Abstract
Aims/hypothesis
Identification of novel biomarkers for type 2 diabetes and their genetic determinants could lead to improved understanding of causal pathways and improve risk prediction. Methods In this study, we used data from non-targeted metabolomics performed using liquid chromatography coupled with tandem mass spectrometry in three Swedish cohorts (Uppsala Longitudinal Study of Adult Men [ULSAM], n = 1138; Prospective Investigation of the Vasculature in Uppsala Seniors [PIVUS], n = 970; TwinGene, n = 1630). Metabolites associated with impaired fasting glucose (IFG) and/or prevalent type 2 diabetes were assessed for associations with incident type 2 diabetes in the three cohorts followed by replication attempts in the Cooperative Health Research in the Region of Augsburg (KORA) S4 cohort (n = 855). Assessment of the association of metabolite-regulating genetic variants with type 2 diabetes was done using data from a meta-analysis of genome-wide association studies.
Results
Out of 5961 investigated metabolic features, 1120 were associated with prevalent type 2 diabetes and IFG and 70 were annotated to metabolites and replicated in the three cohorts. Fifteen metabolites were associated with incident type 2 diabetes in the four cohorts combined (358 events) following adjustment for age, sex, BMI, waist circumference and fasting glucose. Novel findings included associations of higher values of the bile acid deoxycholic acid and monoacylglyceride 18:2 and lower concentrations of cortisol with type 2 diabetes risk. However, adding metabolites to an existing risk score improved model fit only marginally. A genetic variant within the CYP7A1 locus, encoding the rate-limiting enzyme in bile acid synthesis, was found to be associated with lower concentrations of deoxycholic acid, higher concentrations of LDL-cholesterol and lower type 2 diabetes risk. Variants in or near SGPP1, GCKR and FADS1/2 were associated with diabetes-associated phospholipids and type 2 diabetes.
Conclusions/interpretation
We found evidence that the metabolism of bile acids and phospholipids shares some common genetic origin with type 2 diabetes.
Access to research materials
Metabolomics data have been deposited in the Metabolights database, with accession numbers MTBLS93 (TwinGene), MTBLS124 (ULSAM) and MTBLS90 (PIVUS).
Keywords: Genetic, Metabolomics, Prediction, Type 2 diabetes
Introduction
Recent advances in metabolite profiling technology have enabled discovery of novel biomarkers of type 2 diabetes development. It is worthwhile to better characterise these metabolic alterations since they could be of pathogenic importance. Elevated concentrations of branched-chain and aromatic amino acids and lower concentrations of glycine and various lipid species, such as lysophosphatidylcholine (LysoPC) 18:2 are reported to be associated with incident type 2 diabetes, but the causal role of these early aberrations in diabetes pathophysiology is not clear [1–3]. It has been proposed that the identification of genetic determinants of metabolite concentrations would assist in enabling the functional understanding of associations between metabolite concentrations and clinical endpoints [4]. So far, more than 150 associations between genetic variants and various metabolite concentrations are reported from large genome-wide association studies (GWAS), often with large effect sizes [5]. Reported variants affecting metabolite concentrations are often located within genes encoding enzymes or transporters, with a function related to the biochemical nature of the associated metabolites [6]. Some of these genetic variants have recently been used as instrumental variables to study the causal effect of lipid metabolites on cardiovascular risk [7, 8]. The underlying idea of this approach is that a genetic variant determining metabolite concentration could be used as an unbiased proxy to predict the effect of metabolite perturbation on clinical phenotypes of interest.
The primary aim of the present study was to identify metabolites associated with incident type 2 diabetes, using a non-targeted metabolomics approach in four population-based cohort studies, and to investigate whether such metabolites share a common genetic background with type 2 diabetes. A secondary aim was to explore whether the addition of metabolites to the Framingham diabetes risk score [9] would improve prediction of type 2 diabetes.
Methods
Study population
We used data that had been generated previously from non-targeted metabolomics analysis [10] in combination with phenotypic information from fasting individuals from four population-based studies. These studies have all been described in detail previously—the Uppsala Longitudinal Study of Adult Men (ULSAM) [11], the Prospective Investigation of the Vasculature in Uppsala Seniors (PIVUS) [12], a case-cohort subset of the TwinGene study [13] and the Cooperative Health Research in the Region of Augsburg (KORA) [14, 15]. Informed consent was obtained from all participants in the four studies. Details of the cohorts can be found in the ESM Methods.
Outcome definition
Impaired fasting glucose (IFG) at baseline was defined according to the American Diabetes Association criteria as fasting glucose ≥5.6 and <7.0 mmol/l [16]. Type 2 diabetes diagnosis at baseline and during follow-up could be based on biochemical measurement (fasting glucose ≥7.0 mmol/l, HbA1c ≥6.5% (48 mmol/mmol) and/or 2 h post-oral glucose tolerance test glucose ≥11.1 mmol/l) within the study, in addition to health registries and validated medical records. Details of diabetes definitions and analytical methods for glucose for each cohort are given in the ESM Methods. Individuals were censored at date of death or end of study.
Metabolomics analysis
Briefly, plasma samples from the age of 71 years in ULSAM and serum samples from the baseline of PIVUS and TwinGene were treated with methanol to precipitate proteins and dissolve lipids. Non-targeted metabolite profiling was performed using ultra-performance liquid chromatography (Acquity Ultra-Performance Liquid Chromatography) (UPLC) directly coupled to a quadrupole time-of-flight mass spectrometer (Xevo G2 Q-TOF MS) (Waters Corporation, Milford, MA, USA) fitted with an electrospray source operating in positive ion mode. Non-consecutive randomised duplicate samples of 1 μl were injected and separation was performed on a BEH C8 analytical column. Mass analysis was performed in the full scan mode (mass-to-charge ratio, 50–1200).
Data were processed using the open source XCMS package in the R statistical environment [17]. Metabolic feature detection, alignment, grouping, imputation and normalisation were performed separately for each study as previously described [10]. In total, 9755, 10,162 and 7522 metabolic features were detected in the TwinGene, ULSAM and PIVUS cohort, respectively. A metabolic feature is characterised by a unique mass-to-charge ratio and retention time, meaning that a single metabolite can be represented by many metabolic features due to phenomena such as in-source fragmentation, neutral losses, adduct formation and multimer formation. For the present study, only metabolic features present in TwinGene and PIVUS and/or ULSAM were included in the analysis. Since small polar metabolites such as sugars are not well retained by reverse-phase chromatography, all metabolic features with a retention time <35 s were excluded.
