Abstract
Extracellular matrix is a key component of many products in regenerative medicine. Multiple regenerative medicine products currently in the clinic are comprised of human or xenogeneic extracellular matrix. In addition, whole-organ regeneration exploits decellularized native organs as scaffolds for organotypic cell culture. However, precise understanding of the constituents of such extracellular matrix-based implants and scaffolds has sorely lagged behind their use. We present here an advanced protein extraction method using known quantities of proteotypic 13C-labeled peptides to quantify matrix proteins in native and decellularized lung tissues. Using quantitative proteomics that produce picomole-level measurements of a large number of matrix proteins, we show that a mild decellularization technique (“Triton/SDC”) results in near-native retention of laminins, proteoglycans, and other basement membrane and ECM-associated proteins. Retention of these biologically important glycoproteins and proteoglycans is quantified to be up to 27-fold higher in gently-decellularized lung scaffolds compared to scaffolds generated using a previously published decellularization regimen. Cells seeded onto this new decellularized matrix also proliferate robustly, showing positive staining for proliferating cell nuclear antigen (PCNA). The high fidelity of the gently decellularized scaffold as compared to the original lung extracellular matrix represents an important step forward in the ultimate recapitulation of whole organs using tissue-engineering techniques. This method of ECM and scaffold protein analysis allows for better understanding, and ultimately quality control, of matrices that are used for tissue engineering and human implantation. These results should advance regenerative medicine in general, and whole organ regeneration in particular.
Keywords: Quantitative proteomics, Decellularization, Tissue engineering, Regenerative medicine, Extracellular matrix
1. Introduction
Decellularized scaffolds for regenerative medicine are created by removing intact cells from tissues or organs using detergents, nucleases, and other reagents [1]. The resulting scaffold is comprised of extracellular matrix (ECM) proteins that have important physical and biochemical roles in organ function. Use of such scaffolds has rapidly expanded in both the laboratory and in the clinic [2], yet precise knowledge of the components of these matrices remains elusive. Standard proteomics techniques do not render reproducible or quantitative information about crosslinked and highly insoluble matrix elements. Immunostaining, ELISA, and immunoblotting are confounded by antibody cross-reactivity and difficulties in sample preparation, as well as limits on precise quantification. Identification and quantification of the individual components of these bioactive matrix materials is important to understand outcomes following in vitro or in vivo use, and for designing strategies to improve scaffolds for tissue engineering. Using recently developed proteomic techniques [3], we present a detailed, quantitative comparison of the ECM that results from two different decellularization regimens that are used in whole lung engineering. These quantitative proteomics analyses exceed, both in precision and in completeness, all other similar scaffold analyses published to date [4–7] and serve to illustrate the power of this novel method for ECM characterization.
In total, 71 ECM proteins were quantified in native and decellularized lungs. We present the results in 7 categories: proteoglycans; matricellular proteins; other basement membrane and ECM-associated proteins; laminins and fibronectin; elastin-related proteins; cellular proteins; and collagens. In some cases, immunofluorescence and Western blotting were used to corroborate the proteomic results of individual proteins, to visualize the differences between scaffolds, and to add spatial information for key basement membrane proteins. Lungs that are decellularized using the mild method described herein retain basement membrane and basement membrane glycoproteins and proteoglycans to a greater degree than our previously published method (6). We observed near-native retention of a large variety of matrix components that bind cells and support biological activity. However, a trade-off was noted, in that mild decellularization resulted in an increase in remnant cellular proteins that persisted within the scaffold, particularly cytoskeletal proteins. Overall, the results of this study show the feasibility of deriving absolute quantification of a large number of matrix molecules within whole tissues. Armed with this information, it will be possible to tune decellularization and organ culture strategies to better mimic the structural complexities of native tissues.
2. Materials and methods
2.1. Organ procurement
All animal work was performed in accordance with AAALAC guidelines and was approved by the Yale Institutional Animal Care and Use Committee. Lungs were harvested from adult (~3 months old) Fischer 344 male rats [8] and prepared for decellularization.
2.2. Decellularization – Triton/SDC
Lungs were perfused with a series of reagents including antibiotics/antimycotics, phosphate buffered saline (PBS) containing Ca2+ and Mg2+, and 0.0035% Triton X-100. This was followed by a benzonase endonuclease step, wherein lungs were inflated with the enzyme and incubated at room temperature for 30 min a rinse with a 1 M NaCl solution was performed, followed by rinsing with PBS without divalent ions. A series of washes with increasing concentrations of sodium deoxycholate (0.01%, 0.05%, 0.1%) was applied, followed by a 1 h benzonase incubation after perfusion of the enzyme into the lungs via the pulmonary artery. Finally, the lung was perfused with 0.5% Triton X–100, followed by extensive rinsing with PBS and antibiotics/antimycotics for 48 h at room temperature (~25 °C). Lungs were perfused with the antibacterial/antifungal solution for 48 h at 37 °C.
2.3. Decellularization – CHAPS
Lungs were perfused at 37 °C via the pulmonary artery with 500 ml of CHAPS-based decellularization solution [8], pH 12.4. Upon conclusion of detergent application, lungs were treated with benzonase endonuclease (90 U/ml) by inflation of the lungs with the benzonase solution via the trachea and incubated for 1 h at 37 °C. Following benzonase, lungs were rinsed extensively with PBS via the pulmonary artery. Fully rinsed lungs were mounted in a sterile bioreactor and perfused with antibiotics/antimycotics, as described for Triton/SDC.
2.4. Histology and immunofluorescence
Fixed, dehydrated tissues were processed by standard histological techniques at the histological core facilities. Unstained sections were deparaffinized and stained for laminin-γ1, fibronectin, and proliferating cell nuclear antigen (PCNA) by indirect immunofluorescence. Tissue designated for frozen sections was fixed with formalin and embedded in optimal cutting temperature (OCT) gel. The embedded tissue was frozen over dry ice for ~25 min, then transferred to the −80C freezer until sectioned. Sections were thawed to room temperature and rehydrated in PBS for 10 min to stain for collagen IV alpha (α) chains. See Detailed Online Methods.
2.5. Western blot
Whole lower right lobe lung samples were lysed from native and decellularized lungs (both Triton/SDC and CHAPS preparations) with 500–1000 μL of RIPA Buffer (Invitrogen) containing protease inhibitors (Roche), rocked for 1 h at 4 °C, and centrifuged at 14,000 rpm to collect the supernatant. Reducing Laemmli buffer was added to all lysed lungs samples. After normalization for differences in buffer volume for each sample, equal volumes of each lung sample were loaded on a 4–12% SDS-page gel (Bio-Rad) for separation and transferred to polyvinylidene (PVDF) membrane for Western blot analysis. a-actin (Abcam) 1:500, MHC-1 (BD transduction laboratories) 1:500, MHC-2 (Abcam) 1:1000 overnight at 4 °C to probe for proteins of interest.
