Abstract
A kernel-learning based method is proposed to integrate multimodal imaging and genetic data for Alzheimer’s disease (AD) diagnosis. To facilitate structured feature learning in kernel space, we represent each feature with a kernel and then group kernels according to modalities. In view of the highly redundant features within each modality and also the complementary information across modalities, we introduce a novel structured sparsity regularizer for feature selection and fusion, which is different from conventional lasso and group lasso based methods. Specifically, we enforce a penalty on kernel weights to simultaneously select features sparsely within each modality and densely combine different modalities. We have evaluated the proposed method using magnetic resonance imaging (MRI) and positron emission tomography (PET), and single-nucleotide polymorphism (SNP) data of subjects from Alzheimer’s Disease Neuroimaging Initiative (ADNI) database. The effectiveness of our method is demonstrated by both the clearly improved prediction accuracy and the discovered brain regions and SNPs relevant to AD.
1 Introduction
Alzheimer’s disease (AD) is an irreversible and progressive brain disorder. Early prediction of the disease using multimodal neuroimaging data has yielded important insights into the progression patterns of AD [11,16,18]. Among the many risk factors for AD, genetic variation has been identified as an important one [11,17]. Therefore, it is important and beneficial to build prediction models by leveraging both imaging and genetic data, e.g., magnetic resonance imaging (MRI) and positron emission tomography (PET), and single-nucleotide polymorphisms (SNPs). However, it is a challenging task due to the multimodal nature of the data, limited observations, and highly-redundant high-dimensional data.
Multiple kernel learning (MKL) provides an elegant framework to learn an optimally combined kernel representation for heterogeneous data [4,5,10]. When it is applied to the classification problem with multimodal data, data of each modality are usually represented using a base kernel [3,8,12]. The selection of certain sparse regularization methods such as lasso (ℓ1 norm) [13] and group lasso (ℓ2,1 norm) [15], yields different modality selection approaches [3, 8, 12]. In particular, ℓ1-MKL [10] is able to sparsely select the most discriminative modalities. With grouped kernels, group lasso performs sparse group selection, while densely combining kernels within groups. In [8], the group lasso regularized MKL was employed to select the most relevant modalities. In [12], a class of generalized group lasso with the focus on inter-group sparsity was introduced into MKL for channel selection on EEG data, where groups correspond to channels.
In view of the unique and complementary information contained in different modalities, all of them are expected to be utilized for AD prediction. Moreover, compared with modality-wise analysis and then conducting relevant modality selection, integration of feature-level and modality-level analysis is more favorable. However, for some modalities, their features as a whole or individual are weaker than those in other modalities. In these scenarios, as shown in Fig. 1(b), the lasso and group lasso tend to independently select the most discriminative features/groups, making features from weak modalities having less chance to be selected. Moreover, they are less effective to utilize complementary information among modalities with ℓ1 norm penalty [5, 7]. To address these issues, we propose to jointly learn a better integration of multiple modalities and select subsets of discriminative features simultaneously from all the modalities.
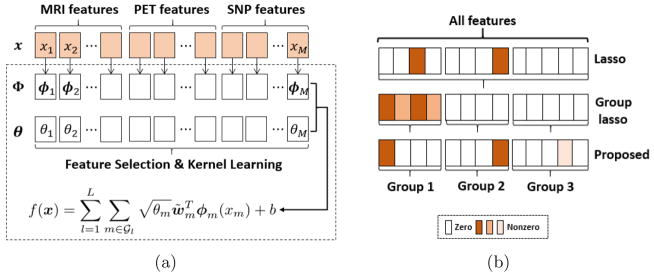
Fig. 1.
Schematic illustration of our proposed framework (a), and different sparsity patterns (b) produced by lasso (ℓ1 norm), group lasso (ℓ2,1 norm) and the proposed structured sparsity (ℓ1,p norm, p > 1). Darker color in (b) indicates larger weights.
Accordingly, we propose a novel structured sparsity (i.e., ℓ1,p norm with p > 1) regularized MKL for heterogeneous multimodal data integration. It is noteworthy that ℓ1,2 norm was considered [6, 7] in settings such as regression, multitask learning etc. Here, we go beyond these studies by considering the ℓ1,p constrained MKL for multimodal feature selection and fusion and its application for AD diagnosis. Moreover, contrary to representing each modality with a single kernel as in conventional MKL based methods [3,4,8], we assign each feature with a kernel and then group kernels according to modalities to facilitate both feature- and group-level analysis. Specifically, we promote sparsity inside groups with inner ℓ1 norm and pursue dense combination of groups with outer nonsparse ℓp norm. Guided by the learning of modality-level dense combination, sparse feature selections in different modalities interact with each other for a better overall performance. This ℓ1,p regularizer is completely different from group lasso [15] and its generalization [9] (i.e., ℓp,1 norm) which gives sparse groups but performs no feature selection within each group [12,15]. An illustration of different sparsity patterns selected by lasso, group lasso and the proposed method is shown in Fig. 1(b). In comparison, the proposed model can not only keep information from each modality with outer nonsparse regularization but also support variable interpretability and scalability with the inner sparse feature selection.
