Abstract
Reproductive traits are economically important for beef cattle production; however, these traits are still a bottleneck in indicine cattle since these animals typically reach puberty at older ages when compared to taurine breeds. In addition, reproductive traits are complex phenotypes, i.e., they are controlled by both the environment and many small-effect genes involved in different pathways. In this study, we conducted genome-wide association study (GWAS) and functional analyses to identify important genes and pathways associated with heifer rebreeding (HR) and with the number of calvings at 53 months of age (NC53) in Nellore cows. A total of 142,878 and 244,311 phenotypes for HR and NC53, respectively, and 2,925 animals genotyped with the Illumina Bovine HD panel (Illumina®, San Diego, CA, USA) were used in GWAS applying the weighted single-step GBLUP (WssGBLUP) method. Several genes associated with reproductive events were detected in the 20 most important 1Mb windows for both traits. Significant pathways for HR and NC53 were associated with lipid metabolism and immune processes, respectively. MHC class II genes, detected on chromosome 23 (window 25-26Mb) for NC53, were significantly associated with pregnancy success of Nellore cows. These genes have been proved previously to be associated with reproductive traits such as mate choice in other breeds and species. Our results suggest that genes associated with the reproductive traits HR and NC53 may be involved in embryo development in mammalian species. Furthermore, some genes associated with mate choice may affect pregnancy success in Nellore cattle.
Introduction
Heifer rebreeding (HR) is the ability of a primiparous cow to rebreed after having the first calving—is an important trait in beef cattle production. Heifers that conceive soon after the first calving permit a fast return on investments. HR has been considered as a selection criterion in some breeding programs, particularly in indicine breeds [1, 2] as cows of these breeds usually present low rebreeding rates [3]. This trait shows a high genetic correlation with other important reproductive traits such as age at first calving [4] and stayability [5].
An alternative selection criterion for HR is the number of calving at 53 mo (NC53) [6]. In addition to discriminating between cows with 2 calves and those that did not rebreed as primiparous (1 calf), NC53 also allows considering in the genetic evaluation heifers that did not get pregnant. Compared with stayability, NC53 allows more emphasis on rebreeding of primiparous cows, one of the main bottlenecks for improving efficiency of beef cows in the tropics, besides allowing anticipating the genetic evaluation of sires using information of their progeny.
Despite the economic importance of HR and NC53, the genetic improvement of these traits is challenging because they present low heritability [5, 6], are expressed relatively late in life and just in females, which present lower selection intensity compared to males. As a consequence, genetic evaluations for these traits using only pedigree and phenotypic information result in expected breeding values with low accuracy. Trying to overcome this, genome-wide association studies (GWAS) have been performed with the hope to detect genomic regions associated to reproductive traits, aiming to use this information as a complementary tool to increase genetic gain.
In general, GWAS of reproductive traits have confirmed the polygenic nature of these traits. Nevertheless, some studies have identified important candidate genes for reproductive traits of beef cattle [7, 8, 9, 10, 11]. Conducting GWAS in independent populations is important to reinforce the association of some genomic regions with the trait of interest. When a genomic region is detected as important by different studies the evidence that this region harbor a true QTL is supposedly increased. Carrying out function analyses with the candidate genes may also help to unravel pathways and key genes involved in the expression of the trait of interest. In this study, we conducted GWAS and functional analyses aiming to identify important candidate genes and pathways associated with HR and NC53 in Bos indicus Nellore cows.
Material and methods
Phenotypes
Phenotypic information of Nellore cows was obtained from the Aliança Nelore database. The cows were born from 1984 to 2010 and were raised on 188 different commercial farms located in the southeastern, western and central regions of Brazil and in Paraguay. The feeding system adopted by these farms basically consists of tropical pastures, mineral salt and water ad libitum. In the dry season, the cows usually receive mineral supplementation.
During the breeding season, which lasts about 90 days and usually occurs in the rainy period, the heifers are either artificially inseminated or naturally mated. Generally, the first mating of heifers occurs at about 26 months of age, although some herds expose the heifers earlier at around 14–18 months of age in an anticipated mating season. In the data analyzed, 46.6% (66,643) of the heifers were exposed early, corresponding to an early pregnancy rate of 21.4% (14,287). Heifers that were exposed early and did not conceive had a second chance at about 26 months of age in the regular mating season, and all heifers that did not get pregnant during this period were culled, including those that were exposed for the first time. During the breeding season, the cows were either artificially inseminated or naturally mated using single or multiple sires (4–5 sires). There were 48.08%, 47.58% and 4.35% of calves born from artificial insemination, multiple sires and single sires, respectively.
Heifer rebreeding was defined as success (1) or failure (0), i.e., heifers that calved or not, respectively, since they had produced the first calf. NC53 was defined as 0, 1 or 2 for those heifers that did not have any calf, one calf or two calves at 53 months of age, given they had the opportunity to reach this age and had performance records until long-yearling age [6].
During editing, heifers with age at first calving less than 21 or greater than 40 months, age at second calving less than 32 or greater than 53 months, and a calving interval less than 11 months were excluded. The remaining number of phenotypes after editing was 142,878 and 244,311 for HR and NC53, respectively. For HR, 45.1% of the heifers failed and 54.9% succeeded to rebreed. For NC53, 38.0%, 28.6% and 33.4% had 0, 1 or 2 calves, respectively. The pedigree file for the two traits contained 369,878 animals distributed over five generations.
Genotypes
A total of 2,925 animals were genotyped with the Illumina Bovine HD panel (Illumina®, San Diego, CA, USA). These animals included 2,212 Nellore heifers/cows from 12 different herds and 713 Nellore sires that had an average of 73.6 offspring evaluated for HR and NC53. The genotyped heifers and sires were born from 2002 to 2009 and from 1965 to 2006, respectively.
Quality control of genotypes was performed excluding single nucleotide polymorphisms (SNPs) from non-autosomal regions, SNPs mapped to the same position, and SNPs with a p-value for Hardy-Weinberg equilibrium < 10−5, a GC score < 0.15, a call rate < 0.95 and a minor allele frequency < 0.02. Samples with a call rate < 0.9 were discarded. The remaining number of SNPs and then samples after quality control was 409,376 and 2,923, respectively.