2.6. Repopulation and culture of seeded scaffolds
Decellularized lung extracellular matrix scaffolds were prepared as described and mounted in biomimetic bioreactors as described previously [9]. After terminal sterilization with antibiotics and antimycotics, 100–150 million rat lung microvascular endothelial cells were introduced into the decellularized lung scaffolds via the pulmonary artery and pulmonary veins in series. Mixed lung cell populations isolated from neonatal rats were subsequently introduced into the scaffold via the trachea [8]. Pulsatile flow via the pulmonary artery was begun 1 h after seeding the mixed lung cell population at ~1 mL/min and constructs were maintained in 50% DMEM + 10% FBS/50% MCDB-131 with 1% penicillin/streptomycin for 4 days. Half the volume of media was changed on days 1 and 3, and the construct was harvested for analysis on day 4.
2.7. Sample preparation for proteomic analysis
Lyophilized native and decellularized lungs were pulverized in liquid nitrogen and processed as previously described [3]. Approximately 5 mgs of each lungtype (Native, CHAPS, Triton/SDC) were processed in quadruplicate. Ten of 2 mm Zirconium oxide beads were added to each tube, and tissue was homogenized by a Bullet Blender® (Next Advance) on power 8 for 3 min. Supernatants were sequentially extracted using high-speed centrifugation following vortexing in CHAPS, Urea, and CNBr buffers.
2.8. Detergent/chaotrope removal & protein digestion
The endogenous protein concentration of each fraction was determined by Bradford assay, prior to proteolytic digestion. Urea and CHAPS were removed from samples through the FASP protocol as previously described [10]. Briefly, 30 μgs of each sample was added to a 10 kD molecular weight cut-off filter. Two levels of 13C6 labeled QconCAT standards (500 amol/μl and 7.5 fmol/μl, 20 μl/injection; See Detailed Online Methods) were spiked into each sample as replicates to increase the quantifiable dynamic range. Samples were then reduced, alkylated, and digested with trypsin. Peptides were eluted into fresh tubes through 3 successive washes with 30% acetonitrile. Peptides were concentrated and acetonitrile removed on a speed-vac and then brought up to final volume (60 μls).
2.9. Liquid chromatography tandem mass spectrometry
Samples were analyzed on the QTRAP®5500 triple quadrupole mass spectrometer (ABSciex) coupled with an Agilent 1200 LC system utilizing methods described previously [3]. A targeted, scheduled Selected Reaction Monitoring (SRM) approach was performed using the QconCAT standards as targets. Transition selection and corresponding elution time, declustering potential, and collision energies were specifically optimized for each peptide of interest using Skyline [11]. Method building and acquisition were performed using the instrument supplied Analyst®Software (Version 1.5.2).
2.10. Protein abundance calculations and statistics
After filtering the data for LTQ and LOD (see Detailed Online Methods), the remaining protein concentrations for 4 lungs in each category (Native, CHAPS, and Triton/SDC) and 2 concentrations of QconCAT spikes were used to calculate a mean and standard deviation for protein concentration. Total lung weight was used to convert protein concentration to total quantity of protein per lung. N values represent the number of biological replicates. Standard error of the mean (SEM) was calculated using the mean, standard deviation, and n values.
2.11. Statistical analysis
Mean, SEM, and n values were entered into GraphPad Prism for statistical analysis. Statistically significant differences were determined using the Holm-Sidak method for multiple comparisons, with alpha = 5%. Proteins were analyzed as one group including 71 proteins. Each row was analyzed individually, without assuming consistent standard deviation (SD). Results, including p-values and whether the differences analyzed were statistically significant are shown in Supplemental Table 5.
3. Results
3.1. Quantitative proteomics
We have previously developed a sequential digestion method that is effective at solubilizing essentially all proteins of the lung extracellular matrix, including those that are insoluble by conventional methods [3]. Of note, urea, cyanogen bromide CNBr, and trifluroacetic acid (TFA) are used to enrich for proteins with varying levels solubility and to chemically digest otherwise insoluble ECM (iECM; Supplemental Fig. 1A) into discrete peptides for detection by liquid chromatography-tandem mass spectrophotometry (LC-MS/MS). Of interest, basement membrane proteins are found in approximately equal quantities in fractions 4 and 5, while fibrillar ECM proteins are located almost exclusively in fraction 5 (Supplemental Fig. 1A); these fractions are not included in most proteomic ECM analyses.
A targeted LC-selected reaction-monitoring (SRM) assay was used to identify and quantify unique peptides in the digested lung-derived peptide mixture. Absolute quantification of the proteins of interest is achieved by adopting the quantitative concatamer approach [12] (QconCAT; where the QconCAT is a concatamer of amino acids that generates a collection of reference peptides upon tryptic cleavage), and by using 13C stable isotope labeled (SIL), “heavy”, concatenated peptides as reference quantities for unlabeled proteins in our samples of interest (Supplemental Fig. 1B, and Hill et al. [3]). Because heavy reference peptides share an amino acid sequence with the endogenous peptides but are increased in weight by 6 Da, they co-elute with the corresponding endogenous peptides in space and time. This generates coincident signals on the intensity-to-time plot obtained by LC-SRM, but offset signals on the signal intensity-vs.-mass/charge ratio plot.
This allows distinction between SIL peptides and the peptides derived from experimental samples. The heavy peptides are spiked into samples at known concentrations, and therefore the integrated peak areas can be used as reference peaks for the naturally derived peptides of known identity but unknown quantity (Supplemental Fig. 1B).
Absolute quantification of peptides, and therefore proteins, in native and decellularized lung samples allows direct comparison between ECM scaffolds. Furthermore, the majority of the peptides used in this study utilize sequences common to multiple species – 76% of the proteins identified utilized sequences shared between three species: rat, pig, and human (Supplemental Tables 1 and 2). This fact facilitates the use of the quantification methods described here to compare the impact of different treatment methods on scaffolds of different origins. More broadly, this fact makes these tools widely applicable to a variety of experimental and clinical model systems.
3.2. Generation of decellularized rat lung extracellular matrices
Decellularization was performed using two series of reagents: 1) a CHAPS-based (0.4%) solution of pH > 12, performed at 37 °C, with washes at room temperature as previously reported ([8,9]; hereafter referred to as “CHAPS”); or 2) a series of low-concentration detergent solutions containing Triton-X-100 (0.0035% and 0.5%) and sodium deoxycholate (SDC; solutions from 0.01 to 0.1%), performed at room temperature with solutions of pH 7.4–8.0 (see Section 2).
3.3. Quantification of proteins in native lung
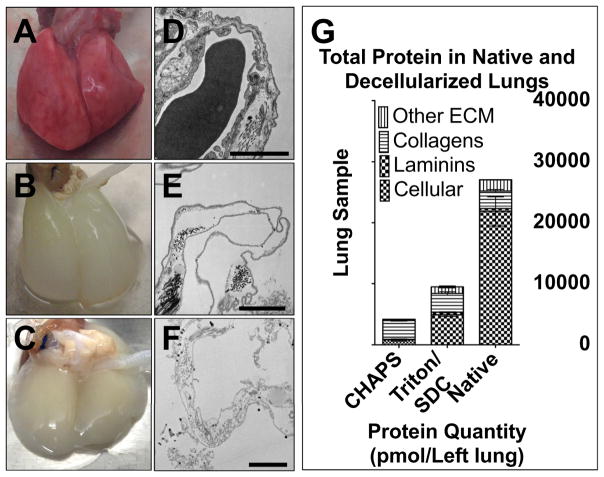
Native and decellularized lungs were grossly structurally intact (Fig. 1A–C). Transmission electron microscopy showed that, in decellularized lungs, cellular components appeared depleted, while basement membranes in the alveolar regions remain intact (Fig. 1D–F). Total protein content in a whole, intact native left lung, based on the 71 proteins assayed here, is 2.7E4 ± 2.4E3 pmol. (Fig. 1G, n = 4 biological replicates per group). Cellular proteins account for 80.7% of total protein in native lung (2.2E4 ± 2.4E3 pmol); collagens make up 10.7% (2.9E3 ± 2.5E2 pmol); laminins make up 1.8% (4.7E2 ± 27 pmol); the other categories of ECM proteins together make up 6.8% (1.8E3 ± 45 pmol).