2 Method
Given a set of N labeled data samples , where , M is the number of all features in all modalities, and yi ∈ {1, −1} is a class label. MKL aims to learn an optimal combination of base kernels, while each kernel describes a different property of the data. To also perform the task of joint feature selection, we assign each feature a base kernel through its own feature mapping. An overview of the proposed framework is illustrated in Fig. 1(a).
2.1 Structured Sparse Feature and Kernel Learning
Let 𝒢 = {1, 2, ···, M } be the feature index set which is partitioned into L non-overlapping groups according to task-specific knowledge. For instance, in our application, we partition 𝒢 into L = 3 groups according to modalities. Let be the M base kernels for the M features respectively, which are induced by M feature mappings . Given the feature space defined by the joint feature mapping Φ(x) = (ϕ1(x1), ϕ2(x2), ···, ϕM (xM))T, we learn a linear discriminant function of the form . Here, we have explicitly written out the group structure in the function f (x), in which w̃m is the normal vector corresponding to ϕm, b encodes the bias, and θ = (θ1, θ2, ···, θM)T contains the weights for the M feature mappings. Therefore, feature mappings with zero weights would not be active in f (x).
In the following, we perform feature selection by enforcing a structured sparsity on weights of the feature mappings. To introduce a more general model, we further introduce (1) M positive weights β = (β1, β2, ···, βM)T for features and (2) L positive weights γ = (γ1, γ2, ···, γL)T for feature groups to encode prior information. If we have no knowledge about group/feature importance, we can set βm = 1 and γl = 1 for each l and m. Accordingly, our generalized MKL model with a structured sparsity inducing constraint can be formulated as below:
| (1) |
where ℒ(t, y) = max(0, 1 − ty) is the hinge loss function, C′ is a trade-off weight, τ controls the sparsity level, and 0 is a vector of all zeros. Similar to the typical MKL [10], this model is equivalent to learning an optimally combined kernel . The inequality constraint employs a weighted ℓ1,p mixed norm (p > 1), i.e., ||·||1,p;β,γ, which simultaneously promotes sparsity inside groups with the inner weighted ℓ1 norm and pursues dense combination of groups with the outer weighted ℓp norm.
The rationale of using this regularization is that, while each individual modality contains redundant high-dimensional features, different modalities can offer unique and complementary information. Owing to the heterogeneity of different modalities, we sparsely select features from each homogenous feature groups, i.e., modalities, and densely integrate different modalities. As has been discussed in [5], with p > 1, the non-sparse ℓp norm has the advantage of better combining complementary features than ℓ1 norm. Moreover, in view of the unequal reliability of different modalities, we take a compromise of ℓ1 lasso and ℓ2 ridge regularization and intuitively set p = 1.5 for inter-group regularization, i.e. ℓ1,1.5. More specifically, due to the geometrical property of the ℓ1.5 contour lines, it results in unequal shrinkage of weights with higher probability than ℓ2 norm, thus allowing the assignment of larger weights for leading groups/modalities.
Further understanding and computation of our model can be achieved with the following lemma and theorem. Let , w = (w1, w2, ···, wM) and also W = (||w1||2, ||w2||2, ···, ||wM ||2)T, we first have the following lemma.
Lemma 1
Given p ≥ 1, positive weights γ and β. We use the convention that 0/0 = 0. For fixed w ≠ 0, the minimal θ in Eq. (1) is attained at
| (2) |
where , and 𝒢lm is the index set containing m
For the fixed w, this lemma gives an explicit solution for θ. The proof can be done by deriving the first order optimality conditions of Eq. (1). Plugging Eq. (2) into the model in Eq. (1) yields the following compact optimization problem.
