Abstract
Congenital heart disease (CHD) represents the most prevalent inborn anomaly. Only a minority of CHD cases are attributed to genetic causes, suggesting a major role of environmental factors. Nonphysiological hypoxia during early pregnancy induces CHD, but the underlying reasons are unknown. Here, we have demonstrated that cells in the mouse heart tube are hypoxic, while cardiac progenitor cells (CPCs) expressing islet 1 (ISL1) in the secondary heart field (SHF) are normoxic. In ISL1+ CPCs, induction of hypoxic responses caused CHD by repressing Isl1 and activating NK2 homeobox 5 (Nkx2.5), resulting in decreased cell proliferation and enhanced cardiomyocyte specification. We found that HIF1α formed a complex with the Notch effector hes family bHLH transcription factor 1 (HES1) and the protein deacetylase sirtuin 1 (SIRT1) at the Isl1 gene. This complex repressed Isl1 in the hypoxic heart tube or following induction of ectopic hypoxic responses. Subsequently, reduced Isl1 expression abrogated ISL1-dependent recruitment of histone deacetylases HDAC1/5, inhibiting Nkx2.5 expression. Inactivation of Sirt1 in ISL1+ CPCs blocked Isl1 suppression via the HIF1α/HES1/SIRT1 complex and prevented CHDs induced by pathological hypoxia. Our results indicate that spatial differences in oxygenation of the developing heart serve as signals to control CPC expansion and cardiac morphogenesis. We propose that physiological hypoxia coordinates homeostasis of CPCs, providing mechanistic explanations for some nongenetic causes of CHD.
Introduction
Cardiac morphogenesis is controlled by a complex morphogenetic program driven by lineage specification, proliferation, differentiation, and migration of cardiac progenitor cells (CPCs). Disruption of the molecular pathways regulating these processes leads to cardiac malformation and congenital heart disease (CHD) (1). Numerous mutations have been identified in familiar and spontaneous forms of CHD, but the majority of CHD cases, which amount to 6 to 8 newborns in every 1,000 live births, cannot be explained by monogenetic causes (2). It is generally believed that both environmental and genetic factors based on variations in many different genes contribute to CHD. Environmental or nongenetic risk factors include diabetes mellitus, obesity, and hypoxic responses, but the molecular events driving CHD have remained enigmatic (3–6). Small observational groups and potential confounding effects have complicated analysis of the contribution of specific environmental effects in human beings. Nevertheless, studies on human populations living at high altitude have associated increased prevalence of CHD with low oxygen levels (7, 8).
Adult multipotent stem/progenitor cells frequently occupy hypoxic niches and respond to low oxygen concentrations by either proliferation or differentiation (5, 9, 10), but studies on embryonic multipotent progenitor cells are critically missing. Currently, it is not known whether CPCs utilize signals that depend on oxygen availability during development. Such processes would most likely involve induction of HIFs, which is one of the foremost cellular reactions to low concentrations of oxygen (11–13). HIF1α has profound effects on different molecules regulating the behavior of stem and/or progenitor cells. Genetic inactivation of Hif1a during early but not late developmental stages causes CHDs, suggesting a critical role of hypoxia responses for normal heart development, although the precise mechanisms of the action of HIF1α and the definition of its targets in this context have not been worked out (4).
In the heart, 2 major populations of CPCs forming the first heart field (FHF) and the second heart field (SHF) drive early cardiac morphogenesis. CPCs of the FHF generate the left ventricle (LV) and parts of the inflow tract, while the right ventricle (RV), the atria, the outflow tract, and parts of the inflow tract are mainly derived from CPCs of the SHF (14, 15). The transcription factors NK2 homeobox 5 (Nkx2.5) and islet 1 (ISL1) play key roles in the complex network, which controls fate decisions and expansion of CPCs (16–18). Isl1 expression marks cells in the SHF with a trilineage potential (cardiomyocytes, endothelial cells, and smooth muscle cells), but Isl1 is also expressed broadly in the coelomic mesoderm, which harbors precursors of both FHF and SHF (19). Nkx2.5 is expressed in the cardiac mesoderm, the adjacent endodermal cells, and cells of the FHF, which have turned off Isl1 expression, indicating that the difference between the FHF and SHF lineages lies primarily in the timing of differentiation (20). Hence, NKX2.5+ISL1– cells in the FHF and particularly in the linear heart tube might be seen as cells that have already acquired a more mature cardiomyocyte fate, a conclusion that is also supported by the direct role of NKX2.5 in repression of progenitor genes (e.g., Isl1 and Fgf10) and the persistent signature of progenitor cell gene expression in the myocardium of Nkx2.5 mutants (19, 21, 22). Isl1 transcription is dynamically regulated in CPCs and becomes silenced when CPCs are incorporated into the heart tube where oxygen concentrations are low (23, 24). Yet nothing is known about the role of hypoxia signaling for Isl1 and Nkx2.5 activity, which determines CPC homeostasis and lineage specification during early cardiogenesis.
In this study, we investigated the role of hypoxia signaling for the regulation of CPCs and the potential crosstalk between Isl1 and Nkx2.5 in this context. We found that Isl1+ CPCs in the SHF are less hypoxic (hereafter referred to as normoxic) compared with the primary linear heart tube. We determined that O2 availability influences the fate of ISL1+ CPCs by controlling Isl1 expression and thereby ISL1-dependent site-specific recruitment of HDAC1/5, which is required for Nkx2.5 silencing, allowing expansion of ISL1+ CPCs. Experimental induction of hypoxia responses during early heart development suppressed proliferation of ISL1+ CPCs and promoted lineage specification to cardiomyocytes. Our study decodes the molecular network that transmits hypoxia responses by identifying SIRT1 and hes family bHLH transcription factor 1 (HES1) as critical components of a repressor complex mediating HIF1α-dependent silencing of Isl1. We provide insights into the molecular mechanism leading to CHDs and demonstrate that inactivation of Sirt1 in ISL1+ CPCs prevents hypoxia-induced cardiac malformations.
Results
Experimental induction of hypoxia responses leads to multiple cardiac defects.
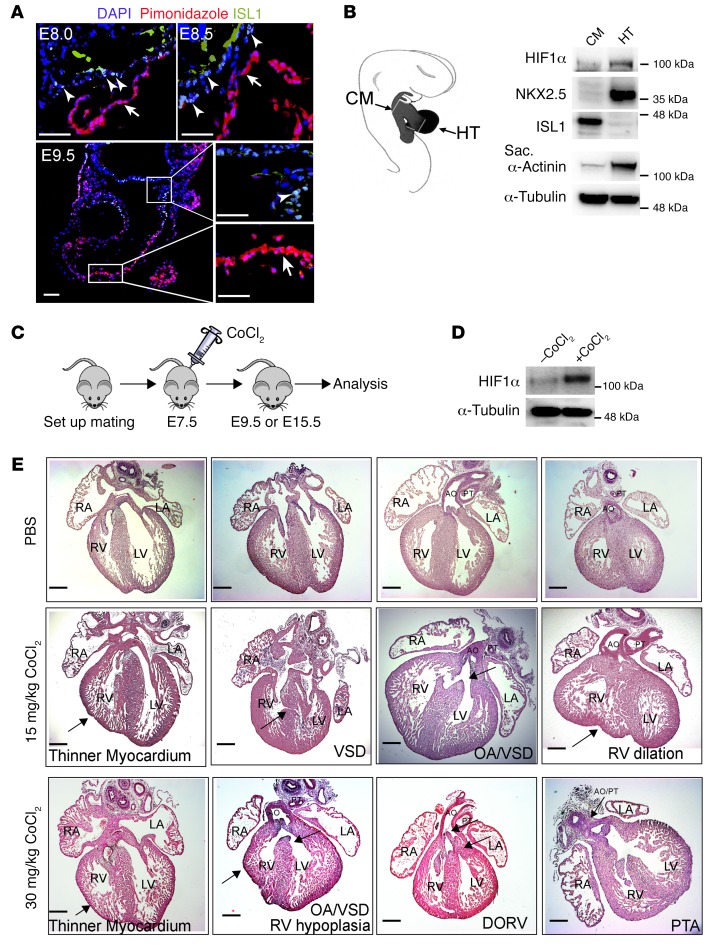
Physiological hypoxia (oxygen concentration <2%) and activation of HIF1α play critical roles for cardiac morphogenesis and function, but it is not clear whether hypoxia signaling is equally important for all parts of the developing heart (25). We therefore monitored the spatial distribution of pimonidazole, a nitroimidazole derivative that incorporates into hypoxic cells when oxygen concentration is below 10 mmHg (<2%) in embryonic mouse hearts between E8.0 and E9.5. Hypoxic cells were mainly localized in the myocardium of the looping heart tube as described previously (25). Surprisingly, however, CPCs in the cardiac mesoderm and the outflow tract, which we defined by expression of Isl1, showed negligible incorporation of pimonidazole, indicating that ISL1+ CPCs are maintained in a nonhypoxic (normoxic) environment (Figure 1A). The spatially distinct distribution of hypoxic cells in the developing heart should invoke hypoxic signaling indicated by HIF1α stabilization in the heart tube, but not in ISL1+ CPCs located in the cardiac mesoderm and the outflow tract. Hence, we separated the cardiac mesoderm and the outflow tract from the heart tube. Western blot analysis revealed lower levels of HIF1α in cardiac mesoderm, including the adjacent outflow/inflow tracts compared with the heart tube, confirming our hypothesis (Figure 1B).
Figure 1. Experimental induction of hypoxia during early pregnancy leads to cardiac defects.
(A) Representative immunofluorescence images of cryosections from E8.0, E8.5, and E9.5 embryos stained for ISL1 and the hypoxia marker pimonidazole. For each time point, 2 embryos from 2 different litters were randomly selected for analysis (n = 4). ISL1+ cells in the cardiac mesoderm (indicated by arrowheads) are nonhypoxic (O2 ≤2%), whereas the myocardium (indicated by arrows) is hypoxic. Scale bars: 50 μm. (B) Western blot analysis of protein levels in the cardiac mesoderm containing adjacent outflow/inflow tract (CM) and the heart tube (HT) microdissected from E9–E9.5 embryos. A schematic depicting the isolation procedure is shown in the left panel. α-Tubulin was used as protein-loading control. Two separate experiments (3 individual litters per experiments) were analyzed, yielding similar results. Sarc, sarcomeric. (C) Schematic of the strategy for chemical induction of hypoxia responses at early embryonic stages. (D) Western blot analysis of HIF1α levels in cardiac mesoderm containing adjacent outflow tract isolated from E9.5 embryos with or without CoCl2 treatment. α-Tubulin was used as protein-loading control. Two separate experiments (3 individual litters per experiments) were analyzed, yielding similar results. (E) H&E staining of E15.5 hearts after experimental induction of hypoxia at E7.5. Fifteen embryos with PBS treatment from 3 litters (upper panels) and 22 embryos with 15 mg/kg (middle panels) or 30 mg/kg CoCl2 (lower panels) treatment from 4 different litters were analyzed. Arrows point to individual cardiac defects named in the figure. AO, aorta; PT, pulmonary trunk; LA, left atrium; RA, right atrium; DORV, double outlet right ventricle; PTA, persistent truncus arteriosus. Scale bars: 200 μm.
