Abstract
Following migration of primordial germ cells to the genital ridge, oogonia undergo several rounds of mitotic division and enter meiosis at approximately E13.5. Most oocytes arrest in the dictyate (diplotene) stage of meiosis circa E18.5. The genes necessary to drive oocyte differentiation in parallel with meiosis are unknown. Here, we have investigated whether expression of spermatogenesis and oogenesis bHLH transcription factor 1 (Sohlh1) and Sohlh2 coordinates oocyte differentiation within the embryonic ovary. We found that SOHLH2 protein was expressed in the mouse germline as early as E12.5 and preceded SOHLH1 protein expression, which occurred circa E15.5. SOHLH1 protein appearance at E15.5 correlated with SOHLH2 translocation from the cytoplasm into the nucleus and was dependent on SOHLH1 expression. NOBOX oogenesis homeobox (NOBOX) and LIM homeobox protein 8 (LHX8), two important regulators of postnatal oogenesis, were coexpressed with SOHLH1. Single deficiency of Sohlh1 or Sohlh2 disrupted the expression of LHX8 and NOBOX in the embryonic gonad without affecting meiosis. Sohlh1-KO infertility was rescued by conditional expression of the Sohlh1 transgene after the onset of meiosis. However, Sohlh1 or Sohlh2 transgene expression could not rescue Sohlh2-KO infertility due to a lack of Sohlh1 or Sohlh2 expression in rescued mice. Our results indicate that Sohlh1 and Sohlh2 are essential regulators of oocyte differentiation but do not affect meiosis I.
Introduction
In contrast to the sex-determining region Y–dependent (SRY-dependent) testis-determining system in early XY gonads, the molecular mechanisms behind XX female gonadal fate are poorly understood. Unlike male germ cells, in which differentiation culminates in meiosis, the oocyte is a unique multitasker, with meiosis and oocyte differentiation proceeding alongside each other. The somatic component plays an important role in XX determination, just as it does in well-characterized XY gonadal differentiation, and forkhead box L2 (Foxl2) expression circa E12 is an important determinant of granulosa cell differentiation and oocyte maintenance (1). However, little is understood regarding oocyte differentiation outside of the somatic sphere of influence.
Previous studies have discovered a group of transcription factors that play a crucial role in postnatal oocyte differentiation; these include spermatogenesis and oogenesis bHLH transcription factor 1 (Sohlh1) (2), Sohlh2 (3, 4), LIM homeobox protein (Lhx8) (5, 6), NOBOX oogenesis homeobox (Nobox) (7), and folliculogenesis-specific basic helix-loop-helix (Figla) (8) genes. Sohlh1 and Sohlh2 are expressed preferentially in the male and female germline and are critical for male and female germline development. Lhx8, Nobox, and Figla are mainly expressed in the female germline, and their KOs disrupt only ovarian development, without affecting male germline differentiation. The postnatal phenotypes in ovaries deficient in these transcriptional regulators share many similarities: embryonic gonads look relatively unaffected on gross histology and oocyte counts, and newborn ovaries are similarly unaffected. Within the first few days of postnatal life, the histology reveals significant disruption in follicle formation, with ensuing oocyte death within a few weeks. However, it is clear from ultrastructural studies of embryonic Nobox-deficient gonads that anatomic abnormalities, such as deficient formation of somatic cell projections, commence as early as E16.5 (7). We therefore focused our studies on the prenatal regulation of oocyte differentiation by oocyte-specific transcriptional regulators. Here, we show that SOHLH1 and SOHLH2 protein expression commence in the embryonic gonad and that Sohlh1 and Sohlh2 gene deficiency disrupts embryonic expression of the oocyte differentiation factors NOBOX and LHX8, without an appreciable effect on meiosis I. We conclude that Sohlh1 and Sohlh2 are critical genes in oocyte differentiation, independent of meiosis. Together with findings from previous studies (9), these results indicate that Sohlh1 and Sohlh2 genes are universal regulators of both male and female germline differentiation, with distinct and sex-specific downstream pathways.
Results
SOHLH2 nucleocytoplasmic translocation is dependent on SOHLH1 expression.
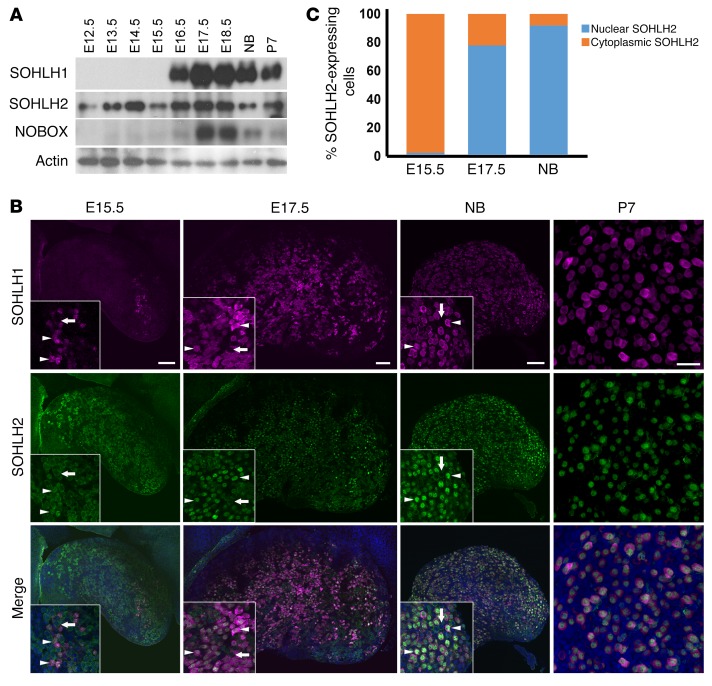
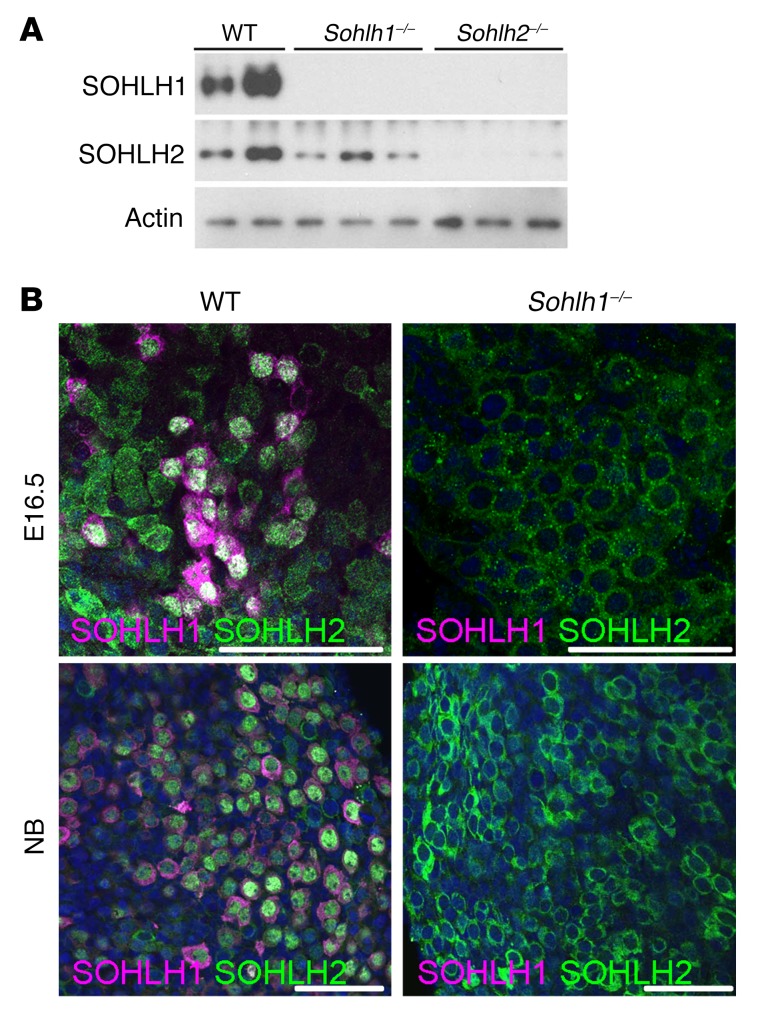
Little is known regarding embryonic expression of SOHLH1, SOHLH2, LHX8, and NOBOX in embryonic gonads. We evaluated the expression of these proteins at different developmental stages of the ovary, using immunofluorescence and Western blotting. SOHLH2 was expressed as early as E12.5, prior to oocyte entry into meiosis (Figure 1A), and preceded SOHLH1 expression. NOBOX protein and RNA expression had an onset similar to that for SOHLH1 expression in the embryonic ovaries (Figure 1A and Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/JCI90281DS1). Interestingly, SOHLH2 expression circa E15.5 and earlier was exclusively cytoplasmic (Figure 1, B and C). We did not detect SOHLH1 expression at E12.5 by immunofluorescence or by composite data derived from the NCBI’s Gene Expression Omnibus (GEO) profiles on embryonic ovarian RNA expression (Supplemental Figure 1). At E15.5, oocytes had entered the zygotene stage of meiosis I. At this time, a small subset of oocytes expressed the SOHLH1 protein, which was located in both the cytoplasm and the nucleus (Figure 1B). Western blotting did not detect SOHLH1 at E15.5 (Figure 1A), while immunofluorescence did, perhaps due to epitope conformation or sensitivity. In oocytes that expressed SOHLH1 at E15.5, we found that SOHLH2 expression was mostly confined to the nucleus (Figure 1, B and C). At E17.5 and beyond, 80% of germ cells coexpressed SOHLH1 and SOHLH2, with SOHLH2 expression mainly confined to the nucleus, while SOHLH1 expression was detected in both the nucleus and cytoplasm (Figure 1, B and C). We found that SOHLH2 translocation from the cytoplasm to the nucleus was closely associated with expression of SOHLH1. SOHLH1 and SOHLH2 proteins were also coexpressed postnatally and were detectable mainly in the oocytes of primordial follicles and a few primary follicles (Figure 1, B and C) (4, 5). Western blotting did not detect SOHLH1 protein in Sohlh2-KO ovaries, while SOHLH2 expression persisted in Sohlh1-KO ovaries (Figure 2A). The Western blot results are consistent with SOHLH2 expression being upstream of SOHLH1. We also assessed whether SOHLH2 localization is disrupted in Sohlh1-deficient embryonic ovaries. Immunofluorescence staining with anti-SOHLH1 and anti-SOHLH2 antibodies on ovaries derived from E16.5 and newborn (E19.5) Sohlh1-deficient animals showed that Sohlh1 deficiency abrogated SOHLH2 translocation into the nucleus (Figure 2B). These results further indicate that SOHLH2 nuclear localization is dependent on SOHLH1 expression.