Annotation of IFG- and diabetes-associated metabolic features was based on spectral matching against an in-house spectral library of authentic standards as well as public databases. The level of confidence was categorised in agreement with the Metabolomics Standard Initiative [18] as level 1–4: 1, match with accurate mass (±5 ppm), overall fragmentation pattern and retention time with the in-house spectral library; 2, match based on accurate mass (±5 ppm) and fragmentation pattern using available spectra in public data bases; 3, match based on a combination of mass spectra and fragmentation pattern knowledge; accurate mass and retention time window to assign the metabolite to a chemical class; 4, unknown.
In KORA, metabolites were extracted using similar methods as for the Swedish cohorts from baseline serum samples and a non-targeted metabolomics analysis was performed by Metabolon (Durham, NC, USA), using three separate analytical methods GC–mass spectrometry (MS), UPLC–MS positive mode and UPLC–MS negative mode. The UPLC–MS platform utilised a Waters Acquity UPLC and a ThermoFisher LTQ mass spectrometer. The methods are described in detail elsewhere [19].
For all metabolite features included in the analysis, peak intensity was transformed to the Log2 scale and then SD-transformed within each of the four cohorts prior to statistical analysis.
Statistical analysis
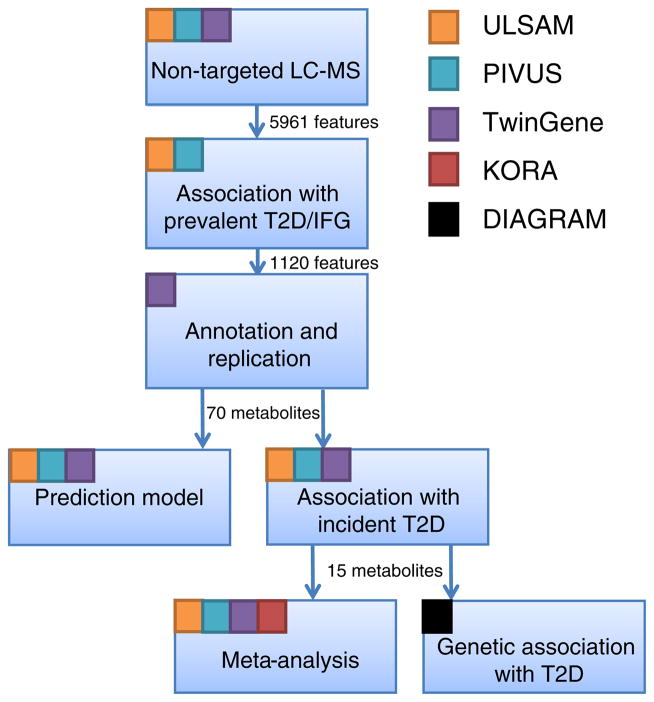
The overall workflow of the study is depicted in Fig. 1. The study was designed assuming that early markers of type 2 diabetes are also altered in individuals with IFG and overt type 2 diabetes. All statistical analysis was done using STATA13 (Stata, College Station, TX, USA) and R v. 3.1.3 (https://www.r-project.org/).
Fig. 1.
Overall workflow of the study. The coloured squares indicate which studies are being used in the different steps in the workflow. T2D, type 2 diabetes
Non-targeted metabolomics of prevalent type 2 diabetes, IFG and incident diabetes
In PIVUS and ULSAM, the association of each metabolic feature was assessed separately with normal fasting glucose vs IFG and normal fasting glucose vs prevalent type 2 diabetes using logistic regression modelling with feature intensity, age, sex, BMI and waist circumference as independent variables. In total, 3276 metabolite features were detected in both PIVUS and ULSAM and here fixed-effects inverse-variance-weighted meta-analysis was performed to pool results; 1622 features were detected only in PIVUS samples and 1063 features were detected only in ULSAM samples. The Benjamini–Hochberg procedure [20] was used to correct for multiple testing (5961 tests) at a false discovery rate (FDR) of 5%. Metabolic features that were identified as being associated with IFG or type 2 diabetes underwent annotation to metabolites and were re-assessed for their association with IFG and prevalent type 2 diabetes in the TwinGene subcohort (n = 1549). Metabolites were excluded if only fragments, but not the parent ion, were found associated with the outcome. A nominal p value cut-off of 0.05 and consistent direction of effect estimates were considered as evidence of replication.
Cox proportional hazard models, adjusted for age and sex, were used to assess the association of IFG- and prevalent type 2 diabetes-associated metabolites with time-to-event to type 2 diabetes in each of the three Swedish cohorts. In TwinGene, models were fitted and re-weighted for the inverse of the sampling probability using the Borgan ‘Estimator II’ [21]. Fixed-effects meta-analysis was used to pool the results and a 5% FDR was applied. We further adjusted models for BMI, waist circumference and fasting glucose concentrations. We assessed the association of metabolites available on the KORA platform with incident type 2 diabetes using the same model specifications and applied fixed-effects meta-analysis of all four cohorts. We tested the probability of binomial probability test (bitest in STATA) for directional replication using the binomial probability test.
Genetic association of metabolic loci with type 2 diabetes
To identify genetic variants regulating the metabolites identified as being associated with incident type 2 diabetes, we extracted results from the GWAS of metabolomics based on the KORA and TwinsUK cohorts with up to 7824 adults [19]. We meta-analysed GWAS results from ULSAM, PIVUS and TwinGene for those metabolites that were not identified or did not have a GWAS signal in KORA and TwinsUK data. A cut-off of p<5×10−8 was used to denote genome-wide significance. To assess the association of these variants with type 2 diabetes, the publicly available data from the GWAS and Metabochip results for type 2 diabetes, including up to 34,840 cases and 114,981 controls from the DIAbetes Genetics Replication and Meta-analysis consortium [22], were accessed and for five genetic variants we used a proxy in linkage disequilibrium with r2>0.8. In additional analysis, we addressed the association of the bile acid-regulating variant within CYP7A1, with other metabolic traits using the MR catalogue (www.mrcatalogue.medschl.cam.ac.uk, accessed 03/03/2016).
Prediction of type 2 diabetes
To determine whether metabolites associated with prevalent type 2 diabetes and IFG could improve type 2 diabetes prediction, we used Lasso penalised Cox regression implemented via the glmnet package in R by setting the overall penalty parameter α to 1 to select those with the highest predictive value. Cohort identity and Framingham diabetes risk score [9] were forced into the model. Model choice was based on tenfold internal cross-validation and the minimum λ achieved by adding exactly five of the 54 metabolite biomarkers that were available in all three cohorts. We used the combined ULSAM/PIVUS cohorts as a training set to derive an additive β coefficient-weighted 5-metabolite risk score. For validation in TwinGene, Cox proportional hazards regression re-weighted for the inverse of the sampling probability was used to assess incremental improvement of adding the metabolite score to the Framingham risk score by likelihood ratio test and C indices [23]. In TwinGene, information on parental history of diabetes was not available to include in the Framingham diabetes risk score; thus, this variable was set to ‘none’.