To analyze whether induction of hypoxia responses in physiologically normoxic ISL1+ cells affects cardiac morphogenesis, we treated pregnant mice carrying E7.5 embryos with cobalt chloride (CoCl2), which elicits hypoxia-like responses (Figure 1, C and D). Morphological analysis of E15.5 embryos treated with CoCl2 (n = 22) at 15 mg/kg i.p. revealed multiple cardiac defects, such as thinner compact myocardium, ventricle septum defects (VSD), overriding aorta (OA), and RV dilation. Increased concentration of CoCl2 (30 mg/kg i.p.) resulted in more severe defects, such as double-outlet RV, persistent truncus arteriosus, and RV hypoplasia (Figure 1E), although such malformations were not seen in all embryos (n = 22) (Supplemental Table 1; supplemental material available online with this article; https://doi.org/10.1172/JCI88725DS1). We concluded that ISL1+ cells require a normoxic niche to contribute normally to cardiogenesis.
Hypoxia responses alter ISL1+ cell homeostatic control and lead to cardiac malformation.
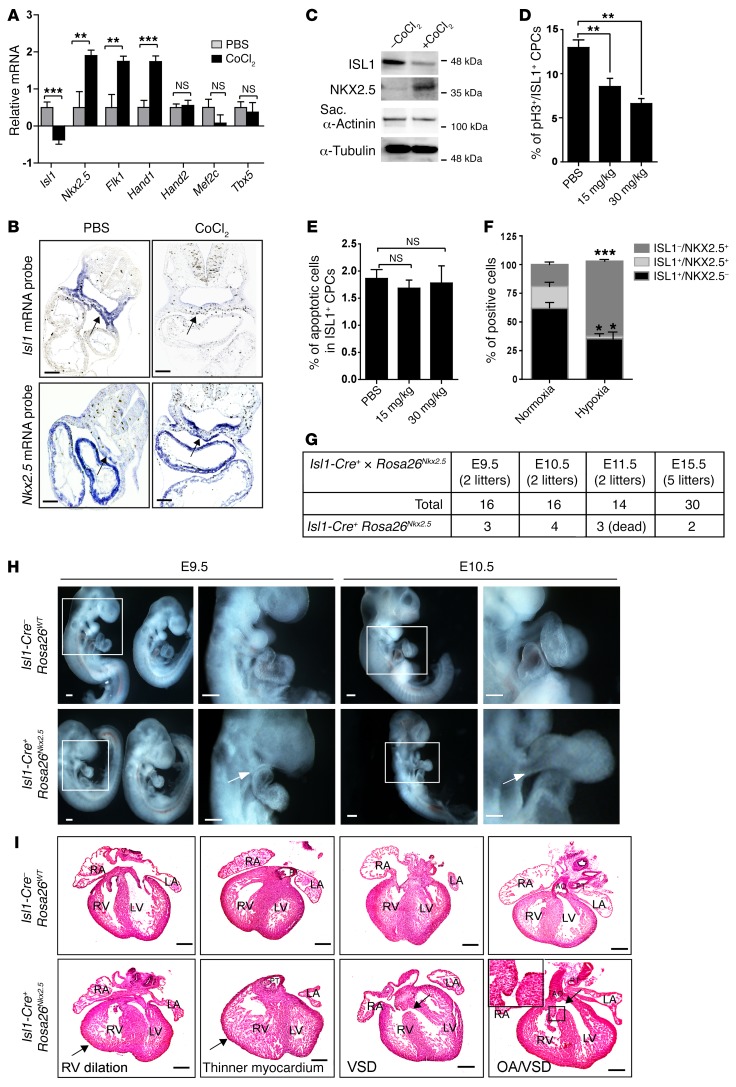
To investigate whether hypoxia responses have a direct impact on the expression of critical CPC regulators, we first performed a quantitative reverse transcription PCR (RT-qPCR) analysis of E9.0 hearts (including the adjacent mesoderm) after CoCl2 treatment. Interestingly, we observed a decrease of Isl1 expression, the master regulator of the SHF. In contrast, expression of several factors characteristic for the FHF increased as, e.g., Hand1 and Nkx2.5, which are expressed in the FHF and at lower levels in the SHF (Figure 2A). To validate these findings and to identify local changes in expression levels, we performed RNA in situ hybridization as well as Western blot analysis of dissected cardiac mesoderm after induction of hypoxia responses. Expression of Isl1 was reduced in the foregut endoderm and cardiac mesoderm of E9.5 embryos after CoCl2 treatment (Figure 2, B and C, and Supplemental Figure 1A). In contrast, expression of Nkx2.5 was upregulated in the cardiac mesoderm (Figure 2, B and C), indicating that experimental induction of hypoxia responses inhibits Isl1 but increases Nkx2.5 expression in the cardiac mesoderm. Additional antibody staining of whole-mount preparations followed by cryosections further confirmed the downregulation of ISL1, but upregulation of NKX2.5, in the cardiac mesoderm after induction of hypoxia responses (Supplemental Figure 1B).
Figure 2. Experimental induction of hypoxia alters the expression of Isl1 and Nkx2.5.
(A) RT-qPCR analysis of Isl1 (n = 8), Nkx2.5 (n = 8), Flk1 (n = 7), Hand1 (n = 6), Hand2 (n = 6), and Tbx5 (n = 6) expression in E9.5 embryonic hearts and the adjacent mesoderm (20–21 somites) after chemical induction of hypoxia responses. PBS-injected mice were used as control. The m34b4 gene was used as a reference for normalization. **P < 0.01; ***P < 0.001, t test. (B) In situ hybridization of C57BL/6 E9.25 embryos (18 somites) for either Isl1 mRNA (upper panel) or Nkx2.5 mRNA (lower panel) after chemical induction of hypoxia responses (30 mg CoCl2/kg body weight). Arrows indicate the NKX2.5+ or ISL1+ cardiogenic region. Representative images from 2 independent experiments are shown. (C) Western blot analysis of ISL1, NKX2.5, and sarcomeric α-actinin levels in cardiac mesoderm containing adjacent outflow tract isolated from E9.5 embryos with or without CoCl2 treatment. α-Tubulin was used as protein-loading control. Two separate experiments (3 individual litters per experiments) were analyzed, yielding similar results. (D) Analysis of ISL1+ cell proliferation in mock (n = 4) or CoCl2-treated (n = 4) E9.5 embryos by immunostaining for ISL1 and phospho-histone H3 (Ser10) (pH3). The percentages of pH3/ISL1 double-positive cells in cardiac mesoderm are shown. At least 6 sections from each embryo were counted. **P < 0.01, ANOVA with Dunnett’s post hoc correction. (E) TUNEL assay of ISL1+ cells after CoCl2 treatment. The percentages of TUNEL-positive ISL1+ CPCs are shown. At least 12 sections from each embryo were counted. ANOVA with Dunnett’s post hoc test was used to calculate significance (n = 3). (F) Immunofluorescence-based quantification of ISL1+NKX2.5–, ISL1+NKX2.5+, and ISL1–NKX2.5+ cells after FACS-based cell sorting of ISL1+ cells from E8.0 ISL1nGFP/+ embryos and 2-day cultivation of isolated cells under either normoxic or hypoxic conditions. Quantification of different cell populations was achieved by counting all immunostained cells in a multiwell dish. *P < 0.05; ***P < 0.001, t test (n = 3). (G) Number of Isl1-Cre+ Rosa26Nkx2.5 embryos after breeding of heterozygous Isl1-Cre+ mice with Rosa26Nkx2.5 mice at different developmental stages. (H) Whole-mount views of E9.5 (left panels) and E10.5 embryos (right panels). Boxed areas are enlarged. Representative images are shown. Arrows indicate shortened outflow tract in Isl1-Cre+ Rosa26Nkx2.5 embryos. Scale bars: 100 μm. (I) H&E staining of E15.5 hearts isolated from either Isl1-Cre– Rosa26WT (upper panel) or Isl1-Cre+ Rosa26Nkx2.5 (lower panel) mice. Thirty embryos from 5 litters including 2 Isl1-Cre+ Rosa26Nkx2.5 embryos were analyzed. Arrows point to individual cardiac defects named in the figure. Scale bars: 200 μm.
To exclude any potential artifacts due to chemical induction of hypoxia responses, we repeated the experiments with E7.5 embryos from pregnant mice housed in a hypoxia chamber at 10% oxygen and with isolated CPCs kept at 1% O2 (Supplemental Figure 1, C and F). Analysis of Isl1 and Nkx2.5 mRNA levels by whole-mount in situ hybridization or RT-qPCR yielded results similar to those after CoCl2 treatment, confirming that hypoxia responses repress Isl1, but promote Nkx2.5, transcription (Supplemental Figure 1, D–G).
Since O2 levels have a profound effect on stem/progenitor cell niches and modulate cell-fate decisions, we analyzed proliferation and apoptosis of ISL1+ cells in CoCl2-treated embryos at E9.5. Induction of hypoxia responses significantly reduced the numbers of ISL1/pH3 double-positive CPCs, but not of NKX2.5/pH3 double-positive cells, relative to mock-treated embryos (Figure 2D and Supplemental Figure 1H), whereas no differences in apoptotic cells were observed (Figure 2E and Supplemental Figure 1I). Similarly, ex vivo culture of FACS-isolated ISL1+ CPCs from E8.0 ISL1nGFP/+ embryos under 1% O2 (Supplemental Figure 2, A–E) reduced the number of ISL1/Ki67 double-positive cells (Supplemental Figure 2F).
The observation that hypoxia attenuates Isl1 but enhances Nkx2.5 expression in cardiac mesoderm raised the intriguing possibility that induction of hypoxia inhibits proliferation of ISL1+ cells at the expense of enhanced cardiomyocyte specification. Indeed, ex vivo culture of freshly isolated ISL1+ cells revealed a striking increase of ISL1–NKX2.5+ cells under hypoxia (63% of the cell population) compared with normoxia conditions (19% of the cell population). In contrast, the relative amounts of ISL1+NKX2.5– and ISL1+NKX2.5+ cells declined from 62% under normoxia to 35% under hypoxia and from 19% under normoxia to 2% under hypoxia, respectively, indicating increased commitment of ISL1+ CPCs to the cardiomyocyte lineage (Figure 2F and Supplemental Figure 2G). We could rule out that the higher numbers of NKX2.5+ cells were caused by augmented proliferation of already committed CPCs, since Nkx2.5 expression was nearly absent in freshly isolated ISL1+ CPCs (Supplemental Figure 2E). Instead, high expression of Nkx2.5 was detected in CPCs that had turned off Isl1 expression, suggesting that Nkx2.5 expression increases after inhibition of Isl1 (Supplemental Figure 2E).
