared to these findings, the rate of 6.1% of women with DRMs at 12 and/or 18 ppm in this study and 8.5% at 12 ppm in Malawi clearly show the benefit over earlier PMTCT recommendations. Nevertheless, the high number of women lost to follow-up in this study (65/124; 52.4% of women) has to be considered. Women lost to follow-up probably also may have been less adherent to the regimen thereby impeding treatment success. Therefore, the true percentage of DRMs is difficulty to assess and is probably underestimated in this study analysing the more adherent women.
Low frequency variants of NNRTI-associated DRMs were detected in two of three women at 12 and 18 ppm. However, their contribution to VF cannot be assessed, as they were found among multiple DRMs. Several studies indicated that low-frequency drug resistant variants are associated with VF[19]. In a recent systematic review of literature it has been shown that individuals with low-frequency drug resistant variants and <95% medication adherence had a 5.1 times higher risk of VF compared to individuals with low-frequency drug resistant variants and a medication adherence of >95%[44]. Kyeyune et al., reported the presence of DRMs in low frequency only for 65% of Ugandan patients failing cART [45]. However, in other studies no association between VF and the presence of low-frequency drug resistant variants was observed[46, 47].
No TDRMs according to the WHO SDRM list were detected in women at baseline in our study cohort. However, based on the algorithm of the IAS-USA, a number of minor PI-resistance mutations were detected at baseline. These mutations are common non-B subtype polymorphisms, some of which have been associated with early treatment failure and higher number of acquired major PI-associated mutations at time of treatment failure under a PI containing regimen[48–50]. This should be kept in mind when switching to a PI containing second-line regimen, as treatment success might me impaired. In total, the prevalence rate of TDRMs in this study was 0%, although highly sensitive NGS was performed. Other studies from Uganda report varying rates of TDRMs between 1.5% and 19% with respect to different populations, regions and surveillance strategies[51, 52]. In pregnant ART-naïve women, two studies between 2006 and 2007 observed TDRM rates ranging from 0% to 5%, respectively[53, 54]. These data are in line with the rate found in our study cohort.
The small sample size is a major limitation of our study as only 49 women with a respective baseline sample could be followed-up until 12/18 ppm. However, limited samples sizes in the context of HIV-1 positive cohort FU are a common problem in these settings, while at the same time, our data are among the first to describe the development of DRMs in HIV-1 positive pregnant women starting Option B+ in a real-life scenario. For a growing number of countries, now implementing the lifelong intake of cART for PMTCT, these data are encouraging with regards to DRMs linked with the initiation of an early lifelong treatment for all HIV-positive pregnant women. For the future, data from large-scale, multicentre studies assessing the emergence of DRMs during Option B+ will be required to confirm the findings of this research.
Supporting information
VL: viral load; wt: wild type; nd: not detected; ns: no sequence from sample; -: absence of sample, or to exclusion according to sample inclusion criterion.
(DOCX)
Acknowledgments
We would like to thank all participating women, midwives, laboratory staff and administration of both The Holy Family Virika Hospital and Fort Portal Regional Referral Hospital. The study was funded and supported by the H.W. & J. Hector Foundation, Germany. The sponsor had no role in study design, in the collection, analysis and interpretation of data, in the writing of the report, or in the decision to submit the article for publication. This research was based on a larger PMTCT study supported by ESTHER Germany, Deutsche Gesellschaft für Internationale Zusammenarbeit (GIZ).
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
https://www.hector-stiftung.de/medizinische-forschung/aidsforschung/projekte/m68/. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. Initials of authors who were receiving funds is not applicable.
References
- 1.WHO HIV/AIDS Fact sheet. Updated July 2016. WHO website.Available from: http://www.who.int/mediacentre/factsheets/fs360/en/.
- 2.UNAIDS The GAP report. 2014. UNAIDS website.Available from: http://www.unaids.org/sites/default/files/media_asset/UNAIDS_Gap_report_en.pdf.