Figure 1. Expression of SOHLH2 and SOHLH1 during embryonic development.
(A) Western blot analysis of protein extracts from embryonic (E12.5–E18.5), newborn (NB), and P7 ovaries. Affinity-purified polyclonal antibodies were used to detect SOHLH1, SOHLH2, NOBOX, and actin expression (n = 3 per group). (B) Whole-mount immunofluorescence staining with SOHLH1 (magenta) and SOHLH2 (green) in ovaries at E15.5, E17.5, the newborn (NB) stage, and P7. White arrows indicate oocytes that lack SOHLH1 expression and show SOHLH2 localization to the cytoplasm, while white arrowheads indicate oocytes expressing both SOHLH1 and SOHLH2 (n = 3 per group). Scale bars: 100 μm. Original magnification: ×40. (C) Quantitation of SOHLH2-expressing cells showing nuclear or cytoplasmic localization (n = 3 per group). Data are representative of at least 3 independent experiments.
Figure 2. Sohlh1 deficiency disrupts SOHLH2 cellular localization.
(A) Western blot of proteins extracted from newborn ovaries show a lack of expression of SOHLH1 and SOHLH2 in Sohlh2–/– ovaries and the presence of SOHLH2 in Sohlh1–/– ovaries (n = 3 per group). (B) Immunofluorescence staining with anti-SOHLH1 (magenta) and anti-SOHLH2 (green) antibodies in E16.5 and newborn ovaries. At both time points, SOHLH1-deficient oocytes did not express SOHLH2 in the nucleus (n = 3 per group). DNA was stained with DAPI (blue). Data are representative of at least 3 independent experiments. Scale bars: 50 μm.
SOHLH1 and SOHLH2 regulate oocyte growth and differentiation.
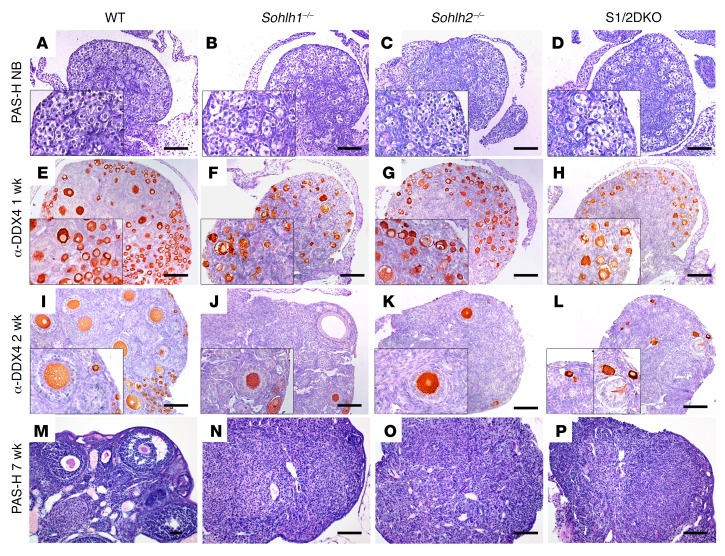
SOHLH1 and SOHLH2 proteins are known to interact in vivo, and we hypothesized that complete elimination of SOHLH1 and SOHLH2 proteins from the ovaries may cause a more severe developmental defect than would single deficiency (9, 10). We generated Sohlh1/Sohlh2 double-deficiency mice (referred to hereafter as S1/2DKO mice) to investigate the effects of combined deficiency on oogenesis. Mice with loss of both Sohlh1 and Sohlh2 in their ovaries showed no histomorphological differences when compared with single-deficiency Sohlh1 and Sohlh2 mice, and both combined and single-deficiency ovaries were substantially smaller when compared with WT ovaries (Figure 3). In newborn mice, we found that Sohlh1- or Sohlh2 single- or double-KO ovarian histology was no different from that of WT littermates (Figure 3, A–D). Sohlh1 or Sohlh2 single-KO ovaries, as well as S1/2DKO ovaries, have remarkably diminished primordial and primary follicles when compared with 1- and 2-week-old WT littermates (Figure 3, E–L) and lose the vast majority of oocytes by the time they reach reproductive maturity at approximately 7 weeks of age (Figure 3, M–P).
Figure 3. Sohlh1 and Sohlh2 double deficiency disrupts folliculogenesis similarly to single deficiency of Sohlh1 or Sohlh2.
(A–P) PAS staining of newborn ovaries. Ovarian phenotype of WT, Sohlh1–/–, Sohlh2–/–, and S1/2DKO mice at the indicated stages of postnatal ovarian development. (E–L) IHC with anti-DDX4 antibody in ovaries harvested from 1- and 2-week-old mice. (M–P) PAS staining of 7-week-old ovaries for the indicated genotypes (n = 3 per group). Data are representative of at least 3 independent experiments. Scale bars: 100 μm. Original magnifcation: ×40. PAS-H, periodic acid–Schiff-hematoxylin.
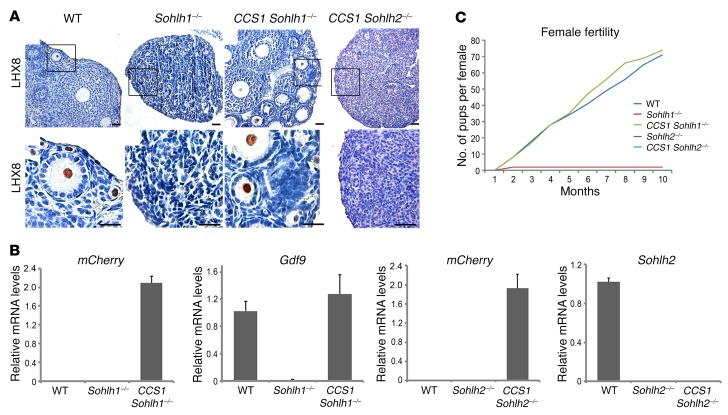
Given that Sohlh1-KO, Sohlh2-KO, and S1/2DKO phenotypes are histologically identical despite a different onset of their embryonic expression (4), that SOHLH2 protein precedes SOHLH1 expression, and that SOHLH2 protein translocation to the nucleus is associated with the appearance of SOHLH1 expression circa E15.5, we further tested the hypothesis that the phenotypes observed in Sohlh2-deficient ovaries are solely due to the disruption of SOHLH1 expression. If this is the case, then activating Sohlh1 expression in Sohlh2-KOs may rescue the Sohlh2-KO phenotype and lead to normal oogenesis. To address this question, we engineered a transgenic mouse that expresses Sohlh1-mCherry under the CMV early enhancer–chicken β-actin hybrid (CAG) promoter, when Cre-mediated excision removes the chloramphenicol acetyltransferase (CAT) gene, which lies between the CAG promoter and Sohlh1 (Supplemental Figure 2) (11). We labeled this conditional Sohlh1-expressing construct CCS1. We used DEAD (Asp-Glu-Ala-Asp) box polypeptide–Cre (Ddx4-Cre) (12), whose expression commences between E15.5 and E18.5, after the onset of meiosis, to activate Sohlh1 expression within superoxide dismutase copper chaperone (CCS1). Moreover, we found that Sohlh1 and Sohlh2 deficiencies did not suppress DDX4 protein expression (Supplemental Figure 3); therefore, Ddx4 promoter–driven Cre will be expressed in the background of Sohlh1- and Sohlh2-deficient ovaries. Ddx4-Cre–driven expression of Sohlh1 from the CCS1 transgene rescued ovarian development and fertility in Sohlh1-KOs (Figure 4A). The expression of mCherry and growth differentiation factor 9 (Gdf9) RNA in the CCS1 Sohlh1–/– ovary indicated successful expression of the Sohlh1-mCherry transgene in Sohlh1–/– mice (Figure 4B). Moreover, SOHLH2 expression in the CCS1 Sohlh1–/– animals was now shifted from the cytoplasm to the nucleus (Supplemental Figure 4). These rescue results are consistent with the appearance of Sohlh1 circa E15.5 and indicate that Sohlh1 does not play a critical role prior to the formation of the embryonic gonad. These results also indicate that Sohlh1 is not essential for meiosis, as meiosis commences around E13.5.