Statistical analysis
The SNP marker effects were estimated using the weighted single-step GBLUP (WssGBLUP) method proposed by Wang et al. [12]. This method was chosen because it permits to combine pedigree, phenotypic and genomic information in a single step, weighting the SNP marker effects according to the variance explained by each SNP, i.e., a higher weight is attributed to the SNP that explains a higher proportion of the genetic variance. This method is particularly appealing when there are many more phenotypes than genotypes as in the present study. Phenotypic information from non-genotyped animals allows predicting more accurately the genetic merit of genotyped animals (ag). As the SNP effect estimates from WssGBLUP are computed as a function of ag, better predictions of ag would ultimately result in better estimation of SNP effects [13]. The WssGBLUP analyses were run using the BLUPF90 family programs [14]. The method first computes the breeding values then the SNP effects, as described below.
Predicted breeding values were obtained with the following threshold animal model [15]:
where y is a vector of underlying liabilities for HR and NC53, β is a vector of fixed effects of the contemporary group, a is a vector of random additive direct genetic effects (breeding values), X and Za are incidence matrices relating elements in β and a to y, respectively, and e is the vector of random residuals. Contemporary groups were defined by concatenating the information of herd, year and season of birth, and weaning and yearling management groups of the heifers. Contemporary groups containing fewer than five heifers and without variability in HR or NC53, i.e., groups consisting of animals with the same categorical response, were excluded from the data. The underlying liabilities for HR and NC53 were defined as follows: HR = 0, if y < t1; HR = 1, if y > t1; NC53 = 0, if y < t1; NC53 = 1, if t1 < y < t2, and NC53 = 2, if y > t2, where t1 and t2 are the thresholds corresponding to the discontinuity in the observed scale of HR and NC53.
The covariance between a and e was assumed to be absent and their variances were equal to and , respectively, where and are the additive direct and residual variances, respectively, H is the matrix which combines pedigree and genomic information [16], and I is an identity matrix. Since the variable in the underlying distribution is not observable, the parameterization was adopted [17].
The parameter estimates of the threshold model were obtained under a Bayesian framework using the THRGIBBS1F90 Gibbs sampling program [18]. Default prior distributions were assumed for the variance components and for the fixed and random effects. The Gibbs sampler was run in a single chain of 500,000 iterations, with a burn-in period of 50,000 and a thinning interval of 50 iterations, totaling 9,000 posterior samples for each parameter to be estimated. The posterior means of the samples were used as the parameter estimates. The convergence of Monte Carlo chains of and heritability was evaluated using the postGibbsf90 software [14] and the Geweke test of the BOA R package [19].
After computing the breeding values, the solutions of SNP effects (û) were then obtained according to VanRaden et al. [20] and Stranden & Garrick [21]: û = DZ′[ZDZ′]−1 âg, where D is a diagonal matrix with weights for SNP effects, Z is a matrix relating genotypes of each locus, and âg is the vector of predicted breeding values of genotyped animals. The SNP effects breeding values, the D matrix and the SNP effects were iteratively recomputed over two iterations, as suggested by Wang et al. [12]. This number of iterations was chosen based on the results of a simulation study [13].
The diagonal elements of D (di) were calculated as: di = ûi2pi(1-pi), where ûi is the allele substitution effect of the ith marker estimated from the previous iteration, and pi is the allele frequency of the second allele of the ith marker [12]. Prior to recomputing û, the D matrix was normalized to enforce the total genetic variance to be constant across iterations.
Detection of significant windows and functional groups
The proportion of variance explained by SNPs within non-overlapping consecutive 1-Mb windows was evaluated for both traits. A total of 2,523 windows spanning all autosomes were considered, with an average density of 162±48 SNPs per window. S1 and S2 Tables show the top 20 significant windows for HR and NC53, respectively. Annotated genes located within these windows were further inspected. The list of genes was provided by the NCBI Map Viewer tool (www.ncbi.nlm.nih.gov/mapview/) using the Bos taurus Annotation Release 103 and Bos taurus UMD 3.1 as reference assembly.
The DAVID software [22, 23], ClueGO program [24] and Cytoscape plug-in [25] were used to group genes according to similarity of the biological processes in which they are involved, aiming to verify if these functional clusters were related to reproductive events.
Results and discussion
Heritability estimates
The Geweke test [26] indicated convergence of the chains (Table 1). The heritability was 0.19 for HR and NC53 (Table 1), in agreement with the estimates reported in other studies on the Nellore breed [2, 5, 6]. This result indicates that, in addition to being highly influenced by the environment, HR and NC53 are affected by an additive genetic component, a finding justifying the execution of GWAS.
Table 1. Estimates of marginal posterior distributions of heritability for heifer rebreeding (HR) and number of calves at 53 months of age (NC53) of Nelore cattle.
| HR | NC53 | |
|---|---|---|
| Heritabilitya | 0.194 ± 0.013 | 0.185 ± 0.008 |
| HPD 95% | 0.168–0.218 | 0.171–0.200 |
| z-scoreb | -0.576 | -0.989 |
| p-valueb | 0.565 | 0.323 |
| ESS | 149.3 | 388.1 |
HPD 95%, 95% highest posterior density interval (inferior—superior); ESS, Effective Sample Size.
aMean±s.d.
bStatistics of Geweke Test.
Significant windows
S1 and S2 Tables show the BTA and position of the top 20 1-Mb significant windows for HR and NC53, respectively, as well as the percentage of genetic additive variance explained by each window and the genes within in each window. All significant genes are described below.
In S1 Table, window 71-72Mb of BTA11 harbors the FOSL2 gene related to endometrial decidualization [27], RBKS associated with oocyte maturation [28], and BRE associated with steroidogenesis in the ovary and maintenance of placental function [29]. The subsequent window of BTA11 (72-73Mb) harbors important genes previously reported to be associated with reproductive events, such as SLC30A3 [30] and CAD [31] described as estrogen receptors, CIB4 associated with male sheep fecundity [32], EMILIN1 associated with placentation and trophoblast invasion in the uterine wall [33], TCF23 which plays an important role in endometrial decidualization [34], UCN associated with steroidogenesis and maintenance of placental function which is expressed in mature mouse spermatozoa and rat epididymis [35, 36, 37], and IFT172, PREB and SNX17 which are associated with neural development [38, 39, 40, 41]. Furthermore, window 49-50Mb of BTA17 has been reported to be associated with age at first calving in Nellore cows [13].