- 3.McIntyre J. Preventing mother-to-child transmission of HIV: successes and challenges. BJOG. 2005. September;112(9):1196–203. doi: 10.1111/j.1471-0528.2005.00716.x [DOI] [PubMed] [Google Scholar]
- 4.Siegfried N, van der Merwe L, Brocklehurst P, Sint TT. Antiretrovirals for reducing the risk of mother-to-child transmission of HIV infection. The Cochrane database of systematic reviews. 2011. (7):CD003510 doi: 10.1002/14651858.CD003510.pub3 [DOI] [PubMed] [Google Scholar]
- 5.Jackson JB, Musoke P, Fleming T, Guay LA, Bagenda D, Allen M, et al. Intrapartum and neonatal single-dose nevirapine compared with zidovudine for prevention of mother-to-child transmission of HIV-1 in Kampala, Uganda: 18-month follow-up of the HIVNET 012 randomised trial. Lancet. 2003. September 13;362(9387):859–68. doi: 10.1016/S0140-6736(03)14341-3 [DOI] [PubMed] [Google Scholar]
- 6.Hauser A, Sewangi J, Mbezi P, Dugange F, Lau I, Ziske J, et al. Emergence of minor drug-resistant HIV-1 variants after triple antiretroviral prophylaxis for prevention of vertical HIV-1 transmission. PloS one. 2012;7(2):e32055 Pubmed Central PMCID: 3285650. doi: 10.1371/journal.pone.0032055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Flys T, Nissley DV, Claasen CW, Jones D, Shi C, Guay LA, et al. Sensitive drug-resistance assays reveal long-term persistence of HIV-1 variants with the K103N nevirapine (NVP) resistance mutation in some women and infants after the administration of single-dose NVP: HIVNET 012. The Journal of infectious diseases. 2005. July 1;192(1):24–9. doi: 10.1086/430742 [DOI] [PubMed] [Google Scholar]
- 8.Coovadia A, Hunt G, Abrams EJ, Sherman G, Meyers T, Barry G, et al. Persistent minority K103N mutations among women exposed to single-dose nevirapine and virologic response to nonnucleoside reverse-transcriptase inhibitor-based therapy. Clinical infectious diseases: an official publication of the Infectious Diseases Society of America. 2009. February 15;48(4):462–72. Pubmed Central PMCID: 2810158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brooks K, Diero L, DeLong A, Balamane M, Reitsma M, Kemboi E, et al. Treatment failure and drug resistance in HIV-positive patients on tenofovir-based first-line antiretroviral therapy in western Kenya. Journal of the International AIDS Society. 2016;19(1):20798 Pubmed Central PMCID: 4882399. doi: 10.7448/IAS.19.1.20798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lockman S, Shapiro RL, Smeaton LM, Wester C, Thior I, Stevens L, et al. Response to antiretroviral therapy after a single, peripartum dose of nevirapine. The New England journal of medicine. 2007. January 11;356(2):135–47. doi: 10.1056/NEJMoa062876 [DOI] [PubMed] [Google Scholar]
- 11.Eshleman SH, Mracna M, Guay LA, Deseyve M, Cunningham S, Mirochnick M, et al. Selection and fading of resistance mutations in women and infants receiving nevirapine to prevent HIV-1 vertical transmission (HIVNET 012). Aids. 2001. October 19;15(15):1951–7. [DOI] [PubMed] [Google Scholar]
- 12.Palombi L, Galluzzo CM, Andreotti M, Liotta G, Jere H, Sagno JB, et al. Drug resistance mutations 18 months after discontinuation of nevirapine-based ART for prevention of mother-to-child transmission of HIV in Malawi. The Journal of antimicrobial chemotherapy. 2015. October;70(10):2881–4. doi: 10.1093/jac/dkv171 [DOI] [PubMed] [Google Scholar]
- 13.Fox Z, Phillips A, Cohen C, Neuhaus J, Baxter J, Emery S, et al. Viral resuppression and detection of drug resistance following interruption of a suppressive non-nucleoside reverse transcriptase inhibitor-based regimen. Aids. 2008. November 12;22(17):2279–89. Pubmed Central PMCID: 2745911. doi: 10.1097/QAD.0b013e328311d16f [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.WHO: Consolidated guidelines on the use of antiretroviral drugs for treating and preventing HIV infection. June 2013. WHO website.Available from: http://apps.who.int/iris/bitstream/10665/85321/1/9789241505727_eng.pdf?ua=1. [PubMed]