Figure 4. Sohlh1 ectopic expression on the background of Sohlh1- and Sohlh2-deficient animals.
(A) Conditional expression of Sohlh1 from the CCS1 transgene (Supplemental Figure 2) with the Ddx4 promoter–driven Cre rescued the Sohlh1–/– phenotype (CCS1 Sohlh1–/–) but not the Sohlh2–/– phenotype (CCS1 Sohlh2–/–). Histology sections were stained with anti-LHX8 antibodies to identify oocyte nuclei (n = 3 per group). WT ovaries show various stages of folliculogenesis, while Sohlh1–/– ovaries lack follicles beyond 3 weeks of postnatal life. Scale bars: 30 μm. Black squares in the upper panel are magnified in the lower panel. Original magnification: ×40. (B) RNA expression of mCherry, Gdf9, and Sohlh2 from the ovaries of mice of various genotypes. mCherry expression shows that the CAG promoter in CCS1 is active in the presence of Ddx4-Cre (n = 6 per group). Error bars indicate the mean ± SD. (C) Fertility data (pups per female) for mice of various genotypes are plotted for a 10-month period (n = 3 per group). Data are representative of at least 3 independent experiments.
We then assessed whether Sohlh1 expression on the Sohlh2-KO background can rescue ovarian development. We assessed fertility parameters such as pups per litter over a 10-month period and performed histomorphometric analyses to determine the presence and types of follicles from 5 pairs of ovaries per genotype (Figure 4, A and C). Sohlh1 expression under the control of Ddx4-Cre did not rescue Sohlh2-KO mice (Figure 4A), despite positive expression of Sohlh1, as evidenced by the presence of the mCherry transcript (Figure 4B). These results indicate that genes other than Sohlh1 are important for Sohlh2 downstream effects or that SOHLH2 expression (absent in the rescue) and heterodimerization (9) with SOHLH1 protein are essential steps for downstream effects and rescue.
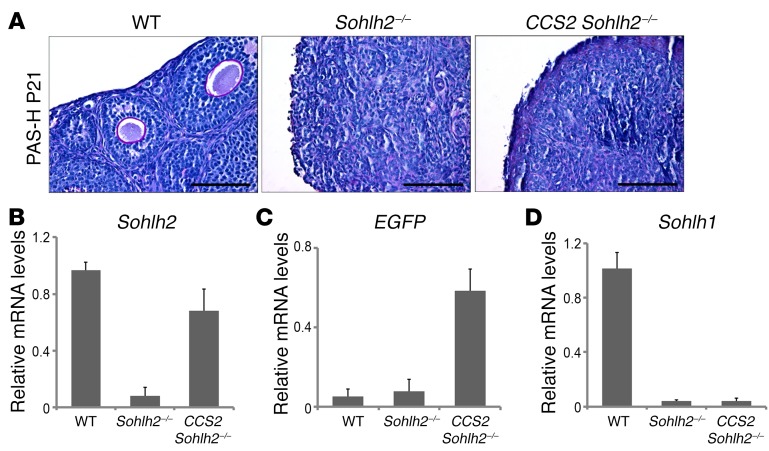
We also attempted to rescue the Sohlh2-KO using the Sohlh2 transgene, whose expression is driven by a floxed CAG promoter (CCS2 transgene line, identical to CCS1, except for Sohlh2 expression). The CCS2 transgene in the presence of Ddx4-Cre and on the background of Sohlh2-KO (CCS2 Sohlh2–/–) did not rescue Sohlh2-KOs (Figure 5A), unlike the ability of CCS1 to rescue Sohlh1-KOs at the same time point (E15.5). One explanation is that, in the Sohlh1-KO rescue with CCS1, there is an ongoing expression of Sohlh2, which allows for SOHLH1:SOHLH2 heterodimers to form and for oogenesis to proceed. In the Sohlh2-KO, even with Sohlh2 rescue with the CCS2 transgene, the expression of Sohlh1 was not restored (Figure 5, B–D), and SOHLH2:SOHLH1 heterodimers were not formed. The lack of Sohlh1 re-expression in CCS2 Sohlh2–/– mice is probably why the fertility of Sohlh2–/– mice fertility was not rescued.
Figure 5. Sohlh2 overexpression with Ddx4 promoter–driven Cre fails to reverse the Sohlh2–/– phenotype.
(A) PAS staining of WT, Sohlh2–/–, and CCS2 Sohlh2–/– ovaries at P21. Hematoxylin was used for counterstaining (n = 3 per group). Scale bars: 50 μm. (B–D) RNA expression of Sohlh2, EGFP, and Sohlh1. Sohlh2 was fused with EGFP, and expression of EGFP showed that the Ddx4-Cre promoter was active in Sohlh2–/– ovaries. However, overexpression of Sohlh2 in Sohlh2–/– ovaries could not rescue Sohlh1 expression (n = 6 per group). Error bars indicate the mean ± SD. Data are representative of at least 3 independent experiments.
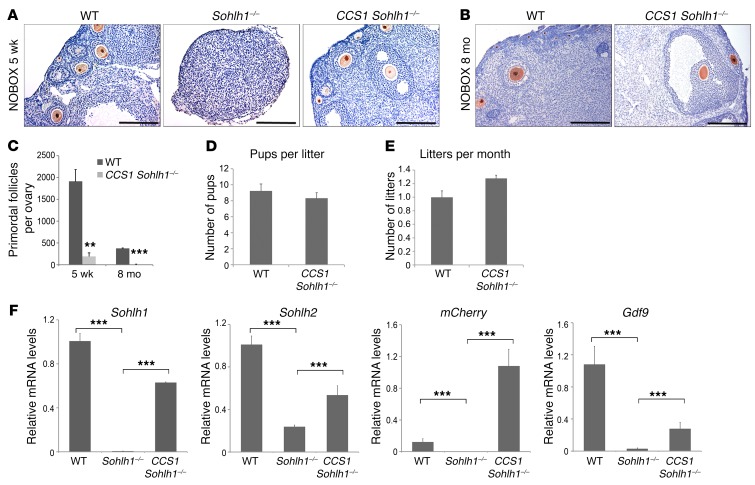
We also wanted to test the hypothesis that Sohlh1-KO rescue will occur when Sohlh1 is expressed postnatally. We used Gdf9-Cre to induce Sohlh1 expression in CCS1 Sohlh1–/– mice. Gdf9-Cre expression commences on P3 in oocytes of primordial follicles (13). To our surprise, CCS1 Sohlh1–/– mice had normal folliculogenesis and were fertile (Figure 6, A, B, D, and E). However, they had a significantly reduced primordial follicle pool size compared with that of WT mice, and expression of Sohlh1, Sohlh2, and Gdf9 was partially rescued (Figure 6, C and F). Nonetheless, these data show that Sohlh1–/– oocytes in newborn ovaries can be rescued and can be fertilized and produce offspring.
Figure 6. Sohlh1 overexpression with Gdf9 promoter–driven Cre rescues Sohlh1–/– fertility but not the primordial follicle reserve.
(A and B) Conditional expression of Sohlh1 from the CCS1 transgene with Gdf9-Cre rescued Sohlh1–/– folliculogenesis. Histology sections were stained with anti-NOBOX antibodies to identify oocyte nuclei. The WT ovary showed various stages of folliculogenesis, while the Sohlh1–/– ovary lacked oocytes at 5 weeks. CCS1 Sohlh1–/– mice showed normal folliculogenesis from primary to large antral follicles at 5 weeks and 8 months, but had an unusually low number of primordial follicles (n = 3 per group). (C) Primordial follicle numbers in WT and CCS1 Sohlh1–/– mice at 5 weeks and 8 months (n = 5 per group). (D and E) Fertility testing of CCS1 Sohlh1–/– mice over a period of 6 months (n = 3 per group). (F) Expression levels of Sohlh1, Sohlh2, mCherry, and Gdf9 in WT, Sohlh1-deficient (Sohlh1–/–), and Sohlh1–/–-rescued ovaries (CCS1 Sohlh1–/–). **P < 0.01 and ***P < 0.001, by 2-tailed Student’s t test. Data represent the mean ± SEM and are representative of at least 3 independent experiments.