As shown in S2 Table, window 6-7Mb of BTA3 harbors the DDR2 gene, which has been associated with first calving and HR in Nellore cattle [10] in a study that used a subset of the present dataset. Window 39-40Mb of BTA9 harbors the AMD1 gene, which is overexpressed in bovine oocyte and cumulus cells [42]. In addition, the AMD1 protein was located in luminal and epithelial cells of bovine endometrium [43]. This window also harbors FYN associated with mouse oocyte maturation [44] and maternal-fetal immune tolerance in humans and mice [45], REV3L which contributes to genome stability during neoplastic transformation and progression in mice—targeted disruption of this gene results in lethality during midgestation [46, 47], and SLC16A10 which is differentially expressed in human placenta at mid-gestation, regulating embryonic development and growth [48]. Window 70-71Mb of BTA 17 was significant for both traits. This window harbors CHEK2 which is down-regulated in human oocytes cryopreserved by slow freezing, reducing oocyte development competence [49], and plays an important role in DNA damage repair in mouse oocyte meiosis [50], and XBP1, a regulator gene associated with endoplasmic reticulum stress, influencing oocyte maturation and embryo development [51]. In pigs, XBP1 expression promoted oocyte maturation, embryonic genome activation and early embryonic development in vitro [52]. In cattle, XBP1 is involved in corpus luteum development and maintenance [53], and is upregulated in large follicles [54]. Window 18-19Mb of BTA26 harbors FRAT1 which is differentially expressed during the secretory phases of the human endometrium [55], and PGAM1 related to spermatogenesis success in mice [56] whose expression was found to be increased in cows with postpartum infection caused by pathogenic bacteria [57].
Some genes (SLIT1, TNR, FAM181B, IFT172, PPM1G, SNX17, and PREB) associated with neural development [38, 39, 40, 41, 58, 59, 60, 61, 62, 63] were also found in the top 20 significant windows for both traits. Costa et al. [10] also detected genes related to neural development that were associated with reproductive traits. Neurulation is an important and one of the first events of embryogenesis [64]. Failure in this stage can result in a non-viable pregnancy, a fact that would explain this association. Genes associated with male fertility [32, 36, 37, 56] were also found within the top 20 windows (CIB4, UCN, and PGAM1), a finding that would explain the genetic correlation between male and female reproductive traits as reported by Irano et al. [11]. Another possible explanation is that sperm quality influences pregnancy success. Thus, these genes may be associated with andrological parameters that also improve conception rates [65].
Functional gene grouping
The top GO terms for the five functional groups with the highest enrichment score for HR and NC53 obtained by DAVID analyses are shown in Tables 2 and 3, respectively. The genes associated with HR were generally distributed in pathways related to metabolism and molecular functions. The top GO term for HR was “intracellular organelle lumen”. Functional group 2 (fatty acid metabolism) contained genes that play a role in the catabolism of short- (ACADS on BTA17 at 65-66Mb) and long-chain (HADHA and HADHB on BTA11 at 73-74Mb) fatty acids. The supply of lipophilic compounds such as fatty acids and some vitamins across the placenta influences growth and fetal fat mass formation [66]. Moreover, fatty acids are precursors of many sex hormones [67] and may influence the estrous cycle and pregnancy maintenance.
Table 2. Gene category and pathway enrichment analysis of HR genes.
| Functional groups | Top Go Term (Id~Name) | Ontology | ES | FDR |
|---|---|---|---|---|
| 1 | GO: 0070013≈ Intracellular Organelle lumen | Cellular Component | 1.17 | 5.4E1 |
| 2 | Kegg: Fatty acid metabolism | 1.08 | 2.0E1 | |
| 3 | Zinc finger region: RING-type | 0.73 | 3.5E1 | |
| 4 | GO:0005509~Calcium ion binding | Molecular Function | 0.60 | 9.4E1 |
| 5 | Microtubule | 0.55 | 8.9E1 |
ES, Enrichment Score; FDR, False Discovery Rate.
Table 3. Gene category and pathway enrichment analysis of NC53 genes.
| Functional groups | Top Go Term (Id~Name) | Ontology | ES | FDR |
|---|---|---|---|---|
| 1 | Go:0042613~MHC class II Protein complex | Cellular Component | 7.81 | 7.3E-2 |
| 2 | Kegg: Asthma | 5.10 | 1.8E-4 | |
| 3 | GO: 0000166~Nucleotide binding | Molecular Function | 0.93 | 7.0E1 |
| 4 | GO: 0046872~Metal ion binding | Molecular Function | 0.52 | 9.8E1 |
| 5 | GO: 0006915~Apoptotic process | Biological Process | 0.40 | 1.0E2 |
ES, Enrichment Score; FDR, False Discovery Rate.
Functional group 5 (microtubule) contained the DYNLL1, KIF3C and TRIM54 genes (BTA17 at 65-66Mb, BTA11 at 73–74 Mb and BTA11 at 72-73Mb, respectively). The DYNLL1 and KIF3C genes encode two motor proteins called kinesins and dyneins that regulate intracellular transport through microtubules. These genes are associated with the mitotic process and play important roles in embryo development and cell migration and differentiation [68, 69, 70, 71]. Microtubules are responsible for many intracellular movements, including chromosome separation during cell division. When a cell enters mitosis, the disassembly rate of microtubules increases about ten-fold and the number of microtubules increases by five- to ten-fold [72]. Since mitosis is also essential for embryonic growth and development [73], these microtubule genes affect the success of embryo development.
For NC53, most genes were associated with immune processes. The top GO term for this trait was “MHC class II protein complex”, which is directly associated with the immune system and pregnancy success [74]. This group of genes will be further discussed below.