Results
Non-targeted metabolomics of prevalent type 2 diabetes, IFG and incident diabetes
Baseline characteristics of the included cohorts and the number of individuals with prevalent diabetes and IFG are shown in Table 1. We found 338 metabolite features to be associated with IFG and 975 features to be associated with prevalent type 2 diabetes in models adjusted for age, sex, BMI and waist circumference in PIVUS and ULSAM combined. In the annotation step, these 1120 features were determined to originate from at least 115 metabolites, of which 69 could be annotated to key adducts of a single unique metabolite and were taken forward to replication in TwinGene. Further, 17 additional metabolites had high-quality spectra but no matching metabolite in our data bases and were labelled as ‘missing retention time’ and taken forward to replication. Of the 86 metabolites taken forward to replication, 70 were associated with at least one of the two outcomes in TwinGene: 13 with both IFG and prevalent type 2 diabetes, 53 with type 2 diabetes only and four with IFG only (ESM Tables 1 and 2).
Table 1.
Baseline characteristics of the four cohorts used in this study
| Characteristic | TwinGenea | ULSAM | PIVUS | KORA S4b |
|---|---|---|---|---|
| N (total) | 1549 (subcohort) 81 (case-cohort) | 1138 | 970 | 855 |
| Prevalent diabetes | 192 (12) | 220 (19) | 113 (12) | – |
| IFG | 444 (29) | 249 (22) | 337 (35) | 325 (38) |
| No. of incident events of type 2 diabetes | 122 | 78 | 70 | 88 |
| Age (years) | 68.0 ± 8.1 | 71.0 ± 0.6 | 70.2 ± 0.2 | 63.1 ± 0.4 |
| % female sex | 42 | 0 | 50 | 49 |
| % current smoker | 13 | 20 | 10 | 49 |
| BMI (kg/m2) | 26.0 ± 3.9 | 26.3 ± 3.4 | 27.1 ± 4.3 | 28.1 ± 4.0 |
| Waist circumference (cm) | 92.8 ± 11.8 | 94.9 ± 9.6 | 91.2 ± 11.6 | 94.5 ± 11.1 |
| Fasting glucose (mmol/l) | 5.7 ± 1.3 | 5.8 ± 1.5 | 5.9 ± 1.8 | 5.5 ± 0.5 |
| HDL-cholesterol (mmol/l) | 1.4 ± 0.4 | 1.3 ± 0.3 | 1.5 ± 0.4 | 1.5 ± 0.4 |
| LDL-cholesterol (mmol/l) | 3.7 ± 1.0 | 3.9 ± 0.9 | 3.4 ± 0.9 | 4.0 ± 1.0 |
| Triacylglycerol (mmol/l) | 1.3 ± 0.7 | 1.5 ± 0.8 | 1.3 ± 0.6 | 1.5 ± 0.8 |
| Systolic blood pressure (mmHg) | 141.0 ± 19.8 | 146.8 ± 18.7 | 149.1 ± 22.6 | 133.0 ± 19 |
| Diastolic blood pressure (mmHg) | 81.7 ± 10.5 | 83.7 ± 9.4 | 78.6 ± 10.2 | 80.0 ± 10 |
| % taking antihypertensive medication | 25 | 35 | 31 | 31 |
| % taking lipid-lowering medication | 17 | 9 | 16 | 11 |
Data are shown as mean ± SD for continuous variables and as n (%) for binary variables
Baseline characteristics are given for subcohort of TwinGene
Replication cohort, individuals with prevalent type 2 diabetes are excluded
There were 78 incident events of type 2 diabetes in the ULSAM cohort, 70 in the PIVUS cohort, 122 in the TwinGene cohort and 88 in the KORA cohort. Of the 70 metabolites found to be associated with prevalent type 2 diabetes and IFG, 36 were also associated with incident type 2 diabetes in the meta-analysis of the three Swedish cohorts in crude models adjusted for age and sex at a 5% FDR and 15 metabolites in ‘fully adjusted models’ additionally adjusted for waist circumference, BMI and fasting glucose (p<0.05) (ESM Table 3). Of those 15, deoxycholic acid, monoacylglyceride 18:2 and cortisol represent a novel finding with the highest level of annotation confidence. The comparison of analytical spectra to standard spectra is shown in ESM Figs 1 and 2.
Five of these 15 compounds (cortisol, γ-glutamyl-leucine, 2-methylbutyroylcarnitine, L-tyrosine and deoxycholic acid) were part of the panel tested in the KORA cohort. The association of 2-methylbutyroylcarnitine and tyrosine with incident type 2 diabetes in the age- and sex-adjusted models was confirmed, although none of the five metabolites were associated in the fully adjusted models (ESM Table 4). For all five metabolites, the directions of effect estimates were the same in KORA as in the Swedish cohorts and, when formally tested, the probability for this distribution was significantly different from the null (binomial probability test for 10/10 to be in the same direction, p=0.002). A post hoc power calculation for replication at an α of 0.05 is shown in ESM Fig. 3 and ESM Table 4.
All five metabolites assessed in KORA showed p < 0.05 in the combined meta-analysis (Table 2). In a sensitivity analysis, we re-ran the meta-analysis excluding the male sex-only cohort ULSAM (ESM Table 5) and obtained similar results.
Table 2.