Oocyte-specific transcriptional regulator expression is dependent on Sohlh1 and Sohlh2.
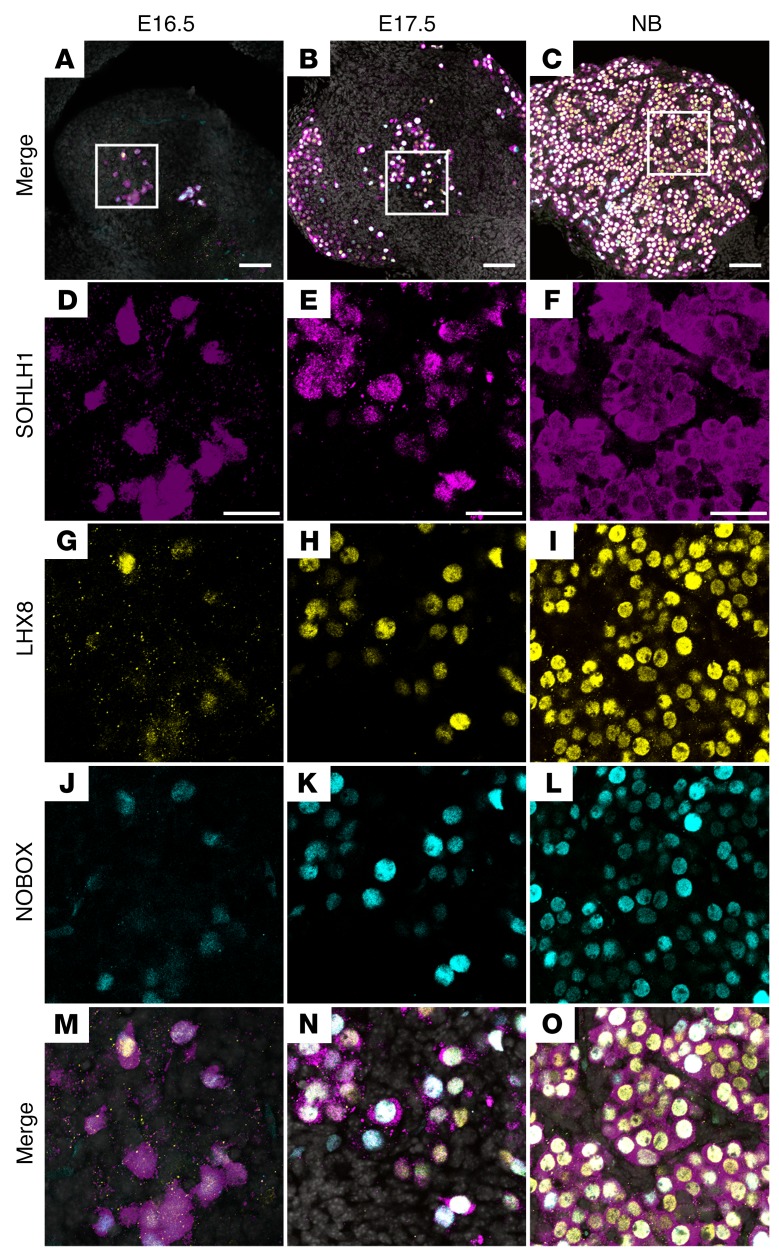
We hypothesized that the appearance of SOHLH1 around E15.5, with concomitant nuclear translocation of SOHLH2, correlates with the embryonic appearance of transcripts that encode genes essential for oocyte differentiation. Previous studies have shown that postnatal deficiency of SOHLH1 and SOHLH2 leads to downregulation of multiple oocyte-specific transcriptional regulators, including Nobox, Figla, KIT proto-oncogene receptor tyrosine kinase (Kit), and Lhx8 (5, 14). We used publicly available GEO profiles of ovarian development to assess the expression of Sohlh1, which correlated very well with our experimental data, and chose Sohlh1 profile neighbors. We discovered that Nobox, Figla, Kit, and Lhx8 overlap Sohlh1 expression (Supplemental Figure 1). Western blot analysis using anti-NOBOX antibodies followed the Nobox RNA GEO profile expression pattern and overlapped with SOHLH1 protein and RNA expression (Figure 1A and Supplemental Figure 1). We analyzed coexpression of SOHLH1, NOBOX, and LHX8 proteins in embryonic ovaries using immunofluorescence with affinity-purified antibodies against SOHLH1, NOBOX, and LHX8. A subset of oocytes within the E16.5 ovary expressed SOHLH1 protein, which was localized in both the nucleus and cytoplasm. We found that NOBOX and LHX8 expression commenced solely in SOHLH1-positive cells in E16.5 ovaries and that more than 90% of the oocytes in E17.5 and newborn ovaries coexpressed SOHLH1 together with LHX8 and NOBOX (Figure 7). These results are consistent with GEO profile expression data showing that essential transcriptional regulators are profile neighbors and are coexpressed in the same subset of germ cells.
Figure 7. SOHLH1, LHX8, and NOBOX are coexpressed in embryonic ovaries.
(A–C) Low-magnification merge images show squared areas that are magnified in D–O. Images show immunofluorescence results from whole-mount ovaries stained with antibodies against SOHLH1 (D–F, magenta), LHX8 (G–I, yellow), and NOBOX (J–L, cyan) at the indicated ages. (M–O) Merge images at higher magnification show coexpression of all 3 transcriptional regulators in the vast majority of cells at the indicated time points. DNA was stained with DAPI (blue) (n = 3 per group). Data are representative of at least 3 independent experiments. Scale bars: (A–C) 100 µm; (D–O) 50 µm.
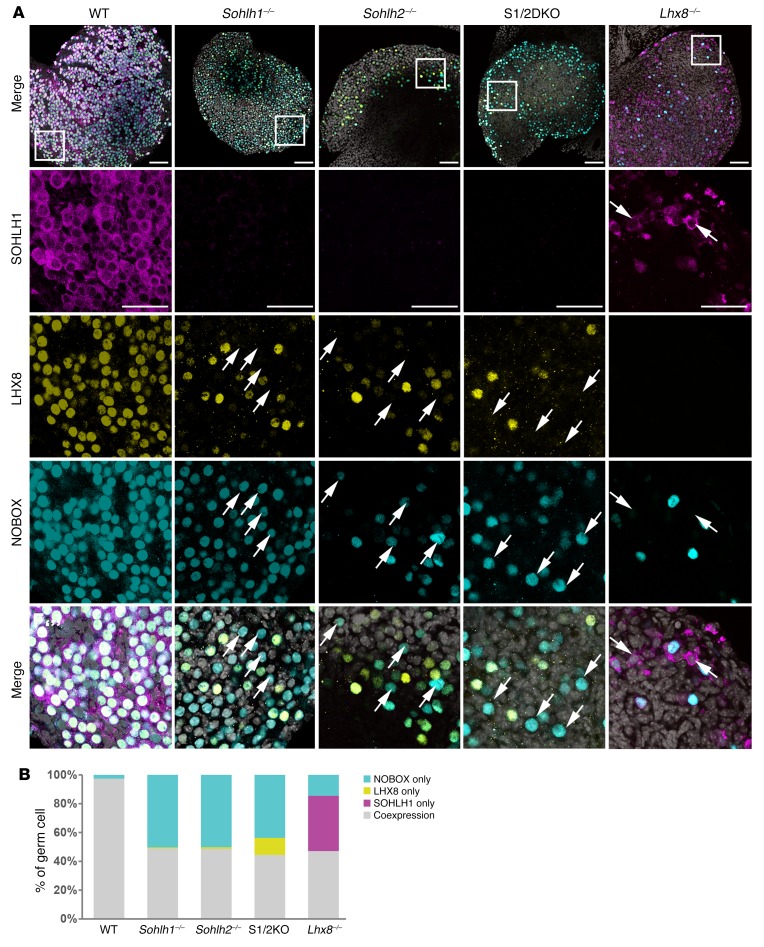
We next examined whether NOBOX and LHX8 protein expression is dependent on SOHLH1 and SOHLH2 expression. In Sohlh1- and Sohlh2-deficient newborn ovaries, we found that expression of NOBOX and LHX8 was reduced but not abolished, as determined by immunofluorescence staining with specific antibodies (Figure 8). Sohlh1 or Sohlh2 single deficiency or S1/2DKO did not affect NOBOX or LHX8 intracellular localizations. However, we observed that LHX8 and NOBOX proteins did not colocalize in 52% and 58% of oocytes, respectively, in the embryonic gonad in either Sohlh1 or Sohlh2 single- or double-KOs (Figure 8, A and B). These results indicate that SOHLH1 and SOHLH2 are required for proper NOBOX and LHX8 expression and colocalization. We also investigated the expression of SOHLH1 protein in Lhx8 global-KO animals (14). SOHLH1 expression was substantially diminished in Lhx8-deficient embryonic ovaries (Figure 8A). Moreover, Sohlh1, Sohlh2, and Lhx8 individual deficiencies markedly downregulated the expression of Kit, a well-characterized receptor in germline differentiation (Supplemental Figure 3). All of these results indicate that SOHLH1 and SOHLH2 and other oocyte-specific transcription factors are codependent on one another for their appropriate expression in the embryonic gonad (4, 5, 7, 14).