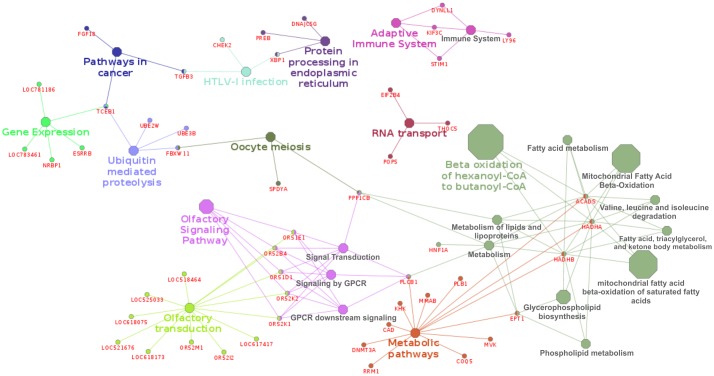
The Cytoscape software was used to cluster the genes into functional groups. For HR, 25 GO terms/groups were generated (Fig 1) and three of them were statistically oversignificant (p < 0.001), namely “beta-oxidation of hexanoyl-CoA to butanoyl-CoA” (the most significant with p = 2.77E-6), “mitochondrial fatty acid beta-oxidation of saturated fatty acids”, and “mitochondrial fatty acid beta-oxidation”. These pathways are associated with lipid metabolism, which was a significant functional group in DAVID analysis. The genes grouped in these pathways were the same as those identified by DAVID analysis (ACADS, HADHA, and HADHB).
Fig 1. Gene network of heifer rebreeding.
Each color represents a functional group that harbored related subgroups and genes.
Metabolism of fatty acids is directly associated with the pregnancy success. Females with no satisfactory corporal condition that do not receive a good nutrition supplementation after calving have higher interval from calving to estrus, decrease conception rate and produce lighter calves [75]. The significant pathways related to lipid metabolism might suggest that there are cows that accumulate more fat in comparison to others. As a consequence, they have a better rebreeding performance.
The “Olfactory signaling pathways” was a significant cluster (0.05 < p-value < 0.01). Genes clustered in this group were: OR51D1, OR51E1, OR52B4, OR52K1, OR52K2 (BTA15, at 51-52Mb). Two of these, OR51D1 and OR52K1 were found to be expressed in female and male primordial germ cells and in unfertilized human oocytes, participating in gamete production [76]. Furthermore, olfactory receptors integrate many metabolic processes and some of them play important role in reproduction [76].
“Oocyte meiosis” is also a pathway extremely related with reproductive events. Despite this pathway was not significant, it clustered three genes associated with HR: FBXW11, PPP1CB, SPDYA. The PPP1CB gene is also associated to “Olfactory” pathway. This gene plays a role in regulation of oocytes chromatin condensation in mouse [77].
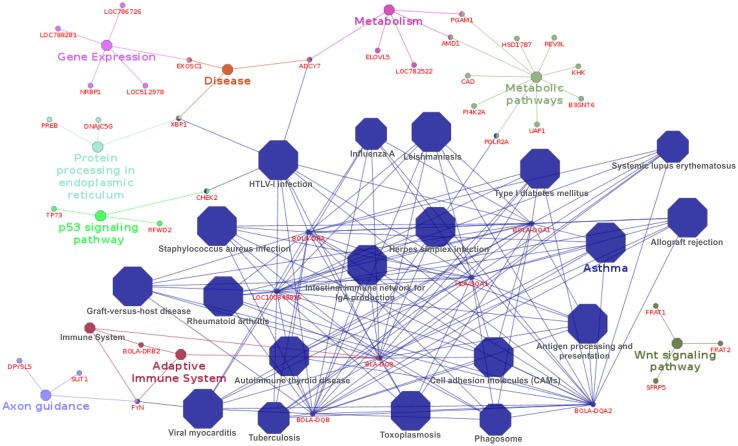
For NC53, 29 GO terms/groups were generated, 19 of them were over significant, that are presented in dark-blue in Fig 2. “Asthma” was the group that presented the higher significance level (p-value = 1.12E-9). The signalized genes from the disease pathways were the ones from the Major Histocompatibility Complex Class II (MHC II). Shortly, the MHC complex codifies cell surface glycoproteins that present the peptide antigen to the T-cells [78]. MHC class II molecules can stimulate the proliferation of some inflammatory cytokines contributing to chronic inflammation responses, as observed in some human autoimmune diseases [79,80]. It is indirectly related to the maternal-fetal tolerance and embryo development [81]. This association between MHC class II and autoimmune diseases could explain why human diseases pathways were found by Cytoscape analyses.
Fig 2. Gene network of number of calving at 53 months of age.
Each color represents a functional group that harbored related subgroups and genes.
MHC is composed of a group of highly polymorphic genes and its variability is important to enable the immune system to recognize and act against pathogens [82]. The genes listed were BLA-DQB, BOLA-DQA1, BOLA-DQA2, BOLA-DQB, BOLA-DRA, and HLA-DQA1, located on BTA23 at 25-26Mb.
In addition to the important immune function of MHC, evidence indicates that males and females choose MHC-dissimilar sexual partners [83], which works as a self/nonself perception to guarantee that their offspring will be immunogenetically viable, consequently ensuring the survival of the species. The more polymorphic the MHC, the higher the probability of fighting efficiently different diseases. Aarnink et al. [74] observed a low survival rate in a Mauritian-origin macaque population with a low level of polymorphism in MHC genes. Negative natural selection acts against offspring whose parents have similar MHC alleles. In beef cattle reproduction, the preference of some bulls for certain cows in single/multiple sire mating might be explained by the action of MHC genes.
The choice of the sexual partner occurs before copulation and is guided by dissimilarity in the MHC genes of the partners that are able to recognize MHC variability based on body odor [83]. Moreover, MHC-based mate choice continues during sperm-oocyte interaction (chemotaxis) after copulation and is mediated by olfactory receptors present on the gametes [76]. The olfactory receptors of the gametes “reflect” the MHC variability of the oocyte/sperm and guide fecundation between the genetically most distant gametes.
The association of MHC genes and olfactory receptors with NC53 allows raising some hypotheses linked to cattle production. For example, artificial insemination does not allow mate choice, suggesting that a dam could be inseminated with semen from a sire with similar MHC alleles. In this case, selection for MHC variability would only be possible during fecundation (gamete level). If MHC genes are similar, fecundation may not occur. Thus, unsuccessful rebreeding may also be explained by the similarity of MHC genes between sexual partners. The detection of some olfactory receptor genes and olfactory pathways associated with HR supports this hypothesis.
