Abstract
Quiescent and proliferating leukemia cells accumulate highly lethal DNA double-strand breaks that are repaired by 2 major mechanisms: BRCA-dependent homologous recombination and DNA-dependent protein kinase–mediated (DNA-PK–mediated) nonhomologous end-joining, whereas DNA repair pathways mediated by poly(ADP)ribose polymerase 1 (PARP1) serve as backups. Here we have designed a personalized medicine approach called gene expression and mutation analysis (GEMA) to identify BRCA- and DNA-PK–deficient leukemias either directly, using reverse transcription-quantitative PCR, microarrays, and flow cytometry, or indirectly, by the presence of oncogenes such as BCR-ABL1. DNA-PK–deficient quiescent leukemia cells and BRCA/DNA-PK–deficient proliferating leukemia cells were sensitive to PARP1 inhibitors that were administered alone or in combination with current antileukemic drugs. In conclusion, GEMA-guided targeting of PARP1 resulted in dual cellular synthetic lethality in quiescent and proliferating immature leukemia cells, and is thus a potential approach to eradicate leukemia stem and progenitor cells that are responsible for initiation and manifestation of the disease. Further, an analysis of The Cancer Genome Atlas database indicated that this personalized medicine approach could also be applied to treat numerous solid tumors from individual patients.
Introduction
Currently available antileukemic treatments often fail to eradicate drug-refractory quiescent leukemia stem cells (LSCs) and drug-resistant proliferating LSCs and leukemia progenitor cells (LPCs). Previous reports suggest that altered DNA repair mechanisms may be responsible for enhanced survival of LSCs and/or LPCs under genotoxic stress caused by reactive oxygen species (ROS) and cytotoxic treatment (1). Thus, leukemia cells may be highly dependent on specific DNA repair mechanisms and targeting these pathways could sensitize LSCs and LPCs to the lethality of DNA damage (2).
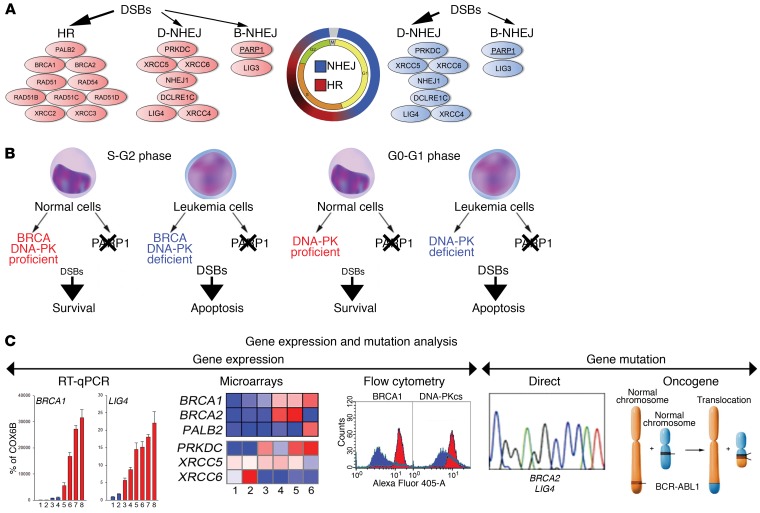
DNA double-strand breaks (DSBs), the most lethal DNA lesions, are usually repaired by BRCA-mediated homologous recombination (HR) and DNA-dependent protein kinase–mediated (DNA-PK–mediated) nonhomologous end-joining (NHEJ) (D-NHEJ) in proliferating cells, whereas D-NHEJ plays a major role in quiescent cells (3). Poly(ADP)ribose polymerase 1–dependent (PARP1-dependent) NHEJ serves as a back-up (B-NHEJ) pathway in both proliferating and quiescent cells (Figure 1A). In addition, PARP1 may decrease or prevent accumulation of potentially lethal DSBs, either by stimulation of base excision repair and single-strand break repair and/or by facilitation of DSB repair protein MRE11-mediated recruitment of the DNA damage marker RAD51 to promote stalled replication fork restart (4, 5).
Figure 1. Proposed model of GEMA-guided dual cellular synthetic lethality triggered by PARP1i in quiescent and proliferating leukemia cells.
(A) DSB repair pathways are cell cycle dependent. (B) The concept of dual cellular synthetic lethality triggered by PARP1i in BRCA- and DNA-PK–deficient proliferating, and in DNA-PK–deficient quiescent leukemia cells. (C) The concept of GEMA.
Cancer-specific defects in DNA repair pathways create the opportunity to employ synthetic lethality, which has been applied against cancer cells harboring mutations in BRCA1 and BRCA2 by using PARP1 inhibitors (6, 7). This finding initiated more than 100 clinical trials, which indicated that biomarkers of the response to PARP1 inhibitors reach beyond that of BRCA1/2 status. In addition, PARP1 inhibitor–mediated synthetic lethality would not eradicate BRCA1/2–mutated quiescent cancer stem cells, including quiescent LSCs, which often are responsible for minimal residual disease and disease relapse (8). We hypothesized that PARP1 inhibition (PARPi) can trigger dual cellular synthetic lethality in proliferating LSCs/LPCs and quiescent LSCs that display quantitative deficiencies in BRCA and DNA-PK pathways (Figure 1B).
Since inactivating mutations in BRCA and DNA-PK pathways (e.g., Fanconi anemia D1 = BRCA2 and LIG4, respectively) are rare in leukemias (9), other strategies for identifying patients with leukemias that display BRCA and DNA-PK (BRCA/DNA-PK) deficiency are needed. We developed a comprehensive gene expression and mutation analysis (GEMA) (Figure 1C) that identifies BRCA/DNA-PK–deficient patients using a combination of gene expression (microarrays, reverse transcription-quantitative PCR [RT-qPCR], and flow cytometry) and gene mutation (rare mutations in BRCA/DNA-PK genes and the presence of oncogenes reducing the expression of these genes) analyses to detect insufficient expression of at least 1 gene in each of the BRCA and DNA-PK pathways.
Results
PARP1i exerted dual cellular synthetic lethality by elimination of BRCA/DNA-PK–deficient proliferating cells and DNA-PK–deficient quiescent cells.
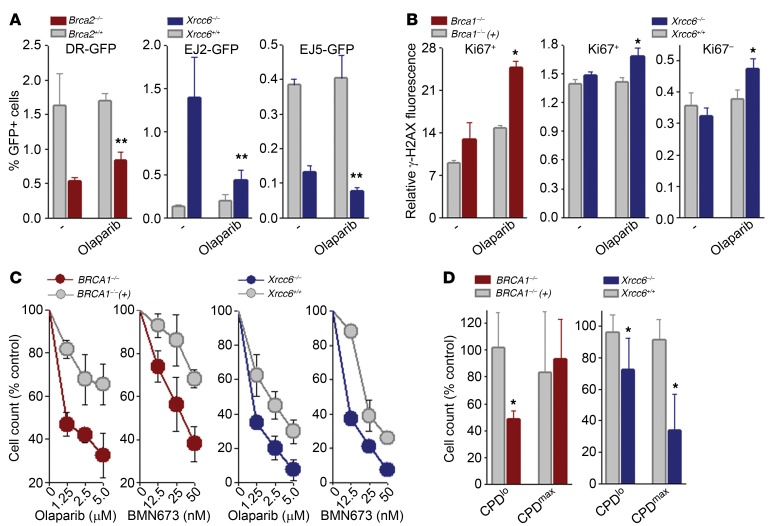
HR activity was strongly reduced in the BRCA2–/– hamster cell line V-C8 in comparison with wild-type V79 cells (Figure 2A; DR-GFP), which was accompanied by abundant elevation of γ-H2AX in PARP1 inhibitor olaparib-treated Ki67+ proliferating cells, indicating accumulation of DSBs (Figure 2B; BRCA1 panel, Ki67+).