Theorem 1
Let . For p > 1, the model in Eq. (1) is equivalent to
| (3) |
The first term is a weighted ℓ1,q norm penalty on W with q ∈ (1, 2). By choosing p = 1.5 and thus q = 1.2, it shares similar group-level regularization property with that in Eq. (1) on θ. Specifically, in each group, only a small number of wm can contribute to the decision function f (x) with nonzero values. Accordingly, few features in each group can be selected. Meanwhile, the sparsely filtered groups are densely combined, while allowing the presence of leading groups.
2.2 Model Computation
After the variable changing, we can optimize the proposed model via a block coordinate descent. For fixed θ, the subproblem of w and b can be computed with any support vector machine (SVM) [2] solver. According to Lemma 1, we can analytically carry out θm with w fixed. θm can be initialized as to satisfy the constraint in Eq. (1). Moreover, from Eq. (3), it is obvious that we can fold τ and C′ into a single trade-off weight C and set τ = 1. In this way, we have single model parameter C which not only acts as the soft margin parameter but also controls the sparsity of θ and W.
3 Experimental Results
3.1 Dataset
We evaluated our method by applying it on a subset of the Alzheimer’s Disease Neuroimaging Initiative (ADNI) dataset1. In total, we used MRI, PET, and SNP data of 189 subjects, including 49 patients with AD, 93 patients with Mild Cognitive Impairment (MCI), and 47 Normal Controls (NC). After preprocessing, the MRI and PET images were segmented into 93 regions-of-interest (ROIs). The gray matter volumes of these ROIs in MRI and the average intensity of each ROI in PET were calculated as features. The SNPs [11] were genotyped using the Human 610-Quad BeadChip. Among all SNPs, only SNPs, belonging to the top AD candidate genes listed on the AlzGene database2 as of June 10, 2010, were selected after the standard quality control and imputation steps. The Illumina annotation information based on the Genome build 36.2 was used to select a subset of SNPs, belonging or proximal to the top 135 AD candidate genes. The above procedure yielded 5677 SNPs from 135 genes. Thus, we totally have 93 + 93 + 5677 = 5863 features from the three modalities for each subject.
3.2 Experimental Settings
For method evaluation, we used the strategy of 10 times repeated 10-fold cross-validation. All parameters were learned by conducting 5-fold inner cross-validation. Three measures including classification accuracy (ACC), sensitivity (SEN), and specificity (SPE) were used. We compared the proposed method with (1) feature selection based methods, i.e., Fisher Score (FS) [2], and Lasso [13], and (2) MKL based methods, i.e., the method of Zhang et al. in [16], and ℓ1-MKL [10]. In the Lasso method, the logistic loss [2] was used. The method in [16] represented each modality with a base kernel and further learned a linearly-combined kernel with cross validation. For FS, Lasso and the method in [16], the linear SVM implemented in LibSVM software3 was used as the classifier. For all methods, we used t-test [2] thresholded by p-value as a feature pre-selection step to reduce feature size and improve computational efficiency. The commonly used p-value < 0.05 was applied for MRI and PET. Considering the large number of SNP features, we selected the p-value from {0.05, 0.02, 0.01}. Therefore, t-test-SVM that combined t-test and SVM was designed for comparison with the same p-value setting as well. For our proposed model, ℓ1-MKL and Zhang’s method, to avoid further kernel parameter selection, each kernel matrix was defined as a linear kernel on a single feature. Furthermore, we simply assumed no knowledge on both feature and group weights and thus we set γ = 1 and β = 1. The soft margin parameter C was selected with grid search from {2−5, 2−4, ···, 25}.
3.3 Results and Discussions
The classification results of AD vs. NC and MCI vs. NC using all the three modalities are listed in Table 1. By taking advantage of the structured feature learning in kernel space, the proposed method outperforms all competing methods in classification rate. For AD vs. NC classification, our method achieves an ACC of 96.1 % with an improvement of 2.1 % over the best performance of other methods. Meanwhile, the standard variance of the proposed method is also lower, demonstrating the stability of the proposed method. For classifying MCI from NC, the improvements by the proposed method is 2.4 % in terms of ACC. In comparison with t-test-SVM, we obtained 4.2 % and 7.6 % improvements in terms of ACC for classifying AD and MCI from NC, respectively. Similar results are obtained for the classification of AD and MCI, which has not listed in Table 1 due to space limit. For example, the ACC of Lasso-SVM, ℓ1-MKL and our method are 70.3 ± 1.5 %, 73.0 ± 1.6 %, and 76.9 ± 1.4 %, respectively. In summary, these results show the improved classification performance by our method.
Table 1.
