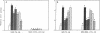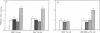Abstract
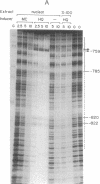
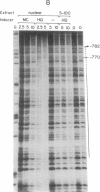
Glutathione S-transferase (GST) Ya subunit gene expression is induced in mammalian tissues by two types of chemical agents: (i) planar aromatic compounds (e.g., 3-methylcholanthrene, beta-naphthoflavone, and 2,3,7,8-tetrachlorodibenzo-p- dioxin) and (ii) electrophiles (e.g., trans-4-phenyl-3-buten-2-one and dimethyl fumarate) or compounds easily oxidized to electrophiles (e.g., tert-butylhydroquinone). To study the mechanism of this induction, we have introduced deletions in the 5' flanking region of a mouse GST Ya subunit gene, fused it to the coding sequence for chloramphenicol acetyltransferase (CAT) activity, and transfected the Ya-CAT genes for expression into hepatoma cells. We show that a single cis-regulatory element, between nucleotides -754 and -713 from the start of transcription, is responsible for the induction by both planar aromatic and electrophilic compounds. Using murine hepatoma cell mutants defective in either the Ah-encoded aryl hydrocarbon receptor (BPrc1 mutant) or in cytochrome P1-450 gene (c1 mutant), we show that induction by planar aromatic but not by electrophilic inducers requires a functional Ah receptor and cytochrome P1-450 activity. From this it is concluded that Ya gene activation by planar aromatic compounds involves metabolism of these inducers by the phase I xenobiotic-metabolizing cytochrome P1-450 system into electrophilic compounds, which is consistent with a recently proposed model [Prochaska, H. J. & Talalay, P. (1988) Cancer Res. 48, 4776-4782]. Therefore, the regulatory sequence of the Ya gene should be considered an electrophile-responsive element (EpRE) activated exclusively by inducers containing an electrophilic center. An EpRE-containing 41-bp oligonucleotide ligated at the -187 site of the Ya gene promoter confers upon it an increase in basal activity and xenobiotic inducibility. The basal activity augments with the number of EpRE copies. DNase I protection patterns show the protection of the EpRE domain by a nuclear factor(s) that becomes more abundant upon exposure of Hepa 1c1c7 cells to tert-butylhydroquinone.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Borgmeyer U., Nowock J., Sippel A. E. The TGGCA-binding protein: a eukaryotic nuclear protein recognizing a symmetrical sequence on double-stranded linear DNA. Nucleic Acids Res. 1984 May 25;12(10):4295–4311. doi: 10.1093/nar/12.10.4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel V., Sharon R., Bensimon A. Regulatory elements controlling the basal and drug-inducible expression of glutathione S-transferase Ya subunit gene. DNA. 1989 Jul-Aug;8(6):399–408. doi: 10.1089/dna.1.1989.8.399. [DOI] [PubMed] [Google Scholar]
- Daniel V., Sharon R., Tichauer Y., Sarid S. Mouse glutathione S-transferase Ya subunit: gene structure and sequence. DNA. 1987 Aug;6(4):317–324. doi: 10.1089/dna.1987.6.317. [DOI] [PubMed] [Google Scholar]
- Daniel V., Tichauer Y., Sharon R. 5'-flanking sequence of mouse glutathione S-transferase Ya gene. Nucleic Acids Res. 1988 Jan 11;16(1):351–351. doi: 10.1093/nar/16.1.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denison M. S., Fisher J. M., Whitlock J. P., Jr Protein-DNA interactions at recognition sites for the dioxin-Ah receptor complex. J Biol Chem. 1989 Oct 5;264(28):16478–16482. [PubMed] [Google Scholar]
- Dignam J. D., Lebovitz R. M., Roeder R. G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983 Mar 11;11(5):1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujisawa-Sehara A., Yamane M., Fujii-Kuriyama Y. A DNA-binding factor specific for xenobiotic responsive elements of P-450c gene exists as a cryptic form in cytoplasm: its possible translocation to nucleus. Proc Natl Acad Sci U S A. 1988 Aug;85(16):5859–5863. doi: 10.1073/pnas.85.16.5859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Israel D. I., Whitlock J. P., Jr Regulation of cytochrome P1-450 gene transcription by 2,3,7, 8-tetrachlorodibenzo-p-dioxin in wild type and variant mouse hepatoma cells. J Biol Chem. 1984 May 10;259(9):5400–5402. [PubMed] [Google Scholar]
- Kimura S., Smith H. H., Hankinson O., Nebert D. W. Analysis of two benzo[a]pyrene-resistant mutants of the mouse hepatoma Hepa-1 P(1)450 gene via cDNA expression in yeast. EMBO J. 1987 Jul;6(7):1929–1933. doi: 10.1002/j.1460-2075.1987.tb02453.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannervik B., Danielson U. H. Glutathione transferases--structure and catalytic activity. CRC Crit Rev Biochem. 1988;23(3):283–337. doi: 10.3109/10409238809088226. [DOI] [PubMed] [Google Scholar]
- Miller A. G., Israel D., Whitlock J. P., Jr Biochemical and genetic analysis of variant mouse hepatoma cells defective in the induction of benzo(a)pyrene-metabolizing enzyme activity. J Biol Chem. 1983 Mar 25;258(6):3523–3527. [PubMed] [Google Scholar]
- Nebert D. W., Jones J. E. Regulation of the mammalian cytochrome P1-450 (CYP1A1) gene. Int J Biochem. 1989;21(3):243–252. doi: 10.1016/0020-711x(89)90182-1. [DOI] [PubMed] [Google Scholar]
- Pickett C. B., Lu A. Y. Glutathione S-transferases: gene structure, regulation, and biological function. Annu Rev Biochem. 1989;58:743–764. doi: 10.1146/annurev.bi.58.070189.003523. [DOI] [PubMed] [Google Scholar]
- Poland A., Knutson J. C. 2,3,7,8-tetrachlorodibenzo-p-dioxin and related halogenated aromatic hydrocarbons: examination of the mechanism of toxicity. Annu Rev Pharmacol Toxicol. 1982;22:517–554. doi: 10.1146/annurev.pa.22.040182.002505. [DOI] [PubMed] [Google Scholar]
- Prochaska H. J., De Long M. J., Talalay P. On the mechanisms of induction of cancer-protective enzymes: a unifying proposal. Proc Natl Acad Sci U S A. 1985 Dec;82(23):8232–8236. doi: 10.1073/pnas.82.23.8232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prochaska H. J., Talalay P. Regulatory mechanisms of monofunctional and bifunctional anticarcinogenic enzyme inducers in murine liver. Cancer Res. 1988 Sep 1;48(17):4776–4782. [PubMed] [Google Scholar]
- Talalay P., De Long M. J., Prochaska H. J. Identification of a common chemical signal regulating the induction of enzymes that protect against chemical carcinogenesis. Proc Natl Acad Sci U S A. 1988 Nov;85(21):8261–8265. doi: 10.1073/pnas.85.21.8261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams R. T. Comparative patterns of drug metabolism. Fed Proc. 1967 Jul-Aug;26(4):1029–1039. [PubMed] [Google Scholar]