Figure 2. PARP1i inhibited B-NHEJ, elevated DSBs, and triggered synthetic lethality in BRCA- or DNA-PK–deficient proliferating cells and in DNA-PK–deficient quiescent cells.
(A) HR, B-NHEJ, and total NHEJ activities were measured in Brca2–/– VC8 cells, Brca2+/+ V79 cells, Xrcc6–/– and Xrcc6+/+ mESCs harboring DR-GFP, EJ2-GFP, or EJ5-GFP reporter cassettes, as indicated, treated or not with 1.25 μM olaparib. (B) Indicated cells were treated or not with 1.25 μM olaparib and DSBs were measured by detecting γ-H2AX in Ki67+ and Ki67– cells. (C and D) Sensitivity of BRCA1-mutated [BRCA1–/–] and BRCA1-reconstituted [BRCA1–/–(+)] MDA-MB-436 breast carcinoma cells, and Xrcc6–/– and Xrcc6+/+ mESCs to 1.25 μM olaparib. (C) Trypan blue exclusion, and (D) CPDmax quiescent cells and CPDlo proliferating cells. Results represent 3 independent experiments. *P < 0.05 in comparison with olaparib-treated BRCA1- and/or DNA-PK–proficient cells; **P < 0.03 in comparison with untreated counterparts using Student’s t test.
While total NHEJ was reduced, B-NHEJ was activated in the absence of XRCC6 (Figure 2A; EJ2-GFP and EJ5-GFP, respectively). Olaparib exerted strong inhibitory activity against B-NHEJ, and also modestly diminished total NHEJ in XRCC6–/– murine embryonic stem cells (mESCs), but did not affect repair in XRCC6+/+ cells. This effect was associated with elevation of γ-H2AX in olaparib-treated XRCC6–/– Ki67+ and Ki67–cells, indicating accumulation of DSBs in proliferating and quiescent cells, respectively (Figure 2B; XRCC6 panels).
PARP1i reduced growth of BRCA1-mutated MDA-MB-436 human breast carcinoma cells (Figure 2C; BRCA1 panels) by elimination of proliferating, but not quiescent cells (Figure 2D; BRCA1 panels). Remarkably, PARP1i reduced the number of XRCC6–/– cells more efficiently than XRCC6+/+ counterparts (Figure 2C; XRCC6 panels), but the effect depended mainly on elimination of quiescent cells, with modest impact on proliferating cells (Figure 2D; XRCC6 panels).
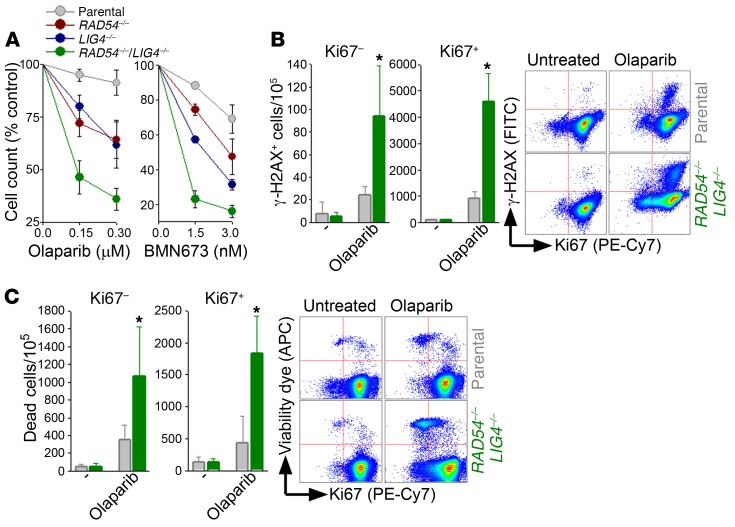
To determine the effect of PARP1i in cells displaying dual BRCA/DNA-PK deficiency we applied 2 previously characterized models: (a) FANCA–/– XRCC6–/– mESCs (10), and (b) the RAD54–/– LIG4–/– human pre-B leukemic cell line Nalm-6 (11). Cells with BRCA/DNA-PK deficiency were equally or more sensitive to olaparib and BMN673, another PARP1 inhibitor, than those displaying individual deficiencies (Figure 3A and Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/JCI90825DS1). Olaparib-treated RAD54–/– LIG4–/– cells and FANCA–/– XRCC6–/– mESCs accumulated DSBs measured by γ-H2AX (Figure 3B and Supplemental Figure 1B), resulting in elevated cell death in both Ki67– quiescent and Ki67+ proliferating subpopulations (Figure 3C and Supplemental Figure 1C). These effects were accompanied by downregulation of poly(ADP-ribose) polymer formation by PARP1i (Supplemental Figure 2).
Figure 3. PARP1i elevated DSBs and triggered dual cellular synthetic lethality in BRCA/DNA-PK–deficient proliferating and quiescent Nalm-6 leukemia cells.
(A) Sensitivity of the indicated cells to olaparib and BMN673. (B and C) Nalm-6 parental cells (gray bars) and RAD54–/– LIG4–/– counterparts (green bars) treated or not with 0.15 μM olaparib for 24 (B) or 48 (C) hours. (B) γ-H2AX–positive Ki67–and Ki67+ cells and (C) dead Ki67– and Ki67+ cells; representative plots are included. Results represent 3 independent experiments. *P < 0.05 in comparison with olaparib-treated parental cells using Student’s t test.
Collectively, these data strongly suggest that PARP1i selectively inhibited B-NHEJ that promoted accumulation of lethal DSBs, resulting in dual cellular synthetic lethality through the eradication of BRCA/DNA-PK–deficient proliferating and quiescent cells.
PARP1i eradicated BRCA/DNA-PK–deficient proliferating and quiescent immature leukemia cells from individual AMLs and ALLs identified by gene expression analysis.
To identify BRCA/DNA-PK–deficient individual leukemias we examined the expression of (a) HR genes, that when absent/mutated caused synthetic lethality with PARP1i (6, 7, 12), and (b) key D-NHEJ genes (13).
Our hypothesis was supported by detection of wide-range expression levels of BRCA1 and PRKDC (encodes for DNA-PK catalytic subunit = DNA-PKcs) in existing databases of individual patient leukemia samples (Supplemental Figure 3, A–C). Moreover, meta-analysis of 166 acute myeloid leukemia (AML) patient samples from The Cancer Genome Atlas (TCGA) RNA-seq database revealed that 15 (9%) samples displayed BRCA/DNA-PK deficiency (Supplemental Figure 3D).