- 15.Kesho Bora Study G, de Vincenzi I. Triple antiretroviral compared with zidovudine and single-dose nevirapine prophylaxis during pregnancy and breastfeeding for prevention of mother-to-child transmission of HIV-1 (Kesho Bora study): a randomised controlled trial. The Lancet Infectious diseases. 2011. March;11(3):171–80. doi: 10.1016/S1473-3099(10)70288-7 [DOI] [PubMed] [Google Scholar]
- 16.Vandamme AM, Van Vaerenbergh K, De Clercq E. Anti-human immunodeficiency virus drug combination strategies. Antiviral chemistry & chemotherapy. 1998. May;9(3):187–203. [DOI] [PubMed] [Google Scholar]
- 17.Paredes R, Marconi VC, Lockman S, Abrams EJ, Kuhn L. Impact of antiretroviral drugs in pregnant women and their children in Africa: HIV resistance and treatment outcomes. The Journal of infectious diseases. 2013. June 15;207 Suppl 2:S93–100. Pubmed Central PMCID: 3657116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gourlay A, Birdthistle I, Mburu G, Iorpenda K, Wringe A. Barriers and facilitating factors to the uptake of antiretroviral drugs for prevention of mother-to-child transmission of HIV in sub-Saharan Africa: a systematic review. Journal of the International AIDS Society. 2013;16:18588 Pubmed Central PMCID: 3717402. doi: 10.7448/IAS.16.1.18588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Metzner KJ, Giulieri SG, Knoepfel SA, Rauch P, Burgisser P, Yerly S, et al. Minority quasispecies of drug-resistant HIV-1 that lead to early therapy failure in treatment-naive and -adherent patients. Clinical infectious diseases: an official publication of the Infectious Diseases Society of America. 2009. January 15;48(2):239–47. [DOI] [PubMed] [Google Scholar]
- 20.Fisher RG, Smith DM, Murrell B, Slabbert R, Kirby BM, Edson C, et al. Next generation sequencing improves detection of drug resistance mutations in infants after PMTCT failure. Journal of clinical virology: the official publication of the Pan American Society for Clinical Virology. 2015. January;62:48–53. Pubmed Central PMCID: 4279108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Quinones-Mateu ME, Avila S, Reyes-Teran G, Martinez MA. Deep sequencing: becoming a critical tool in clinical virology. Journal of clinical virology: the official publication of the Pan American Society for Clinical Virology. 2014. September;61(1):9–19. Pubmed Central PMCID: 4119849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Samuel R, Paredes R, Parboosing R, Moodley P, Gordon M. Minority HIV-1 drug-resistant mutations and prevention of mother-to-child transmission: perspectives for resource-limited countries. AIDS reviews. 2014. Oct-Dec;16(4):187–98. [PubMed] [Google Scholar]
- 23.UNAIDS: The HIV and AIDS Uganda Progress Report 2014. June 2015. UNAIDS website.Available from: http://www.unaids.org/sites/default/files/country/documents/UGA_narrative_report_2015.pdf.
- 24.Palombi L, Pirillo MF, Marchei E, Jere H, Sagno JB, Luhanga R, et al. Concentrations of tenofovir, lamivudine and efavirenz in mothers and children enrolled under the Option B-Plus approach in Malawi. The Journal of antimicrobial chemotherapy. 2016. April;71(4):1027–30. doi: 10.1093/jac/dkv435 [DOI] [PubMed] [Google Scholar]
- 25.Hauser A, Hofmann A, Hanke K, Bremer V, Bartmeyer B, Kuecherer C, et al. National molecular surveillance of recently acquired HIV infections in Germany, 2013–2014. Euro Surveillance 2016;accepted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cleland A, Davis C, Adams N, Lycett C, Jarvis LM, Holmes H, et al. Development of multiplexed nucleic acid testing for human immunodeficiency virus type 1 and hepatitis C virus. Vox sanguinis. 2001;81(2):93–101. [DOI] [PubMed] [Google Scholar]
- 27.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014. August 1;30(15):2114–20. Pubmed Central PMCID: 4103590. doi: 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Magoc T, Salzberg L. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics btr507. [DOI] [PMC free article] [PubMed]
- 29.LI H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. [q-bioGN]. 2013;arXiv:1303.3997
- 30.Walter H, Schmidt B, Korn K, Vandamme AM, Harrer T, Uberla K. Rapid, phenotypic HIV-1 drug sensitivity assay for protease and reverse transcriptase inhibitors. Journal of clinical virology: the official publication of the Pan American Society for Clinical Virology. 1999. June;13(1–2):71–80. [DOI] [PubMed] [Google Scholar]