Evidently, these hypotheses need to be confirmed, but some changes in cattle handling could be proposed until the influence of MHC has been established. The replacement of sire semen after pregnancy failure and the use of natural mating for part of the females might promote higher pregnancy rates since these practices tend to better respect the natural MHC selection process. MHC genes are not the only reason for pregnancy failure, but the present results suggest that they may influence pregnancy rates and therefore deserve further attention.
Conclusion
Genes that play important roles in embryo development, lipid metabolism and sexual partner choice were associated with reproductive traits in Nellore cattle. Further studies may confirm the biological role of these genes behind the pregnancy success in mammals.
Supporting information
(DOCX)
(DOCX)
Acknowledgments
The financial support from Coordenação de Aperfeiçoamento de Pessoal de nível superior (CAPES) and Fundação de Amparo à Pesquisa (FAPESP), process n° 2014/09603-2. Also financial supports from CNPq (n° 559631/2009-0) and FAPESP (n° 2009/16118-5) are acknowledged. DeltaGen® breeding program for providing the data used in this study.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
Financial support provided by Coordenação de Aperfeiçoamento de Pessoal de nível superior (CAPES) and Fundação de Amparo à Pesquisa (FAPESP), process no 2014/09603-2 to (TPM), CNPq (no 559631/2009-0) to (LGA), and FAPESP (no 2009/16118-5) to (LGA). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Luna-Nevarez P, Rincon G, Medrano JF, Riley DG, Chase CC, Coleman SW, et al. Single nucleotide polymorphisms in the growth hormone—insulin-like growth factor axis in straightbred and crossbred Angus, Brahman, and Romosinuano heifers: Population genetic analyses and association of genotypes with reproductive phenotypes. J Anim Sci. 2011;89(4):926–34. 10.2527/jas.2010-3483 [DOI] [PubMed] [Google Scholar]
- 2.Boligon AA, de Albuquerque LG. Genetic parameters and relationships between heifers rebreeding and hip height in Nellore cattle. R. Bras. Zootec. 2012; 41(3):598–602. [Google Scholar]
- 3.Corrêa ES, Euclides Filho K, Alves RGO, Vieira A. Desempenho reprodutivo em um sistema de produção de gado de corte. Brazil, Campo Grande: EMBRAPA/CNPGC, 2001. http://www.infoteca.cnptia.embrapa.br/handle/doc/325111. [Google Scholar]
- 4.Boligon AA, Baldi F, de Albuquerque LG. Genetic correlations between heifer subsequent rebreeding and age at first calving and growth traits in Nellore cattle by Bayesian inference. Genet Mol Res. 2012; 11(4): 4516–4524. 10.4238/2012.October.17.2 [DOI] [PubMed] [Google Scholar]
- 5.Guarini AR, Neves HHR, Schenkel FS, Carvalheiro R, Oliveira JA, Queiroz SA. Genetic relationship among reproductive traits in Nellore cattle. Animal. 2014; 9(5). [DOI] [PubMed] [Google Scholar]
- 6.Neves HHR, Carvalheiro R, Queiroz SA. Genetic parameters for an alternative criterion to improve productive longevity of Nellore cows. J Anim Sci. 2012; 90:4209–4216. 10.2527/jas.2011-4766 [DOI] [PubMed] [Google Scholar]
- 7.Fortes MRS, Lehnert SA, Bolormaa S, Reich C, Fordyce G, Corbet NJ, et al. Finding genes for economically important traits: Brahman cattle puberty. Anim Prod Sci, 2012; 52: 143–150. [Google Scholar]
- 8.Haweken RJ, Zhang YD, Fortes MRS, Collis E, Barris WC, Corbet NJ, et al. Genome-wide association studies of female reproduction in tropically adapted beef cattle. J Anim Sci. 2012; 90:1398–1410. 10.2527/jas.2011-4410 [DOI] [PubMed] [Google Scholar]
- 9.Fortes MRS, Suhaimi AHMS, Porto-Neto LR, McWillian SM, Flatscher-Bader T, Moore SS, et al. Post-partum anoestrus in tropical beef cattle: a systems approach combining gene expression and genome-wide association results. Livest Sci. 2014; 166:158–166. [Google Scholar]
- 10.Costa RB, Camargo GM, Diaz ID, Irano N, Dias MM, Carvalheiro R, et al. Genome-wide association study of reproductive traits in Nellore heifers using Bayesian inference. GSE. 2015;47(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Irano N, de Camargo GMF, Costa RB, Terakado APN, Magalhães AFB, Silva RMO, et al. Genome Wide Association Study for Indicator Traits of Sexual Precocity in Nellore Cattle. PLoS ONE. 2016. August; 11(8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang H, Misztal I, Aguilar I, Legarra A, Muir WM. Genome-wide association mapping including phenotypes from relatives without genotypes. Genet Res. 2012; 94:73–83. [DOI] [PubMed] [Google Scholar]
- 13.Melo TP, Takada L, Baldi F, Oliveira HN, Dias MM, Neves HHR, et al. Assessing the value of phenotypic information from non-genotyped animals for QTL mapping of complex traits in real and simulated populations. BMC Genet. 2016; 17–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Misztal I. BLUPF90—a flexible mixed model program in Fortran 90. User Manual. Animal and Dairy Science. Athens, GA, USA: University of Georgia; 2012. http://nce.ads.uga.edu/wiki/lib/exe/fetch.php?media=blupf90.pdf document.
- 15.Gianola D, Foulley JL. Sire evaluation for ordered categorical data with a threshold model. Genet Sel Evol. 1983; 15(2):201–224. 10.1186/1297-9686-15-2-201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aguilar I, Misztal I, Johnson DL, Legarra A, Tsuruta S, Lawlor TJ. Hot topic: A unified approach to utilize phenotypic, full pedigree, and genomic information for genetic evaluation of Holstein final score. J Dairy Sci. 2010; 93:743–52. 10.3168/jds.2009-2730 [DOI] [PubMed] [Google Scholar]
- 17.Sorensen D; Gianola D. Likelihood, Bayesian, and MCMC Methods in Quantitative Genetics. New York: Springer; 2002. [Google Scholar]
- 18.Tsuruta S, Misztal I. THRGIBBS1F90 for estimation of variance component with threshold and linear models. In: World congress on genetics applied to livestock production, 2006, Belo Horizonte. Proceedings…Belo Horizonte: UFMG, 2006: 27–31.