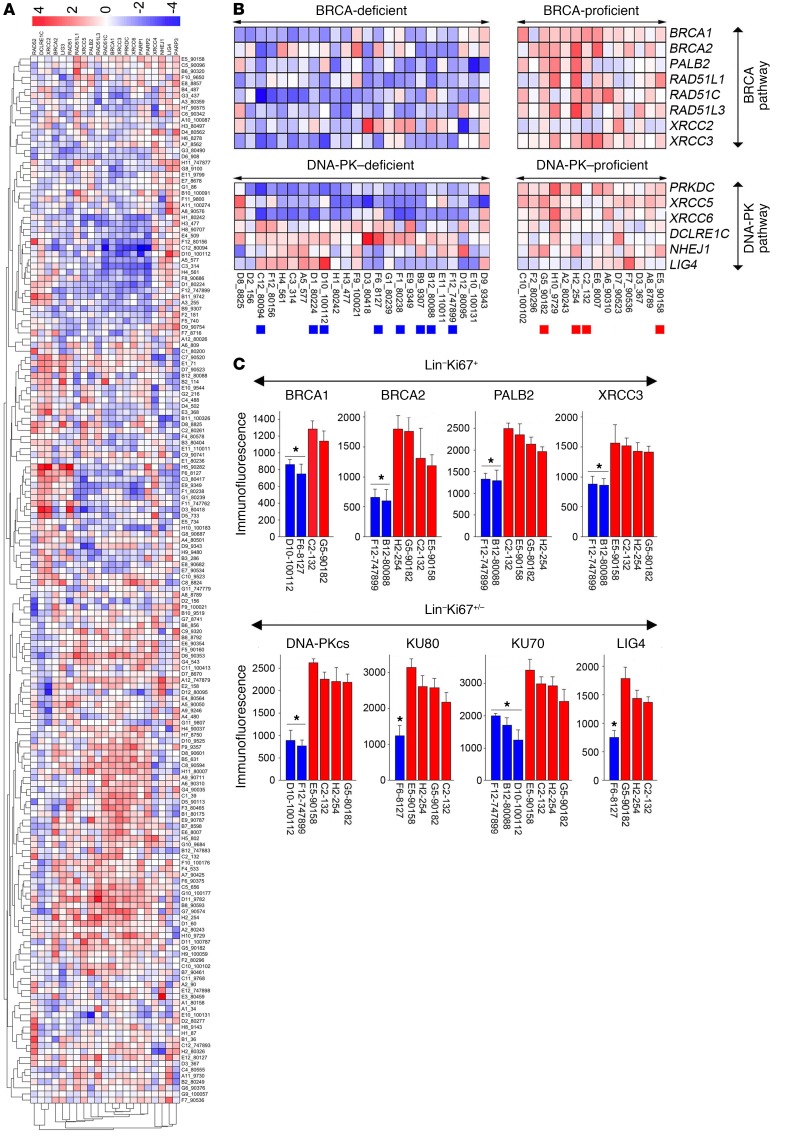
Gene expression analysis of our set of 172 cytogenetically normal AML samples revealed a wide range of expression levels of HR and D-NHEJ genes (Figure 4A). AML samples were assigned to the BRCA/DNA-PK–deficient group if a row Z-score value of at least 1 gene in HR (BRCA1, BRCA2, PALB2, RAD51L1 [RAD51B], RAD51C, RAD51L3 [RAD51D], XRCC2, and XRCC3) and D-NHEJ (PRKDC [encoding DNA-PKcs], XRCC5 [KU80], XRCC6 [KU70], DCLRE1C, NHEJ1, LIG4, and XRCC4) was less than –1.5. Conversely, BRCA/DNA-PK–proficient samples displayed row Z-score values greater than –0.75 for all genes in HR and D-NHEJ. Using this approach we identified 26 BRCA/DNA-PK–deficient and 16 BRCA/DNA-PK–proficient AMLs (Figure 4B). This finding was supported by detection of BRCA/DNA-PK–deficient AML samples in another microarray database (Supplemental Figure 4).
Figure 4. mRNA microarrays identified BRCA/DNA-PK–deficient AMLs.
(A) Gene expression analysis of DSB repair genes in 172 AML samples. (B) BRCA/DNA-PK–deficient and –proficient samples; those used in further analyses are marked by colored squares at the bottom. (C) Flow cytometric analysis of DSB repair proteins in Lin–Ki67+/– cells from individual AMLs predicted to be BRCA/DNA-PK deficient (blue) or proficient (red). Each AML sample was measured 3–6 times.*P < 0.001 in comparison with all BRCA/DNA-PK–proficient samples using Student’s t test.
Downregulation of at least 1 protein in each of the BRCA and DNA-PK pathways was detected in Lin–Ki67+ proliferating cells and Lin–Ki67+/– proliferating/quiescent cells, respectively, from individual BRCA/DNA-PK–deficient AMLs, compared with BRCA/DNA-PK–proficient samples (Figure 4C). The BRCA/DNA-PK–deficient/proficient status of preselected samples was further validated by direct measurement of HR and NHEJ activities using specific reporter cassettes (Supplemental Figure 5A) (14). Both HR and NHEJ activities were downregulated in representative BRCA/DNA-PK–deficient samples when compared with BRCA/DNA-PK–proficient counterparts (Supplemental Figure 5B).
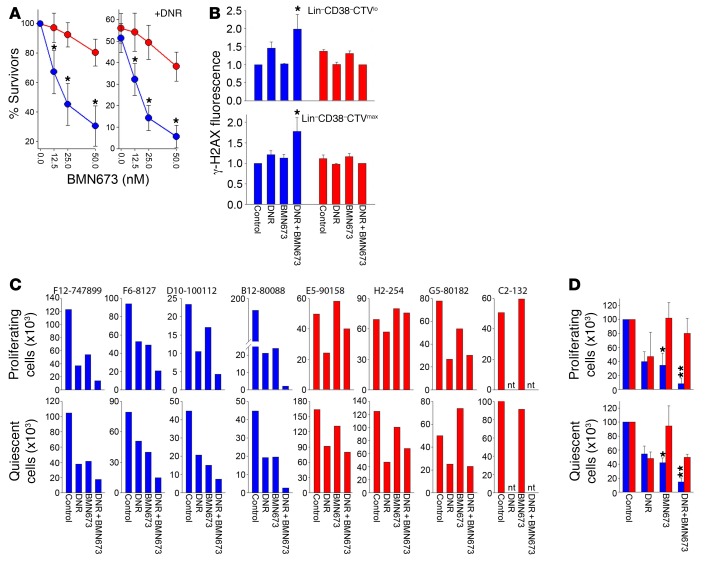
BRCA/DNA-PK–deficient Lin– AML primary cells from individual patients were more sensitive to BMN673, and BMN673 plus daunorubicin (DNR), in comparison with BRCA/DNA-PK–proficient counterparts (Figure 5A). LSC-enriched Lin–CD38– AML patient cells were used to study the role of PARP1i in quiescent and proliferating LSCs because this subpopulation was the most enriched in LSCs in NSG mice (15). The BRCA/DNA-PK–deficient phenotype was accompanied by accumulation of DSBs in both Lin–CD38–CTVlo proliferating cells and Lin–CD38–CTVmax quiescent cells treated with suboptimal doses of BMN673 and DNR (Figure 5B). These cell subpopulations were much more sensitive to BMN673 alone and in combination with DNR in comparison with BRCA/DNA-PK–proficient counterparts (Figure 5, C and D). Moreover, BMN673 + DNR exerted a synergistic effect against BRCA/DNA-PK–deficient cells.
Figure 5. BRCA/DNA-PK–deficient AMLs were sensitive to dual cellular synthetic lethality exerted by BMN673.