Performance comparison of different methods in terms of “mean ± standard deviation” for AD vs. NC and MCI vs. NC classifications, using MRI, PET and SNPs. The superscript “*” indicates statistically significant difference (p-value < 0.05) compared with the proposed method
| Methods | AD vs. NC (%) | MCI vs. NC (%) | ||||
|---|---|---|---|---|---|---|
| ACC | SEN | SPE | ACC | SEN | SPE | |
| t-test-SVM | 91.9 ± 1.9* | 92.7 ± 2.0 | 91.1 ± 3.0 | 72.7 ± 2.1* | 85.4 ± 2.9 | 47.7 ± 5.2 |
| FS-SVM | 92.4 ± 1.3* | 93.5 ± 2.7 | 91.3 ± 1.9 | 76.1 ± 1.4* | 84.3 ± 2.2 | 59.8 ± 3.1 |
| Zhang et al. | 92.6 ± 1.4* | 92.7 ± 1.4 | 92.6 ± 2.7 | 75.1 ± 2.2* | 82.8 ± 3.0 | 59.8 ± 2.9 |
| Lasso-SVM | 93.5 ± 1.4* | 94.9 ± 1.7 | 92.1 ± 1.8 | 76.3 ± 2.3* | 85.2 ± 2.3 | 58.7 ± 4.3 |
| ℓ1-MKL | 94.0 ± 1.4* | 94.3 ± 2.5 | 93.6 ± 2.0 | 77.9 ± 1.4* | 85.7 ± 1.4 | 62.6 ± 4.0 |
| Proposed | 96.1 ± 1.0 | 97.3 ± 1.0 | 94.9 ± 1.8 | 80.3 ± 1.6 | 85.6 ± 1.9 | 69.8 ± 3.7 |
To further investigate the benefit of SNP data and multimodality fusion, in Table 2 we illustrate the performance of the proposed method w.r.t different modality combinations. First of all, the performance of any single modality is much lower than that of their combinations. Among the three modalities, the SNP data shows the lowest performance. However, when combined with other modalities, genetic data can obviously help improve predictions. For example, in AD and NC classification, the performances using MRI+SNP and PET+SNP demonstrate 2.7 % and 5.7 % improvements in terms of ACC over the cases of only using MRI and PET, respectively; the improvement with MRI+PET+SNP over that with MRI+PET is 3.8 %. Similar results are obtained for MCI vs. NC.
Table 2.
Comparison of our proposed method in the cases of using different modality combinations. “*” indicates statistically significant difference with MRI+PET+SNP
| Modalities | AD vs. NC (%) | MCI vs. NC (%) | ||||
|---|---|---|---|---|---|---|
| ACC | SEN | SPE | ACC | SEN | SPE | |
| MRI | 88.4* | 84.1 | 93.0 | 71.6 | 83.9 | 47.2 |
| PET | 86.3* | 84.5 | 88.1 | 68.8 | 85.5 | 35.7 |
| SNP | 76.0* | 69.8 | 82.6 | 66.2 | 75.4 | 48.1 |
| MRI+PET | 92.3* | 91.9 | 91.7 | 76.4 | 83.9 | 61.5 |
| MRI+SNP | 91.1* | 89.8 | 92.6 | 74.9 | 84.5 | 55.7 |
| PET+SNP | 92.0* | 90.8 | 93.2 | 71.3 | 81.4 | 51.3 |
| MRI+PET+SNP | 96.1 | 97.3 | 94.9 | 80.3 | 85.6 | 69.8 |
The most selected brain regions and SNPs in our algorithm can also be the potential biomarkers used in clinical diagnosis. In MRI, hippocampal formation and uncus in parahippocampal gyrus are recognized in both AD vs. NC and MCI vs. NC classifications, as well as multiple temporal gyrus regions. This is in line with the findings of the most affected regions in AD in previous neuro-studies [3,8,16,18]. Amygdala, one of the subcortical regions, is the integrative center for emotions, is also identified as AD. In PET, angular gyri, precuneus, and entorhinal cortices are the regions identified, which are also among the altered regions in AD reported in prior studies [16,18]. As to the genetic information, the most selected SNPs for AD and NC classification are from APOE gene, VEGFA gene, and SORCS1 gene. For MCI prediction, the most selected SNPs are from KCNMA1 gene, APOE gene, VEGFA gene and CTNNA3 gene. Generally, our results are consistent with the existing results [11,17]. For instance, APOE and SORCS1 genes are the well-known top candidate genes related to AD and MCI [11]. VEGFA, the expression of vascular endothelial growth factor, represents a potential mechanism where vascular and AD pathologies are related [1].