- 19.Smith BJ. Bayesian Output Analysis Program (BOA) for MCMC version 1.1.7–2 user’s manual. [S.I.: s.n.], 2015.
- 20.Vanraden PM, Van Tassell CP, Wiggans GR, Sonstegard TS, Schnabel RD, Taylor JF, et al. Invited review: Reliability of genomic predictions for North American Holstein bulls. J Dairy Sci. 2009; 92(1):16–24. 10.3168/jds.2008-1514 [DOI] [PubMed] [Google Scholar]
- 21.Stranden I, Garrick DJ. Technical note: Derivation of equivalent computing algorithms for genomic predictions and reliabilities of animal merit. J Dairy Sci. 2009; 92(6):2971–2975. 10.3168/jds.2008-1929 [DOI] [PubMed] [Google Scholar]
- 22.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID Bioinformatics Resources. Nature Protoc. 2009a;4(1):44–57. [PubMed] [DOI] [PubMed] [Google Scholar]
- 23.Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009b;37(1):1–13. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bindea G, Mlecnik B. ClueGo, a Cytoscape plug-in to decifer biological networks v2.0.0. User’s manual, 2012.
- 25.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003; 13:2498–2504. 10.1101/gr.1239303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Geweke J. Evaluating the Accuracy of Sampling-Based Approaches to the Calculation of Posterior Moments In Bernardo JM, Berger JO, Dawid AP, Smith AFM (Eds). Bayesian statistics. 1992; 4:169–194. Oxford: Oxford University Press, Clarendon Press. [Google Scholar]
- 27.Mazur EC, Vasquez YM, Li X, Kommagani R, Jiang L, Chen R, Lanz RB, et al. Progesterone receptor transcriptome and cistrome in decidualized human endometrial stromal cells. Endocrinology. 2015;156(6):2239–53. 10.1210/en.2014-1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim Y, Kim E-Y, Seo Y-M, Yoon TK, Lee W-S, Lee K-A. Function of the pentose phosphate pathway and its key enzyme, transketolase, in the regulation of the meiotic cell cycle in oocytes. Clin Exp Reprod Med 2012;39(2):58–67. 10.5653/cerm.2012.39.2.58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miao J, Chan K-W, Chen GG, Chun S-Y, Chan JYH, et al. Blocking BRE expression in Leydig cells inhibits steroidogenesis by down-regulating 3β-hydroxysteroid dehydrogenase. J Endocrinol. 2005. June 1;185(3):507–17. 10.1677/joe.1.06079 [DOI] [PubMed] [Google Scholar]
- 30.Fong CJ, Burgoon LD, Williams KJ, Forgacs AL, Zacharewski TR. Comparative temporal and dose-dependent morphological and transcriptional uterine effects elicited by tamoxifen and ethynylestradiol in immature, ovariectomized mice. BMC Genomics. 2007; 8:151 10.1186/1471-2164-8-151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Khan S, Barhoumi R, Burghardt R, Liu S, Kim K, Safe S. Molecular Mechanism of Inhibitory Aryl Hydrocarbon Receptor-Estrogen Receptor/Sp1 Cross Talk in Breast Cancer Cells. Mol Endocrinol. 2006; 20(9):2199–2214. 10.1210/me.2006-0100 [DOI] [PubMed] [Google Scholar]
- 32.Yu Y, Zhang Y, Song X, Jin M, Guan Q, Zhang Q, et al. Alternative splicing and tissue expression of CIB4 gene in sheep testis. Anim Reprod Sci. 2010; 120: 1–9. 10.1016/j.anireprosci.2010.01.004 [DOI] [PubMed] [Google Scholar]
- 33.Spessotto P, Bulla R, Danussi C, Radillo O, Cervi M, Monami G. EMILIN1 represents a major stromal element determining human trophoblast invasion of the uterine wall. J Cell Sci. 2006; 119:4574–4584. 10.1242/jcs.03232 [DOI] [PubMed] [Google Scholar]
- 34.Kommagani R, Szwarc MM, Kovanci E, Creighton CJ, O’Malley BW, DeMayo FJ, et al. A Murine Uterine Transcriptome, Responsive to Steroid Receptor Coactivator-2, Reveals Transcription Factor 23 as Essential for Decidualization of Human Endometrial Stromal Cells. Biol Reprod. 2014; 90(4):75, 1–11. 10.1095/biolreprod.114.117531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oki Y, Sasano H. Localization and physiological roles of urocortin. Peptides. 2004;25(10):1745–9. 10.1016/j.peptides.2004.06.023 [DOI] [PubMed] [Google Scholar]
- 36.Tao J, Lin M, Sha J, Tan G, Soong TW, Li S. Separate locations of urocortin and its receptors in mouse testis: function in male reproduction and the relevant mechanisms. Cell Physiol Biochem. 2007;19(5–6):303–12. 10.1159/000100649 [DOI] [PubMed] [Google Scholar]
- 37.De Luca A, Liguori G, Squillacioti C, Paino S, Germano G, Alì S, et al. Expression of urocortin and its receptors in the rat epididymis. Reprod Biol. 2014;14(2):140–7. 10.1016/j.repbio.2014.01.007 [DOI] [PubMed] [Google Scholar]
- 38.Huangfu D, Liu A, Rakeman AS, Murcia NS, Niswander L, Anderson KV. Hedgehog signalling in the mouse requires intraflagellar transport proteins. Nature. 2003;426(6962):83–7. 10.1038/nature02061 [DOI] [PubMed] [Google Scholar]
- 39.Gorividsky M, Mukhopadhyay M, Wilsch-Braeuninger M, Phillips M, Teufel A, Kim C, et al. Intraflagellar transport protein 172 is essential for primary cilia formation and plays a vital role in patterning the mammalian brain. Dev Biol. 2009;325(1):24–32. 10.1016/j.ydbio.2008.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Clelland CLT, Craciun L, Bancroft C, Lufkin T. Mapping and Developmental Expression Analysis of the WD-Repeat Gene Preb. Genomics. 2000;63(3):391–9. 10.1006/geno.1999.6089 [DOI] [PubMed] [Google Scholar]