(A) Lin– cells from BRCA/DNA-PK–deficient (blue, n = 8) and BRCA/DNA-PK–proficient (red, n = 4) samples treated with BMN673 (left) and DNR + BMN673 (right). (B) γ-H2AX immunofluorescence in BRCA/DNA-PK–deficient (blue) and BRCA/DNA-PK–proficient (red) cells (n = 3–5) untreated (control) and treated with 1 μg/ml DNR, 12.5 nM BMN673, or DNR + BMN673. *P < 0.05 in comparison with DNR-treated counterparts by Student’s t test. (C) Number of proliferating Lin–CD38–CTVlo and quiescent Lin–CD38–CTVmax cells from individual BRCA/DNA-PK–deficient (blue) and –proficient (red) samples treated with DNR, BMN673, or DNR + BMN673. (D) Cumulative percentages from samples examined in C. *P < 0.02, **P < 0.05 in comparison with corresponding BRCA/DNA-PK–proficient samples using Student’s t test. **P < 0.03 in comparison with DNR- or BMN673-treated BRCA/DNA-PK–deficient samples using 2-way ANOVA. nt, not tested.
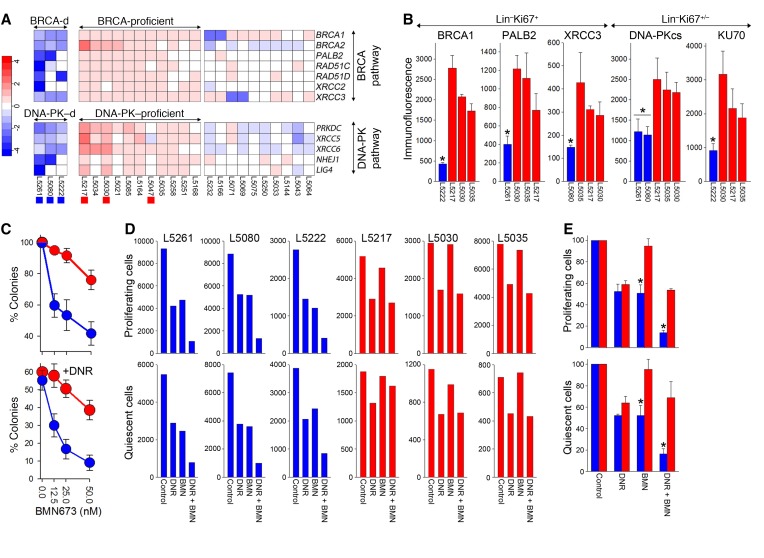
Next, Fluidigm RT-qPCR analysis of 26 therapy-related myelodysplastic syndrome (t-MDS)/AML samples revealed that 3 samples (12%) displayed BRCA/DNA-PK deficiency (Z score < –1.5 of at least 1 gene in BRCA and DNA-PK pathways) and 11 were BRCA/DNA-PK proficient (Z score > –0.5 of all genes in BRCA and DNA-PK pathways) (Figure 6A). Immunofluorescent staining followed by flow cytometric analysis of BRCA/DNA-PK–deficient and BRCA/DNA-PK–proficient samples (Figure 6A; marked with blue and red squares, respectively) confirmed downregulation of at least 1 protein in each BRCA and DNA-PK pathway in BRCA/DNA-PK–deficient proliferating Lin–Ki67+ cells and proliferating/quiescent Lin–Ki67+/– cells, respectively, compared with BRCA/DNA-PK–proficient samples (Figure 6B).
Figure 6. RT-qPCR identified BRCA/DNA-PK–deficient t-MDS/AMLs sensitive to dual synthetic lethality exerted by BMN673.
(A) BRCA/DNA-PK–deficient and –proficient t-MDS/AML samples; those used in further analyses are marked by colored squares at the bottom. Three samples (12%) displayed BRCA/DNA-PK deficiency (Z score < –1.5 of at least 1 gene in BRCA and DNA-PK pathways) and 11 were BRCA/DNA-PK proficient (Z score > –0.5 of all genes in BRCA and DNA-PK pathways). (B) Flow cytometric analysis of the indicated DSB repair proteins in BRCA/DNA-PK–deficient (blue) proliferating Lin–Ki67+ cells and proliferating/quiescent Lin–Ki67+/– cells, respectively, compared with BRCA/DNA-PK–proficient (red) samples. Each t-MDS/AML sample was measured 2–3 times. *P < 0.04 in comparison with all BRCA/DNA-PK–proficient samples. (C) Colonies from Lin–CD34+ BRCA/DNA-PK–deficient (blue, n = 3) and BRCA/DNA-PK–proficient (red, n = 3) samples treated with BMN673 (top graph) or DNR + BMN673 (bottom graph). (D) Number of proliferating Lin–CTVlo and quiescent Lin–CTVmax cells from individual BRCA/DNA-PK–deficient (blue) and –proficient (red) samples treated with DNR, BMN673, and DNR + BMN673. (E) Cumulative percentages from samples examined in panel D. *P < 0.003 in comparison with corresponding BRCA/DNA-PK–proficient samples using Student’s t test. *P = 0.08 or 0.11 in comparison with DNR- or BMN673-treated BRCA/DNA-PK–deficient samples, respectively, using 2-way ANOVA.
BRCA/DNA-PK–deficient Lin–CD34+ t-MDS/AML primary cells from 3 patients were more sensitive to BMN673 and BMN673 + DNR in comparison with BRCA/DNA-PK–proficient counterparts from 3 patients (Figure 6C). Consistent with this finding, immature Lin–CD34+CTVlo proliferating cells and Lin–CD34+CTVmax quiescent cells from BRCA/DNA-PK–deficient t-MDS/AMLs were much more sensitive to BMN673 alone and in combination with DNR in comparison with the counterparts from BRCA/DNA-PK–proficient samples (Figure 6, D and E). BMN673 + DNR exerted a borderline synergistic effect against BRCA/DNA-PK–deficient cells.
To design a more diagnostically oriented approach for predicting BRCA/DNA-PK deficiency we employed RT-qPCR to quantify expression of DSB repair genes compared with a housekeeping gene such as COX6B. Our data suggest that BRCA1 and LIG4 mRNA levels lower than 1,100% and 2%, respectively, of that encoded by COX6B is predictive of BRCA/DNA-PK deficiency in 2 of 11 (18%) samples, resulting in the highest sensitivity to BMN673-driven synthetic lethality (Supplemental Figure 6, A–C). Meta-analysis of the B cell acute lymphoblastic leukemia (B-ALL) transcriptome database supported our observation that a cohort of B-ALLs were BRCA/DNA-PK deficient and that BRCA1 and LIG4 were frequently downregulated in these cases (Supplemental Figure 6D).
Altogether, gene expression profiling by mRNA microarrays and by RT-qPCR followed by flow cytometry identified individual cases of BRCA/DNA-PK–deficient AMLs/ALLs, in which immature quiescent and proliferating cells were highly sensitive to PARP1i alone or in combination with the approved first-line AML therapy drug, DNR.
PARP1i exerted an antileukemic effect against BRCA/DNA-PK–deficient primary leukemia xenografts.