Figure 8. Transcriptional regulator deficiencies disrupt coexpression of oocyte-specific transcriptional regulators.
(A) Immunofluorescence staining with SOHLH1 (magenta), LHX8 (yellow), and NOBOX (cyan) in ovaries from WT, Sohlh1–/–, Sohlh2–/–, S1/2DKO, and Lhx8–/– mice. Scale bars: 100 μm (top) and 50 μm (enlargements). Arrows indicate oocytes expressing only LHX8 or NOBOX but not both (n = 3 per group). DNA was stained with DAPI (gray). (B) Quantitation of NOBOX-only, LHX8-only, SOHLH1-only, and LHX8/NOBOX-coexpressed oocytes in A. Data are representative of at least 3 independent experiments.
Sohlh1 and Sohlh2 deficiencies do not disrupt meiosis I.
Meiosis in mouse ovaries commences circa E13.5 and arrests in the diplotene stage before birth. By E16.5, most oocytes show zygotene and pachytene stages of meiosis, whereas at E18.5, pachytene and diplotene stages predominate. The substages of meiosis I prophase are defined by the following chromosome configurations and structures: pairing, which occurs during the leptotene and zygotene stages; synapsis, which is completed at the onset of the pachytene stage; and desynapsis, which occurs during the diplotene stage (15). Rapid oocyte loss occurs when meiosis I components are perturbed, as exemplified by KO models for mutS homolog 5 (Msh5) (16), REC8 meiotic recombination protein (Rec8) (17), and DNA meiotic recombinase 1 (Dmc1) (18). SOHLH2 expression precedes SOHLH1 expression in the embryonic gonad and overlaps the initiation of meiosis I. SOHLH1 and SOHLH2 ovaries lose oocytes rapidly after birth, at the time when most oocytes have entered the diplotene stage of meiosis I. We studied whether gross perturbations in meiosis account for rapid oocyte loss in Sohlh1- and Sohlh2-deficient ovaries.
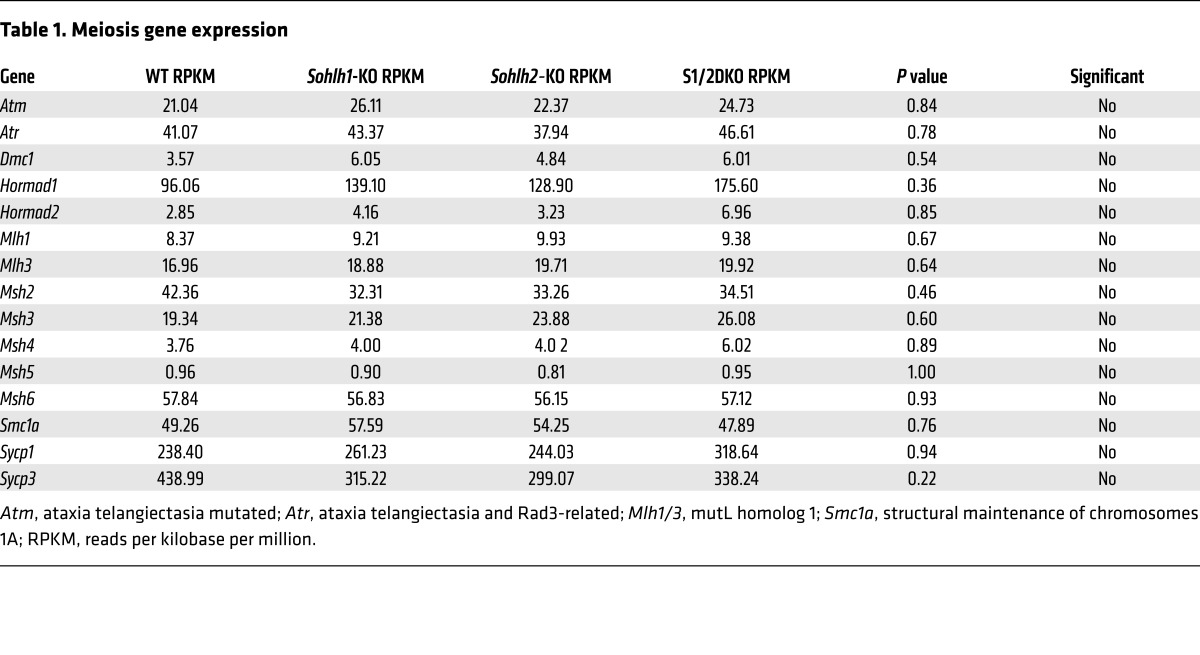
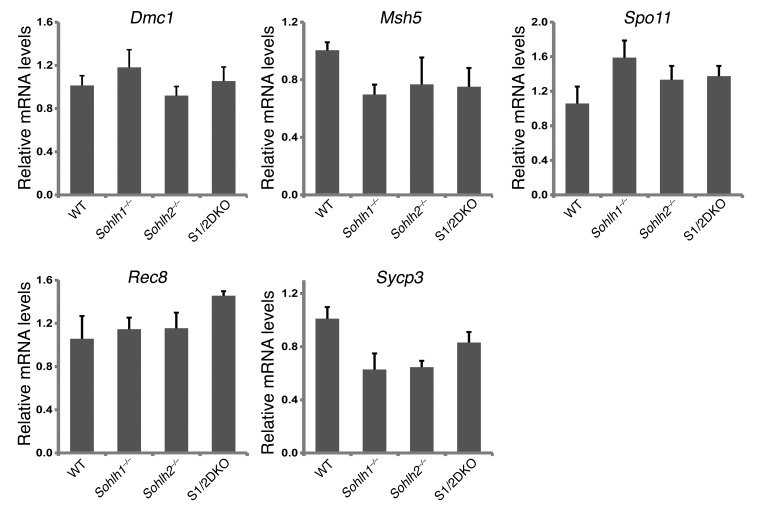
We performed RNA-sequencing (RNA-seq) on Sohlh1 and Sohlh2 single-KOs as well as on S1/2DKO newborn ovaries (Table 1) to assess the expression profiles of genes encoding well-characterized meiotic factors. The relative transcript levels of Dmc1, Msh5, SPO11 meiotic protein covalently bound to DSB (Spo11), Rec8, and other well-characterized meiotic genes were not significantly different between the WT and Sohlh1, Sohlh2, or S1/2DKO ovaries (Figure 9). These results indicate that meiotic components currently known to disrupt early oogenesis are not affected by Sohlh1 or Sohlh2 deficiency and that oocyte differentiation can be governed by factors independent of meiosis.
Table 1. Meiosis gene expression.
Figure 9. Sohlh1 and Sohlh2 deficiency and meiosis.
Quantitative PCR analysis was conducted for Dmc1, Msh5, Spo11, Rec8, and synaptonemal complex protein 3 (Sycp3) transcripts in WT, Sohlh1–/–, Sohlh2–/–, and S1/2DKO mice (n = 6 per group). Gapdh was used for normalization, and the ΔΔCt was calculated for the fold change. Error bars indicate the mean ± SD. A 2-tailed Student’s t test was used to calculate P values (compared with WT). Data are representative of at least 3 independent experiments. A P value of less than 0.05 was considered statistically significant. No significant differences in expression were found between different genotypes (P value was greater than 0.05).
SOHLH1- and SOHLH2-dependent gene expression profile in the ovary.