Metabolites associated with incident diabetes mellitus in the combined analysis with TwinGene, ULSAM, PIVUS and KORA S4
| Metabolitea | Annotation confidence | Adduct form | HR (95% CI) for age- and sex-adjusted models | p value | I2 | HR (95% CI) for fully adjusted models | p value | No. of cohorts | I2 |
|---|---|---|---|---|---|---|---|---|---|
| Cortisol | 1 | M + H | 0.85 (0.77, 0.94) | 1.2 × 10−3 | 80 | 0.84 (0.76, 0.92) | 4.1 × 10−4 | 4 | 78 |
| γ-Glutamyl-leucine | 2 | M + H | 1.48 (1.32, 1.67) | 3.5 × 10−11 | 69 | 1.25 (1.10, 1.41) | 4.1 × 10−4 | 4 | 68 |
| LysoPC/PC(O-16:1/0:0) | 3 | M + H | 0.69 (0.61, 0.78) | 2.3 × 10−9 | 66 | 0.81 (0.71, 0.93) | 2.8 × 10−3 | 3 | 0 |
| 2-Methylbutyroylcarnitine | 2 | M + H | 1.38 (1.24, 1.53) | 2.5 × 10−9 | 0 | 1.20 (1.06, 1.35) | 3.7 × 10−3 | 4 | 30 |
| Barogenin | 2 | M + H | 1.38 (1.22, 1.57) | 2.4 × 10−7 | 0 | 1.21 (1.05, 1.38) | 6.4 × 10−3 | 3 | 0 |
| L-Tyrosine | 1 | M + H | 1.46 (1.30, 1.64) | 3.4 × 10−10 | 3 | 1.17 (1.04, 1.32) | 8.4 × 10−3 | 4 | 0 |
| SM (33:1) | 2 | M + Na | 0.83 (0.74, 0.92) | 4.2 × 10−4 | 0 | 0.87 (0.77, 0.97) | 0.01 | 3 | 0 |
| LysoPC (20:2) | 2 | M + H | 0.78 (0.70, 0.88) | 2.8 × 10−5 | 19 | 0.85 (0.74, 0.97) | 0.01 | 3 | 0 |
| Monoacylglycerol (18:2) | 1 | M + Na | 1.43 (1.24, 1.65) | 8.2 × 10−7 | 62 | 1.23 (1.04, 1.46) | 0.02 | 2 | 0 |
| CerPE (38:2) | 2 | M + H | 0.87 (0.77, 0.97) | 1.3 × 10−2 | 0 | 0.87 (0.77, 0.99) | 0.03 | 3 | 0 |
| missing@tg43 | 4 | 1.55 (1.31, 1.83) | 2.7 × 10−7 | 0 | 1.21 (1.01, 1.45) | 0.03 | 2 | 0 | |
| SM (d18:2/18:1) | 2 | M + H | 0.86 (0.77, 0.97) | 1.0 × 10−2 | 0 | 0.88 (0.78, 0.99) | 0.04 | 3 | 0 |
| SM (34:2) | 2 | M + H | 0.88 (0.79, 0.98) | 2.2 × 10−2 | 0 | 0.89 (0.80, 1.00) | 0.04 | 3 | 0 |
| Deoxycholic acid | 1 | M + Na | 1.27 (1.14, 1.41) | 1.1 × 10−5 | 31 | 1.13 (1.00, 1.27) | 0.04 | 4 | 0 |
| PC (42:7) | 2 | M + H | 0.80 (0.72, 0.90) | 1.5 × 10−4 | 30 | 0.87 (0.77, 1.00) | 0.04 | 3 | 0 |
HR per SD-unit of Log2-transformed metabolite increase and 95% CI are given for age- and sex-adjusted and fully adjusted (age, sex, BMI, waist circumference and fasting glucose at baseline) models for 1-SD increase of log2-scaled metabolite increase. Only metabolites with p < 0.05 in the fully adjusted models are shown
For metabolites annotated at level 4, the metabolite is named ‘missing@retention time’ measured in TwinGene
Genetic association of metabolic loci with type 2 diabetes
Using published GWAS from KORA and TwinsUK [19], as well as from a meta-analysis from ULSAM, PIVUS and TwinGene, we identified a total of 12 metabolite-regulating genetic variants for eight of the 15 metabolites at a genome-wide significance level (p<5×10−8). The association of these genetic variants with type 2 diabetes was assessed using published summary statistics from a large meta-analysis of GWAS for type 2 diabetes [22]. Four of the 12 genetic variants were found to be associated with type 2 diabetes at a nominal p value threshold (Table 3). First, a variant in the gene encoding cholesterol 7α-hydroxylase (CYP7A1) was found to be associated with both decreased concentrations of the bile acid deoxycholic acid and decreased risk of type 2 diabetes. We further investigated the association of CYP7A1 with other metabolic traits using the largest available GWAS results and found associations with higher LDL-cholesterol and higher triacylglycerol levels (Table 4). Second, genetic variants associated with lower concentrations of sphingomyelin (SM) 33:1 (a variant within SYNE2 [upstream SGPP1]) and ceramide phosphoethanolamine (CerPE) 38:2 (a variant within GCKR), respectively, identified in ULSAM, PIVUS and TwinGene), were found to be associated with lower risk of type 2 diabetes. Third, a variant in MYRF (upstream of FADS2) identified in ULSAM, PIVUS and TwinGene was found to be associated with lower LysoPC 20:2 and increased risk of type 2 diabetes.
Table 3.
Genetic variants associated with candidate metabolites and their association with type 2 diabetes
| Metabolite | SNP
|
SNP–metabolite
|
SNP–type 2 diabetes
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rsIDa | Closest gene | Chr. | EA | β | SE | p value | Source | β | SE | p value | Sourceb | |
| 2-Methylbutyroylcarnitine | rs662138 | SLC22A1 | 6 | C | 0.02 | 0.003 | 3.7 × 10−8 | Shin et al [19] | 0.01 | 0.016 | 0.48 | GWAS + Metabochip |
| 2-Methylbutyroylcarnitine | rs272893 | SLC22A4 | 5 | T | 0.24 | 0.024 | 1.8 × 10−23 | ULSAM/PIVUS/TG | 0.010 | 0.021 | 0.70 | GWAS |
| SM (33:1) | rs12879919 (rs12889954) | SYNE2, SGPP1 | 14 | A | −0.23 | 0.040 | 1.0 × 10−8 | ULSAM/PIVUS/TG | −0.04 | 0.017 | 0.04 | GWAS + Metabochip |
| CerPE (38:2) | rs1260326 | GCKR | 2 | T | −0.16 | 0.025 | 2.1 × 10−10 | ULSAM/PIVUS/TG | −0.06 | 0.012 | 1.6 × 10−6 | GWAS + Metabochip |
| Deoxycholic acid | rs8192870 | CYP7A1 | 8 | T | −0.04 | 0.007 | 4.0 × 10−8 | Shin et al [19] | −0.03 | 0.012 | 8.1 × 10−3 | GWAS + Metabochip |
| L-Tyrosine | rs9400467 | SLC16A10 | 6 | T | −0.01 | 0.002 | 6.5 × 10−14 | Shin et al [19] | 0.03 | 0.02 | 0.17 | GWAS |
| L-Tyrosine | rs172650 (rs4788817) | TAT | 16 | A | 0.01 | 0.002 | 2.8 × 10−10 | Shin et al [19] | −0.003 | 0.012 | 0.80 | GWAS + Metabochip |
| L-Tyrosine | rs12728678 | KCNN3 | 1 | T | −0.01 | 0.002 | 2.2 × 10−8 | Shin et al [19] | −0.04 | 0.025 | 0.09 | GWAS |
| LysoPC(20:2) | rs174536 (rs174535) | MYRF, FADS1/2 | 11 | A | −0.24 | 0.024 | 1.3 × 10−22 | ULSAM/PIVUS/TG | 0.03 | 0.012 | 0.03 | GWAS + Metabochip |
| SM (34:2) | rs174583 | FADS2 | 11 | T | −0.14 | 0.024 | 1.1 × 10−8 | ULSAM/PIVUS/TG | −0.02 | 0.012 | 0.08 | GWAS + Metabochip |
| SM (d18:2/18:1) | rs12529505 | AK9 | 6 | A | −0.15 | 0.027 | 2.7 × 10−8 | ULSAM/PIVUS/TG | −0.03 | 0.02 | 0.16 | GWAS |
SNP identifiers in parenthesis indicates that a proxy (r2 > 0.8) was used for SNP–type 2 diabetes association
Data derived from the public repository accompanying Morris et al [22] at http://diagram-consortium.org/downloads.html
EA, effect allele; SNP, single-nucleotide polymorphism; TG, TwinGene
Table 4.