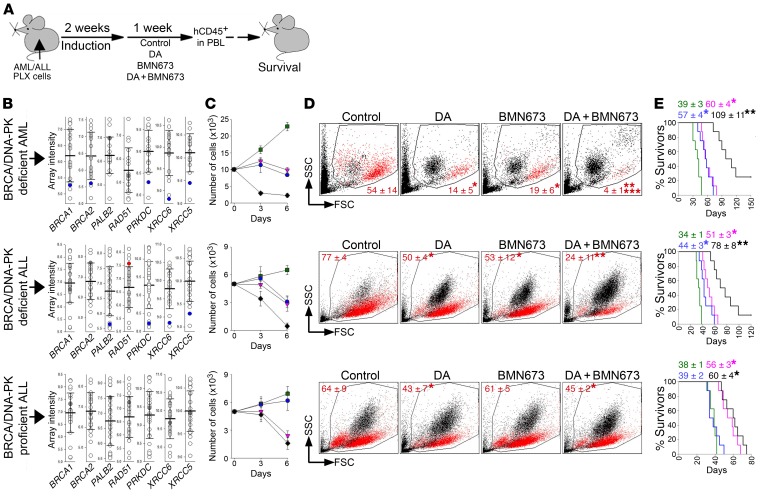
NSG mice bearing BRCA/DNA-PK–deficient or –proficient primary leukemia xenografts (PLXs) were left untreated or treated with vehicle (control), doxorubicin (DA), BMN673, or DA + BMN673 (Figure 7A). Therapeutic effect was measured by detection of hCD45+ cells in peripheral blood and by median survival time (MST). BRCA/DNA-PK–deficient and –proficient PLXs were preselected based on microarray analysis (Figure 7B), by in vitro sensitivity to BMN673 alone or in combination with DNR (Figure 7C), and by nuclear foci formation or flow cytometric detection of DSB repair proteins (Supplemental Figure 5, C and D).
Figure 7. PARP1i exerted a therapeutic effect in mice bearing BRCA/DNA-PK–deficient PLXs.
(A) Experimental design: NSG mice were injected with PLX cells and treated with diluents (control), doxorubicin + ara-C (DA), BMN673, or DA + BMN673 (8 mice/group). Human CD45+ (hCD45+) cells were detected in peripheral blood leukocytes (PBLs) 1 week after the treatment, and survival was determined. (B) Microarray profile of the indicated genes in AML (top plot) and B-ALL (middle and bottom plots) PLXs. Each circle represents an individual PLX; error bars show SD of the mean. Data from PLXs used for further experiments are marked in blue, gray, and red, indicating lower than average, average, and higher than average expression levels, respectively, of the indicated genes in BRCA/DNA-PK–deficient (BRCA/DNA-PK-D) AML and B-ALL (top and middle plots, respectively) and in BRCA/DNA-PK–proficient (BRCA/DNA-PK-p) B-ALL (bottom plot) cells. (C) Living cells from individual PLXs treated in vitro (triplicate experiment) with diluents (green), DA (purple), BMN673 (blue), or DA + BMN673 (black). (D) Representative plots of PBLs from treated mice (n = 6/group); percentage of hCD45+ cells (red dots) is indicated. *P ≤ 0.001, **P ≤ 0.001 in comparison with control and DA or BMN673, respectively, using Student’s t test adjusted for multiple comparisons. ***P < 0.03 in comparison with DA- or BMN673-treated mice using 2-way ANOVA. (E) Survival curves and MST of PLX mice treated with diluents (green), DA (purple), BMN673 (blue), or DA + BMN673 (black). * P < 0.003, **P < 0.003 in comparison with control and DA or BMN673, respectively, using Kaplan-Meier log-rank test.
Control mice succumbed to leukemias characterized by splenomegaly (3- to 15-fold enlarged spleen) and the presence of hCD45+ cells in peripheral blood. Approximately 60% and 79% of these hCD45+ cells were also CD33+ or CD19+, respectively, thus confirming the development of myeloid and lymphoid malignancies. BMN673 alone and a combination of DA + BMN673 reduced the number of BRCA/DNA-PK–deficient hCD45+ AML and B-ALL cells, but did not affect BRCA/DNA-PK–proficient B-ALL cells (Figure 7D). Moreover, DA + BMN673 exerted synergistic and additive effects against BRCA/DNA-PK–deficient AML and ALL, respectively. Selective elimination of BRCA/DNA-PK–deficient hCD45+ PLX cells by BMN673 and DA + BMN673 was associated with extended disease latency (Figure 7E). A cohort of mice treated with DA + BMN673 survived more than 150 (AML) and 120 (ALL) days and hCD45+ cells were not detected in these mice.
In addition, 2 × 106 bone marrow cells (BMCs) from untreated and DA + BMN673–treated individual mice bearing BRCA/DNA-PK–deficient PLX (AML and ALL) were transplanted to secondary recipient mice. All mice injected with cells from untreated donors died within 27 (AML) and 21 (ALL) days, whereas 6 of 8 (AML) and 4 of 8 (ALL) recipients of cells from DA + BMN673–treated donors survived 100 days or more (Supplemental Figure 7). Recipients of AML/ALL cells from mice treated with DA + BMN673 demonstrated significantly prolonged survival, suggesting that LSCs were targeted.
PARP1i eradicated BRCA1/DNA-PK–deficient proliferating and quiescent immature leukemia cells identified by gene mutation analysis.
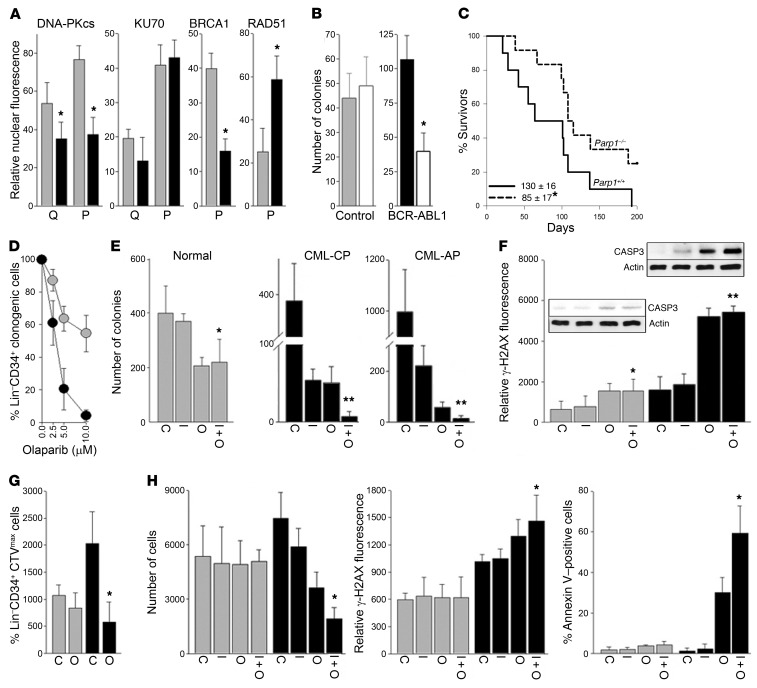
BCR-ABL1 oncogenic tyrosine kinase causes downregulation of BRCA1 and DNA-PKcs protein expression (16, 17). In concordance, we detected downregulation of DNA-PKcs and/or BRCA1 in BCR-ABL1–positive chronic myeloid leukemia (CML) in chronic phase (CML-CP) Lin–CD34+Ki67– quiescent and Lin–CD34+Ki67+ proliferating immature cells, respectively (Figure 8A). These observations combined with high levels of ROS-induced DSBs in untreated and tyrosine kinase inhibitor (TKI)–treated CML-CP quiescent and proliferating LSCs and LPCs (18) create an opportunity to trigger dual cellular synthetic lethality by PARP1i.
Figure 8. PARP1i inhibited BCR-ABL1–mediated transformation and eliminated BRCA1/DNA-PKcs–deficient quiescent and proliferating immature CML cells.