- 41.Sotelo P, Farfán P, Benitez ML, Bu G, Marzolo MP. Sorting nexin 17 regulates ApoER2 recycling and reelin signaling. PLoS One. 2014;9(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Regassa A, Rings F, Hoelker M, Cinar U, Tholen E, Looft C, et al. Transcriptome dynamics and molecular cross-talk between bovine oocyte and its companion cumulus cells. BMC Genomics. 2011;12(1):1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ramos Rdos S, Mesquita FS, D’Alexandri FL, Gonella-Diaza AM, Papa Pde C, Binelli M. Regulation of the polyamine metabolic pathway in the endometrium of cows during early diestrus. Mol Reprod Dev. 2014;81(7):584–94. 10.1002/mrd.22323 [DOI] [PubMed] [Google Scholar]
- 44.Luo J, McGinnis LK, Kinsey WH. Role of Fyn kinase in oocyte developmental potential. Reproduction, fertility, and development. 2010;22(6):966–76. 10.1071/RD09311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu Q, Tian FJ, Xie QZ, Zhang J, Liu L, Yang J. Fyn Plays a Pivotal Role in Fetomaternal Tolerance Through Regulation of Th17 Cells. Am J Reprod Immunol. 2016;75(5):569–79. 10.1111/aji.12498 [DOI] [PubMed] [Google Scholar]
- 46.Wittschieben JP, Reshmi SC, Gollin SM, Wood RD. Loss of DNA Polymerase ζ Causes Chromosomal Instability in Mammalian Cells. Cancer Res. 2006. January 5;66(1):134–42. 10.1158/0008-5472.CAN-05-2982 [DOI] [PubMed] [Google Scholar]
- 47.Esposito G, Godin† I, Klein U, Yaspo M-L, Cumano A, Rajewsky K. Disruption of the Rev3l-encoded catalytic subunit of polymerase ζ in mice results in early embryonic lethality. Current Biology. 2000. October 1;10(19):1221–4. [DOI] [PubMed] [Google Scholar]
- 48.Uusküla L, Männik J, Rull K, Minajeva A, Kõks S, Vaas P, et al. Mid-Gestational Gene Expression Profile in Placenta and Link to Pregnancy Complications. PLoS ONE. 2012. November 7;7(11):e49248 10.1371/journal.pone.0049248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Monzo C, Haouzi D, Roman K, Assou S, Dechaud H, Hamamah S. Slow freezing and vitrification differentially modify the gene expression profile of human metaphase II oocytes. Human Reproduction. 2012. July 1;27(7):2160–8. 10.1093/humrep/des153 [DOI] [PubMed] [Google Scholar]
- 50.Bolcun-Filas E, Rinaldi VD, White ME, Schimenti JC. Reversal of female infertility by Chk2 ablation reveals the oocyte DNA damage checkpoint pathway. Science (New York, NY). 2014. January 31;343(6170):533–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Michalak M, Gye MC. Endoplasmic reticulum stress in periimplantation embryos. Clinical and Experimental Reproductive Medicine. 2015. March;42(1):1–7. 10.5653/cerm.2015.42.1.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang JY, Diao YF, Oqani RK, Han RX, Jin DI. Effect of Endoplasmic Reticulum Stress on Porcine Oocyte Maturation and Parthenogenetic Embryonic Development In Vitro. Biology of Reproduction. 2012. April 1;86(4):128 10.1095/biolreprod.111.095059 [DOI] [PubMed] [Google Scholar]
- 53.Park H-J, Park S-J, Koo D-B, Kong I-K, Kim MK, Kim J-M, et al. Unfolding protein response signaling is involved in development, maintenance, and regression of the corpus luteum during the bovine estrous cycle. Biochem. Biophys. Res. Commun. 2013. November 15;441(2):344–50. 10.1016/j.bbrc.2013.10.056 [DOI] [PubMed] [Google Scholar]
- 54.Hatzirodos N, Irving-Rodgers HF, Hummitzsch K, Harland ML, Morris SE, Rodgers RJ. Transcriptome profiling of granulosa cells of bovine ovarian follicles during growth from small to large antral sizes. BMC Genomics. 2014;15(1):1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.DeSouza L, Diehl G, Yang EC, Guo J, Rodrigues MJ, Romaschin AD, et al. Proteomic analysis of the proliferative and secretory phases of the human endometrium: protein identification and differential protein expression. Proteomics. 2005;5(1):270–81. 10.1002/pmic.200400920 [DOI] [PubMed] [Google Scholar]
- 56.Zhang S, Zhao Y, Lei B, Li C, Mao X. PGAM1 is Involved in Spermatogenic Dysfunction and Affects Cell Proliferation, Apoptosis, and Migration. Reprod Sci. 2015. October 1;22(10):1236–42. 10.1177/1933719115572485 [DOI] [PubMed] [Google Scholar]
- 57.Ledgard AM, Smolenski GA, Henderson H, Lee RS. Influence of pathogenic bacteria species present in the postpartum bovine uterus on proteome profiles. Reprod Fertil Dev. 2015;27(2):395–406. 10.1071/RD13144 [DOI] [PubMed] [Google Scholar]
- 58.Dickinson RE, Hryhorskyj L, Tremewan H, Hogg K, Thomson AA, McNeilly AS, et al. Involvement of the SLIT/ROBO pathway in follicle development in the fetal ovary. Reproduction (Cambridge, England). 2010. February;139(2):395–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yeh ML, Gonda Y, Mommersteeg MTM, Barber M, Ypsilanti AR, Hanashima C, et al. Robo1 Modulates Proliferation and Neurogenesis in the Developing Neocortex. J Neurosci. 2014. April; 34(16):5717–5731. 10.1523/JNEUROSCI.4256-13.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Meganathan K, Jagtap S, Srinivasan SP, Wagh V, Hescheler J, Hengstler J, et al. Neuronal developmental gene and miRNA signatures induced by histone deacetylase inhibitors in human embryonic stem cells. Cell Death Dis. 2015. May 7; 6: e1756 10.1038/cddis.2015.121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xu J-C, Xiao M-F, Jakovcevski I, Sivukhina E, Hargus G, Cui Y-F, et al. The extracellular matrix glycoprotein tenascin-R regulates neurogenesis during development and in the adult dentate gyrus of mice. J Cell Sci. 2014. January 30;127(3):641. [DOI] [PubMed] [Google Scholar]