- 31.Rhee SY, Gonzales MJ, Kantor R, Betts BJ, Ravela J, Shafer RW. Human immunodeficiency virus reverse transcriptase and protease sequence database. Nucleic acids research. 2003. January 1;31(1):298–303. Pubmed Central PMCID: 165547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wensing AM, Calvez V, Günthard HF, Johnson VA, Paredes R, Pillay D, et al. 2015 Update of the Drug Resistance Mutations in HIV-1. Top Antivir Med. 2015;23(4):132–41. [PMC free article] [PubMed] [Google Scholar]
- 33.Bennett DE, Camacho RJ, Otelea D, Kuritzkes DR, Fleury H, Kiuchi M, et al. Drug resistance mutations for surveillance of transmitted HIV-1 drug-resistance: 2009 update. PloS one. 2009;4(3):e4724 Pubmed Central PMCID: 2648874. doi: 10.1371/journal.pone.0004724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pineda-Pena AC, Faria NR, Imbrechts S, Libin P, Abecasis AB, Deforche K, et al. Automated subtyping of HIV-1 genetic sequences for clinical and surveillance purposes: performance evaluation of the new REGA version 3 and seven other tools. Infection, genetics and evolution: journal of molecular epidemiology and evolutionary genetics in infectious diseases. 2013. October;19:337–48. doi: 10.1016/j.meegid.2013.04.032 [DOI] [PubMed] [Google Scholar]
- 35.Schnack A, Rempis E, Decker S, Braun V, Rubaihayo J, Busingye P, et al. Prevention of Mother-to-Child Transmission of HIV in Option B+ Era: Uptake and Adherence During Pregnancy in Western Uganda. AIDS patient care and STDs. 2016. March;30(3):110–8. doi: 10.1089/apc.2015.0318 [DOI] [PubMed] [Google Scholar]
- 36.WHO. World Health Organization Global Strategy for the Surveillance and Monitoring of HIV Drug Resistance. 2012.
- 37.Mancinelli S, Galluzzo CM, Andreotti M, Liotta G, Jere H, Sagno JB, et al. Virological Response and Drug Resistance 1 and 2 Years Post-Partum in HIV-Infected Women Initiated on Life-Long Antiretroviral Therapy in Malawi. AIDS research and human retroviruses. 2016. May 5. [DOI] [PubMed] [Google Scholar]
- 38.Ngarina M, Kilewo C, Karlsson K, Aboud S, Karlsson A, Marrone G, et al. Virologic and immunologic failure, drug resistance and mortality during the first 24 months postpartum among HIV-infected women initiated on antiretroviral therapy for life in the Mitra plus Study, Dar es Salaam, Tanzania. BMC infectious diseases. 2015;15:175 Pubmed Central PMCID: 4392730. doi: 10.1186/s12879-015-0914-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.WHO: Consolidated guidelines on the use of antiretroviral drugs for treating and preventing HIV infection. Recommendations for a public health approach—Second edition. June 2016. WHO website.Available from: http://www.who.int/hiv/pub/arv/arv-2016/en/. [PubMed]
- 40.Kalembo FW, Zgambo M. Loss to Followup: A Major Challenge to Successful Implementation of Prevention of Mother-to-Child Transmission of HIV-1 Programs in Sub-Saharan Africa. Isrn Aids. 2012. July 31;2012:589817 Pubmed Central PMCID: 3767368. doi: 10.5402/2012/589817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Geretti AM, Fox Z, Johnson JA, Booth C, Lipscomb J, Stuyver LJ, et al. Sensitive assessment of the virologic outcomes of stopping and restarting non-nucleoside reverse transcriptase inhibitor-based antiretroviral therapy. PloS one. 2013;8(7):e69266 Pubmed Central PMCID: 3715458. doi: 10.1371/journal.pone.0069266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cortez KJ, Maldarelli F. Clinical management of HIV drug resistance. Viruses. 