(A) Expression of indicated nuclear proteins in Lin–CD34+Ki67– quiescent (Q) and Lin–CD34+Ki67+ proliferating (P) cells from CML-CP patients (black bars, n = 3) and normal donors (gray bars, n = 3). *P < 0.05 using Student’s t test. (B) Number of colonies from BCR-ABL1–transformed or nontransformed (control) Parp1+/+ and Parp1–/– mBMCs (triplicate experiment). *P = 0.006 using Student’s t test. (C) Survival curves and MST of the tet-off SCLtTA p210BCR-ABL1 Parp1–/– (n = 12) and SCLtTA p210BCR-ABL1 Parp1+/+ (n = 10) mice. *P = 0.04 using Kaplan-Meier log-rank test. (D–H) Lin–CD34+ cells from CML-CP and CML-AP patients (black symbols; n = 3–10) and healthy donors (gray symbols; n = 3) were (D) treated with the indicated concentrations of olaparib or (E–H) left untreated (C) or treated with 1 μM imatinib (I), 5 μM olaparib (O), or a combination (I + O) for 5 days. (D and E) Lin–CD34+ clonogenic cells. *P < 0.05, **P < 0.001 in comparison with I using Student’s t test. **P ≤ 0.005 in comparison with I or O using 2-way ANOVA. (F) γ-H2AX immunofluorescence. *P < 0.05, **P < 0.001 in comparison with I. Active caspase 3 (CASP3) and β-actin detected by Western blot on parallel gels. (G) Lin–CD34+CTVmax quiescent cells/105 cells. *P < 0.05 in comparison with C using Student’s t test. (H) Lin–CD34+CD38–CTVmax quiescent cells. (Left) Number of cells/106 cells. *P < 0.05 in comparison with I. (Middle) γ-H2AX immunofluorescence. *P = 0.007 in comparison with I. (Right) Apoptotic cells. *P = 0.03 in comparison with I using Student’s t test.
BCR-ABL1–transformed Parp1–/– murine BMCs (mBMCs) formed approximately 2.5-fold fewer colonies than their Parp1+/+ counterparts (Figure 8B). To determine the role of PARP1 in disease-mimicking settings, a tet-off SCLtTA p210BCR-ABL1–inducible mouse model of CML-CP was employed (19). CML-CP–like disease latency was prolonged in SCLtTA p210BCR-ABL1 Parp1–/– mice when compared with SCLtTA p210BCR-ABL1 Parp1+/+ counterparts (Figure 8C). Altogether, it appears that PARP1 plays an important role in BCR-ABL1–mediated leukemogenesis.
Olaparib selectively reduced clonogenic activity of Lin–CD34+ CML-CP immature cells in a dose-dependent manner (Figure 8D). The combination of TKI imatinib and PARP1 inhibitor olaparib synergistically abrogated clonogenic potential of Lin–CD34+ immature cells from CML-CP and also from more aggressive CML accelerated phase (CML-AP) patient samples (Figure 8E); the effect was preceded by the accumulation of DSBs measured by γ-H2AX and induction of apoptosis detected by activation of caspase 3 (Figure 8F).
Olaparib also eliminated Lin–CD34+CTVmax quiescent CML-CP cells (Figure 8G) and imatinib plus olaparib reduced the number of LSC-enriched Lin–CD34+CD38–CTVmax quiescent CML-CP cells (Figure 8H). The latter effect was accompanied by an accumulation of DSBs and annexin V–positive apoptotic cells (Figure 8H). Moreover, olaparib combined with imatinib or ponatinib exerted synergistic effects against BCR-ABL1–positive and BCR-ABL1(T315I)–positive B-ALL, respectively (Supplemental Figure 8).
Meta-analyses of transcriptome databases and other reports suggested that the fusion oncoprotein AML1-ETO, but not the oncogenic partners HOXA9 and MEIS1 or the oncogenic mutant FLT3/ITD, may negatively modulate BRCA and DNA-PK pathways (20–22). In fact, AML1-ETO–transformed mBMCs display decreased expression of BRCA1, BRCA2, and/or KU70 proteins in comparison with HOXA9 + MEIS1–transformed cells (Supplemental Figure 9A). Parp1–/– mBMCs expressing AML1-ETO, but not HOXA9 + MEIS1, demonstrated reduced clonogenic activity in comparison with Parp1+/+ counterparts (Supplemental Figure 9B). AML1-ETO–transformed Parp1+/+ mBMCs were highly sensitive to olaparib in comparison with cells expressing HOXA9 + MEIS1 or GFP only (Supplemental Figure 9C). In addition, olaparib eliminated Lin–CD34+ immature leukemia cells, including LSC-enriched Lin–CD34+CD38–CFSElo proliferating and Lin–CD34+CD38–CFSEmax quiescent cell populations from AML patient samples carrying t(8;21)(q22;q22) encoding for AML1-ETO (Supplemental Figure 9D). Moreover, the combination of olaparib and low doses of first-line therapy drug DNR exerted anticlonogenic effects against cells expressing AML1-ETO, but not against those expressing HOXA9-MEIS1 or GFP (Supplemental Figure 9E).
PARP1i exerted an antileukemic effect in vivo against BCR-ABL1–positive CML.
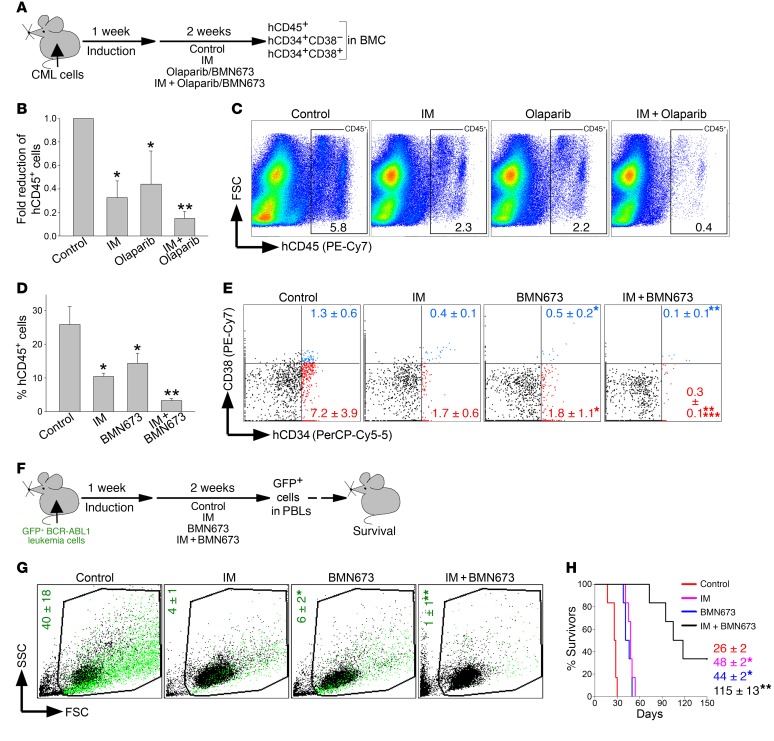
To test the therapeutic potential of PARP1i in CML, sublethally irradiated NSG mice were injected with 2.5 × 106 Lin–CD34+ CML-CP cells obtained from 4 individual patients followed by treatment with vehicle, imatinib, olaparib, or imatinib + olaparib (Figure 9A). Since CML-CP cells usually do not induce lethal disease, we monitored leukemia cells in bone marrow 7 days after treatment. Imatinib or olaparib reduced the number of hCD45+ leukemia cells by 2- to 3-fold; in contrast, combination of these drugs caused a greater than 5-fold reduction of leukemia cells (Figure 9, B and C).
Figure 9. PARP1i exerted a therapeutic effect in mice bearing BCR-ABL1–positive leukemia.