Fig. 1.
Structure and composition of native and decellularized lungs. A) Photograph of native rat lungs. B) Photograph of lungs decellularized with Triton/SDC. C) Photograph of lungs decellularized with pH 12.4, CHAPS-based decellularization fluid (CHAPS). D) Transmission electron micrograph (TEM) of native rat lungs. E) TEM of rat lungs decellularized with Triton/SDC. F) TEM of rat lungs decellularized with CHAPS. D–F scale bars = 2 μm. G) Bar chart showing the relative compositions of native and decellularized lungs, as assessed by quantitative ECM proteomics.
When ordered by abundance, the majority of the 10 most abundant non-cellular proteins in native lung are collagens (Supplemental Table 3). This class of protein provides the majority of the bulk and structural framework for all types of ECM. Collagen I is a ubiquitous structural component of all tissues and organs, while collagen VI shows greater temporal and tissue-specific expression; it is especially abundant in lung [13]. The 10 least abundant non-cellular proteins (Supplemental Table 4) are largely associated with the fibrillar or with the basement membrane components of the ECM, and likely serve functions in mediating ECM-ECM and cell-ECM interactions.
Quantification of these matrix proteins in an intact organ provides a benchmark for comparison of subsequently decellularized scaffolds. In addition, this information can be used as a point of comparison for other in vitro studies wherein tissue or organ specific cells are producing matrix, and for the selection of culture substrates for stem cell or other cell culture work.
3.4. Analysis of protein content in decellularized lung scaffolds
Triton/SDC lung scaffolds contain more total protein than CHAPS lungs (9.5E3 ± 4.4E2 pmol vs. 4.2E3 ± 2.1E2 pmol protein, respectively p < 0.0001; Fig. 1G). Of note, the absolute quantities of collagens and laminins are similar in Triton/SDC lungs and native lungs (2.9E3 ± 2.5E2 pmol vs. 3.0E3 ± 2.9E2 pmol, p = 0.7133; and 4.7E2 ± 27 pmol vs. 4.1E2 ± 71 pmol, p = 0.4524, respectively). The remaining ECM proteins are present in Triton/SDC lungs in similar proportion to native lung (11.7% vs. 6.8% of total protein; 24.3% vs. 35% of ECM protein) but are significantly reduced in quantity (1.1E3 ± 1.6E2 pmol vs. 1.8E3 ± 45 pmol, p = 0.0043). Overall, Triton/SDC lungs show greater overall preservation of ECM components found in native lung than CHAPS lungs.
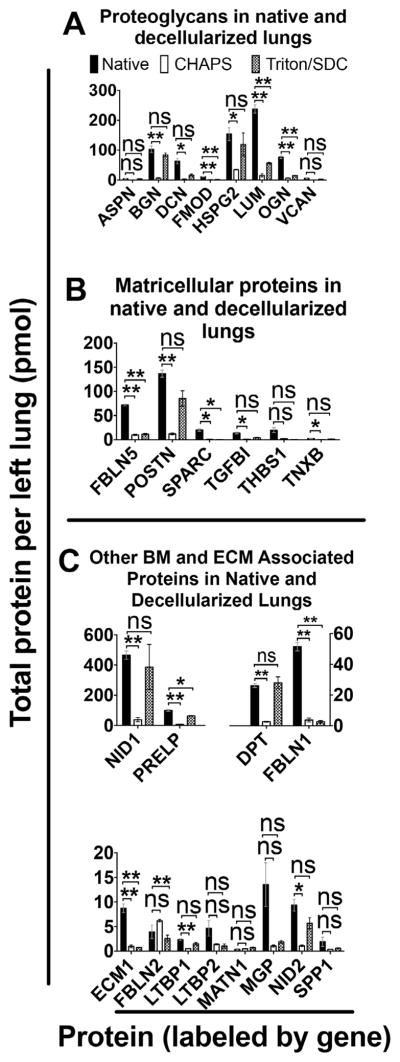
3.5. Proteoglycans
Proteoglycans confer structure to matrix in part by organizing collagen and elastin fibrils in space [14,15] and contribute to lung elasticity and to alveolar stability at low and medium lung volumes [16], thereby enabling gas exchange. We quantified 8 proteoglycans in native and decellularized lungs (Fig. 2A). Of the proteoglycans assayed here, lumican is the most abundant in native lung (2.4E2 ± 14 pmol per left lung). Lumican is also one of only 3 proteoglycans that exhibits significant loss in Triton/SDC lungs as compared to native lung.
Fig. 2.
Quantification of key protein categories. A) Proteoglycans found in native and decellularized lung. B) Matricellular protein quantifications. C) Other basement membrane and ECM-associated proteins. Error bars are ± SEM, n = 4 biological replicates, with two different concentrations of QconCAT peptides. (Genes displayed in Supplemental Table 7.) Single asterisk (*) indicates α = 0.05; (**) indicates α = 0.01 (see Supplemental Table 5). ns = non significant difference.
The remaining 5 of the 8 proteoglycans assayed here are present in Triton/SDC scaffolds in quantities that are not significantly different from native lung: asporin, biglycan, decorin, heparan sulfate proteoglycan 2 (perlecan), and versican (see Supplemental Tables 5 and 6 for statistical analysis and absolute quantities, respectively). In addition, all 8 proteoglycans investigated here are more abundant in Triton/SDC lungs than in CHAPS lungs. This may be due to the fact that the ionic interactions between the charged sugar moieties on glycoproteins and proteoglycans and the other ECM components are more easily abrogated by the zwitterionic CHAPS than by non-ionic Triton-X-100. Indeed, 2 of the proteoglycans, asporin and versican, are nearly absent in CHAPS lung scaffolds, with a protein content of less than 1 pmol per left lung. In total, when comparing CHAPS-based decellularization to the Triton/SDC-based method, there is a significant net 4-fold increase in proteoglycans, as measured by protein mass (68 ± 6.0 pmol in CHAPS vs. 3.0E2 ± 39 pmol in Triton/SDC; p = 0.0089).
3.6. Matricellular proteins, basement membrane and ECM-associated proteins
We investigated 6 matricellular proteins in native lungs and decellularized scaffolds (Fig. 2B). The presence of matricellular proteins is important to promote cell migration, differentiation, and function. Among these proteins in native lung, there is the greatest quantity of periostin (1.4E2 ± 7.5 pmol per left lung), followed by fibulin-5 (72 ± 1.6 pmol).