Association of the T allele CYP7A1 variant rs8192870 or its corresponding C allele of the proxy rs2326077 (r2 = 0.881) with metabolic traits
| Phenotype | Study | Year | β | SE | p value | No. of controls | No. of cases | Unit |
|---|---|---|---|---|---|---|---|---|
| Type 2 diabetesa | Morris et al (2012) [22] | 2012 | −0.033 | 0.012 | 8.1 × 10−3 | 114,981 | 34,840 | log(OR) |
| Fasting glucosea | Dupuis et al (2010) [36] | 2010 | −0.004 | 0.002 | 0.07 | 133,010 | 0 | mmol/l |
| Log(fasting insulin)a | Dupuis et al (2010) [36] | 2010 | −0.002 | 0.003 | 0.41 | 108,557 | 0 | pmol/l |
| 2 h fasting glucosea | Dupuis et al (2010) [36] | 2010 | −0.010 | 0.011 | 0.37 | 42,854 | 0 | mmol/l |
| LDL-cholesterola | Global Lipids Genetics Consortium (2013) [41] | 2013 | 0.034 | 0.004 | 5.0 × 10−17 | 172,996 | 0 | IVNT |
| Triacylglycerolsa | Global Lipids Genetics Consortium (2013) [41] | 2013 | 0.018 | 0.003 | 5.4 × 10−7 | 177,766 | 0 | IVNT |
| HDL-cholesterola | Global Lipids Genetics Consortium (2013) [41] | 2013 | 0.004 | 0.004 | 0.22 | 187,069 | 0 | IVNT |
| BMI | Locke et al (2015) [42] | 2015 | 0.001 | 0.004 | 0.88 | 235,991 | 0 | INVT |
| Waist-to-hip ratio | Shungin et al (2015) [43] | 2015 | 0.006 | 0.004 | 0.19 | 144,548 | 0 | INVT |
Results are extracted from the largest available GWAS datasets
Metabochip proxy rs2326077
IVNT, inverse normal transformed trait
Prediction of type 2 diabetes
In 1763 individuals comprising the PIVUS and ULSAM cohorts (70 and 78 incident events, respectively), a LASSO predictor selection adjusted for cohort and Framingham diabetes risk score resulted in a five-metabolite score that included tyrosine, barogenin, LysoPC/phosphatidylcholine (PC)(O-16:1/0:0), PC(O-18:1/0:0)/PC(P-18:0/0:0) and LysoPC(20:2). In the validation sample of 1394 fasting individuals without prevalent diabetes and 122 incident events in TwinGene, the metabolite score improved the Framingham diabetes risk model’s fitting (χ2=7.371, p=0.007) and marginally improved discrimination of incident diabetes events (C index for the Framingham diabetes risk score of 0.848 [95% CI 0.793, 0.903] improved to 0.855 [95% CI 0.800, 0.910]). One SD increase in the five-metabolite score, when added to the Framingham diabetes model, increased the 10 year risk of type 2 diabetes by 29% (HR 1.294, 95% CI 1.071, 1.564).
Discussion
Using a non-targeted metabolomics approach, our study confirmed several known metabolites to be associated with incident type 2 diabetes and also identified novel associations for three compounds annotated with the highest level of confidence—deoxycholic acid, monoacylglyceride 18:2 and the steroid hormone cortisol. For four metabolites, we identified genetic variants associated with both metabolite concentrations (at a genome-wide significance level) and type 2 diabetes (at a nominal level).
Bile acid synthesis
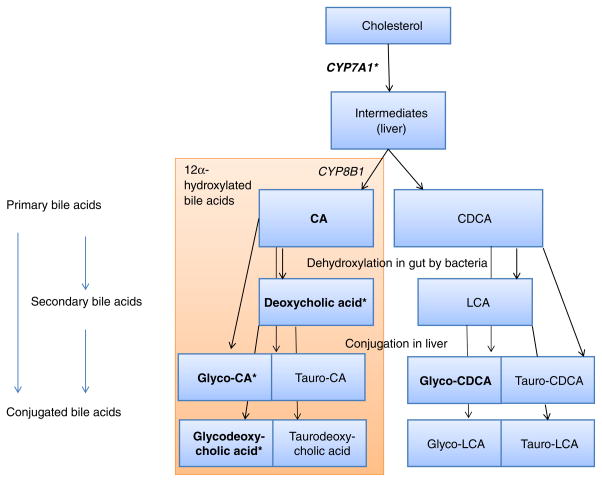
The main finding of our study is the phenotypic and genetic correlation of bile acid concentrations with type 2 diabetes. In the present study, increased concentrations of three 12α-hydroxylated bile acids (deoxycholic acid, glycocholic acid and glycodeoxycholic acid) were associated with incident diabetes in the age- and sex-adjusted models. One of these, deoxycholic acid, remained significant in the model adjusted for BMI, waist circumference, age, sex and concentration of fasting glucose. In a previous study, increased 12α-hydroxylated bile acid concentrations were linked to worse insulin resistance [24]. Another study found elevated concentrations of deoxycholic acid, but lower concentrations of cholic acid, when persons with prevalent diabetes were compared with healthy controls [25]. We note that out of four 12α-hydroxylated bile acids captured on our metabolomics platform, three were associated with prevalent and incident diabetes. The results from the current study highlight the complex interactions between lipid metabolism, type 2 diabetes and bile acid concentrations. In the liver, the enzyme cholesterol 7α-hydroxylase (encoded by CYP7A1) is the rate-limiting enzyme in the conversion of cholesterol to primary bile acids (Fig. 2). Using a genome-wide approach, we found that a genetic variant within CYP7A1 was associated with decreased deoxycholic acid concentrations, decreased risk of type 2 diabetes and increased concentrations of LDL-cholesterol and triacylglycerols, which supports our observational findings.