(A) Experimental design: sublethally irradiated NSG mice were injected with Lin–CD34+ CML-CP cells (B and C) or Lin–CD34+ CML-AP cells (D and E) followed by treatment with vehicle (control), imatinib (IM), BMN673, olaparib, IM + BMN673, or IM + olaparib for 14 consecutive days. Leukemia burden was assessed by detection of indicated human cells in mBMCs. (B) hCD45+ CML-CP cells (n = 4–8/group). *P ≤ 0.003, **P < 0.02 in comparison with control and IM or olaparib, respectively, using Student’s t test adjusted for multiple comparisons. (C) Percentage of hCD45+ CML-CP cells (framed) in representative plots. (D) hCD45+ CML-AP cells (n = 4/group). *P < 0.01, **P ≤ 0.001 in comparison with control and IM or olaparib, respectively, using Student’s t test adjusted for multiple comparisons. (E) Percentage of Lin–hCD34+CD38– (red dots) and Lin–hCD34+CD38+ (blue dots) CML-AP cells; representative plots from 4 mice/group are shown. *P < 0.02, **P < 0.03 in comparison with control and IM, respectively, using Student’s t test. ***P = 0.08 in comparison with individually treated mice by 2-way ANOVA. (F) Experimental design: sublethally irradiated SCID mice were injected with GFP+BCR-ABL1 leukemia cells and 1 week later treated with vehicle (control), IM, BMN673, or IM + BMN673 for 14 consecutive days. GFP+ leukemia cells in peripheral blood leukocytes (PBLs) and survival were scored. (G) Percentages of GFP+BCR-ABL1 leukemia cells (green dots) in representative plots from 3–4 mice/group. *P = 0.01 in comparison with control using Student’s t test. **P < 0.02 in comparison with individually treated mice by 2-way ANOVA. (H) Survival curves and MST (n = 6 mice/group). * P < 0.001, **P < 0.001 in comparison with control and IM or BMN673, respectively, using Kaplan-Meier log-rank test.
The above experiments were repeated using 1 × 107 Lin–CD34+ cells from a more advanced CML-AP patient; 1 week following injection mice were treated with vehicle, imatinib, BMN673, or imatinib + BMN673 (Figure 9A). Imatinib or BMN673 diminished the number of hCD45+ cells by 2- to 2.5-fold, whereas imatinib + BMN673 exerted a 7-fold reduction (Figure 9D). Moreover, imatinib or BMN673 applied individually reduced the number of LSC-enriched Lin–CD34+CD38– cells and Lin–CD34+CD38+ LPCs (23) by 2.5- to 4-fold, whereas the combination reduced the LSC-enriched population and LPCs by 24- and 13-fold, respectively (Figure 9E). Imatinib + BMN673 exerted borderline synergistic effects against LSC-enriched Lin–CD34+CD38– cells.
In addition, sublethally irradiated SCID mice were injected with GFP+ BCR-ABL1–positive murine leukemia cells followed by imatinib and/or BMN673 treatment (Figure 9F). When applied individually, imatinib or BMN673 reduced the leukemia burden in peripheral blood by 7- to 10-fold, whereas the combination exerted a synergistic effect and caused a 40-fold reduction (Figure 9G). This effect was associated with prolongation of the MST of leukemic mice treated with imatinib or BMN673 by 1.7- to 1.8-fold and by 4.4-fold of those treated with a combination (Figure 9H). Two out of 6 mice treated with imatinib + BMN673 survived more than 150 days and GFP+ cells were not detected in these animals.
Discussion
Quiescent and proliferating tumor cells accumulate spontaneous and treatment-induced DSBs, which are eventually repaired and thereby tolerated (18, 24, 25). Inhibition of DSB repair has emerged as a promising antitumor treatment, especially in the context of synthetic lethality, which targets specific pathways to eradicate tumor cells while sparing normal cells (26).
The success of the PARP1 inhibitor olaparib in BRCA1- and BRCA2-deficient breast tumors has established a proof of concept for personalized cancer therapy utilizing synthetic lethality (6, 7). Tumors from individual patients predicted to be sensitive to PARP1i are usually identified either by detecting mutations in genes encoding BRCA pathway proteins, epigenetic-mediated downregulation of BRCA pathway protein expression, or by functional tests uncovering inefficient RAD51 foci formation (26). While these approaches may be effective at targeting proliferating cells, they are not able to simultaneously eradicate quiescent therapy-refractory cancer stem cells (3). However, quiescent LSCs are often responsible for minimal residual disease and disease relapse (8). Therefore, we postulated that a precision medicine–guided PARP1i-mediated dual cellular synthetic lethality approach that simultaneously targets quiescent and proliferating cancer stem and progenitor cells could be curative.
As expected, PARP1 inhibitors (olaparib and BMN673) elevated lethal DSBs and promoted synthetic lethality in BRCA-deficient (BRCA1–/–, FANCA–/–, RAD54–/–) proliferating tumor cells. This effect is likely caused by the abrogation of PARP1-mediated stalled replication fork restart and/or inhibition of PARP1-mediated B-NHEJ (27, 28). In addition, we show that PARP1i elevated DSBs, resulting in synthetic lethality also in DNA-PK–deficient (XRCC6–/–, LIG4–/–) cells; this phenomenon is supported by previous reports that combined use of DNA-PK and PARP1 inhibitors radiosensitized cells (29) and that PARP1 inhibitor–treated Lig4–/– and Dclre1c–/– mouse embryonic fibroblasts accumulated DSBs, thereby reducing cell survival (30).
DNA-PK–deficient proliferating cells repair DSBs by HR and/or B-NHEJ (31); thus, PARP1i would abrogate B-NHEJ activity and also delay stalled replication fork restart and HR (32), which is consistent with a modest but significant degree of synthetic lethality. Interestingly, the lethal effect of PARP1i was more pronounced in DNA-PK–deficient quiescent rather than proliferating cells. Quiescent cells accumulate potentially lethal DSBs, which are repaired by NHEJ (D-NHEJ and/or B-NHEJ) (25, 31); thus, targeting PARP1 in DNA-PK–deficient quiescent cells is synthetically lethal. This idea is supported by the observation that radiation-induced DSBs in Lig4–/– cells reduced cell survival independently of DNA replication (30).
We report here that olaparib and BMN673 induced synthetic lethality in both proliferating and quiescent BRCA/DNA-PK–deficient cells. This dual cellular synthetic lethality effect results from the accumulation of lethal DSBs in PARP inhibitor–treated BRCA/DNA-PK–deficient cells in S-G2/M of the cell cycle, and in PARP1 inhibitor–treated DNA-PK–deficient cells in G0/G1 phase. However, it is plausible that BRCA/DNA-PK–deficient cells with PARP1i-induced DNA damage/DSBs can continue through the G2/M checkpoint and/or spindle-assembly checkpoint, leading to the accumulation of excessive lethal DNA damage/DSBs in DNA-PK–deficient G0/G1 cells, thus resulting in cell death (33, 34).
It has been reported that BRCA-deficient tumors with low D-NHEJ activity might be less responsive to PARPi (35). These studies, however, relied on short-term disabling of the DNA-PK pathway by using genetic or pharmacologic approaches. BRCA/DNA-PK–deficient cells tested here harbored long-term DNA-PK deficiency, presumably enforcing substantial remodeling of DSB repair pathways, which promotes sensitivity to PARP1i. This inference is supported by this work and other reports that BCR-ABL1 leukemia cells, which display BRCA1 and DNA-PKcs deficiencies (16, 17), are highly sensitive to PARP1i, and that hyperactive NHEJ caused resistance to PARP1i in BRCA-deficient Fanca–/– and Fancc–/– hematopoietic stem and progenitor cells (36).