Triton/SDC produces lungs that retain four of the 6 proteins in this category at near-native levels (periostin; transforming growth factor, beta-induced; thrombospondin-1; and tenascin X), while CHAPS lungs only retain a single protein in this category (thrombospondin-1) at a level similar to native lung (See Supplemental Tables 5 and 6). The increased loss of proteins in this category using the CHAPS method is likely due to the non-covalent interactions of matricellular proteins with other ECM, which confers a greater relative solubility during the decellularization process. The difference in decellularization methods is greatest for periostin. Retention of periostin is encouraging for the purposes of reseeding with respiratory epithelium, since periostin is a ligand for the αvβ3 and αvβ5 integrins that enable adhesion and migration of other types of epithelial cells [17] and may provide a similar function for pulmonary epithelium.
We assayed 12 additional basement membrane and ECM-associated proteins in native and decellularized lungs (Fig. 2C) and found that 75% of these proteins (9 out of 12) are retained at near-native levels in Triton/SDC lung scaffolds. Of the proteins retained by Triton/SDC, nidogen-1 is by far the most abundant protein in native lung (native = 4.6E2 ± 28 pmol, followed by prolargin (1E2 ± 5.5 pmol), and fibulin-1 (51 ± 2.8 pmol). Nidogen-2 is also retained in Triton/SDC lungs (see Supplemental Tables 5 and 6). Since nidogens-1 and -2 and other proteins in this category provide structural support as linkers between other ECM proteins, these proteins are likely to confer improved structural stability in Triton/SDC scaffolds [18]. Preservation of linker proteins also likely indicates diminished or minimal damage to the interstitial fibrillar proteins and basement membrane proteins. Overall, 67% of the matricellular proteins and 75% of the basement membrane and other ECM-associated proteins assayed here are preserved at native levels in Triton/SDC lungs, compared to 17% and 42% in CHAPS scaffolds, respectively. This represents a substantial improvement with respect to maintenance of native proteins in acellular lung scaffolds.
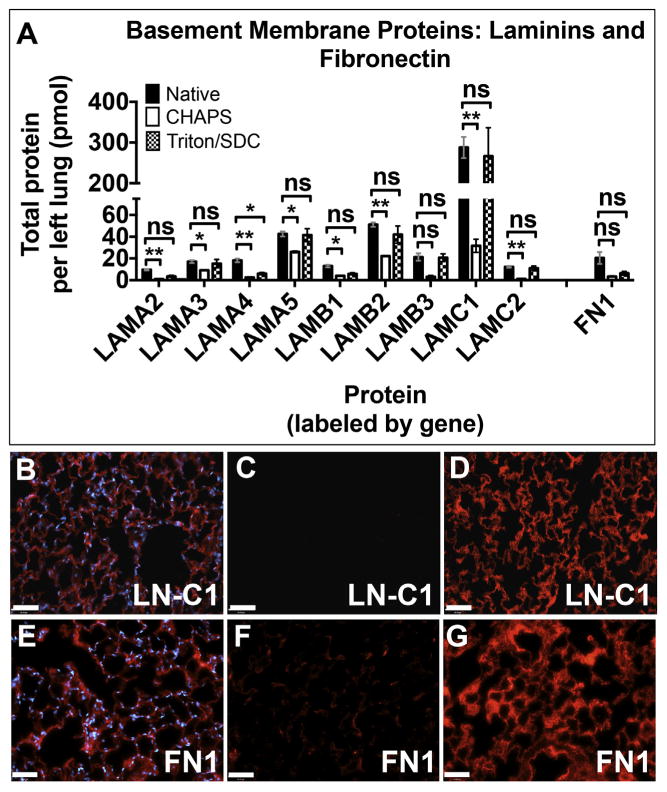
3.7. Adhesion proteins: laminins, fibronectin, and collagen IV
Laminins are an essential component of all alveolar, airway, and vascular basement membranes [19]. They promote cell adhesion and support organ-specific cell phenotypes. Therefore, we quantified 9 laminin subunits (Fig. 3A). Only one was diminished in Triton/SDC lungs (laminin α4, p = 7.301E-4, Triton compared to native). In contrast, only one of the laminin subunits assayed here was retained in CHAPS lungs (laminin β3, p = 2.911E-4). Comparison of Triton/SDC lung with native lung shows that >90% of native laminin α-5, β-3, and ϒ1 were present in Triton/SDC lungs after decellularization. (see Supplemental Table 6). In contrast, the greatest retention of a laminin subunit in CHAPS lungs was that of α-5, which was present at 61% (26 ± 0.83 pmol) of native levels.
Fig. 3.
Assessment of the basement membrane proteins laminins and fibronectin. A) Quantitative proteomics for nine laminin subunits and fibronectin. Error bars are ± SEM, n = 4 biological replicates, with two different concentrations of QconCAT peptides. (Genes displayed in Supplemental Table 7.) Single asterisk (*) indicates α = 0.05; (**) indicates α = 0.01 (see Supplemental Table 5). ns = non-significant difference. Immunofluorescence for laminin γ1) in (B) native lung; (C) CHAPS lung; (D) Triton/SDC lung. Immunofluorescence for fibronectin in (E) native lung; (F) CHAPS lung; (G) Triton/SDC lung. Antigen of interest is stained in red. Nuclei are stained with DAPI (blue). Scale bars = 50 μm. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
We also assessed the amount of fibronectin, a basement membrane component that promotes adhesion and migration, in our scaffolds. Very little (3.3 ± 0.55 pmol) was found in CHAPS lungs, while Triton/SDC lungs contain 6.3 ± 1.7 pmol – just over 30% of the amount of fibronectin found in native lungs. The presence of fibronectin is expected to enhance cell adhesion and proliferation of alveolar epithelium [20,21] and club cells, as well as vascular endothelium [22].
Immunofluorescence indicated that the overall distribution and abundance of these proteins is comparable to native lung. While staining for laminin γ1suggests faithful retention of the delicate alveolar structure (Fig. 3B–D), the pattern of fibronectin indicated that the protein may be somewhat damaged (Fig. 3E–G). Overall, these results indicate that the total amount, structural integrity, location, and cell adherence domains in laminin in Triton/SDC lungs are relatively intact.
Bronchiolar and alveolar epithelium also rely on collagen IV for adhesion, differentiation, and migration [23]. To quantify this basement membrane component, we evaluated α2 and α5 chains of collagen IV. For both of these proteins, there was a positive trend in the abundance of these proteins in Triton/SDC lungs, as compared to CHAPS lungs (Supplemental Fig. 2A and Supplemental Table 6). Both proteins were retained with appropriate distribution in Triton/SDC lungs, as indicated by immunofluorescence with antibodies specific to the non-collagenous (NC1) cell-binding domains of these proteins (Supplemental Fig. 2B–G). Many of the fibrillar collagens (I, V, XIV, and XVII) and fibril-associated collagen XII are also retained at native levels in Triton/SDC lungs when compared to native lung by quantitative proteomics (Supplemental Fig. 3A, B) and trichrome staining (Supplemental Fig. 3C–E).