So far, our experiments essentially suggested that hypoxia results in a “switch” of ISL1+ cells to NKX2.5+ cells, eventually resulting in cardiac malformations. To directly address this possibility and to investigate whether untimely expression of Nkx2.5 in ISL1+ cells recapitulates hypoxia-induced cardiac defects, we inserted the Nkx2.5 cDNA into the Rosa26 locus behind a loxP-stop cassette (Supplemental Figure 3A). Activation of Nkx2.5 expression in ISL1+ cells using Isl1-Cre mice (Isl1-Cre+ Rosa26Nkx2.5 mice) (Supplemental Figure 3, B and C) caused obvious heart malformations (i.e., shortened outflow tract, cardiac looping defects) in all double-heterozygous animals at E9.5 and E10.5 (Figure 2H). The majority of Isl1-Cre+ Rosa26Nkx2.5 embryos died around E11, although a few survived until E15.5 (Figure 2G), showing cardiac malformations similar to those seen after induction of hypoxia responses, including thinner compact myocardium, VSD, OA, and RV dilation (Figure 2I). Taken together, our results indicate that temporal and spatial oxygen availability critically regulates ISLl1+ cell proliferation and Nkx2.5 expression in the developing heart, while reduced oxygen availability at the wrong place and time leads to CHDs.
The SIRT1-HES1 complex silences Isl1 gene expression in a hypoxia-dependent manner.
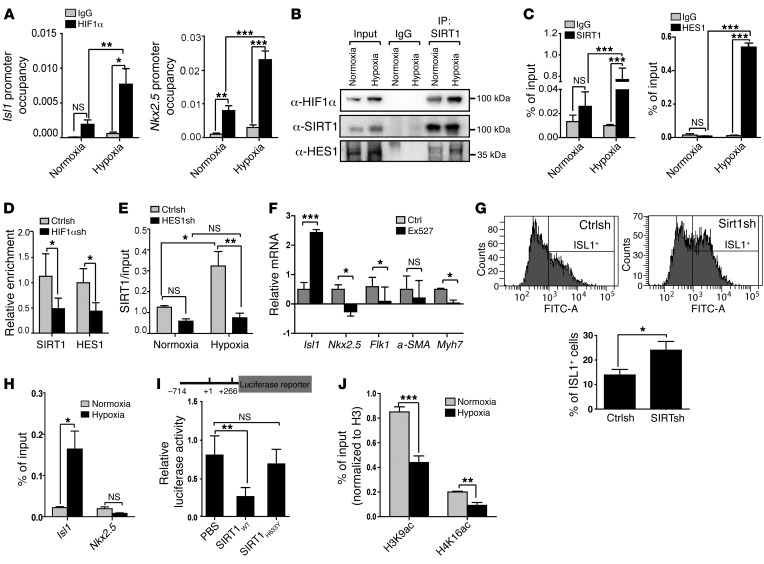
Due to the strong effects of hypoxia on Isl1 and Nkx2.5 expression, we wondered whether both genes are direct targets of hypoxia signaling. Bioinformatics analysis uncovered the presence of hypoxia regulatory elements (HREs) in the regulatory regions of Isl1 and Nkx2.5 genes. ChIP analysis detected binding of HIF1α to these regulatory elements, which was further enhanced in response to hypoxia (Figure 3A and Supplemental Figure 4, A and B).
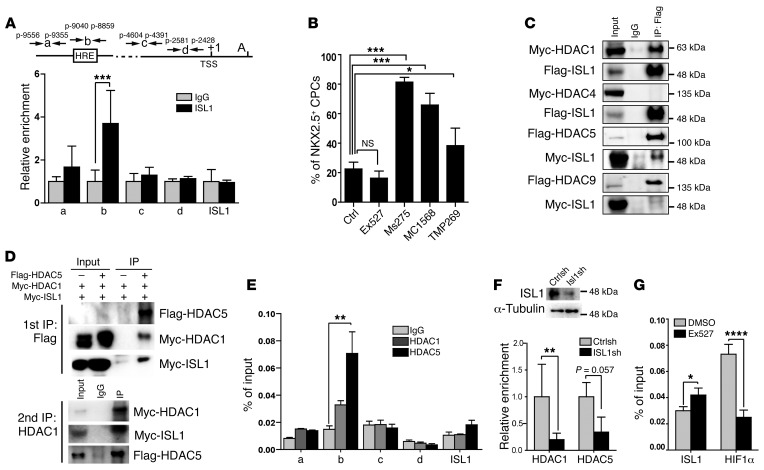
Figure 3. A SIRT1-HES1–containing complex represses Isl1 expression in a hypoxia-dependent manner.
(A) ChIP analysis of HIF1α at Isl1 (positions –468 to –285) (n = 4) and Nkx2.5 (positions –9040 to –8859) (n = 3) promoters after hypoxia treatment of day-6 EBs (1% O2, 16 hours). *P < 0.05; **P < 0.01; ***P < 0.001, ANOVA with Tukey’s post hoc correction. Enrichment of HIF1α was normalized to input DNA. (B) Co-IP assay of SIRT1 with HIF1α or HES1 under hypoxia. 5% input was used as loading control. 2 independent experiments were performed, generating similar results. (C) ChIP analyses of SIRT1 and HES1 binding to the Isl1 promoter (positions –468 to –285) under hypoxia (1% O2, 12 hours) and normoxia (21% O2) in E8.5 embryos. Enrichment of SIRT1 or HES1 was normalized to input DNA. ***P < 0.001, ANOVA with Tukey’s post hoc correction (n = 3). (D) ChIP analyses of SIRT1 (n = 8) and HES1 (n = 6) binding to the Isl1 promoter (positions –468 to –285) after lentivirus-mediated Hif1a knockdown in CoCl2–treated differentiating ESCs. Enrichment of SIRT1 or HES1 was normalized to input DNA. *P < 0.05, ANOVA with Tukey’s post hoc correction. (E) ChIP analysis of SIRT1 binding to the promoter of Isl1 (positions –468 to –285) in CPCs after Hes1 knockdown. *P < 0.05; **P < 0.01, ANOVA with Tukey’s post hoc correction (n = 3). (F) RT-qPCR analysis of Isl1, Nkx2.5, Flk1 (EB at E6), a-SMA, and Myh7 (EB at E8) expression in differentiating ES cells after treatment with the SIRT1 inhibitor (1 μM EX527, 24 hours; Isl1, Nkx2.5, a-SMA, Myh7: n = 3; Flk1: n = 5). The m34b4 gene was used as a reference for normalization. *P < 0.05; ***P < 0.001, t test. SIRT1 inhibitor treatment increases Isl1, but decreases Nkx2.5 and Flk1 expression. (G) FACS analyses of ISL1+ cells in EBs 6 days after Sirt1 knockdown. Left panels, histograms of ISL1+ cells. Right panel, quantification of ISL1+ cells. *P < 0.05, t test (n = 3). (H) ChIP analysis of SIRT1 binding to Isl1 (positions –468 to –285) and Nkx2.5 promoters (positions –9040 to –8859) in embryonic hearts after induction of hypoxia responses. *P < 0.05, t test (n = 3). (I) Luciferase reporter assays of the proximal Isl1 promoter with WT and H633Y mutant SIRT1. *P < 0.05; **P < 0.01, ANOVA with Dunnett’s post hoc correction (n = 3). (J) ChIP analysis of H3K9ac and H4K16ac at the Isl1 promoter in NKX2.5–EmGFP+ cells under hypoxia conditions. Enrichment of H3K9ac and histone H4K16ac was normalized to histone H3. **P < 0.01; ***P < 0.001, t test (n = 3).
HIF1α often induces transcription by recruiting coactivators such as histone acetyltransferase (HAT) p300 (26, 27), but sometimes also functions as a transcriptional repressor (28). We noticed that the HRE in the regulatory region of Isl1 was localized next to an HES1-binding site (N-box motif). HES1 is a Notch target and forms a repressor complex with the class III histone deacetylase SIRT1 (29). Importantly, inactivation of Hes1 leads to defects in SHF development (30). Therefore, we reasoned that HES1 and SIRT1 might form a ternary complex with HIF1α to represses the Isl1 promoter in CPCs during hypoxia. Western blot and co-IP experiments revealed that hypoxia increased protein levels and promoted formation of a complex containing SIRT1, HES1, and HIF1α, as indicated by IP with either SIRT1 or HIF1α antibodies (Figure 3B and Supplemental Figure 4C). In addition, ChIP experiments indicated strongly increased binding of SIRT1 and HES1 to the Isl1 proximal promoter harboring the N-box motif in CPCs under hypoxia conditions (Figure 3C and Supplemental Figure 4, D–G), which was significantly decreased under hypoxia when HIF1α was depleted by shRNAs (Figure 3D and Supplemental Figure 4H). Similarly, depletion of HES1 by shRNAs prevented binding of SIRT1 to the Isl1 promoter, suggesting that HES1 recruits SIRT1 to the Isl1 promoter to repress transcription during hypoxia (Figure 3E and Supplemental Figure 5A).
To gain further evidence for the role of HES1 and SIRT1 in mediating hypoxia-dependent gene repression, we mutated the N-box within the Isl1 promoter, which increased activity of an Isl1-luciferase construct and prevented Isl1-promoter repression by hypoxia (Supplemental Figure 5B). Furthermore, inactivation of SIRT1 in proliferating CPCs either by shRNA knockdown or by treatment with the SIRT1-specific inhibitor Ex527 increased Isl1 gene expression (Figure 3F and Supplemental Figure 5, C and D) as well as the number of ISL1+ cells (Figure 3G), but suppressed other cardiac-specific genes, including Nkx2.5 and Myh7 (Figure 3F and Supplemental Figure 5, C and D). Accordingly, we found that SIRT1 binds specifically to the Isl1 but not to the Nkx2.5 promoter (Figure 3H and Supplemental Figure 4F) and that Sirt1 knockdown abrogates hypoxia-mediated silencing of the Isl1 promoter (Supplemental Figure 5E). Enzymatically inactive SIRT1 (SIRT1H633Y) did not repress Isl1 promoter activity (Figure 3I), and induction of hypoxia responses decreased H3K9 and H4K16 acetylation at the Isl1 promoter in CPCs, indicating that SIRT1-mediated hypoacetylation leads to Isl1 gene silencing (Figure 3J).
Interestingly, hypoxia responses did not change Sirt1 mRNA and NAD+ concentrations (Supplemental Figure 6, A–C), but caused accumulation of ROS and activation of JNK, which results in enhanced activity of SIRT1 (31) reflected by deacetylation of histone H3K9 (Supplemental Figure 6, D and E). We concluded that hypoxia-induced Isl1 gene silencing is mediated by HIF1α/HES1-dependent recruitment of SIRT1 specifically to the Isl1 promoter. Increased SIRT1 activity induced by activation of JNK might also contribute to Isl1 gene silencing after hypoxia, although we cannot exclude a mere association. This mechanism allows inverse transcriptional regulation of Isl1 and Nkx2.5 by HIF1α following differential recruitment of cofactors such as HES1 and SIRT1.