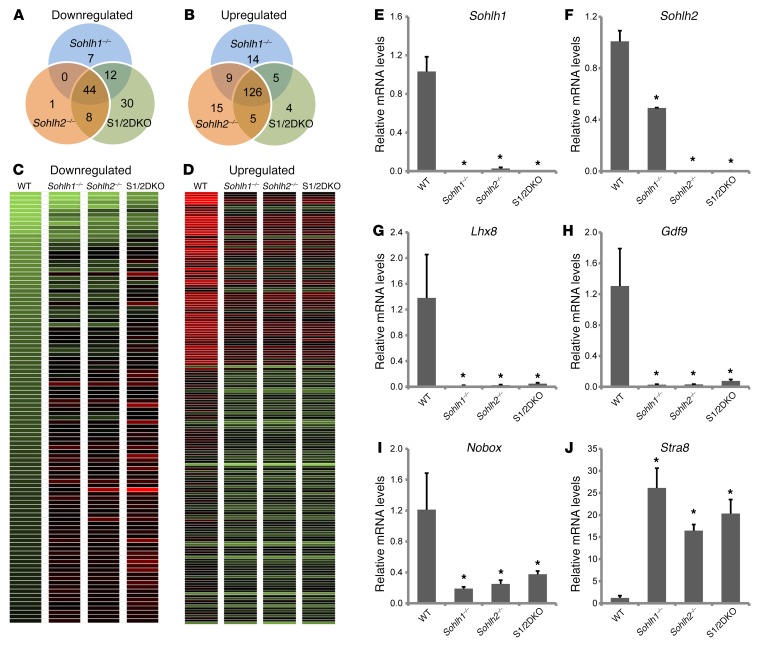
We performed RNA-seq analysis on single-KO or S1/2DKO newborn mouse ovaries to determine the molecular perturbations that precede a frank pathology in double-KO versus single-KO mouse ovaries. SOHLH1 and SOHLH2 were coexpressed in the vast majority of oocytes in newborn ovaries, and the histology of newborn ovaries was equivalent between WT and KOs. Analysis of the RNA-seq data revealed that 63 genes were significantly downregulated and 154 genes were markedly upregulated in Sohlh1–/– mice; 53 genes were downregulated and 155 were upregulated in Sohlh2–/– mice; and 94 genes were downregulated and 140 were upregulated in double-KO ovaries (Figure 10). Sohlh1- and Sohlh2-regulated networks were mainly repressed in the presence of Sohlh1 and Sohlh2, but a substantial number of genes were also activated (downregulated in KOs). There was a significant overlap in genes misexpressed between Sohlh1 and Sohlh2 single-KO ovaries. Among downregulated genes, Sohlh1 and Sohlh2 single-KOs shared 44 genes of a total of 63 among Sohlh1-KOs (70%) and 53 genes among Sohlh2-KOs (83%). Among upregulated genes, Sohlh1 and Sohlh2 shared 135 genes (90%). S1/2DKO ovaries showed a substantially higher number of downregulated genes, 94, while the number of upregulated genes was not significantly different. The significantly higher number of genes downregulated in S1/2DKOs argues that Sohlh1 and Sohlh2 may have synergistic effects at the molecular level, despite a lack of effect on the histology phenotype. SOHLH1 and SOHLH2 proteins are known to heterodimerize (9), and heterodimers may have additional effects on gene expression.
Figure 10. Sohlh1 and Sohlh2 deficiencies show significant overlap in affected genes.
Venn diagrams show overlap and differences in the number of downregulated (A) and upregulated (B) transcripts as well as transcript heatmaps (C and D) derived from Sohlh1–/–, Sohlh2–/–, and S1/2DKO mice when compared with the WT newborn ovary transcriptomes. Transcript data were derived from RNA-seq, and genes that showed a greater-than 2-fold change compared with WT were included. (E–J) Quantitative PCR analyses derived from RNA-seq of select genes such as Sohlh1 (E), Sohlh2 (F), Lhx8 (G), Nobox (H), Gdf9 (I), and Stra8 (J) in WT, Sohlh1–/–, Sohlh2–/–, and S1/2DKO mice (n = 6 per group). Gapdh was used for normalization, and ΔΔCt was calculated for the fold change. Error bars indicate the mean ± SD. *P < 0.05, by 2-tailed Student’s t test (compared with WT). Data are representative of at least 3 independent experiments.
The downregulated genes include well-known oocyte-specific genes, such as Lhx8, Nobox, Zp3, and Kit (Figure 10 and Supplemental Table 1). Thirty genes were exclusively downregulated in S1/2DKO ovaries (Figure 10 and Supplemental Table 2). The downregulated gene list is enriched in the histone transcripts H2afx, Hist2h4, Hist2h3c2, Hist1h4m, Hist2h2ac, and Hist1h2ag, although the significance of this is unclear. One hundred twenty-six genes were upregulated in both single- and double-KO ovaries (Figure 10 and Supplemental Table 3) and include stimulated by retinoic acid gene 8 (Stra8) as well as testis-specific genes such as histone cluster 1, H1t (Hist1h1t); histone cluster 1, H2aa (Hist1h2aa); histone cluster 1, H2ba (Hist1h2ba); testis-expressed gene 16 (Tex16); and testis-expressed gene 101 (Tex101). Multiple members of the reproductive homeobox cluster (Rhox) family of homeobox genes, important in male infertility (19), are also upregulated and, in part, account for the preponderance of misregulated X chromosome genes (39%) in Sohlh1- and Sohlh2-deficient ovaries. The overexpression of testis-specific genes has been previously observed in other oocyte-specific KOs of Nobox (20) and Figla (21) genes and signifies the importance of oocytes in repressing male germline differentiation.
Discussion
Oocyte differentiation is closely intertwined with the onset of meiosis I, but the drivers of oocyte differentiation are unknown. The role of somatic factors is well established in embryonic male gonadal differentiation. Moreover, we and others have previously shown that Sohlh1 and Sohlh2, two unique and germline-specific basic helix-loop-helix transcriptional regulators, coordinate male spermatogonial differentiation (9, 10, 22, 23). Much less is known regarding female gonadal differentiation, though somatic proteins encoded by Wnt4 and Foxl2 genes occupy a prominent role in the female pathway. Even less is known regarding oocyte-expressed genes that drive oocyte differentiation independently of meiosis. We and others have previously shown the importance of the transcriptional regulators SOHLH1 (2, 5), SOHLH2 (3, 4), LHX8 (5, 6), NOBOX (7), FIGLA (8), and TATA-box–binding protein–associated factor 4b (TAF4B) (24) in postnatal oocyte differentiation. Postnatal studies have shown that these factors are critical in primordial follicle formation, primordial follicle–to–primary follicle transition, and oocyte survival. However, the embryonic onset of their expression and action in the embryonic ovary is poorly understood. SOHLH1 and SOHLH2 are exclusively expressed in the male and female germline (2, 3). We hypothesized that SOHLH1 and SOHLH2 are important factors in driving oocyte differentiation in the embryonic ovary.
The Sohlh2 gene was initially identified as a homolog of Sohlh1 (3), with their limited homology confined to the bHLH domains (~50%). Our data presented here, as well as the GEO expression profile data (Supplemental Figure 1), show that Sohlh2 RNA and protein expression commences relatively early, as early as E12.5, and precedes Sohlh1 expression. Interestingly, we observed that SOHLH2 protein was predominantly located in the cytoplasm prior to the appearance of SOHLH1 protein (Figure 1). SOHLH1 protein’s appearance, circa E15.5, led to the translocation of SOHLH2 into the nucleus and coincided with the appearance of LHX8 and NOBOX proteins (Figure 7). Sohlh1–/– ovaries, examined at E16.5 and in newborns, showed that SOHLH2 protein expression was confined to the cytoplasm (Figure 2), reinforcing the conclusion that SOHLH2 cellular localization depends on SOHLH1 expression. Single deficiency of Sohlh1, Sohlh2, Lhx8, or Nobox disrupts the pattern of expression of other genes, implying an interdependence of these factors (Figure 8).
We do not know whether cytoplasmic SOHLH2 has a specific function. Transcription factors like p53 have posttranscriptional roles and bind transcripts to regulate their translation (25, 26). We hypothesize that cytoplasmic SOHLH2 is inactive and that translocation and heterodimerization with SOHLH1 are necessary for activity, however, further studies are needed to address this hypothesis.
Because SOHLH1 and SOHLH2 show limited homology, we originally hypothesized that these 2 proteins play independent roles and that double deficiency will produce synergistic effects in the ovary. However, the onset and extent of pathology in mice lacking both Sohlh1 and Sohlh2 is identical to that in single-KO mice (Figure 3). The lack of a synergistic effect is consistent with the interpretation that Sohlh1 and Sohlh2 share a common pathway that is enabled at E15.5, when the appearance of SOHLH1 induces SOHLH2 nuclear translocation. Previous studies have shown that SOHLH1 and SOHLH2 physically interact and can form homo- and heterodimers (9, 10). Consistent with KO studies, which indicate that SOHLH1 and SOHLH2 could regulate common pathways, RNA-seq analyses of gene expression in Sohlh1- and Sohlh2-deficient ovaries showed a large overlap in misexpressed genes. Double mutants had more profound changes in gene expression, consistent with Sohlh1 and Sohlh2 synergism (Figure 10). SOHLH1 and SOHLH2 heterodimers may regulate genes involved in later steps of oocyte differentiation rather than those involved in the earlier steps of development.