- 62.Marks M, Pennimpede T, Lange L, Grote P, Herrmann BG, Wittler L. Analysis of the Fam181 gene family during mouse development reveals distinct strain-specific expression patterns, suggesting a role in nervous system development and function. 2015;575(2 Pt 2):438–51. [DOI] [PubMed] [Google Scholar]
- 63.Foster WH, Langenbacher A, Gao C, Chen J, Wang Y. Nuclear phosphatase PPM1G in cellular survival and neural development. Dev Dyn. 2013. September;242(9):1101–9. 10.1002/dvdy.23990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hyttel P, Sinowatz F, Vejlsted M, Betteridge K. Essentials of Domestic Animal Embryology. 1st ed China: Elsevier Limited; 2009. [Google Scholar]
- 65.Fortes MR, Kemper K, Sasazaki S, Reverter A, Pryce JE, Barendse W, et al. Evidence for pleiotropism and recent selection in the PLAG1 region in Australian Beef cattle. Anim Genet. 2013;44(6):636–47. 10.1111/age.12075 [DOI] [PubMed] [Google Scholar]
- 66.Herrera E, Ortega-Senovilla H. Lipid Metabolism During Pregnancy and its Implications for Fetal Growth. Curr Pharm Biotechnol. 2014;15(1):24–31. [DOI] [PubMed] [Google Scholar]
- 67.Nelson DL, Cox MM. Princípios de Bioquímica de Lehninger. 6th ed Porto Alegre: Artmed; 2014. 1328 p. [Google Scholar]
- 68.Dunsch AK, Hammond D, Lloyd J, Schermelleh L, Gruneberg U, Barr FA. Dynein light chain 1 and a spindle-associated adaptor promote dynein asymmetry and spindle orientation. J Cell Bio. 2012. September 17;198(6):1039–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zaytseva O, Tenis N, Mitchell N, Kanno S, Yasui A, Heierhorst J, et al. The Novel Zinc Finger Protein dASCIZ Regulates Mitosis in Drosophila via an Essential Role in Dynein Light-Chain Expression. Genetics. 2014. February;196(2):443–53. 10.1534/genetics.113.159541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Navone F, Consalez GG, Sardella M, Caspani E, Pozzoli O, Frassoni C, et al. Expression of KIF3C kinesin during neural development and in vitro neuronal differentiation. J Neurochem. 2001;77(3):741–53. [DOI] [PubMed] [Google Scholar]
- 71.Spencer JA, Eliazer S, Ilaria RL, Richardson JA, Olson EN. Regulation of Microtubule Dynamics and Myogenic Differentiation by Murf, a Striated Muscle Ring-Finger Protein. J Cell Biol. 2000. August 21;150(4):771–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Carvalho HF, Recco-Pimentel SM. A célula. 2nd ed Barueri, SP: Manole; 2007. [Google Scholar]
- 73.McGeady TA, Quinn PJ, Fitzpatrick ES, Ryan MT, Cahalan S. Veterinary Embriology. 1st ed Victoria, Australia: Wiley-Blackwell; 2006. [Google Scholar]
- 74.Aarnink A, Mee ET, Savy N, Congy-Jolivet N, Rose NJ, Blancher A. Deleterious impact of feto-maternal MHC compatibility on the success of pregnancy in a macaque model. Immunogenetics. 2014;66(2):105–13. 10.1007/s00251-013-0752-3 [DOI] [PubMed] [Google Scholar]
- 75.Ciccioli NH, Wettemann RP, Spicer LJ, Lents CA, White FJ, Keisler DH. Influence of body condition at calving and postpartum nutrition on endocrine function and reproductive performance of primiparous beef cows. J Anim Sci. 2003;81(12):3107–20. [DOI] [PubMed] [Google Scholar]
- 76.Diedrichs F, Mlody B, Matz P, Fuchs H, Chavez L, Drews K, et al. Comparative molecular portraits of human unfertilized oocytes and primordial germ cells at 10 weeks of gestation. Int J Dev Biol. 2012;56(10–12):789–97. 10.1387/ijdb.120230ja [DOI] [PubMed] [Google Scholar]
- 77.Swain JE, Ding J, Brautigan DL, Villa-Moruzzi E, Smith GD. Proper Chromatin Condensation and Maintenance of Histone H3 Phosphorylation During Mouse Oocyte Meiosis Requires Protein Phosphatase Activity. Biology of Reproduction. 2007. April 1;76(4):628–38. 10.1095/biolreprod.106.055798 [DOI] [PubMed] [Google Scholar]
- 78.Janeway CA, Travers P, Walport M, Shlomchik MJ. Imunobiologia: O sistema imune na saúde e na doença. 6th ed Porto Alegre: Artmed; 2007. [Google Scholar]
- 79.Abbas AK, Lichtman AHH, Pillai S. Cellular and Molecular Immunology. 8th ed Philadelphia, PA: Elsevier; 2015. [Google Scholar]
- 80.Liu Y, Yin H, Zhao M, Lu Q. TLR2 and TLR4 in Autoimmune Diseases: a comprehensive Review. Clin Rev Allergy Immunol. 2014;47(2):136–47. 10.1007/s12016-013-8402-y [DOI] [PubMed] [Google Scholar]
- 81.Fu B, Li X, Sun R, Tong X, Ling B, Tian Z, et al. Natural killer cells promote immune tolerance by regulating inflammatory T(H)17 cells at the human maternal—fetal interface. Proceedings of the National Academy of Sciences of the United States of America. 2013. January 15;110(3): E231–40. 10.1073/pnas.1206322110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nicholas FW. Introduction to Veterinary Genetics. New York: Oxford University Press; 1996. [Google Scholar]
- 83.Ziegler A, Santos PSC, Kellermann T, Uchanska-Ziegler B. Self/nonself perception, reproduction and the extended MHC. Self Nonself. 2010;1(3):176–9. 10.4161/self.1.3.12736 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.