4 Conclusion
We developed a kernel-based multimodal feature selection and integration method, and further applied it on imaging and genetic data for AD diagnosis. Instead of independently selecting features from each modality and then combining them together [16] or performing most relevant modality selection [8,14], we integrated the multimodal feature selection and combination in a novel structured sparsity regularized kernel learning framework. A block coordinate descent algorithm was derived to solve our general ℓ1,p (p ≥ 1) constrained non-smooth objective function. Comparisons by various experiments have shown better AD diagnosis performance by our proposed method. In future work, we will incorporate prior knowledge about feature/group importance into the proposed framework.
Acknowledgments
J. Peng was partially supported by NSFC (11401231) and NSFFC (2015J01254).
Footnotes
References
- 1.Chiappelli M, Borroni B, Archetti S, et al. VEGF gene and phenotype relation with Alzheimer’s disease and mild cognitive impairment. Rejuvenation Res. 2006;9(4):485–493. doi: 10.1089/rej.2006.9.485. [DOI] [PubMed] [Google Scholar]
- 2.Duda R, Hart P, Stork D. Pattern Classification. Wiley; New York: 2012. [Google Scholar]
- 3.Hinrichs C, Singh V, Xu G, Johnson S. MKL for robust multi-modality AD classification. In: Yang G-Z, Hawkes D, Rueckert D, Noble A, Taylor C, editors. MICCAI 2009. LNCS. Vol. 5762. Springer; Heidelberg: 2009. pp. 786–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jin Y, Wee CY, Shi F, et al. Identification of infants at high-risk for autism spectrum disorder using multiparameter multiscale white matter connectivity networks. Hum Brain Mapp. 2015;36(12):4880–4896. doi: 10.1002/hbm.22957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kloft M, Brefeld U, Sonnenburg S, et al. Lp-norm multiple kernel learning. J Mach Learn Res. 2011;12:953–997. [Google Scholar]
- 6.Kong D, Fujimaki R, Liu J, et al. Exclusive feature learning on arbitrary structures via L12-norm. NIPS. 2014:1655–1663. [Google Scholar]
- 7.Kowalski M. Sparse regression using mixed norms. Appl Comput Harmon Anal. 2009;27(3):303–324. [Google Scholar]
- 8.Liu F, Zhou L, Shen C, et al. Multiple kernel learning in the primal for multimodal Alzheimer’s disease classification. IEEE J Biomed Health Inform. 2014;18(3):984–990. doi: 10.1109/JBHI.2013.2285378. [DOI] [PubMed] [Google Scholar]
- 9.Liu J, Ye J. Efficient l1/lq norm regularization. 2010 arXiv:1009.4766. [Google Scholar]
- 10.Rakotomamonjy A, Bach F, Canu S, et al. Simple MKL. J Mach Learn Res. 2008;9:2491–2521. [Google Scholar]
- 11.Shen L, Thompson P, Potkin S, et al. Genetic analysis of quantitative phenotypes in AD and MCI: imaging, cognition and biomarkers. Brain Imaging Behav. 2014;8(2):183–207. doi: 10.1007/s11682-013-9262-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Szafranski M, Grandvalet Y, Rakotomamonjy A. Composite kernel learning. Mach Learn. 2010;79(1):73–103. [Google Scholar]
- 13.Tibshirani R. Regression shrinkage and selection via the lasso. J R Stat Soc Series B Stat Methodol. 1996;58(1):267–288. [Google Scholar]
- 14.Wang H, Nie F, Huang H. ICML. 2013. Multi-view clustering and feature learning via structured sparsity; pp. 352–360. [Google Scholar]
- 15.Yuan M, Lin Y. Model selection and estimation in regression with grouped variables. J R Stat Soc Series B Stat Methodol. 2006;68(1):49–67. [Google Scholar]
- 16.Zhang D, Wang Y, Zhou L, et al. Multimodal classification of Alzheimer’s disease and mild cognitive impairment. NeuroImage. 2011;55(3):856–867. doi: 10.1016/j.neuroimage.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Z, Huang H, Shen D. Integrative analysis of multi-dimensional imaging genomics data for Alzheimer’s disease prediction. Front Aging Neuros. 2013;6:260. doi: 10.3389/fnagi.2014.00260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhu X, Suk HI, Lee SW, et al. Subspace regularized sparse multi-task learning for multilclass neurodegenerative disease identification. IEEE Trans Biomed Eng. 2015;63(3):607–618. doi: 10.1109/TBME.2015.2466616. [DOI] [PMC free article] [PubMed] [Google Scholar]