Inactivation of Sirt1 in ISL1+ cells enhances Isl1 expression and rescues hypoxia-induced CHDs.
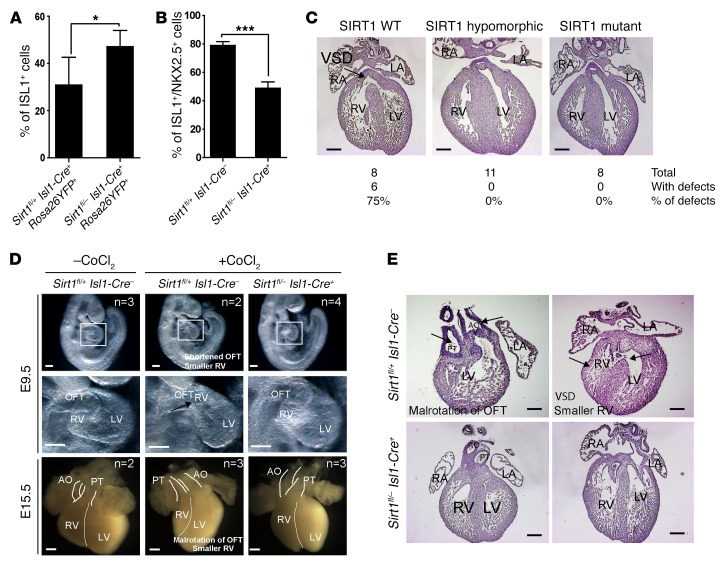
To elucidate the physiological role of SIRT1 for silencing Isl1 expression in ISL1+ cells during hypoxia, we deleted exon 4, which encodes the conserved SIRT1 catalytic domain in mice. Consistent with a previous study (32), germline Sirt1–/– mutants showed multiple CHDs (Supplemental Figure 7, A–C). In addition, we found a strong upregulation of Isl1 mRNA in E8.0 Sirt1–/– embryos (5-somite stage) (Supplemental Figure 7D). We next specifically inactivated Sirt1 in ISL1+ cells using Isl1-Cre+ mice and tagged the ISL1+ lineage with a Rosa26YFP+ reporter (thereafter referred to as Sirt1fl/– Isl1-Cre+ RosaYFP+) (Supplemental Figure 8, A and B). Sirt1fl/– Isl1-Cre+ RosaYFP+ mice were viable and fertile, but exhibited slightly impaired RV function and reduced body weight (data not shown). In contrast with germline Sirt1 mutants, we did not detect major cardiac malformations, although we noted a thinner myocardial compact layer (Supplemental Table 2). The rather minor phenotype of Sirt1fl/– Isl1-Cre+ compared with germline Sirt1 mutants might indicate that the loss of Sirt1 can be mostly compensated in the SHF, thereby preventing CHDs, but not in other parts of the developing heart, such as FHF or the cardiac neural crest. Importantly, inactivation of Sirt1 in ISL1+ cells increased Isl1 expression, reduced Nkx2.5 expression, and generated more ISL1+ cells (Figure 4, A and B, and Supplemental Figure 8C), validating the results of our in vitro analysis.
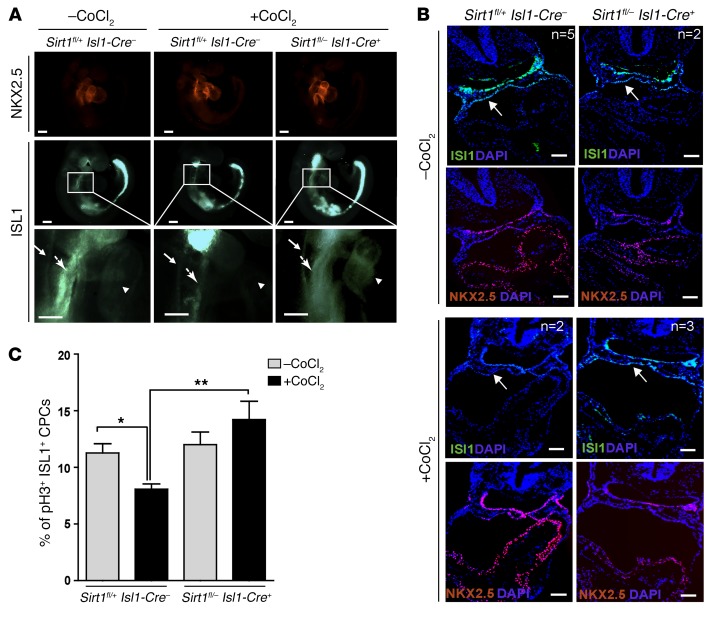
Figure 4. Inactivation of Sirt1 in ISL1+ cells increases the number of ISL1+ cells and rescues hypoxia-induced CHDs.
(A) FACS analysis of ISL1+ cells and their derivatives in E9.5 embryonic hearts (20–24 somites) after Sirt1 inactivation. *P < 0.05, t test (n = 3). (B) Quantification of ISL1+NKX2.5+ cells in the cardiac mesoderm by immunostaining of E9.5 embryos (20–24 somites). ***P < 0.001, t test (n = 3). (C) H&E staining of E15.5 hearts of control (Sirt1 WT), Sirt1fl/+ Isl1-Cre+ (Sirt1 hypomorphic), and Sirt1fl/– Isl1-Cre+ (Sirt1 mutant) embryos after chemical induction of hypoxia responses (15 mg CoCl2/kg body weight) at E7.5. Note the rescue of CHDs in Sirt1fl/– Isl1-Cre–, Sirt1fl/+ Isl1-Cre+, and Sirt1fl/– Isl1-Cre+ embryos compared with controls. Twenty-seven embryos from 4 litters, including 8 Sirt1fl/– Isl1-Cre+ embryos, were analyzed. Numbers of specific CHDs are listed in Supplemental Table 2. Scale bars: 200 μm. (D) Whole-mount views of E9.5 embryos (upper panels) and E15.5 embryonic hearts (lower panels) without and with chemical induction of hypoxia responses (30 mg CoCl2/kg body weight). For each time point, 1 litter was analyzed. Numbers of analyzed embryos for each condition are indicated in the figure. Representative images are shown. Inactivation of Sirt1 in the SHF ameliorates CHDs. OFT, outflow tract. Scale bars: 100 μm. (E) H&E staining of severe cardiac malformations in control, but not in Sirt1fl/– Isl1-Cre+, embryos at E15.5 after chemical induction of hypoxia responses (30 mg CoCl2/kg body weight). Arrows point to individual cardiac defects named in the figure. Scale bars: 100 μm.
Since experimental hypoxia causes CHDs and SIRT1 mediates repression of Isl1 transcription in response to hypoxia, we hypothesized that reduced expression of Sirt1 might rescue the adverse effects of acute hypoxia episodes during pregnancy. Induction of hypoxia responses by injection of 15 mg/kg body weight CoCl2 at E7.5 caused CHDs (e.g., VSD, OA) in approximately 75% of Sirt1fl/+ Isl1-Cre– and C57BL/6 control hearts, which increased to nearly 100% the incidence of CHD when a CoCl2 dosage of 30 mg/kg body weight was used (Figure 4C and Supplemental Table 1). In stark contrast, mutant mice receiving the same treatment, but lacking one (Sirt1fl/– Isl1-Cre–, Sirt1fl/+ Isl1-Cre+) or both alleles of Sirt1 (Sirt1fl/– Isl1-Cre+) showed no signs of CHD, such as thinner myocardium, VSD, smaller RV with shortened outflow tract, or malrotation of the outflow tract (Figure 4, C–E), suggesting that reduction of Sirt1 expression is sufficient to rescue the cardiac defects. To investigate whether the prevention of cardiac malformations caused by induction of hypoxia responses in Sirt1fl/– Isl1-Cre–, Sirt1fl/+ Isl1-Cre+, and Sirt1fl/– Isl1-Cre+ embryos correlated with increased expression of Isl1, we compared ISL1 levels in WT and mutant embryos after CoCl2 treatment. Immunofluorescence staining revealed a strong attenuation of ISL1, but not NKX2.5, expression in WT embryos after induction of hypoxia responses (Figure 5, A and B). Importantly, however, inactivation of Sirt1 in the SHF normalized expression of Isl1 (Figure 5, A and B) and restored proliferation of ISL1+ cells (Figure 5C), emphasizing the critical role of SIRT1 in mediating Isl1 suppression during hypoxia responses.
Figure 5. Inactivation of Sirt1 in ISL1+ cells enhances Isl1 expression and promotes proliferation of ISL1+ cells after induction of hypoxia responses.
(A) Whole-mount immunostaining of E9.5 control and Sirt1 mutant embryos for NKX2.5 and ISL1 with and without chemical induction of hypoxia responses (30 mg/kg body weight CoCl2 treatment). The pale green staining in the head and tail is due to unspecific autofluorescence. Neural tube (arrows with small arrowheads), cardiac mesoderm (arrows with big arrowheads), and heart tubes (arrowheads) are indicated. Embryos with or without chemical induction of hypoxia responses were obtained from 3 different litters. Scale bars: 100 μm. (B) Representative images of sections from control and Sirt1 mutant E9.5 embryos following whole-mount immunostaining for ISL1 and NKX2.5 with and without chemical induction of hypoxia responses (15 mg/kg body weight CoCl2 treatment). Embryos with (Sirt1fl/+ Isl1-Cre–: n = 2 Sirt1fl/– Isl1-Cre+: n = 3) or without (Sirt1fl/+ Isl1-Cre–: n = 5; Sirt1fl/– Isl1-Cre+: n = 2) chemical induction of hypoxia responses were obtained from 2 different litters. Note reduced ISL1 levels in the cardiac mesoderm (white arrows) in control, but not in Sirt1fl/– Isl1-Cre+, littermates after chemical induction of hypoxia responses. Scale bars: 200 μm. (C) Quantification of ISL1+ cell proliferation by pH3 immunostaining in control (–CoCl2: n = 4; +CoCl2: n = 5) and Sirt1fl/– Isl1-Cre+ E9.0 embryos (n = 3) after chemical induction of hypoxia responses. Note that inactivation of Sirt1 in the SHF rescues reduced proliferation of ISL1+ cells. *P < 0.05; **P < 0.01, ANOVA with Dunnett’s post hoc correction.
ISL1 forms a complex with HDAC1/HDAC5 and silences Nkx2.5 gene expression.