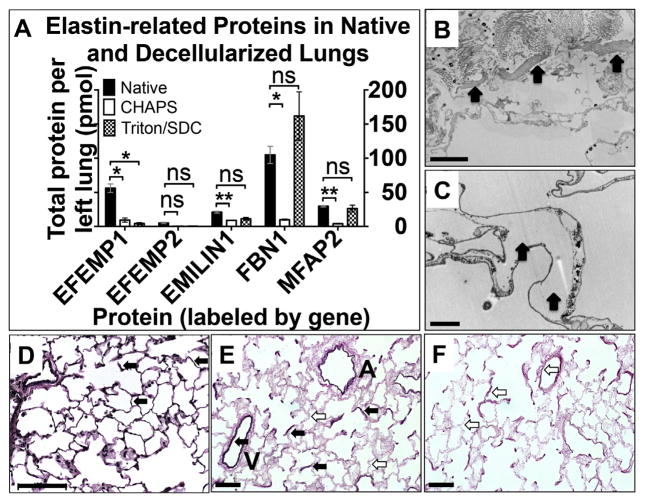
3.8. Elastin and elastin-associated protein content
Elastin, a critical component for ECM mechanics, remains challenging to extract, digest, and quantify. Therefore, we probed for proteins physically associated with elastin instead. Fig 4A shows that fibrillin-1 (1.0E2 ± 12 pmol), fibulin-3 (56 ± 6.2 pmol), and fibulin-4 (30 ± 1.0 pmol) are the most abundant in native lung. All elastin-associated proteins except fibulin-3 (i.e. 4/5) were found in Triton/SDC lungs in amounts equivalent to native lung. In contrast, 4 out of 5 of these proteins were significantly depleted in lungs treated with CHAPS (Supplemental Tables 5 and 6).
Fig. 4.
Elastin and elastin-associated proteins. A) Proteomic quantification of elastin-related proteins in native and decellularized lungs. Error bars are ± SEM, n = 4 biological replicates, with two different concentrations of QconCAT peptides. (Genes displayed in Supplemental Table 7.) Single asterisk (*) indicates α = 0.05; (**) indicates α = 0.01 (see Supplemental Table 5). ns = non significant difference. B) Transmission electron micrograph (TEM) of Triton/SDC-decellularized lung. Arrows indicate intraseptal space where elastin might usually be found. (C) TEM of CHAPS-decellularized lung. Arrows indicate elastin. Scale bars in (B,C) = 2 μm. VVG stain of native rat lung (D), Triton/SDC-treated lung (E) and CHAPS-treated lung (F). Black arrows indicate elastic fibers in arterial walls and throughout the sepatae. White arrows indicate the absence of elastin in locations similar to those highlighted in native lung (panel D). A = airway. V = vessel. Scale bars = 100 μm (D); 50 μm (E, F).
Consistent with these findings, relatively large islands of amorphous, electron-dense material typical of interstitial elastin is present throughout Triton/SDC lungs in images acquired using TEM (Fig 4B). These are not readily visible in CHAPS-treated lungs (Fig. 4C). Histological evaluation of these samples by Verhoeffvan Gieson staining indicates elastin fibers in the arterial walls and throughout the septae of native lung (Fig. 4D; black arrows) and both airways and vessels of Triton/SDC lungs (Fig. 4E). CHAPS lungs are fairly depleted of elastic fibers (Fig. 4F; white arrows).
3.9. Cellular proteins in acellular lung scaffolds
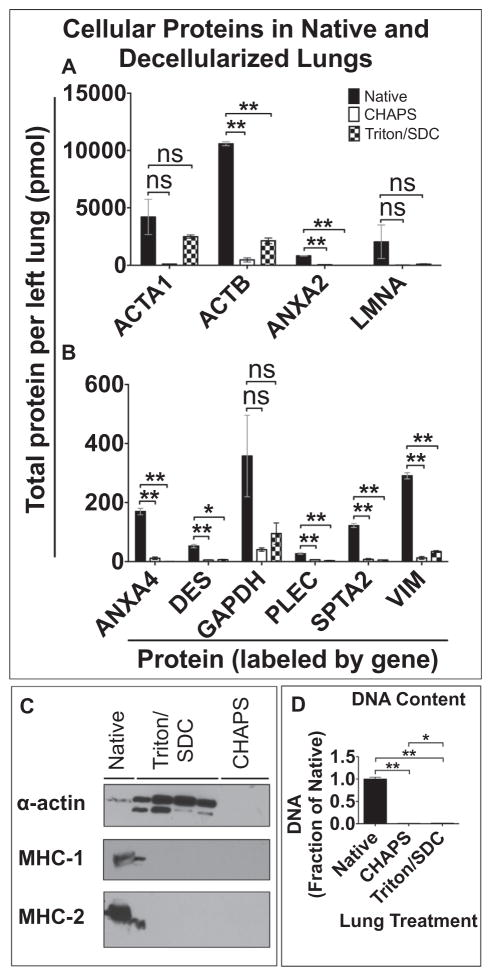
In addition to extracellular matrix proteins, we quantified 17 cellular proteins in native lung and acellular lung scaffolds: 9 cytoskeletal proteins and protein glyceraldehyde-3-phosphate dehydrogenase (GAPDH) are shown (Fig. 5A, B; additional 7 listed in Supplemental Table 8). In the context of decellularized scaffolds, cellular protein can serve as readout of efficiency of cell removal. Of the proteins assayed here, the three most abundant proteins in native lung are β-actin (1.1E4 ± 1.7E2 pmol), α-actin (4.2E3 ± 1.5E3 pmol), and lamin A/C (2.0E3 ± 1.5E3). Of note, 6 of the 17 cellular proteins assayed show no statistically significant difference in abundance between native lung and Triton/SDC lungs. Overall, cellular proteins constitute 52% of the total protein in Triton/SDC lungs (Fig. 1G) that is represented by the peptides described here. In comparison, cellular proteins account for 19% of the total protein quantified in CHAPS lungs (Fig. 1G).
Fig. 5.
Quantification of cellular components. Proteomic quantification of (A) high-abundance proteins and (B) low-abundance proteins in native and decellularized lungs. Error bars are ± SEM, n = 4 biological replicates, with two different concentrations of QconCAT peptides. (Genes displayed in Supplemental Table 7.) Single asterisk (*) indicates α = 0.05; (**) indicates α = 0.01 (see Supplemental Table 5). ns = non significant difference. C) Western blot for α-actin, MHC-1 and -2 molecules in native (lane 1) and decellularized lung samples. (Bleed-over is noted in lane 2, from the adjacent lane 1, in MHC-1 and -2 blots.) D) DNA quantification by PicoGreen Assay. Single asterisk (*) indicates statistically significant difference, p < 0.05; (**) indicates statistical significance, p < 0.01.
Western blot did not reveal a large amount of α-actin, however, perhaps due to protein conformational changes imparted by the harsh CHAPS treatment. In contrast, probing for α-actin by immunoblotting in Triton/SDC lungs does indicate that there is a substantial amount of α-actin in these scaffolds (Fig. 5C). Triton X-100 has been used for decades to separate cytoskeletal proteins (detergent insoluble) from other cytosolic components (detergent soluble) when used in low concentrations [24].
Both MHC-1 and MHC-2 molecules appear to be completely removed by both decellularization methods on immunoblotting (Fig. 5C), based on the antibodies used here. Hence, both methods may be effective at eliminating highly immunogenic MHC molecules. The immunological implications of implanting a scaffold with remnant cytoskeletal elements is unclear, though the highly conserved nature of cytoskeletal proteins between individuals likely means that their immunogenicity is negligible. Evaluation of DNA content in native lung and decellularized scaffolds with PicoGreen indicates that >98% of the DNA present in native lung is removed by either Triton/SDC or CHAPS decellularization (quantities of DNA in scaffolds shown as fraction of native lung, which = 1; Fig. 5D).