Meiosis is a big event in the female embryonic ovary, commencing circa E13.5. Meiosis is dissociable from oocyte differentiation, as shown in studies of postnatal phenotypes in Stra8-deficient ovaries (27). Stra8, one of the genes stimulated by retinoic acid, is essential for oocyte entry into meiosis. Stra8 has a narrow window of expression around E14.5, after which it rapidly declines (Supplemental Figure 1). SOHLH1 has been proposed, in somatic cell lines, to play a role in Stra8 downregulation (28). Despite the lack of a meiotic prophase, a subset of Stra8-deficient oocytes survive fetal life, can synthesize zona pellucida, can organize surrounding somatic cells into follicles, are ovulated in response to hormonal stimulation, undergo asymmetric cell division to produce a polar body, and cleave to form 2-cell embryos upon fertilization (29). Nonetheless, Stra8-deficient ovaries are highly abnormal, with a large loss of oocytes commencing in fetal ovaries and oocyte depletion being completed by 6 weeks’ postnatal age, with resulting sterility (30).
Deficiency of Sohlh1 and Sohlh2 is associated with persistent expression of Stra8 (Figure 10J and Supplemental Table 3). Because Stra8 expression persists in both Sohlh1- and Sohlh2-KOs, it is likely that SOHLH1:SOHLH2 heterodimers are important in regulating Stra8 expression. Despite persistent Stra8 expression, meiosis I is grossly normal, and expression of other meiotic gene transcripts is unaffected (Figure 9). Other meiotic gene KOs support the notion that abnormal meiosis does not disrupt oocyte differentiation. For example, HORMA domain–containing 1 (Hormad1) is a major pachytene checkpoint, and our previous work revealed that Hormad1-deficient oocytes have unsynapsed chromosomes and lack pachynema, yet have a normal number of primordial follicles with normal oogenesis and folliculogenesis (31, 32). Hormad1 deficiency does not affect oocyte differentiation (33), nor does it perturb the expression of SOHLH1 (Supplemental Figure 3).
The expression of many oocyte-specific transcriptional regulators, such as LHX8, NOBOX, and FIGLA, is initiated in the embryonic ovaries and overlaps with SOHLH1 expression and SOHLH2 nuclear translocation. Moreover, Sohlh1 and Sohlh2 gene deficiencies disrupt LHX8 and NOBOX protein embryonic expression without disrupting meiosis. The loss of oocytes at the very early ages in single and combined Sohlh1 and Sohlh2 deficiencies is therefore unlikely to be due to meiosis but rather to disruption in the regulation of genes such as Lhx8 and Nobox (5, 7, 14). Our results are consistent with the interpretation that SOHLH1 and SOHLH2 are important regulators of oocyte differentiation, independent of meiosis, whose molecular interplay begins circa E15.5 with the appearance of SOHLH1 and nuclear translocation of SOHLH2.
Recent human studies have shown that SOHLH1 and SOHLH2 loss-of-function mutations are involved in a subset of XX and XY gonadal dysgenesis cases (34, 35) and show the importance of this pathway in human gonadal differentiation (34, 35). Our results show a dynamic interaction between oocyte-specific transcriptional regulators in the embryonic gonad and the importance of this interaction in orchestrating postnatal primordial follicle formation and oocyte differentiation independently of meiosis. Our previous studies in testes showed that Sohlh1 and Sohlh2 also coordinate spermatogonial differentiation (2, 9). Sohlh1 and Sohlh2 are therefore universal regulators of male and female germline differentiation. The sex-specific interplay of SOHLH1 and SOHLH2 is not well understood, but must be sexually dimorphic at the molecular level. For example, LHX8 and NOBOX determine ovarian development; their ovarian expression is dependent on SOHLH1 and SOHLH2, yet LHX8 and NOBOX deficiencies have no effect on male gonadal development or male infertility. The identity of somatic as well as germline factors that induce and extinguish Sohlh1 and Sohlh2 expression and orchestrate sex-specific germline differentiation remain to be determined.
Methods
Animal breeding and transgenic lines.
Pups were weaned at 3 weeks of age, and breeding pairs were set up at 6 weeks of age. One mating pair was placed per cage and inspected daily for the presence of litters. Sohlh1, Sohlh2, Lhx8, and Hormad1 mice used in the current studies were previously described (2, 3, 14, 31). The CCS1- and CCS2-transgenic mice were generated by integrating the CAG-loxp-CAT-loxp Sohlh1-mCherry (CCS1) transgene or the CAG-loxp-CAT-loxp Sohlh2-EGFP (CCS2) transgene into the pronuclei of zygotes of FVB × FVB mice. Founder CCS1 or CCS2 mice were bred with FVB mice to expand the colony. Ddx4-Cre mice were purchased from The Jackson Laboratory, and Gdf9-Cre mice were a gift of Austin Cooney (University of Texas at Austin, Austin, Texas, USA) (13).
Histology and whole-mount immunostaining.
Ovaries were fixed in 10% buffered formalin (Sigma-Aldrich). Fixed tissues were embedded in paraffin, serially sectioned (5-μm thickness), and stained with hematoxylin (Sigma-Aldrich) and periodic acid–Schiff (PAS). Germ cell cysts and primordial, primary, and secondary follicles were defined as described previously (5). WT and mutant oocytes were stained concurrently with the same mixture of antibodies.
For whole mounting, ovaries were fixed in 4% paraformaldehyde and serially washed with 25%, 50%, 75%, and 100% methanol. Oocytes were labeled with anti-SOHLH1 (2); anti-SOHLH2 (3); anti-DDX4 (ab13840; Abcam); anti–c-KIT (a gift of Tasumi Hirata, National Institute of Genetics, Mishima, Japan) (36); anti-NOBOX (7); and anti-LHX8 antibodies (14). Confocal microscopy (Nikon A1) was used to detect immunofluorescence.
RNA-seq.
RNA-seq was performed in WT and Sohlh1- and Sohlh2 single- or double-KO mouse ovaries. Total RNA was isolated from pools of 5 newborn ovaries, using the RNeasy Mini Kit (QIAGEN) and following the manufacturer’s protocol. A cDNA library was made using a SMARTer Stranded Total RNA Sample Prep Kit (Clontech), and next-generation sequencing was performed on an Illumina NextSeq 500 (Illumina) with the Mid Output Kit (Illumina). Approximately 40 million reads were generated per sample (WT control and single and double knockouts cDNA). Sohlh1- and Sohlh2 single- or double-KO RNA-seq reads were mapped and analyzed using TopHat (37), Cufflinks (38), and Cuffdiff (39). The raw RNA-seq data from this study have been deposited in the NCBI’s Sequence Read Archive (accession no. PRJNA293873). RNA-seq data for specific transcripts were validated by quantitative PCR as previously described (4).
Statistics.
Data are presented as the mean ± SEM. Results were analyzed using a 2-tailed Student’s t test. A P value of less than 0.05 was considered statistically significant.
Study approval.
All experimental and surgical procedures complied with the NIH Guide for the Care and Use of Laboratory Animals (National Academies Press, 2011) and were approved by the IACUC of the University of Pittsburgh, under protocol number 14094558.
Author contributions
YHS, KJG, VM, HS, and HWA conducted the experiments. YHS, KJG, YR, and AR analyzed and interpreted the data. YHS, YR, and AR wrote the manuscript. YR and AR reviewed and revised the manuscript. AR supervised the study.
Supplementary Material
Acknowledgments
We thank Huaiyang Jiang (Magee-Womens Research Institute, Pittsburgh, PA, USA) for his help with the RNA-seq data analysis. This study was supported by the Magee-Womens Research Foundation and National Institutes of Health grants HD044858 and HD070647.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information:J Clin Invest. 2017;127(6):2106–2117.https://doi.org/10.1172/JCI90281.
See the related Commentary at The SO(H)L(H) “O” drivers of oocyte growth and survival but not meiosis I.
Contributor Information
Yong-Hyun Shin, Email: shinyon@iu.edu.
Yu Ren, Email: yur4@pitt.edu.
Hitomi Suzuki, Email: suzukih.arc@cmn.tmd.ac.jp.
Hyo won Ahn, Email: hwahn@uga.edu.
Vasil Mico, Email: VAM39@pitt.edu.
Aleksandar Rajkovic, Email: rajkovic@upmc.edu.