Prompted by the inverse transcriptional regulation of Isl1 and Nkx2.5 in CPCs, we searched for a putative ISL1-binding site in the Nkx2.5 gene. ChIP disclosed binding of ISL1 to a site located in close proximity to the HRE motif at position –9040/–8859 (Figure 6A), which might be used by ISL1 to recruit a corepressor. To identify such a potential corepressor, we treated ISL1+ cells with inhibitors specific for class I and IIa HDACs, which dramatically increased the number of NKX2.5+ cells during differentiation, whereas treatment of CPCs with Ex527, a class III inhibitor that increases Isl1 expression, had the opposite effect (Figure 6B). Protein co-IP assays revealed that ISL1 interacts with HDAC1 and HDAC5, but not with HDAC4 or HDAC9 (Figure 6C). Furthermore, sequential co-IPs revealed that ISL1, HDAC1, and HDAC5 form a tripartite complex (Figure 6D), which was corroborated by ChIP experiments in ISL1+ cells, indicating concomitant binding of HDAC1 and HDAC5 to the ISL1-binding site in the Nkx2.5 gene (Figure 6E). These findings, which are in line with previous studies in P19 cells suggesting an involvement of class I and class II HDACs in Nkx2.5 gene silencing (33), indicate that ISL1 forms a complex with HDAC1 and HDAC5 and thereby represses Nkx2.5 expression when Isl1 expression is high. To exclude the possibility that HDACs repress Nkx2.5 independently of ISL1, we analyzed the binding of HDAC1 and HDAC5 to the Nkx2.5 promoter in the absence of ISL1. ChIP experiments in Isl1 knockdown cells revealed that depletion of ISL1 impairs binding of HDAC1 and HDAC5 to the Nkx2.5 promoter (Figure 6F). The close vicinity of the ISL1-binding sites to the HRE motif (Figure 6A) further indicates that ISL1 and HIF1α might regulate Nkx2.5 transcription in a mutually exclusive manner, depending on their activities. In fact, inhibition of SIRT1 enhanced binding of ISL1 in ChIP experiments, but prevented binding of HIF1α (Figure 6G), strongly arguing for a decisive role of the ISL1/HDAC1/HDAC5 complex in Nkx2.5 gene repression.
Figure 6. ISL1 represses Nkx2.5 transcription by recruitment of HDACs.
(A) ChIP analysis of ISL1 binding to the Nkx2.5 promoter. ***P < 0.001, t test (amplicons a–c, n = 4; amplicon d and ISL1, n = 3). Both the HRE motif and the putative ISL1-binding site are located at –9040 to –8859 (amplicon b). (B) Quantification of NKX2.5hi cells following in vitro differentiation of ISL1+ cells after treatment with HDAC inhibitors. *P < 0.05; ***P < 0.001, ANOVA with Dunnett’s post hoc correction (control, n = 6; Ex527, n = 5; Ms275 and MC1568, n = 3; TMP269, n = 5). (C) Co-IP analysis of the interaction of ISL1 with different HDACs in HEK293T cells after transfection. Two independent experiments were performed, generating similar results. (D) Sequential Co-IP analysis of the interaction of ISL1 with HDAC1 and HDAC5 in HEK293T cells. (E) ChIP analysis of HDAC1 and HDAC5 binding to the ISL1 site in Nkx2.5 promoter. **P < 0.01, ANOVA with Dunnett’s post hoc correction (amplicons a, c, d, ISL1, n = 3; amplicon b, n = 6). (F) ChIP analysis showing HDAC1 and HDAC5 binding to the Nkx2.5 promoter (positions –9040 to –8859) after lentiviral-mediated expression of ISL1 shRNA. **P < 0.01, t test (n = 3). (G) ChIP analysis showing ISL1 and HIF1α binding to the Nkx2.5 promoter (positions –9040 to –8859) after inhibition of SIRT1 enzymatic activity by Ex527 (1 μM). Note that enzymatic inhibition of SIRT1 enhances binding of ISL1, but reduces binding of HIF1α. *P < 0.05; ****P < 0.0001, t test (n = 3).
Discussion
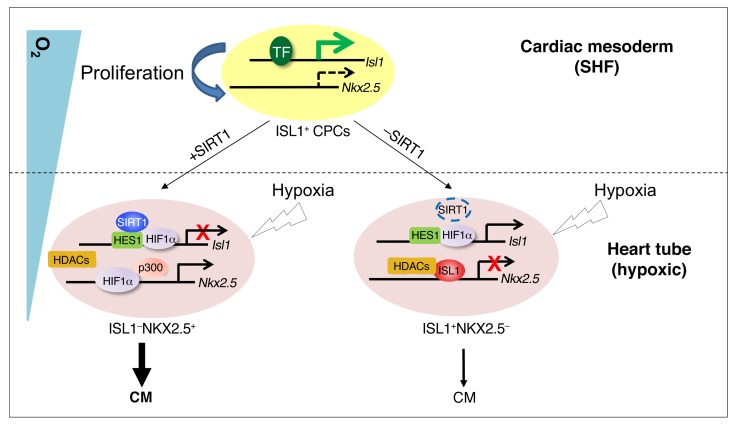
The effect of oxygen availability varies depending on the cell type. Stem/progenitor cell populations usually show enhanced proliferation or self-renewal and suppression of differentiation under hypoxia conditions (5, 9, 10). Here, we describe an unexpected scenario, in which ISL1+ CPCs proliferate in a nonhypoxia niche, but cease proliferation and succumb to precocious myocyte specification when exposed to a hypoxic environment. Challenge of the nonhypoxic niche by experimental hypoxia causes CHD and disrupts the inverse transcriptional control of Isl1 and Nkx2.5, two key cardiac transcription factors that are both direct targets of HIF1α. We demonstrate that HIF1α, which is a well-known transcriptional activator, represses Isl1 by recruitment of SIRT1, thereby inhibiting proliferation of ISL1+ CPCs and inducing myocyte specification. Prevention of precocious myocyte specification enables self-renewal of ISL1+ cells, which requires formation of an ISL1/HDAC1/HDAC5-containing corepressor complex that restricts Nkx2.5 expression. Correspondingly, directed expression of Nkx2.5 in ISL1+ cells recapitulated several aspects of nonphysiological hypoxia, including formation of CHDs, thereby validating our model (Figure 7). According to our model, Nkx2.5 is “on” in the presence of SIRT1 under pathological hypoxic conditions because Isl1 is “off” due to the repressive activity of the HIF1α/HES1/SIRT1 complex, which prevents ISL1/HDAC-mediated inhibition of Nkxk2.5. Nkx2.5 is “off” in the absence of SIRT1 under pathological hypoxic conditions, because Isl1 is “on” due to disruption of the repressive HIF1α/HES1/SIRT1 complex. In this setting, the continued expression of Isl1 allows the ISL1/HDAC1/5 complex to turn Nkx2.5 “off.”
Figure 7. Schematic model of the regulation of ISL1+ CPC behavior by hypoxia, transcription factors, and epigenetic modifiers during early heart development.
ISL1+ CPCs maintain high Isl1 expression directed by transcription activators such as β-catenin/LEF, FOXO1, and GATA4 in response to inductive signals, but low Nkx2.5 expression levels in the cardiac mesoderm, where O2 concentrations are relatively high, favoring self-renewal and expansion. ISL1 recruits HDACs to the Nkx2.5 promoter in CPCs to repress Nkx2.5 expression and prevent precocious myocyte specification. Migration of ISL1+ cells into the growing physiologically hypoxic (O2 ≤2%) heart tube results in HIF1α-SIRT1-HES1–dependent silencing of Isl1 expression and HIF1α-p300–dependent stimulation of Nkx2.5 expression, promoting cardiomyocyte specification. In the absence of SIRT1, repression of Isl1 transcription is relieved, resulting in increased Isl1 levels and enhanced proliferation of CPCs. As a consequence, ISL1-mediated Nkx2.5 repression is augmented and the transition of CPCs from an ISL1+NKX2.5– to an ISL1–NKX2.5+ state is hindered, compromising cardiomyocyte specification. TF, transcription factors.
Previous studies demonstrated differential temporal requirements of hypoxia signaling responses for normal cardiac morphogenesis (25). Notably, inactivation of Hif1α, one of the main transducers of hypoxia signaling, causes septation and conotruncal heart defects after inactivation at E10.5, but not at E13.5 (4). The temporal requirement of hypoxia signaling is interwoven with a precise spatial control determined by the existence of highly hypoxic areas within the developing embryo. We found that under physiological conditions, the SHF is considerably less hypoxic when compared with the primary heart tube, which allows proper expansion of CPCs and explains why nonphysiological shortage of oxygen at the wrong location and time causes CHDs (6). Once the contribution of the SHF has expired, transient pathological hypoxia has less severe effects. This conclusion fits well with the remarkable ability of fetal hearts to cope with hypoxia due to the adaption of fetal cardiomyocytes to low oxygen levels (34).
The histone deacetylase SIRT1 is well known as conferring resistance to metabolic and hypoxia stress by deacetylating key signaling molecules controlling cell metabolism, survival, and proliferation (35). Lack of Sirt1 in the cardiovascular system renders Sirt1 mutants more susceptible to cell death induced by ischemia/reperfusion injury, but has only minor effects at baseline conditions (36–38). In the developing embryo, SIRT1 also mediates responses to hypoxia, but serves a different purpose, not only orchestrating stress responses, but also cellular fate decisions. The reason for this distinction probably lies in the requirement of physiological hypoxia for embryonic heart development, while hypoxia is an unfavorable condition in most adult organs. Interestingly, expression of Sirt1 in the heart declines dramatically between E12.5 and E13.5, when Isl1 expression is turned off and the newly formed cardiac vasculature increases oxygen supply (our unpublished data and ref. 39). The disappearance of hypoxic areas in the heart and the decline of Sirt1 expression might mark a transition of the function of SIRT1 from a morphogen to a stress-response gene that serves different functions, but employs similar mechanisms. This dual function of SIRT1 might also partially explain the phenotypic differences between germline Sirt1 (32) and Sirt1fl/– Isl1-Cre+ mutants, which showed only a minor phenotype characterized by a thinner myocardial compact layer. Interestingly, SIRT1 seems rather to play an adverse role in the SHF, acting as a cellular stress-response gene, when the SHF becomes hypoxic. The untimely employment of SIRT1 might make a bad situation worse by triggering adverse responses, such as the precocious repression of Isl1, a scenario that mimics the gain-of-function phenotype in Isl1-Cre+ Rosa26Nkx2.5 mutants. Such a concept might explain why inactivation of Sirt1 in the ISL1+ lineage rescues the phenotype caused by hypoxia responses, but otherwise has little effect. In contrast, SIRT1 might serve a critical role in the FHF, giving rise to the hypoxic primary heart tube, probably by participating in repression of Isl1 gene expression, although we did not investigate this possibility. At present, we do not know why the FHF is more sensitive to the loss of SIRT1 than the SHF, but future insights into the complex network governed by different sirtuins might provide answers to the differential requirement of SIRT1 in different cell types.