2011. April;3(4):347–78. Pubmed Central PMCID: 3185705. doi: 10.3390/v3040347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kyeyune F, Nankya I, Metha S, Akao J, Ndashimye E, Tebit DM, et al. Treatment failure and drug resistance is more frequent in HIV-1 subtype D versus subtype A-infected Ugandans over a 10-year study period. Aids. 2013. July 31;27(12):1899–909. Pubmed Central PMCID: 4494684. doi: 10.1097/QAD.0b013e3283610ec7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li JZ, Paredes R, Ribaudo HJ, Svarovskaia ES, Metzner KJ, Kozal MJ, et al. Low-frequency HIV-1 drug resistance mutations and risk of NNRTI-based antiretroviral treatment failure: a systematic review and pooled analysis. Jama. 2011. April 6;305(13):1327–35. Pubmed Central PMCID: 3325645. doi: 10.1001/jama.2011.375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kyeyune F, Gibson RM, Nankya I, Venner C, Metha S, Akao J, et al. Low-Frequency Drug Resistance in HIV-Infected Ugandans on Antiretroviral Treatment Is Associated with Regimen Failure. Antimicrobial agents and chemotherapy. 2016. June;60(6):3380–97. Pubmed Central PMCID: 4879357. doi: 10.1128/AAC.00038-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zoufaly A, Jochum J, Hammerl R, Nassimi N, Raymond Y, Burchard GD, et al. Virological failure after 1 year of first-line ART is not associated with HIV minority drug resistance in rural Cameroon. The Journal of antimicrobial chemotherapy. 2015. March;70(3):922–5. doi: 10.1093/jac/dku470 [DOI] [PubMed] [Google Scholar]
- 47.Boltz VF, Bao Y, Lockman S, Halvas EK, Kearney MF, McIntyre JA, et al. Low-frequency nevirapine (NVP)-resistant HIV-1 variants are not associated with failure of antiretroviral therapy in women without prior exposure to single-dose NVP. The Journal of infectious diseases. 2014. March 1;209(5):703–10. Pubmed Central PMCID: 3923545. doi: 10.1093/infdis/jit635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Perno CF, Cozzi-Lepri A, Balotta C, Forbici F, Violin M, Bertoli A, et al. Secondary mutations in the protease region of human immunodeficiency virus and virologic failure in drug-naive patients treated with protease inhibitor-based therapy. The Journal of infectious diseases. 2001. October 15;184(8):983–91. doi: 10.1086/323604 [DOI] [PubMed] [Google Scholar]
- 49.Perno CF, Cozzi-Lepri A, Forbici F, Bertoli A, Violin M, Stella Mura M, et al. Minor mutations in HIV protease at baseline and appearance of primary mutation 90M in patients for whom their first protease-inhibitor antiretroviral regimens failed. The Journal of infectious diseases. 2004. June 1;189(11):1983–7. doi: 10.1086/386307 [DOI] [PubMed] [Google Scholar]
- 50.Nadembega WM, Giannella S, Simpore J, Ceccherini-Silberstein F, Pietra V, Bertoli A, et al. Characterization of drug-resistance mutations in HIV-1 isolates from non-HAART and HAART treated patients in Burkina Faso. Journal of medical virology. 2006. November;78(11):1385–91. doi: 10.1002/jmv.20709 [DOI] [PubMed] [Google Scholar]
- 51.Lee GQ, Bangsberg DR, Muzoora C, Boum Y, Oyugi JH, Emenyonu N, et al. Prevalence and virologic consequences of transmitted HIV-1 drug resistance in Uganda. AIDS research and human retroviruses. 2014. September;30(9):896–906. Pubmed Central PMCID: 4151058. doi: 10.1089/AID.2014.0043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Price MA, Wallis CL, Lakhi S, Karita E, Kamali A, Anzala O, et al. Transmitted HIV type 1 drug resistance among individuals with recent HIV infection in East and Southern Africa. AIDS research and human retroviruses. 2011. January;27(1):5–12. Pubmed Central PMCID: 3045073. doi: 10.1089/aid.2010.0030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Galluzzo CM, Germinario EA, Bassani L, Mancini MG, Okong P, Vyankandondera J, et al. Antiretroviral resistance mutations in untreated pregnant women with HIV infection in Uganda and Rwanda. AIDS research and human retroviruses. 2007. November;23(11):1449–51. doi: 10.1089/aid.2007.0109 [DOI] [PubMed] [Google Scholar]
- 54.Ndembi N, Lyagoba F, Nanteza B, Kushemererwa G, Serwanga J, Katongole-Mbidde E, et al. Transmitted antiretroviral drug resistance surveillance among newly HIV type 1-diagnosed women attending an antenatal clinic in Entebbe, Uganda. AIDS research and human retroviruses. 2008. June;24(6):889–95. doi: 10.1089/aid.2007.0317 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
VL: viral load; wt: wild type; nd: not detected; ns: no sequence from sample; -: absence of sample, or to exclusion according to sample inclusion criterion.
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.