Fig. 2.
Overview of bile acid metabolism. Metabolites with name in bold indicates that these were measured on the platform. *p < 0.05 for incident type 2 diabetes in sex- and age-adjusted models. CA, cholic acid; CDCA, chenodeoxycholic acid; LCA, lithocholic acid
The higher level of LDL-cholesterol in carriers of the bile acid-increasing variant is likely due to a lower activity of the cholesterol 7α-hydroxylase, which will clear less cholesterol from the circulation. The effect of the CYP7A1 variant on LDL-cholesterol and type 2 diabetes is consistent with recent findings that LDL-increasing variants in the gene encoding 3-hydroxy-3-methylglutaryl-CoA reductase (HMGCR) and a polygenic LDL-cholesterol risk score are both associated with lower risk of diabetes [26, 27]. A variant in CYP7A1 decreasing LDL-cholesterol has previously been linked to lower fasting glucose [28]. The direction of effects, with higher levels of bile acids in the circulation linked to increased risk of diabetes, seems however counterintuitive, as bile acids are increasingly being recognised as hormones that regulate various metabolic processes in beneficial ways, including increasing incretin secretion in the gut [29], although different classes of bile acids affect downstream receptor signalling in different ways, not all of which may promote glucose homeostasis [30]. However, with regards to pharmaceutical applications, bile acid sequestrants such as colesevelam (approved for lipid-lowering purposes) bind to bile acids in the gut and thus increase CYP7A1 expression through feedback systems. The drug results in lowered LDL-cholesterol through increased bile acid production and has been approved for glucose-lowering treatment in hyperglycaemia [31], although the underlying mechanism for this effect is little explored and stands in contrast to our results. To our knowledge, we present the largest human sample establishing a possible common genetic origin between dyslipidaemia, reduced 12α-hydroxylated bile acid synthesis and lower risk of type 2 diabetes.
Phospholipid metabolism
Circulating concentrations of different LysoPC species have been found to be reduced in diabetes, impaired glucose tolerance and coronary heart disease [2, 3, 8, 32, 33]. In the present study, lower LysoPC(20:2) and its associated genetic variant near FADS1/2 were found to be associated with higher risk of type 2 diabetes. Fatty acid desaturases (encoded by fatty acid desaturases gene family) introduce double bonds into saturated fatty acids and variants in this locus has previously been linked to blood lipid concentrations [34], fatty acid concentrations [35] and fasting glucose [36]. In our genetic analysis, the direction of the effect was consistent with the observational analysis, where an increased level of LysoPC(20:2) was associated with a lower risk of type 2 diabetes. We speculate that decreased expression of FADS genes likely increases the concentrations of saturated fatty acids in different lipids, which may affect insulin sensitivity and insulin secretion and hence diabetes risk.
SMs have also previously been linked to type 2 diabetes [2]; however, to the best of our knowledge, their analogues, CerPEs, have not. In our study, SM d18:2/18:2, SM 34:2, SM (33:1) and CerPE 38:2 were all found to be inversely associated with incident type 2 diabetes. CerPEs are produced in trace amounts together with SMs and are located in the plasma membrane, but their functions are largely unknown [37]. We found a genetic variant within SYNE2 just upstream of the sphingosine-1-phosphate phosphatase 1 gene (SGPP1) that was associated with SM(33:1) and type 2 diabetes, but in a direction different from that revealed by the observational results. The sphingosine-1-phosphate phosphatase 1 protein regulates sphingosine and long-chain ceramide metabolism [38] and has previously been associated with SM concentrations [39] and may play a role in insulin secretion [40]. We further found that a variant in the glucokinase regulator gene (GCKR) was associated with lower CerPE 38:2 levels and lower risk of type 2 diabetes. The encoded protein regulates the activity of glucokinase (a key enzyme in glucose homeostasis) in the liver. Variants within this locus are well-known markers for diabetes and lipid traits.
Prediction
Addition of five metabolites to the established Framingham risk score for diabetes did increase model fit significantly but added very little (less than 1%) to discrimination. Future studies including also the monosaccharides and polar amino acids that could be detected by GC–MS would have the potential to define a larger set of metabolites that also might increase discrimination.
Strengths and limitations
Strengths of the present study include the use of a non-targeted metabolomics approach in four prospective cohorts and its integration with genetics data to provide evidence for shared causal pathways between several metabolites and type 2 diabetes. However, since some of the genetic variants (e.g. GCKR, FADS2) were commonly associated with several metabolites, a basic assumption for a Mendelian randomisation study (non-pleiotropic effects of genetic instruments) was violated, precluding analysis for causal directions. For CYP7A1 and its association with our main findings on bile acids, although its encoded enzyme is specific to bile acid biosynthesis, it not suitable to disentangle the effect of bile acids from those of their immediate precursor, cholesterol.
Only five of the 15 candidate metabolites could be analysed in KORA due to different analysis methods. The KORA sample had limited power to detect true effect sizes, especially in the fully adjusted models. Nevertheless, the magnitudes and directions of the associations found in the Swedish meta-analysis and in KORA were similar, supporting the validity of the results. The KORA S4 cohort with targeted metabolite profiles analysed on a different metabolomics platform from that used in the present study was previously used to assess the association of a limited number of metabolites (3 and 14, respectively) with incident type 2 diabetes [2, 3], but the metabolites did not overlap with the five assessed in KORA in the current study.
A limitation concerning generalisability is the inclusion of mostly elderly white persons. Another limitation is that ULSAM is a male sex-only cohort and this could have biased the results if there were different concentrations of metabolites in men and women. However, our sensitivity analysis where we exclude ULSAM from the meta-analysis shows similar results. Degradation of analytes is likely to reduce the power to detect differences between groups but as long as there are no differences in degradation among diabetes controls and those with events, there will be no bias causing false-positive findings. Again, results from the meta-analysis without ULSAM (which was the study with the longest freezer storage time) were similar to those of the full meta-analysis. Further, only liquid chromatography was used for separation of metabolites in the three Swedish cohorts; this limits the correct detection and identification of monosaccharides and polar amino acids, which have been highlighted in type 2 diabetes [1, 3]. It is therefore likely that a combination with other methods such as GC–MS would have increased the number of metabolites discovered, especially from glucose-related pathways, which indeed are of great interest for the present research topic. Finally, we were not able to include family history of type 2 diabetes in the Framingham diabetes risk score, which may have overestimated the contribution of the metabolite risk score.