During heart development, SIRT1 closely interacts with HIF1α to control expansion of ISL1+ CPCs. So far, SIRT1 has been described as an amplifier of HIF1α activity during hypoxia (40–42), deacetylating and stabilizing HIF1α, thereby leading to enhanced hypoxia responses. Here, we demonstrate that SIRT1 also functions as negative coregulator of HIF1α, which silences Isl1 expression by deacetylation of histones. In addition, SIRT1 might also deacetylate HIF1α, thereby boosting the effects of hypoxia on Isl1 gene silencing, although this remains to be shown. HIF1α itself has been mainly described as a transcriptional activator forming a complex with the HAT p300. Hence, it was intriguing that interaction of HIF1α with the HES1/SIRT1 complex converted HIF1α from an activator into a repressor. This scenario resembles increased binding of SIRT1 to MASH1 in adult neuronal stem cells during oxidative stress and hence might indicate a general principle (29). Repression of Isl1 expression might not rely solely on hypoxia responses, since inactivation of Nkx2.5 causes persistent expression of Isl1 in the forming heart tube (19). Furthermore, a recent finding demonstrated direct repression of Isl1 by NKX2.5 for proper development of the ventricular myocardium (21). However, we assume that hypoxia is the decisive initial step to silencing Isl1 expression in SHF CPCs migrating into the linear heart tube, which relieves suppression of Nkx2.5 by ISL1, subsequently enabling repression of Isl1 by NKX2.5.
Various combinations of cardiac transcription factors drive development of the SHF (14, 15), in which ISL1 plays a central role. Similarly to what occurs in Nkx2.5, mutations in the human Isl1 gene are associated with a diverse range of cardiac malformations (43–45). Therefore, it is not surprising that a complex network rules the activity of Isl1 (46, 47), to which we added a decisive component. We demonstrated that Isl1 is repressed by hypoxia responses through HES1/HIF1α-dependent recruitment of SIRT1 to the Isl1 gene and by activation of Sirt1. Inhibition of Isl1 expression occurs when ISL1+ CPCs migrate from the normoxic cardiac mesoderm to the hypoxic heart tube, where Isl1 gene expression is low, but Nkx2.5 expression is high.
The presence of an ISL1-binding site in the Nkx2.5 gene close to the HRE establishes another regulatory layer. The high level of Isl1 expression in normoxic conditions attenuates Nkx2.5 gene expression, which is required for preventing precocious myocyte specification of CPCs. Suppression of Nkx2.5 is best achieved by a factor that defines the SHF, such as Isl1. Since Sirt1 does not get activated in the normoxic SHF and the regulatory region of the Nkx2.5 lacks an N-box next to the HRE element, which is required for recruitment of SIRT1, other HDACs, such as HDAC1/HDAC5, are necessary to suppress Nkx2.5 gene expression. The direct regulation of Nkx2.5 by ISL1 and HDAC1/HDAC5 represents what we believe to be a novel facet of SHF development and reflects the necessity of a proper balance of Isl1 and Nkx2.5 levels to control proliferation and differentiation of CPCs (22). Exacerbation of hypoxic responses under pathological or experimental conditions disrupts this balance and results in a “switch” of ISL1+ to NKX2.5+ cells due to SIRT1-dependent downregulation of Isl1, which relieves repression of Nkx2.5. To prove the validity of this model, we expressed NKX2.5 specifically in ISL1+ cells, which essentially recapitulated many hypoxia-induced heart defects. However, it should be mentioned that not all severe heart defects seen in hypoxia-treated embryos were apparent in Isl1-Cre+ Rosa26Nkx2.5 embryos at E15.5. We assume that this phenomenon is caused by early embryonic lethality, since the number of Isl1-Cre+ Rosa26Nkx2.5 embryos at E15.5 was substantially lower than the expected Mendelian ratio.
Taken together, our data support a model in which hypoxia signaling–induced crosstalk between transcription factors and epigenetic modulation determines the fate of ISL1+ cells during early cardiogenesis and prevents CHD. We demonstrate that hypoxia and SIRT1 tie Isl1 and Nkx2.5 into a negative regulatory loop that coordinates expansion of SHF CPCs and ensures proper acquisition of myocyte subtype identity, thereby shaping the heart.
Methods
Animals.
Sirt1-floxed mice were generated by flanking exon 4 with loxP sites (Supplemental Figure 6A). Sirt1-floxed mice were crossed to the CMV-Cre deleter strain (The Jackson Laboratories) to obtain heterozygous Sirt1 mutant mice (Sirt1+/–). Rosa26YFP mice were obtained from Frank Constantini (Columbia University, New York, New York, USA) (48). Rosa26Nkx2.5 mice were generated by insertion of an HA-NKX2.5cDNA-V5 PCR fragment into the Rosa26 locus by homologous recombination in V6.5 embryonic stem (ES) cells obtained from Rudolph Jaenisch (Whitehead Institute for Biomedical Research, Cambridge, Massachusetts, USA) (49) (Supplemental Figure 3A). Isl1-Cre transgenic mice were provided by Sylvia Evans (UCSD, San Diego, California, USA) (50). Isl1nGFP/+ mice (Supplemental Figure 2A) were generated by the dRMEC method using Isl1tm1a(KOMP)Wtsi ES (R1 ES cell line) cells obtained from EUCOMM (51). All mouse strains were backcrossed and maintained on a C57BL/6 genetic background. C57BL/6 mice were obtained from Charles River Laboratories. Primers used for genotyping are listed in Supplemental Table 3. To induce hypoxic responses, pregnant females were placed in their home cages in a hypoxia chamber with 10% O2/90% N2. Oxygen in the chamber was measured using an oxygen analyzer (Engineered Systems and Designs). During hypoxia, humidity and temperature were monitored in accordance with Max Planck Institute Institutional Animal Care and Use Committee procedures. Gas flow was maintained at 0.1–0.2 l per minute. For chemical induction of hypoxic responses, CoCl2 (Sigma-Aldrich) was administered intraperitoneally at 15 or 30 mg/kg body weight per injection. Pimonidazole was administered intraperitoneally at 60 mg/kg body weight per injection 90 minutes before analysis. Each time point or treatment modality was covered by analysis of at least 2 randomly selected embryos from at least 2 litters.
Plasmids.
pcDNA3-Flag-ISL1 and pcDNA3-Myc-IS:1 were described previously (46); pcDNA3-Flag-HDAC9 was described previously (52); pcDNA-Flag-HDAC5 was purchased from Addgene (catalog 33209); pcDNA3-Myc-HDAC1, pcDNA3-Myc-HDAC4, pcDNA3-Flag-SIRT1, and pcDNA3-Flag-SIRT1H633Y have been described elsewhere (53, 54). The Isl1 promoter was cloned from WT mouse genomic DNA (C57BL/6) in pGEM-T Easy Vector (Promega) and subsequently subcloned in pTA-Luciferase plasmid (Invitrogen). The primers used for cloning are listed in Supplemental Table 3.
Embryo isolation, cell sorting, and FACS analysis.
For isolation of embryonic ISL1+ CPCs (ISL1nGFP/+) and cells derived from the ISL1+ lineage (Isl1-Cre RosaYFP+), E8.0 (5 pairs of somites) and E9.0 (14 pairs of somites) embryos or individual embryonic hearts from E15.5 embryos were dissected and dissociated into single cells as described previously (55). ISL1+ or NKX2.5+ CPCs were isolated from embryoid bodies (EB) by treatment with 0.05% trypsin/EDTA for 3 minutes at 37°C. Dissociated cells were filtered through a 40-μm cell strainer and fluorescence sorted using a FACSAriaIII (BD Biosciences). Sorted ISL1+ CPCs were further cultured within differentiation or proliferation medium on cardiac fibroblast feeder cells as described (56). For FACS analysis, cells were fixed in methanol followed by washing (PBS) and blocking steps (1% BSA in PBS) in a volume of 100 μl for 10 minutes at room temperature. Afterwards, cells were incubated with antibodies listed in Supplemental Table 5, washed in PBS, and resuspended in sorting buffer (PBS containing 25 mM Hepes, pH 7.0, 2 mM EDTA, 1% FBS) for FACS analysis. Data were acquired on an LSRII flow cytometer (BD) and analyzed using FlowJo software. To induce hypoxia in CPCs, cells were grown in a humidified atmosphere at 37°C at 1% O2/5% CO2 to induce hypoxic responses. Cells cultivated in a 21% O2/5% CO2 incubator were used as normoxic controls.
ES cell differentiation, NAD+/NADH measurement, and ROS measurement.
Undifferentiated ES cells (V6.5 ES cell line) were maintained on mouse embryonic fibroblast (MEF) feeder cells in DMEM supplemented with 15% FCS (Invitrogen), 1 mM sodium pyruvate, 0.1 mM nonessential amino acids, 0.1 mM 2-mercaptoethanol (Sigma-Aldrich), 0.1 mM nonessential amino acids (Invitrogen), 4.5 g/ml d-glucose, and 1,000 U/ml of leukemia inhibitory factors (LIF). To induce EB formation, 1 × 106 ES cells/10 cm dish were cultured in 10 ml ES medium without LIF. Intracellular NAD concentrations were determined using the NAD/NADH Quantification Kit (BioVision Inc.). Briefly, 2 × 105 cells were sonicated in the NAD/NADH extraction buffer and passed through 10-kDa cut-off filters. One half of the lysate was used to determine total NAD concentration; the other half was heated to 60°C for 30 minutes and used to determine NADH concentration. Reactions were prepared in triplicate in 96-well plates and read at 450 nm. NAD+ concentration was determined by subtracting the NADH from the total NAD concentration. Intracellular ROS levels were determined in freshly sorted ISL1+ CPCs cultured for 16 hours under normoxia or hypoxia using the CellROX probe (0.5 mM) (Thermo Fisher Scientific), which was directly added for 30 minutes to the culture medium. After 3 rinses with PBS at 37°C, cells were fixed for 5 minutes in PFA and rinsed 3 times in PBS. Cells were taken up in PBS and with an LSRII FACS (BD Biosciences). Data were analyzed using FlowJo software.
Cell culture and plasmid transfection.
HEK293T and C2C12 cells (ATTC) were grown in DMEM (Sigma-Aldrich) supplemented with 10% FCS (Sigma-Aldrich), 2 mM l-glutamine, 100 U penicillin, and 100 μg /ml streptomycin at 37°C, 5% CO2. HEK293T cells were transfected with 10 μg DNA using calcium phosphate precipitation at a density of 2 × 106/10 cm dish.
Luciferase reporter assay.
A 1.0-kb genomic DNA fragment upstream of the Isl1 translational start site was amplified and cloned into pTA-Luc (Promega). Mutations in the conserved HES1-binding site (N-box) of the Isl1 5′ promoter region were introduced using the QuikChange Site-Directed Mutagenesis Kit (Agilent) according to the manufacturer’s protocol. Primers are listed in Supplemental Table 3. Firefly luciferase and renilla activities were determined using the Dual-Luciferase Reporter Assay (Promega) with a Mithras LB940 plate reader (Berthold) 48 hours after Lipofectamine 2000–based transfections (Invitrogen) into HEK293T or C2C12 cells. Firefly luciferase activities were normalized to renilla. Each transfection was done in triplicate
shRNA-mediated knockdown by lentiviral infection.