References
- 1.Schmidt D, et al. The murine winged-helix transcription factor Foxl2 is required for granulosa cell differentiation and ovary maintenance. Development. 2004;131(4):933–942. doi: 10.1242/dev.00969. [DOI] [PubMed] [Google Scholar]
- 2.Ballow D, Meistrich ML, Matzuk M, Rajkovic A. Sohlh1 is essential for spermatogonial differentiation. Dev Biol. 2006;294(1):161–167. doi: 10.1016/j.ydbio.2006.02.027. [DOI] [PubMed] [Google Scholar]
- 3.Ballow DJ, Xin Y, Choi Y, Pangas SA, Rajkovic A. Sohlh2 is a germ cell-specific bHLH transcription factor. Gene Expr Patterns. 2006;6(8):1014–1018. doi: 10.1016/j.modgep.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 4.Choi Y, Yuan D, Rajkovic A. Germ cell-specific transcriptional regulator sohlh2 is essential for early mouse folliculogenesis and oocyte-specific gene expression. Biol Reprod. 2008;79(6):1176–1182. doi: 10.1095/biolreprod.108.071217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pangas SA, et al. Oogenesis requires germ cell-specific transcriptional regulators Sohlh1 and Lhx8. Proc Natl Acad Sci U S A. 2006;103(21):8090–8095. doi: 10.1073/pnas.0601083103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qin Y, Zhao H, Kovanci E, Simpson JL, Chen ZJ, Rajkovic A. Analysis of LHX8 mutation in premature ovarian failure. Fertil Steril. 2008;89(4):1012–1014. doi: 10.1016/j.fertnstert.2007.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rajkovic A, Pangas SA, Ballow D, Suzumori N, Matzuk MM. NOBOX deficiency disrupts early folliculogenesis and oocyte-specific gene expression. Science. 2004;305(5687):1157–1159. doi: 10.1126/science.1099755. [DOI] [PubMed] [Google Scholar]
- 8.Soyal SM, Amleh A, Dean J. FIGalpha, a germ cell-specific transcription factor required for ovarian follicle formation. Development. 2000;127(21):4645–4654. doi: 10.1242/dev.127.21.4645. [DOI] [PubMed] [Google Scholar]
- 9.Suzuki H, et al. SOHLH1 and SOHLH2 coordinate spermatogonial differentiation. Dev Biol. 2012;361(2):301–312. doi: 10.1016/j.ydbio.2011.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Toyoda S, Yoshimura T, Mizuta J, Miyazaki J. Auto-regulation of the Sohlh1 gene by the SOHLH2/SOHLH1/SP1 complex: implications for early spermatogenesis and oogenesis. PLoS ONE. 2014;9(7):e101681. doi: 10.1371/journal.pone.0101681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kawamoto S, et al. A novel reporter mouse strain that expresses enhanced green fluorescent protein upon Cre-mediated recombination. FEBS Lett. 2000;470(3):263–268. doi: 10.1016/S0014-5793(00)01338-7. [DOI] [PubMed] [Google Scholar]
- 12.Gallardo T, Shirley L, John GB, Castrillon DH. Generation of a germ cell-specific mouse transgenic Cre line, Vasa-Cre. Genesis. 2007;45(6):413–417. doi: 10.1002/dvg.20310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lan ZJ, Xu X, Cooney AJ. Differential oocyte-specific expression of Cre recombinase activity in GDF-9-iCre, Zp3cre, and Msx2Cre transgenic mice. Biol Reprod. 2004;71(5):1469–1474. doi: 10.1095/biolreprod.104.031757. [DOI] [PubMed] [Google Scholar]
- 14.Choi Y, Ballow DJ, Xin Y, Rajkovic A. Lim homeobox gene, lhx8, is essential for mouse oocyte differentiation and survival. Biol Reprod. 2008;79(3):442–449. doi: 10.1095/biolreprod.108.069393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moens PB, Kolas NK, Tarsounas M, Marcon E, Cohen PE, Spyropoulos B. The time course and chromosomal localization of recombination-related proteins at meiosis in the mouse are compatible with models that can resolve the early DNA-DNA interactions without reciprocal recombination. J Cell Sci. 2002;115(Pt 8):1611–1622. doi: 10.1242/jcs.115.8.1611. [DOI] [PubMed] [Google Scholar]
- 16.de Vries SS, et al. Mouse MutS-like protein Msh5 is required for proper chromosome synapsis in male and female meiosis. Genes Dev. 1999;13(5):523–531. doi: 10.1101/gad.13.5.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bannister LA, Reinholdt LG, Munroe RJ, Schimenti JC. Positional cloning and characterization of mouse mei8, a disrupted allelle of the meiotic cohesin Rec8. Genesis. 2004;40(3):184–194. doi: 10.1002/gene.20085. [DOI] [PubMed] [Google Scholar]
- 18.Bannister LA, et al. A dominant, recombination-defective allele of Dmc1 causing male-specific sterility. PLoS Biol. 2007;5(5):e105. doi: 10.1371/journal.pbio.0050105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Richardson ME, Bleiziffer A, Tüttelmann F, Gromoll J, Wilkinson MF. Epigenetic regulation of the RHOX homeobox gene cluster and its association with human male infertility. Hum Mol Genet. 2014;23(1):12–23. doi: 10.1093/hmg/ddt392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Choi Y, Qin Y, Berger MF, Ballow DJ, Bulyk ML, Rajkovic A. Microarray analyses of newborn mouse ovaries lacking Nobox. Biol Reprod. 2007;77(2):312–319. doi: 10.1095/biolreprod.107.060459. [DOI] [PubMed] [Google Scholar]
- 21.Joshi S, Davies H, Sims LP, Levy SE, Dean J. Ovarian gene expression in the absence of FIGLA, an oocyte-specific transcription factor. BMC Dev Biol. 2007;7:67. doi: 10.1186/1471-213X-7-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hao J, et al. Sohlh2 knockout mice are male-sterile because of degeneration of differentiating type A spermatogonia. Stem Cells. 2008;26(6):1587–1597. doi: 10.1634/stemcells.2007-0502. [DOI] [PubMed] [Google Scholar]
- 23.Toyoda S, et al. Sohlh2 affects differentiation of KIT positive oocytes and spermatogonia. Dev Biol. 2009;325(1):238–248. doi: 10.1016/j.ydbio.2008.10.019. [DOI] [PubMed] [Google Scholar]
- 24.Falender AE, Shimada M, Lo YK, Richards JS. TAF4b, a TBP associated factor, is required for oocyte development and function. Dev Biol. 2005;288(2):405–419. doi: 10.1016/j.ydbio.2005.09.038. [DOI] [PubMed] [Google Scholar]
- 25.Tournillon AS, et al. p53 binds the mdmx mRNA and controls its translation. Oncogene. 2017;36(5):723–730. doi: 10.1038/onc.2016.236. [DOI] [PubMed] [Google Scholar]
- 26.Cassiday LA, Maher LJ. Having it both ways: transcription factors that bind DNA and RNA. Nucleic Acids Res. 2002;30(19):4118–4126. doi: 10.1093/nar/gkf512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dokshin GA, Baltus AE, Eppig JJ, Page DC. Oocyte differentiation is genetically dissociable from meiosis in mice. Nat Genet. 2013;45(8):877–883. doi: 10.1038/ng.2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Desimio MG, Campolo F, Dolci S, De Felici M, Farini D. SOHLH1 and SOHLH2 directly down-regulate STIMULATED BY RETINOIC ACID 8 (STRA8) expression. Cell Cycle. 2015;14(7):1036–1045. doi: 10.1080/15384101.2015.1007721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koubova J, et al. Retinoic acid activates two pathways required for meiosis in mice. PLoS Genet. 2014;10(8):e1004541. doi: 10.1371/journal.pgen.1004541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paredes A, et al. Loss of synaptonemal complex protein-1, a synaptonemal complex protein, contributes to the initiation of follicular assembly in the developing rat ovary. Endocrinology. 2005;146(12):5267–5277. doi: 10.1210/en.2005-0965. [DOI] [PubMed] [Google Scholar]
- 31.Shin YH, et al. Hormad1 mutation disrupts synaptonemal complex formation, recombination, and chromosome segregation in mammalian meiosis. PLoS Genet. 2010;6(11):e1001190. doi: 10.1371/journal.pgen.1001190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Daniel K, et al. Meiotic homologue alignment and its quality surveillance are controlled by mouse HORMAD1. Nat Cell Biol. 2011;13(5):599–610. doi: 10.1038/ncb2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shin YH, McGuire MM, Rajkovic A. Mouse HORMAD1 is a meiosis i checkpoint protein that modulates DNA double- strand break repair during female meiosis. Biol Reprod. 2013;89(2):29. doi: 10.1095/biolreprod.112.106773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bayram Y, et al. Homozygous loss-of-function mutations in SOHLH1 in patients with nonsyndromic hypergonadotropic hypogonadism. J Clin Endocrinol Metab. 2015;100(5):E808–E814. doi: 10.1210/jc.2015-1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qin Y, et al. Novel variants in the SOHLH2 gene are implicated in human premature ovarian failure. Fertil Steril. 2014;101(4):1104–1109.e6. doi: 10.1016/j.fertnstert.2014.01.001. [DOI] [PubMed] [Google Scholar]
- 36.Yamatani H, Sato Y, Fujisawa H, Hirata T. Chronotopic organization of olfactory bulb axons in the lateral olfactory tract. J Comp Neurol. 2004;475(2):247–260. doi: 10.1002/cne.20155. [DOI] [PubMed] [Google Scholar]
- 37.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25(9):1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Trapnell C, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28(5):511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Trapnell C, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7(3):562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.