3.10. Recellularization of gently decellularized lung
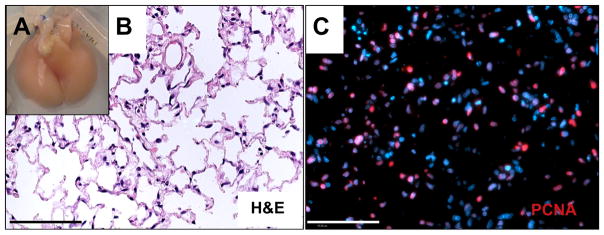
To evaluate cellular viability on gently decellularized scaffolds, we repopulated Triton/SDC scaffolds with neonatal epithelium and microvascular endothelium. Triton/SDC scaffolds populated with cells and cultured in a bioreactor with vascular perfusion for 4 days are structurally intact grossly (Fig. 6A) and by histological evaluation (Fig. 6B). Alveolar spaces are open and cells populate the alveolar septae widely and evenly throughout the parenchyma (Fig. 6B). Staining for proliferating cell nuclear antigen (PCNA) is positive in many cells (estimated at >50%, based on immunofluorescence images; Fig. 6C). Taken together, these data suggest that the gently decellularized Triton/SDC scaffold is hospitable to cell adhesion, survival, and proliferation despite the abundance of cellular material detected in the scaffold prior to introducing cells for tissue regeneration. Even more encouragingly, cells cultured within the Triton/SDC scaffold express protein markers of several key native lung cell phenotypes, including type II alveolar epithelial cells, club cells, endothelial cells, and interstitial cells, as indicated by staining for pro-SPC, CCSP, CD31 and VE-cadherin, and α-actin, respectively (Supplemental Fig. 4). Direct correlation between the appearance and abundance of these proteins and the presence or abundance of any specific ECM proteins is beyond the scope of this work, but is a tantalizing avenue for future research.
Fig. 6.
Repopulation of Triton/SDC scaffolds. A) Gross image of repopulated Triton/SDC scaffold after culture at 37 °C with 50% DMEM + 10% FBS/50% MCDB-131 Complete containing antibiotics/antimycotics. B) Hematoxylin and eosin image of repopulated lung after 4 days in culture. C) Immunofluorescent staining with anti-PCNA antibody on a paraffin section of cultured lung. PCNA is red; nuclei are stained with DAPI (blue). Scale bars = 100 μm. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
4. Discussion
In tissue engineering, various detergents and protocols have been compared with semi-quantitative proteomics [5,25,26] and by cellular repopulation of the resulting scaffolds [5,25,27] or sheets of ECM [28]. However, many published accounts of lung ECM components may be missing key components in their inventories of scaffold proteins due to insufficient extraction; prior reports use fewer than 5 fractions and CNBr is generally not used to dissociate the matrix [4–7,25,29]. In this and a prior study [3], we observed that the first 3 fractions contain almost no proteoglycans, basement membrane proteins, or fibrillar collagens, the last of which are well represented in the matrix overall (Supplemental Fig. 3). This suggests that, while the identification and quantification of subunits of proteins such as collagens III and additional α chains of collagen IV are limited in this study by probe selection and synthesis, many of the semi-quantitative assessments of lung ECM components may have inventories that are either missing or under-representing key ECM components due to insufficient extraction.
The decellularization protocol used to prepare the scaffold may also impact the readout of our studies and others, as demonstrated in Supplemental Fig. 3. Here, the apparent increase in collagen I α1 and α2 chains and the statistically significant increase in collagen V α2 may be attributable to solubilization of the extracellular matrix by the detergent used to render the scaffold acellular in advance of preparation for proteomic analysis. This “pre-solubilization” could allow for greater extraction later, when the scaffolds are prepared for proteomic analysis. This effect would be augmented by more aggressive decellularization regimens (i.e. pH 12.4 CHAPS), compared to Triton/SDC; and would be reflected in a greater increase in protein content reported for CHAPS decell, compared to native, than for Triton/SDC compared to native.
Of the matrix components quantified here, the retention of many proteoglycans and laminin subunits in the Triton/SDC scaffolds is particularly encouraging with respect to organ specific cell differentiation. Proteoglycans are likely to improve both the biochemical and mechanical cues received by cells cultivated within the ECM scaffold. They provide ligand-binding sites for integrins [30,31] and assist in the sequestration of a variety of growth factors [32] that may promote tissue and organ-specific phenotypes. As such, a matrix containing proteoglycans dispersed among structural components such as fibrillar collagens may better support epithelial and mesenchymal phenotypes that more closely recapitulate native lung.
This study also demonstrated the persistence of cellular proteins in both scaffolds, which raises concern over the potential for immunogenicity when implanted in vivo. Limited evidence suggests, however, that the presence of residual cellular proteins may not be detrimental to the survival or sustainability of a tissue engineered graft. Subcutaneous implantation of decellularized lung that contained β-actin actually invoked a biological response that was less damaging to the structure of the scaffold than the response evoked by a scaffold that was more devoid of cellular proteins [33]. In balancing the complete removal of cells and cellular material with the retention of ECM components, the preservation of matrix may supersede the importance of removal of residual proteins.
Thus far, we have demonstrated that, at a minimum, the additional remnant cellular proteins that accompany gains in ECM proteins do not appear to interfere with initial cell-matrix interactions. That is, despite the increased quantities of cellular proteins, cells introduced to the decellularized scaffolds are able to adhere to and proliferate on the Triton/SDC decellularized ECM scaffold. Future work should additionally evaluate the function of the cells seeded onto and cultured within this scaffold over time. In addition, a direct comparison of cells seeded onto CHAPS scaffolds with cells seeded onto Triton/SDC scaffolds would be interesting and informative with regard to whether the molecular differences quantified in these scaffolds correlate with observable differences in cell behavior or function. Ultimately, it may be possible to use the methods presented here to correlate the in vitro and in vivo outcomes of cell seeding with the protein profiles of respective scaffolds.
Finally, the use of this technology to analyze other ECM, for example, at different stages in development will advance our understanding of tissue and organ formation [34]. Evaluation of both developmental ECM, and ECM produced by stem cells in vitro [35] may help generate better intermediate cells (i.e. endoderm) from which to derive organ-specific cell types such as terminally differentiated, functional pulmonary epithelium, which has remained an elusive target to date [36].
In summary, the methods presented here can be used to characterize current tissue engineering efforts, and allow for refinement of decellularization procedures based on protein profiles, both with respect to extent of antigen removal and conservation of ECM proteins. Future work overall will endeavor to correlate protein quantity information with cell localization and function, with the aim of bringing rational design and greater mechanistic understanding to the next generation of engineered organs.
5. Conclusions
In conclusion, there is a need for more precise information regarding the composition of extracellular matrix products and biologically-derived scaffolds. Using absolutely quantitative proteomics, enabled by the use of heavy peptides as independent references, it is possible, for the first time, to quantify extracellular matrix and other proteins in native rat lung and decellularized lung scaffolds prepared by two different methods. There is a net increase in proteoglycans, laminins, and other basement membrane and basement membrane-associated proteins when lung scaffolds are prepared using a “gentle” decellularization protocol that uses Triton/SDC, compared to high pH CHAPS solution. At the same time, though no intact cells or nuclei are present, cellular proteins do persist in the acellular scaffolds – a phenomenon noted by Li et al. as well [26]. Ultimately, the ability to correlate precise information about scaffold constitution with overall biological outcome should prove useful in advancing the field of tissue engineering and regenerative medicine overall.