Lentivirus-mediated shRNA knockdown of Sirt1 (Sigma-Aldrich MISSION shRNA library, SHCLNG-NM_019812.1), HIF1α (Sigma-Aldrich MISSION shRNA library, SHCLNG-NM_010431.1), Isl1 (CGGCAATCAAATTCACGACCA), and Hes1 (Sigma-Aldrich MISSION shRNA library, SHCLNG-XM_192801.2) was accomplished using previously described protocols (57). Nonconfluent cells were incubated for 24 hours in lentivirus-containing medium with 8 μg/ml polybrene, which was replaced with growth media containing 2 μg/ml puromycin for another 2 days before further analysis. Efficient knockdown of target genes was confirmed by both Western blot and RT-qPCR analysis.
Immunoprecipitation and Western blot analysis.
Embryonic hearts or FACS-isolated CPCs were washed in cold PBS and lysed in buffer containing 10 mM Tris (pH 7.4), 200 mM NaCl, 2 mM EDTA, 2 mM EGTA, and 1% Triton X-100 with protease inhibitors. Either 2 μg antibodies (listed in Supplemental Table 5) or IgG control was added to the lysate. Lysates were gently rotated for 4 hours before protein A agarose beads (Roche) were added for 2 hours at 4°C. After extensive washing with lysis buffer, beads were heated in SDS sample buffer or eluted with 3× Flag peptide (200 ng/ml) for second immunoprecipitation. Immunoprecipitated proteins were fractionated, blotted, and analyzed with different antibodies as indicated. For Western blots, tissue or cells were incubated in lysis buffer and resolved by SDS-PAGE before transfer to nitrocellulose filters. Protein expression was visualized using an enhanced chemiluminescence detection system (GE Healthcare) and quantified using a ChemiDoc gel documentation system (Bio-Rad). Antibodies are listed in Supplemental Table 5.
ChIP.
Tissue samples or freshly sorted cells were crosslinked with 1% formaldehyde (Sigma-Aldrich), and chromatin DNA was sheared to an average size of 200–500 bp by sonication with a Biorupter (Diagenode). Protein-DNA complexes were immunoprecipitated with control IgG or specific antibodies, followed by incubation with Protein A/G agrarose beads (Roche). After washing and elution, protein-DNA complexes were purified using chelex-100 (Bio-Rad) as described (58). Immunoprecipitated chromatin was analyzed by qPCR using SYBR green with primers specific for promoter regions of Isl1 and Nkx2.5 (see Supplemental Tables 3 and 4). All experiments were performed at least in triplicate.
Gene expression analyses.
Total RNA from FACS-isolated CPCs, embryonic hearts, or embryos was extracted using the TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. RNA was reverse transcribed with Superscript II (Invitrogen) following standard procedures. Real-time PCR was performed with 3 technical replicates using the iCycler (Bio-Rad) and the Absolute QPCR SYBR Green Fluorescein Mix (Abgene). Relative quantitation of mRNA gene expression was performed using either a standard curve–based data processing method as described (59) or the ΔCt method. The Ct values of the target genes were normalized to the m36b4 housekeeping gene using the equation ΔCt = Ctreference – Cttarget and expressed as ΔCt. Relative mRNA expressions are shown, with the average from control samples set as 0.5. Primers and PCR conditions are listed in Supplemental Table 4.
Whole-mount and histological analysis.
Embryos or embryonic hearts of different developmental stages were isolated and immediately fixed in 4% PFA. For paraffin sections, samples were dehydrated following standard protocols, embedded into paraffin, sectioned at 10 μm, and stained with H&E (Chroma). For cryosections, fixed tissues were equilibrated in 30% sucrose/PBS and frozen on dry ice. Sections of 8 μm were mounted on Superfrost slides for immunofluorescence staining. For in vitro CPC differentiation, purified ISL1+ CPCs were cultured and differentiated on fibronectin-coated glass chamber slides (fibronectin from BD Biosciences; glass chamber slides from Greiner) as described previously (60). HDAC inhibitors were added for 48 hours to cultured CPCs before fixation with 4% PFA and subsequent immunofluorescence staining. The following final concentrations were used: 1 μM for EX527 (Cayman), 5 μM for Ms257, 1 μM for MC1568, and 1 μM for TMP269 (Selleckchem). Antibodies for immunofluorescence are listed in Supplemental Table 5. Image acquisition and analysis were acquired with an ImageXpress microscope equipped with MetaXpress software (Molecular Devices). TUNEL assays to monitor apoptosis were carried out using the In Situ Cell Death Detection Kit (Roche) according to the manufacturer’s protocol. Whole-mount in situ hybridizations for Isl1 mRNA and Nkx2.5 mRNA were performed as described previously (55) using dual DIG-labeled Isl1 or Nkx2.5 antisense probes synthesized from ISL1- and NKX2.5-cDNA-pCR-BluntII-TOPO clones (21).
Statistics.
For all quantitative analyses, a minimum of 3 biological replicates were analyzed. Statistical tests were selected based on the assumption that sample data come from a population following a probability distribution based on a fixed set of parameters. To determine statistical significance of differences between 2 groups, t tests were used, and for multiple groups, ANOVA with Dunnett’s or Tukey’s post hoc correction was used, as indicated. P < 0.05 was considered statistically significant. Calculations were done using the GraphPad Prism 5 software package. Error bars represent SEM. No statistical method was used to predetermine sample size.
Study approval.
All animal experiments were done in accordance with the Guide for the Care and Use of Laboratory Animals (NIH publication no. 85-23, 1996) and were reviewed and approved by the Committee for Animal Rights Protection of the State of Hessen (Regierungspraesidium Darmstadt, Darmstadt, Germany, project number B2/1010).
Author contributions
XY and TB conceived and designed experiments and wrote the manuscript. XY performed most of the experiments, analyzed the data, and prepared figures. HQ and FW performed cell culture and mouse studies. XL performed shRNA knockdown and ChIP experiments. JF and EB generated essential reagents. YZ and GD contributed to experimental design, data analysis, and manuscript writing.
Supplementary Material
Acknowledgments
We thank Marion Wiesnet for technical help and Stefan Günther for FACS data analysis. This work is supported by the Deutsche Forschungsgemeinschaft (DFG) Excellence Cluster Cardio-Pulmonary System (ECCPS and SFB TR 81 TPA2), the LOEWE Center for Cell and Gene Therapy, the German Center for Cardiovascular Research, and the Foundation Leducq (13 CVD 01).
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information:J Clin Invest. 2017;127(6):2235–2248.https://doi.org/10.1172/JCI88725.
Contributor Information
Xuejun Yuan, Email: Xuejun.Yuan@mpi-bn.mpg.de.
Hui Qi, Email: Hui.Qi@mpi-bn.mpg.de.
Xiang Li, Email: xiang.li@mpi-bn.mpg.de.
Fan Wu, Email: fan.wu@mpi-bn.mpg.de.
Jian Fang, Email: Jian.Fang@mpi-bn.mpg.de.
Eva Bober, Email: Eva.Bober@mpi-bn.mpg.de.
Gergana Dobreva, Email: Gergana.Dobreva@mpi-bn.mpg.de.
Yonggang Zhou, Email: t.braun@kerckhoff.mpg.de.
Thomas Braun, Email: thomas.braun@mpi-bn.mpg.de.
References
- 1.Kelly RG, Buckingham ME, Moorman AF. Heart fields and cardiac morphogenesis. Cold Spring Harb Perspect Med. 2014;4(10):a015750. doi: 10.1101/cshperspect.a015750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fahed AC, Gelb BD, Seidman JG, Seidman CE. Genetics of congenital heart disease: the glass half empty. Circ Res. 2013;112(4):707–720. doi: 10.1161/CIRCRESAHA.112.300853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Patel SS, Burns TL. Nongenetic risk factors and congenital heart defects. Pediatr Cardiol. 2013;34(7):1535–1555. doi: 10.1007/s00246-013-0775-4. [DOI] [PubMed] [Google Scholar]
- 4.Kenchegowda D, Liu H, Thompson K, Luo L, Martin SS, Fisher SA. Vulnerability of the developing heart to oxygen deprivation as a cause of congenital heart defects. J Am Heart Assoc. 2014;3(3):e000841. doi: 10.1161/JAHA.114.000841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dunwoodie SL. The role of hypoxia in development of the Mammalian embryo. Dev Cell. 2009;17(6):755–773. doi: 10.1016/j.devcel.2009.11.008. [DOI] [PubMed] [Google Scholar]
- 6.Webster WS, Abela D. The effect of hypoxia in development. Birth Defects Res C Embryo Today. 2007;81(3):215–228. doi: 10.1002/bdrc.20102. [DOI] [PubMed] [Google Scholar]
- 7.Miao CY, Zuberbuhler JS, Zuberbuhler JR. Prevalence of congenital cardiac anomalies at high altitude. J Am Coll Cardiol. 1988;12(1):224–228. doi: 10.1016/0735-1097(88)90378-6. [DOI] [PubMed] [Google Scholar]
- 8.Hasan A. Relationship of high altitude and congenital heart disease. Indian Heart J. 2016;68(1):9–12. doi: 10.1016/j.ihj.2015.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Simon MC, Keith B. The role of oxygen availability in embryonic development and stem cell function. Nat Rev Mol Cell Biol. 2008;9(4):285–296. doi: 10.1038/nrm2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mohyeldin A, Garzón-Muvdi T, Quiñones-Hinojosa A. Oxygen in stem cell biology: a critical component of the stem cell niche. Cell Stem Cell. 2010;7(2):150–161. doi: 10.1016/j.stem.2010.07.007. [DOI] [PubMed] [Google Scholar]
- 11.Majmundar AJ, Wong WJ, Simon MC. Hypoxia-inducible factors and the response to hypoxic stress. Mol Cell. 2010;40(2):294–309. doi: 10.1016/j.molcel.2010.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaelin WG, Ratcliffe PJ. Oxygen sensing by metazoans: the central role of the HIF hydroxylase pathway. Mol Cell. 2008;30(4):393–402. doi: 10.1016/j.molcel.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 13.Semenza GL. Hypoxia-inducible factors in physiology and medicine. Cell. 2012;148(3):399–408. doi: 10.1016/j.cell.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Buckingham M, Meilhac S, Zaffran S. Building the mammalian heart from two sources of myocardial cells. Nat Rev Genet. 2005;6(11):826–835. doi: 10.1038/nrg1710. [DOI] [PubMed] [Google Scholar]
- 15.Meilhac SM, Esner M, Kelly RG, Nicolas JF, Buckingham ME. The clonal origin of myocardial cells in different regions of the embryonic mouse heart. Dev Cell. 2004;6(5):685–698. doi: 10.1016/S1534-5807(04)00133-9. [DOI] [PubMed] [Google Scholar]