Conclusions
We identified novel metabolites that were associated with incident type 2 diabetes. A genetic variant linked to bile acid metabolism was associated with type 2 diabetes and LDL-cholesterol, suggesting shared causal pathways. Non-targeted metabolomics linked with genetic data is a powerful approach to discover new pathophysiological mechanisms linked to type 2 diabetes development.
Acknowledgments
Funding This study was supported by the Swedish Research Council (VR; grant no. 2012-1397), the Swedish Heart-Lung Foundation (20140422), the Swedish Diabetes Foundation (Diabetesfonden; grant no. 2013-024), the Knut och Alice Wallenberg Foundation, the European Research Council (ERC Starting Grant), the Family Ernfors Foundation and Uppsala University. Genotyping was performed by the SNP&SEQ Technology Platform in Uppsala. The computations were partly performed on resources provided by the Swedish National Infrastructure for Computing (SNIC) at UPPMAX.
The KORA study was initiated and financed by the Helmholtz Zentrum München – German Research Center for Environmental Health, which is funded by the German Federal Ministry of Education and Research (BMBF) and by the State of Bavaria. Furthermore, KORA research was supported within the Munich Center of Health Sciences (MC-Health), Ludwig-Maximilians-Universität, as part of LMUinnovativ. Part of this project was supported by EU FP7 grants HEALTH-2009-2.2.1-3/242114 (Project OPTiMiSE) and HEALTH-2013-2.4.2-1/602936 (Project CarTarDis) and by a grant from the BMBF to the German Center for Diabetes Research (DZD e. V.).
Abbreviations
- CerPE
Ceramide phosphoethanolamine
- FDR
False discovery rate
- GWAS
Genome-wide association study
- IFG
Impaired fasting glucose
- KORA
Cooperative Health Research in the Region of Augsburg
- LysoPC
Lysophosphatidylcholine
- PC
Phosphatidylcholine
- PIVUS
Prospective Investigation of the Vasculature in Uppsala Seniors
- SM
Sphingomyelin
- ULSAM
Uppsala Longitudinal Study of Adult Men
- UPLC
Ultra-performance liquid chromatography
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s00125-016-4041-1) contains peer-reviewed but unedited supplementary material, which is available to authorised users.
Contribution statement TF, EI and LL conceived the study. TF, SB, SG and CN implemented, and TF, AG, EI and LL planned, the statistical analysis. SS, CDB, CB, JEP, GK, AP, PKM, RWS, VG, CG, NLP, EI and LL participated in data acquisition. TF and LL wrote the draft manuscript and SS, SB, CB, CN, AG, SG, CDB, JEP, GK, AP, PKM, RWS, VG, CG, NLP and EI revised it. All authors approved the final version. TF is the guarantor of the study.
Duality of interest The authors declare that there is no duality of interest associated with this manuscript.
References
- 1.Wang TJ, Larson MG, Vasan RS, et al. Metabolite profiles and the risk of developing diabetes. Nat Med. 2011;17:448–453. doi: 10.1038/nm.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Floegel A, Stefan N, Yu Z, et al. Identification of serum metabolites associated with risk of type 2 diabetes using a targeted metabolomic approach. Diabetes. 2013;62:639–648. doi: 10.2337/db12-0495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang-Sattler R, Yu Z, Herder C, et al. Novel biomarkers for pre-diabetes identified by metabolomics. Mol Syst Biol. 2012;8:615. doi: 10.1038/msb.2012.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burgess S, Timpson NJ, Ebrahim S, Davey Smith G. Mendelian randomization: where are we now and where are we going? Int J Epidemiol. 2015;44:379–388. doi: 10.1093/ije/dyv108. [DOI] [PubMed] [Google Scholar]
- 5.Suhre K, Raffler J, Kastenmuller G. Biochemical insights from population studies with genetics and metabolomics. Arch Biochem Biophys. 2016;589:168–176. doi: 10.1016/j.abb.2015.09.023. [DOI] [PubMed] [Google Scholar]
- 6.Kastenmuller G, Raffler J, Gieger C, Suhre K. Genetics of human metabolism: an update. Hum Mol Genet. 2015;24:R93–R101. doi: 10.1093/hmg/ddv263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kettunen J, Demirkan A, Wurtz P, et al. Genome-wide study for circulating metabolites identifies 62 loci and reveals novel systemic effects of LPA. Nat Commun. 2016;7:11122. doi: 10.1038/ncomms11122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ganna A, Salihovic S, Sundstrom J, et al. Large-scale metabolomic profiling identifies novel biomarkers for incident coronary heart disease. PLoS Genet. 2014;10:e1004801. doi: 10.1371/journal.pgen.1004801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wilson PW, Meigs JB, Sullivan L, Fox CS, Nathan DM, D’Agostino RB., Sr Prediction of incident diabetes mellitus in middle-aged adults: the Framingham Offspring Study. Arch Intern Med. 2007;167:1068–1074. doi: 10.1001/archinte.167.10.1068. [DOI] [PubMed] [Google Scholar]
- 10.Ganna A, Fall T, Salihovic S, et al. Large-scale non-targeted metabolomic profiling in three human population-based studies. Metabolomics. 2015;12:1–13. [Google Scholar]
- 11.Hedstrand H. A study of middle-aged men with particular reference to risk factors for cardiovascular disease. Ups J Med Sci Suppl. 1975;19:1–61. [PubMed] [Google Scholar]
- 12.Lind L, Fors N, Hall J, Marttala K, Stenborg A. A comparison of three different methods to evaluate endothelium-dependent vasodilation in the elderly: the Prospective Investigation of the Vasculature in Uppsala Seniors (PIVUS) study. Arterioscler Thromb Vasc Biol. 2005;25:2368–2375. doi: 10.1161/01.ATV.0000184769.22061.da. [DOI] [PubMed] [Google Scholar]
- 13.Magnusson PK, Almqvist C, Rahman I, et al. The Swedish Twin Registry: establishment of a biobank and other recent developments. Twin Res Hum Genet. 2013;16:317–329. doi: 10.1017/thg.2012.104. [DOI] [PubMed] [Google Scholar]
- 14.Holle R, Happich M, Lowel H, Wichmann HE, Group MKS. KORA—a research platform for population based health research. Gesundheitswesen. 2005;67(Suppl 1):S19–S25. doi: 10.1055/s-2005-858235. [DOI] [PubMed] [Google Scholar]
- 15.Rathmann W, Haastert B, Icks A, et al. High prevalence of undiagnosed diabetes mellitus in Southern Germany: target populations for efficient screening. The KORA survey 2000. Diabetologia. 2003;46:182–189. doi: 10.1007/s00125-002-1025-0. [DOI] [PubMed] [Google Scholar]
- 16.American Diabetes A. Diagnosis and classification of diabetes mellitus. Diabetes Care. 2005;28(Suppl 1):S37–S42. doi: 10.2337/diacare.28.suppl_1.s37. [DOI] [PubMed] [Google Scholar]