Supplementary Material
Statement of Significance.
The extracellular matrix (ECM) in large part defines the biochemical and mechanical properties of tissues and organs; these inherent cues make acellular ECM scaffolds potent substrates for tissue regeneration. As such, they are increasingly prevalent in the clinic and the laboratory. However, the exact composition of these scaffolds has been difficult to ascertain. This paper uses targeted proteomics to definitively quantify 71 proteins present in acellular lung ECM scaffolds. We use this technique to compare two decellularization methods and demonstrate superior retention of ECM proteins important for cell adhesion, migration, proliferation, and differentiation in scaffolds treated with low-concentration detergent solutions. In the long term, the ability to acquire quantitative biochemical data about biological substrates will facilitate the rational design of engineered tissues and organs based on precise cell-matrix interactions.
Acknowledgments
Thanks to Pavlina Baevova for assistance in preparing lung extracellular matrix scaffolds and to Kevin Boehm for assistance with literature searches. We thank Dr. Feng Dai for assistance with statistical analysis. We gratefully acknowledge the gift of anticollagen IV alpha chain antibodies from Dr. Yoshikazu Sado, Shigei Medical Research Institute, Okayama, Japan. This work was funded by the National Institutes of Health (1U01HL111016-01, Niklason) and supported by the Medical Scientist Training Program (NIH/NIGMS T32 GM007205, KLL).
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.actbio.2016.09.043.
Footnotes
Disclosure
LEN has a founder and shareholder in Humacyte, Inc, which is a regenerative medicine company. Humacyte produces engineered blood vessels from allogeneic smooth muscle cells for vascular surgery. LEN’s spouse has equity in Humacyte, and LEN serves on Humacyte’s Board of Directors. LEN is an inventor on patents that are licensed to Humacyte and that produce royalties for LEN. LEN has received an unrestricted research gift to support research in her laboratory at Yale. Humacyte did not fund these studies, and Humacyte did not influence the conduct, description or interpretation of the findings in this report.
R.C.H and K.C.H have a financial interest in Matriqs Biology, LLC, a proteomic analysis contract research organization. Matriqs Biology did not fund these studies, and did not affect the design, interpretation, or reporting of any of the experiments herein.
References
- 1.Crapo PM, Gilbert TW, Badylak SF. An overview of tissue and whole organ decellularization processes. Biomaterials. 2011:3233–3243. doi: 10.1016/j.biomaterials.2011.01.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Macchiarini P, Jungebluth P, Go T, Asnaghi M, Rees L, Cogan T, Dodson A, Martorell J, Bellini S, Parnigotto P, Dickinson S, Hollander A, Mantero S, Conconi M, Birchall M. Clinical transplantation of a tissue-engineered airway. Lancet. 2008 doi: 10.1016/S0140-6736(08)61598-6. [DOI] [PubMed] [Google Scholar]
- 3.Hill RC, Calle EA, Dzieciatkowska M, Niklason LE, Hansen KC. Quantification of extracellular matrix proteins from a rat lung scaffold to provide a molecular readout for tissue engineering. Mol Cell Proteomics. 2015;14:961–973. doi: 10.1074/mcp.M114.045260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bonvillain RW, Danchuk S, Sullivan DE, Betancourt AM, Semon JA, Eagle ME, Mayeux JP, Gregory AN, Wang G, Townley IK, Borg ZD, Weiss DJ, Bunnell BA. A nonhuman primate model of lung regeneration: detergent-mediated decellularization and initial in vitro recellularization with mesenchymal stem cells. Tissue Eng. 2012:2437–2452. doi: 10.1089/ten.tea.2011.0594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Elizabeth Gilpin S, Guyette JP, Gonzalez G, Ren X, Asara JM, Mathisen DJ, Vacanti JP, Ott HC. Perfusion decellularization of human and porcine lungs: bringing the matrix to clinical scale. J Heart Lung Transplant. 2013 doi: 10.1016/j.healun.2013.10.030. [DOI] [PubMed] [Google Scholar]
- 6.Booth AJ, Hadley R, Cornett AM, Dreffs AA, Matthes SA, Tsui JL, Weiss K, Horowitz JC, Fiore VF, Barker TH, Moore BB, Martinez FJ, Niklason LE, White ES. Acellular normal and fibrotic human lung matrices as a culture system for in vitro investigation. Am J Resp Crit Care Med. 2012 doi: 10.1164/rccm.201204-0754OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wagner DE, Bonenfant NR, Sokocevic D, Desarno MJ, Borg ZD, Parsons CS, Brooks EM, Platz JJ, Khalpey ZI, Hoganson DM, Deng B, Lam YW, Oldinski RA, Ashikaga T, Weiss DJ. Three-dimensional scaffolds of acellular human and porcine lungs for high throughput studies of lung disease and regeneration. Biomaterials. 2014;35:2664–2679. doi: 10.1016/j.biomaterials.2013.11.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Petersen TH, Calle EA, Zhao L, Lee EJ, Gui L, Raredon MB, Gavrilov K, Yi T, Zhuang ZW, Breuer C, Herzog E, Niklason LE. Tissue-engineered lungs for in vivo implantation. Science. 2010 doi: 10.1126/science.1189345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Petersen TH, Calle EA, Colehour MB, Niklason LE. Bioreactor for the long-term culture of lung tissue. Cell Transplant. 2011:1117–1126. doi: 10.3727/096368910X544933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Green JM. Peer reviewed: a practical guide to analytical method validation. Anal Chem. 1996 [Google Scholar]
- 11.Mani DR, Abbatiello SE, Carr SA. Statistical characterization of multiple-reaction monitoring mass spectrometry (MRM-MS) assays for quantitative proteomics. BMC Bioinformatics. 2012;13(Suppl 16):S9. doi: 10.1186/1471-2105-13-S16-S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pratt JM, Simpson DM, Doherty MK, Rivers J, Gaskell SJ, Beynon RJ. Multiplexed absolute quantification for proteomics using concatenated signature peptides encoded by QconCAT genes. Nat Protoc. 2006;1:1029–1043. doi: 10.1038/nprot.2006.129. [DOI] [PubMed] [Google Scholar]
- 13.Fitzgerald J, Rich C, Zhou FH, Hansen U. Three novel collagen VI chains, 4 (VI), 5(VI), and 6(VI) J Biol Chem. 2008:20170–20180. doi: 10.1074/jbc.M710139200. [DOI] [PubMed] [Google Scholar]
- 14.Bristow J, Carey W, Egging D, Schalkwijk J. Tenascin-X, collagen, elastin, and the Ehlers–Danlos syndrome. Am J Med Genet C Semin Med Genet. 2005;139C:24–30. doi: 10.1002/ajmg.c.30071. [DOI] [PubMed] [Google Scholar]