- 16.Evans SM, Yelon D, Conlon FL, Kirby ML. Myocardial lineage development. Circ Res. 2010;107(12):1428–1444. doi: 10.1161/CIRCRESAHA.110.227405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stainier DY. Zebrafish genetics and vertebrate heart formation. Nat Rev Genet. 2001;2(1):39–48. doi: 10.1038/35047564. [DOI] [PubMed] [Google Scholar]
- 18.Zhou Y, Kim J, Yuan X, Braun T. Epigenetic modifications of stem cells: a paradigm for the control of cardiac progenitor cells. Circ Res. 2011;109(9):1067–1081. doi: 10.1161/CIRCRESAHA.111.243709. [DOI] [PubMed] [Google Scholar]
- 19.Prall OW, et al. An Nkx2-5/Bmp2/Smad1 negative feedback loop controls heart progenitor specification and proliferation. Cell. 2007;128(5):947–959. doi: 10.1016/j.cell.2007.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dyer LA, Kirby ML. The role of secondary heart field in cardiac development. Dev Biol. 2009;336(2):137–144. doi: 10.1016/j.ydbio.2009.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dorn T, et al. Direct nkx2-5 transcriptional repression of isl1 controls cardiomyocyte subtype identity. Stem Cells. 2015;33(4):1113–1129. doi: 10.1002/stem.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Watanabe Y, et al. Fibroblast growth factor 10 gene regulation in the second heart field by Tbx1, Nkx2-5, and Islet1 reveals a genetic switch for down-regulation in the myocardium. Proc Natl Acad Sci U S A. 2012;109(45):18273–18280. doi: 10.1073/pnas.1215360109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cai CL, et al. Isl1 identifies a cardiac progenitor population that proliferates prior to differentiation and contributes a majority of cells to the heart. Dev Cell. 2003;5(6):877–889. doi: 10.1016/S1534-5807(03)00363-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kwon C, Qian L, Cheng P, Nigam V, Arnold J, Srivastava D. A regulatory pathway involving Notch1/beta-catenin/Isl1 determines cardiac progenitor cell fate. Nat Cell Biol. 2009;11(8):951–957. doi: 10.1038/ncb1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krishnan J, et al. Essential role of developmentally activated hypoxia-inducible factor 1alpha for cardiac morphogenesis and function. Circ Res. 2008;103(10):1139–1146. doi: 10.1161/01.RES.0000338613.89841.c1. [DOI] [PubMed] [Google Scholar]
- 26.Arany Z, et al. An essential role for p300/CBP in the cellular response to hypoxia. Proc Natl Acad Sci U S A. 1996;93(23):12969–12973. doi: 10.1073/pnas.93.23.12969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ema M, et al. Molecular mechanisms of transcription activation by HLF and HIF1alpha in response to hypoxia: their stabilization and redox signal-induced interaction with CBP/p300. EMBO J. 1999;18(7):1905–1914. doi: 10.1093/emboj/18.7.1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eltzschig HK, et al. HIF-1-dependent repression of equilibrative nucleoside transporter (ENT) in hypoxia. J Exp Med. 2005;202(11):1493–1505. doi: 10.1084/jem.20050177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Prozorovski T, et al. Sirt1 contributes critically to the redox-dependent fate of neural progenitors. Nat Cell Biol. 2008;10(4):385–394. doi: 10.1038/ncb1700. [DOI] [PubMed] [Google Scholar]
- 30.Rochais F, Dandonneau M, Mesbah K, Jarry T, Mattei MG, Kelly RG. Hes1 is expressed in the second heart field and is required for outflow tract development. PLoS ONE. 2009;4(7):e6267. doi: 10.1371/journal.pone.0006267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nasrin N, et al. JNK1 phosphorylates SIRT1 and promotes its enzymatic activity. PLoS ONE. 2009;4(12):e8414. doi: 10.1371/journal.pone.0008414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cheng HL, et al. Developmental defects and p53 hyperacetylation in Sir2 homolog (SIRT1)-deficient mice. Proc Natl Acad Sci U S A. 2003;100(19):10794–10799. doi: 10.1073/pnas.1934713100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Karamboulas C, et al. HDAC activity regulates entry of mesoderm cells into the cardiac muscle lineage. J Cell Sci. 2006;119(Pt 20):4305–4314. doi: 10.1242/jcs.03185. [DOI] [PubMed] [Google Scholar]
- 34.Breckenridge RA, et al. Hypoxic regulation of hand1 controls the fetal-neonatal switch in cardiac metabolism. PLoS Biol. 2013;11(9):e1001666. doi: 10.1371/journal.pbio.1001666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guarani V, Potente M. SIRT1 - a metabolic sensor that controls blood vessel growth. Curr Opin Pharmacol. 2010;10(2):139–145. doi: 10.1016/j.coph.2010.01.001. [DOI] [PubMed] [Google Scholar]
- 36.Hsu CP, et al. Silent information regulator 1 protects the heart from ischemia/reperfusion. Circulation. 2010;122(21):2170–2182. doi: 10.1161/CIRCULATIONAHA.110.958033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Potente M, et al. SIRT1 controls endothelial angiogenic functions during vascular growth. Genes Dev. 2007;21(20):2644–2658. doi: 10.1101/gad.435107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Della-Morte D, Dave KR, DeFazio RA, Bao YC, Raval AP, Perez-Pinzon MA. Resveratrol pretreatment protects rat brain from cerebral ischemic damage via a sirtuin 1-uncoupling protein 2 pathway. Neuroscience. 2009;159(3):993–1002. doi: 10.1016/j.neuroscience.2009.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sakamoto J, Miura T, Shimamoto K, Horio Y. Predominant expression of Sir2alpha, an NAD-dependent histone deacetylase, in the embryonic mouse heart and brain. FEBS Lett. 2004;556(1–3):281–286. doi: 10.1016/s0014-5793(03)01444-3. [DOI] [PubMed] [Google Scholar]
- 40.Nath KA. The role of Sirt1 in renal rejuvenation and resistance to stress. J Clin Invest. 2010;120(4):1026–1028. doi: 10.1172/JCI42184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dioum EM, et al. Regulation of hypoxia-inducible factor 2alpha signaling by the stress-responsive deacetylase sirtuin 1. Science. 2009;324(5932):1289–1293. doi: 10.1126/science.1169956. [DOI] [PubMed] [Google Scholar]
- 42.Yoon H, Shin SH, Shin DH, Chun YS, Park JW. Differential roles of Sirt1 in HIF-1α and HIF-2α mediated hypoxic responses. Biochem Biophys Res Commun. 2014;444(1):36–43. doi: 10.1016/j.bbrc.2014.01.001. [DOI] [PubMed] [Google Scholar]
- 43.Stevens KN, et al. Common variation in ISL1 confers genetic susceptibility for human congenital heart disease. PLoS ONE. 2010;5(5):e10855. doi: 10.1371/journal.pone.0010855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Friedrich FW, et al. A novel genetic variant in the transcription factor Islet-1 exerts gain of function on myocyte enhancer factor 2C promoter activity. Eur J Heart Fail. 2013;15(3):267–276. doi: 10.1093/eurjhf/hfs178. [DOI] [PubMed] [Google Scholar]
- 45.Osoegawa K, Schultz K, Yun K, Mohammed N, Shaw GM, Lammer EJ. Haploinsufficiency of insulin gene enhancer protein 1 (ISL1) is associated with d-transposition of the great arteries. Mol Genet Genomic Med. 2014;2(4):341–351. doi: 10.1002/mgg3.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Witzel HR, Jungblut B, Choe CP, Crump JG, Braun T, Dobreva G. The LIM protein Ajuba restricts the second heart field progenitor pool by regulating Isl1 activity. Dev Cell. 2012;23(1):58–70. doi: 10.1016/j.devcel.2012.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Caputo L, et al. The Isl1/Ldb1 complex orchestrates genome-wide chromatin organization to instruct differentiation of multipotent cardiac Progenitors. Cell Stem Cell. 2015;17(3):287–299. doi: 10.1016/j.stem.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Srinivas S, et al. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Eggan K, et al. Hybrid vigor, fetal overgrowth, and viability of mice derived by nuclear cloning and tetraploid embryo complementation. Proc Natl Acad Sci USA. 2001;98(11):6209–6214. doi: 10.1073/pnas.101118898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang L, et al. Isl1Cre reveals a common Bmp pathway in heart and limb development. Development. 2006;133(8):1575–1585. doi: 10.1242/dev.02322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Osterwalder M, Galli A, Rosen B, Skarnes WC, Zeller R, Lopez-Rios J. Dual RMCE for efficient re-engineering of mouse mutant alleles. Nat Methods. 2010;7(11):893–895. doi: 10.1038/nmeth.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Petrie K, et al. The histone deacetylase 9 gene encodes multiple protein isoforms. J Biol Chem. 2003;278(18):16059–16072. doi: 10.1074/jbc.M212935200. [DOI] [PubMed] [Google Scholar]
- 53.Zhou Y, Santoro R, Grummt I. The chromatin remodeling complex NoRC targets HDAC1 to the ribosomal gene promoter and represses RNA polymerase I transcription. EMBO J. 2002;21(17):4632–4640. doi: 10.1093/emboj/cdf460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhou Y, Schmitz KM, Mayer C, Yuan X, Akhtar A, Grummt I. Reversible acetylation of the chromatin remodelling complex NoRC is required for non-coding RNA-dependent silencing. Nat Cell Biol. 2009;11(8):1010–1016. doi: 10.1038/ncb1914. [DOI] [PubMed] [Google Scholar]
- 55.Wystub K, Besser J, Bachmann A, Boettger T, Braun T. miR-1/133a clusters cooperatively specify the cardiomyogenic lineage by adjustment of myocardin levels during embryonic heart development. PLoS Genet. 2013;9(9):e1003793. doi: 10.1371/journal.pgen.1003793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bu L, et al. Human ISL1 heart progenitors generate diverse multipotent cardiovascular cell lineages. Nature. 2009;460(7251):113–117. doi: 10.1038/nature08191. [DOI] [PubMed] [Google Scholar]
- 57.Zhang T, et al. Prmt5 is a regulator of muscle stem cell expansion in adult mice. Nat Commun. 2015;6:7140. doi: 10.1038/ncomms8140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nelson JD, Denisenko O, Bomsztyk K. Protocol for the fast chromatin immunoprecipitation (ChIP) method. Nat Protoc. 2006;1(1):179–185. doi: 10.1038/nprot.2006.27. [DOI] [PubMed] [Google Scholar]
- 59.Larionov A, Krause A, Miller W. A standard curve based method for relative real time PCR data processing. BMC Bioinformatics. 2005;6:62. doi: 10.1186/1471-2105-6-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Domian IJ, et al. Generation of functional ventricular heart muscle from mouse ventricular progenitor cells. Science. 2009;326(5951):426–429. doi: 10.1126/science.1177350. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.