- 17.Smith CA, Want EJ, O’Maille G, Abagyan R, Siuzdak G. XCMS: processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal Chem. 2006;78:779–787. doi: 10.1021/ac051437y. [DOI] [PubMed] [Google Scholar]
- 18.Sumner LW, Amberg A, Barrett D, et al. Proposed minimum reporting standards for chemical analysis Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI) Metabolomics. 2007;3:211–221. doi: 10.1007/s11306-007-0082-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shin SY, Fauman EB, Petersen AK, et al. An atlas of genetic influences on human blood metabolites. Nat Genet. 2014;46:543–550. doi: 10.1038/ng.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Benjamini Y, Hochberg Y. Controlling the false discovery rate—a practical and powerful approach to multiple testing. J R Stat Soc Ser B Methodol. 1995;57:289–300. [Google Scholar]
- 21.Ganna A, Reilly M, de Faire U, Pedersen N, Magnusson P, Ingelsson E. Risk prediction measures for case-cohort and nested case-control designs: an application to cardiovascular disease. Am J Epidemiol. 2012;175:715–724. doi: 10.1093/aje/kwr374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morris AP, Voight BF, Teslovich TM, et al. Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat Genet. 2012;44:981–990. doi: 10.1038/ng.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harrell FE, Jr, Califf RM, Pryor DB, Lee KL, Rosati RA. Evaluating the yield of medical tests. JAMA. 1982;247:2543–2546. [PubMed] [Google Scholar]
- 24.Haeusler RA, Astiarraga B, Camastra S, Accili D, Ferrannini E. Human insulin resistance is associated with increased plasma levels of 12alpha-hydroxylated bile acids. Diabetes. 2013;62:4184–4191. doi: 10.2337/db13-0639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Suhre K, Meisinger C, Doring A, et al. Metabolic footprint of diabetes: a multiplatform metabolomics study in an epidemiological setting. PLoS One. 2010;5:e13953. doi: 10.1371/journal.pone.0013953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fall T, Xie W, Poon W, et al. Using genetic variants to assess the relationship between circulating lipids and type 2 diabetes. Diabetes. 2015;64:2676–2684. doi: 10.2337/db14-1710. [DOI] [PubMed] [Google Scholar]
- 27.Swerdlow DI, Preiss D, Kuchenbaecker KB, et al. HMG-coenzyme A reductase inhibition, type 2 diabetes, and bodyweight: evidence from genetic analysis and randomised trials. Lancet. 2014;385:351–361. doi: 10.1016/S0140-6736(14)61183-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li N, van der Sijde MR, Study LC, et al. Pleiotropic effects of lipid genes on plasma glucose, HbA1c and HOMA-IR levels. Diabetes. 2014;63:3149–3158. doi: 10.2337/db13-1800. [DOI] [PubMed] [Google Scholar]
- 29.Meier JJ, Nauck MA. Incretin-based therapies: where will we be 50 years from now? Diabetologia. 2015;58:1745–1750. doi: 10.1007/s00125-015-3608-6. [DOI] [PubMed] [Google Scholar]
- 30.Lew JL, Zhao A, Yu J, et al. The farnesoid X receptor controls gene expression in a ligand- and promoter-selective fashion. J Biol Chem. 2004;279:8856–8861. doi: 10.1074/jbc.M306422200. [DOI] [PubMed] [Google Scholar]
- 31.Ooi CP, Loke SC. Colesevelam for type 2 diabetes mellitus. Cochrane Database Syst Rev. 2012;12:CD009361. doi: 10.1002/14651858.CD009361.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Drogan D, Dunn WB, Lin W, et al. Untargeted metabolic profiling identifies altered serum metabolites of type 2 diabetes mellitus in a prospective, nested case control study. Clin Chem. 2015;61:487–497. doi: 10.1373/clinchem.2014.228965. [DOI] [PubMed] [Google Scholar]
- 33.Barber MN, Risis S, Yang C, et al. Plasma lysophosphatidylcholine levels are reduced in obesity and type 2 diabetes. PLoS One. 2012;7:e41456. doi: 10.1371/journal.pone.0041456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Teslovich TM, Musunuru K, Smith AV, et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466:707–713. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu JH, Lemaitre RN, Manichaikul A, et al. Genome-wide association study identifies novel loci associated with concentrations of four plasma phospholipid fatty acids in the de novo lipogenesis pathway: results from the Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) consortium. Circ Cardiovasc Genet. 2013;6:171–183. doi: 10.1161/CIRCGENETICS.112.964619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dupuis J, Langenberg C, Prokopenko I, et al. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat Genet. 2010;42:105–116. doi: 10.1038/ng.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bickert A, Ginkel C, Kol M, et al. Functional characterization of enzymes catalyzing ceramide phosphoethanolamine biosynthesis in mice. J Lipid Res. 2015;56:821–835. doi: 10.1194/jlr.M055269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Le Stunff H, Galve-Roperh I, Peterson C, Milstien S, Spiegel S. Sphingosine-1-phosphate phosphohydrolase in regulation of sphingolipid metabolism and apoptosis. J Cell Biol. 2002;158:1039–1049. doi: 10.1083/jcb.200203123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Demirkan A, van Duijn CM, Ugocsai P, et al. Genome-wide association study identifies novel loci associated with circulating phospho- and sphingolipid concentrations. PLoS Genet. 2012;8:e1002490. doi: 10.1371/journal.pgen.1002490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cantrell Stanford J, Morris AJ, Sunkara M, Popa GJ, Larson KL, Ozcan S. Sphingosine 1-phosphate (S1P) regulates glucose-stimulated insulin secretion in pancreatic beta cells. J Biol Chem. 2012;287:13457–13464. doi: 10.1074/jbc.M111.268185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Global Lipids Genetics C. Willer CJ, Schmidt EM, et al. Discovery and refinement of loci associated with lipid levels. Nat Genet. 2013;45:1274–1283. doi: 10.1038/ng.2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Locke AE, Kahali B, Berndt SI, et al. Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;518:197–206. doi: 10.1038/nature14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shungin D, Winkler TW, Croteau-Chonka DC, et al. New genetic loci link adipose and insulin biology to body fat distribution. Nature. 2015;518:187–196. doi: 10.1038/nature14132. [DOI] [PMC free article] [PubMed] [Google Scholar]