- 15.Lee JH, Ki CS, Chung ES, Chung TY. A novel decorin gene mutation in congenital hereditary stromal dystrophy: a Korean family. Korean J Ophthalmol. 2012;26:301–305. doi: 10.3341/kjo.2012.26.4.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cavalcante FS, Ito S, Brewer K, Sakai H, Alencar AM, Almeida MP, Andrade JS, Jr, Majumdar A, Ingenito EP, Suki B. Mechanical interactions between collagen and proteoglycans: implications for the stability of lung tissue. J Appl Physiol. 1985;2005(98):672–679. doi: 10.1152/japplphysiol.00619.2004. [DOI] [PubMed] [Google Scholar]
- 17.Gillan L, Matei D, Fishman DA, Gerbin CS, Karlan BY, Chang DD. Periostin secreted by epithelial ovarian carcinoma is a ligand for alpha(V)beta(3) and alpha(V)beta(5) integrins and promotes cell motility. Cancer Res. 2002;62:5358–5364. [PubMed] [Google Scholar]
- 18.Willem M, Miosge N, Halfter W, Smyth N, Jannetti I, Burghart E, Timpl R, Mayer U. Specific ablation of the nidogen-binding site in the laminin gamma1 chain interferes with kidney and lung development. Development. 2002;129:2711–2722. doi: 10.1242/dev.129.11.2711. [DOI] [PubMed] [Google Scholar]
- 19.Sannes PL, Burch KK, Khosla J, McCarthy KJ, Couchman JR. Immunohistochemical localization of chondroitin sulfate, chondroitin sulfate proteoglycan, heparan sulfate proteoglycan, entactin, and laminin in basement membranes of postnatal developing and adult rat lungs. Am J Respir Cell Mol Biol. 1993:245–251. doi: 10.1165/ajrcmb/8.3.245. [DOI] [PubMed] [Google Scholar]
- 20.Koval M, Ward C, Findley MK, Roser-Page S, Helms MN, Roman J. Extracellular matrix influences alveolar epithelial claudin expression and barrier function. Am J Respir Cell Mol Biol. 2010:172–180. doi: 10.1165/rcmb.2008-0270OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dunsmore SE, Lee YC, Martinez-Williams C, Rannels DE. Synthesis of fibronectin and laminin by type II pulmonary epithelial cells. Am J Physiol. 1996:L215–L223. doi: 10.1152/ajplung.1996.270.2.L215. [DOI] [PubMed] [Google Scholar]
- 22.Atkinson JJ, Adair-Kirk TL, Kelley DG, Demello D, Senior RM. Clara cell adhesion and migration to extracellular matrix. Respir Res. 2008;1 doi: 10.1186/1465-9921-9-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hinenoya N, Naito I, Momota R, Sado Y, Kumagishi K, Ninomiya Y, Ohtsuka A. Type IV collagen alpha chains of the basement membrane in the rat bronchioalveolar transitional segment. Arch Histol Cytol. 2008:185–194. doi: 10.1679/aohc.71.185. [DOI] [PubMed] [Google Scholar]
- 24.Nebe B, Bohn W, Pommerenke H, Rychly J. Flow cytometric detection of the association between cell surface receptors and the cytoskeleton. Cytometry. 1997;28:66–73. doi: 10.1002/(sici)1097-0320(19970501)28:1<66::aid-cyto8>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 25.Wallis JM, Borg ZD, Daly AB, Deng B, Ballif BA, Allen GB, Jaworski DM, Weiss DJ. Comparative assessment of detergent-based protocols for mouse lung de-cellularization and re-cellularization. Tissue Eng Part C Methods. 2012:420–432. doi: 10.1089/ten.tec.2011.0567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Q, Uygun BE, Geerts S, Ozer S, Scalf M, Gilpin SE, Ott HC, Yarmush ML, Smith LM, Welham NV, Frey BL. Proteomic analysis of naturally-sourced biological scaffolds. Biomaterials. 2016;75:37–46. doi: 10.1016/j.biomaterials.2015.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nichols JEE, Niles J, Riddle M, Vargas G, Schilagard T, Ma L, Edward K, Lafrancesca S, Sakamoto J, Vega S, Ogedegbe M, Mlcak R, Deyo D, Woodson L, McQuitty C, Lick S, Beckles D, Melo E, Cortiella J. Production and assessment of decellularized pig and human lung scaffolds. Tissue Eng. 2013 doi: 10.1089/ten.tea.2012.0250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Faulk DM, Carruthers CA, Warner HJ, Kramer CR, Reing JE, Zhang L, D’Amore A, Badylak SF. The effect of detergents on the basement membrane complex of a biologic scaffold material. Acta Biomater. 2013 doi: 10.1016/j.actbio.2013.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Daly AB, Wallis JM, Borg ZD, Bonvillain RW, Deng B, Ballif BA, Jaworski DM, Allen GB, Weiss DJ. Initial binding and recellularization of decellularized mouse lung scaffolds with bone marrow-derived mesenchymal stromal cells. Tissue Eng. 2012:1–16. doi: 10.1089/ten.tea.2011.0301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goyal A, Pal N, Concannon M, Paul M, Doran M, Poluzzi C, Sekiguchi K, Whitelock JM, Neill T, Iozzo RV. Endorepellin, the angiostatic module of perlecan, interacts with both the alpha2beta1 integrin and vascular endothelial growth factor receptor 2 (VEGFR2): a dual receptor antagonism. J Biol Chem. 2011;286:25947–25962. doi: 10.1074/jbc.M111.243626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zeltz C, Brezillon S, Kapyla J, Eble JA, Bobichon H, Terryn C, Perreau C, Franz CM, Heino J, Maquart FX, Wegrowski Y. Lumican inhibits cell migration through alpha2beta1 integrin. Exp Cell Res. 2010;316:2922–2931. doi: 10.1016/j.yexcr.2010.08.002. [DOI] [PubMed] [Google Scholar]
- 32.Hopf M, Gohring W, Kohfeldt E, Yamada Y, Timpl R. Recombinant domain IV of perlecan binds to nidogens, laminin-nidogen complex, fibronectin, fibulin-2 and heparin. Eur J Biochem. 1999;259:917–925. doi: 10.1046/j.1432-1327.1999.00127.x. [DOI] [PubMed] [Google Scholar]
- 33.Tsuchiya T, Balestrini JL, Mendez J, Calle EA, Zhao L, Niklason LE. Influence of pH on extracellular matrix preservation during lung decellularization. Tissue Eng Part C Methods. 2014 doi: 10.1089/ten.tec.2013.0492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kessels MY, Huitema LFA, Boeren S, Kranenbarg S, Schulte-Merker S, van Leeuwen JL, de Vries SC. Proteomics analysis of the zebrafish skeletal extracellular matrix. PLoS ONE. 2014:e90568. doi: 10.1371/journal.pone.0090568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hughes C, Radan L, Chang WY, Stanford WL, Betts DH, Postovit LM, Lajoie GA. Mass spectrometry-based proteomic analysis of the matrix microenvironment in pluripotent stem cell culture. Mol Cell Proteomics. 2012;11:1924–1936. doi: 10.1074/mcp.M112.020057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Villegas SN, Rothová M, Barrios-Llerena ME, Pulina M, Hadjantonakis A-K, Le Bihan T, Astrof S, Brickman JM. eLife. eLife Sciences Publications Limited; 2013. PI3K/Akt1 signalling specifies foregut precursors by generating regionalized extra-cellular matrix. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.