PARP1i-induced dual cellular synthetic lethality in BRCA/DNA-PK–deficient cell lines prompted us to test PARP1i in BRCA/DNA-PK–deficient individual tumors identified by our personalized medicine strategy referred to as GEMA. Leukemias were used as testing models because their hierarchical compartments of quiescent LSCs and proliferating LSCs/LPCs are well characterized (37). Our report suggests that PARP1i eliminated not only proliferating LSCs/LPCs, but also therapy-refractory quiescent LSCs if they displayed BRCA/DNA-PK–deficient status detected by GEMA. The lethal effect of PARP1i was associated with the accumulation of DSBs and apoptosis, and was selective for BRCA/DNA-PK–deficient leukemia cells, because normal cells and BRCA/DNA-PK–proficient leukemia cells were mostly spared.
Gene mutation profiling showed that leukemias expressing BCR-ABL1 (100% of CMLs and 2%–7% of ALLs in pediatric age and 20%–40% of ALLs in adults) and AML1-ETO (4%–12% of adult and 12%–30% of pediatric AMLs) are BRCA/DNA-PK deficient. The mechanisms responsible for the downregulation of genes in the BRCA/DNA-PK pathways in leukemia cells expressing BCR-ABL1 and AML1-ETO are characterized only partially. For example, BCR-ABL1 kinase inhibits BRCA1 and DNA-PKcs protein expression by suppression of translation and/or enhanced protein degradation (16, 17). While expression of these proteins is partially restored in imatinib-treated cells, other key members of HR (RAD51) and D-NHEJ (LIG4, DCLRE1C, NHEJ1) are strongly downregulated by imatinib, thus promoting PARP1i-mediated synthetic lethality (38, 39). The BRCA2-deficient phenotype of AML1-ETO–positive cells may depend on lack of stimulation of HOXA9 (22).
GEMA, with emphasis on gene mutation analysis, may also be applied to identify additional oncogenes that sensitize cells to PARP1 inhibitor treatment, for example TMPRSS2-ERG–positive prostate carcinomas (DNA-PKcs deficiency), EGFR mutant–positive lung carcinomas (Fanconi anemia deficiency), EWSR1-FLI1–positive Ewing’s sarcomas, and KRAS mutant–positive leukemias (40–43).
Moreover, using gene expression profiling we determined that 9%–25% of AMLs/ALLs that did not contain these oncogenes also displayed BRCA/DNA-PK deficiency. It is also plausible that additional BRCA/DNA-PK–deficient AMLs/ALLs could be identified if the list of target genes were to be expanded. For example, inclusion of Fanconi anemia genes (FANCA, FANCC, FANCG, FANCI), in addition to FANCD1 (also known as BRCA2), increased the pool of predicted BRCA/DNA-PK–deficient samples in TCGA database from 9% to 12%.
BRCA/DNA-PK deficiency in individual AMLs/ALLs detected by gene expression profiling may be due to epigenetic alterations. Analysis of methylome arrays revealed distinct BRCA and DNA-PK pathway methylome profiles in individual patients, which could be associated with reduced expression of methylated genes, resulting in sensitivity to PARP1i (44). Differences in expression of heterogeneous nuclear ribonucleoproteins (e.g., hnRPN C) and miRNAs (e.g., miR-101 and miR-182) may also contribute to a BRCA/DNA-PK–deficient phenotype of individual AMLs/ALLs (45–47).
In conclusion, we postulate that PARP1i-mediated dual cellular synthetic lethality may simultaneously eradicate BRCA/DNA-PK–deficient quiescent and proliferating LSCs/LPCs (Supplemental Figure 10). In addition, we have developed a potentially novel clinically oriented personalized medicine approach, which we refer to as GEMA, for identifying patients with AMLs/ALLs displaying BRCA/DNA-PK deficiency, which could benefit from PARP1i-induced dual cellular synthetic lethality. GEMA-guided PARP1i-mediated dual cellular synthetic lethality may have a broad application for cancer treatment considering that numerous individual samples from a variety of tumor types in TCGA database may display BRCA/DNA-PK deficiency (Supplemental Figure 11).
Methods
Primary cells. Peripheral blood and bone marrow samples from patients with newly diagnosed CML-CP, CML-AP, ALL, AML, and t-MDS/AML were obtained from: (a) Stem Cell and Leukemia Core Facility of the University of Pennsylvania, Philadelphia, Pennsylvania, USA; (b) Department of Internal Medicine I, Division of Hematology & Hemostaseology, Medical University of Vienna; (c) Princess Margaret Cancer Centre; (d) Department of Hematology, Institute of Hematology and Blood Transfusion, Warsaw, Poland; and (e) Division of Hematopoietic Stem Cell and Leukemia Research, City of Hope National Medical Center. Clinical annotations for these samples are listed in Supplemental Tables 1–4. Samples of normal hematopoietic cells were purchased from Cambrex Bio Science. Lin–CD34+ cells were obtained from mononuclear fractions by magnetic sorting using the EasySep negative selection human progenitor cell enrichment cocktail followed by Human CD34 Positive Selection Cocktail (StemCell Technologies). PLXs containing greater than 80% and greater than 90% of AML and B-ALL cells, respectively, were used.
Cell lines. BRCA1-mutated and BRCA1-reconstituted MDA-MB-436 human breast carcinoma cells were obtained from Neil Johnson (Fox Chase Cancer Center). BRCA2–/– V-C8 and BRCA2+/+ V79 hamster cell lines carrying the DR-GFP reporter cassette were obtained from Maria Jasin (Memorial Sloan Kettering Cancer Center, New York, New York, USA) (48). XRCC6–/– and parental mESC lines with EJ2-GFP and EJ5-GFP reporters and FANCA–/– and FANCA–/– XRCC6–/– mESC lines were obtained from Jeremy Stark (Beckman Research Institute of the City of Hope) (10). Nalm-6 parental cells and RAD54–/–, LIG4–/–, and RAD52–/– LIG4–/– isogenic lines were purchased from Horizon.
Transgenic/knockout mice. SCLtTA p210BCR-ABL1 mice (19) were cross-bred with Parp1–/– mice (provided by Roberto Caricchio, Temple University School of Medicine) to generate SCLtTA p210BCR-ABL1 Parp1–/– and SCLtTA p210BCR-ABL1 Parp1+/+ animals. Transgenic/knockout mice were identified by PCR of tail snip DNA. DNA isolation and purification from mice tails were performed using the REDExtract-N-Amp Tissue PCR Kit (Sigma-Aldrich). Genotyping for the SCLtTA and p210BCR-ABL1 transgenes and Parp1 was performed using transgene/knockout–specific primers (Operon) and 2× GoTaq polymerase Master Mix (Promega). BCR-ABL1-specific primers (forward, 5′-GAGCGTGCAGAGTGGAGGGAGAACA-3′; reverse, 5′-GGTACCAGGAGTGTTTCTCCAGACTG-3′) amplified a 500-bp fragment using amplification conditions of 40 cycles at 94°C for 45 seconds, 55°C for 1 minute, and 72°C for 1 minute. SCLtTA-specific primers (tTA, 5′-TTTCGATCTGGACATGTTGG-3′; SCL,5′-AGAACAGAATTCAGGGTCTTCCTT-3′) yielded a 750-bp product using amplification conditions consisting of 40 cycles at 94°C for 40 seconds, 60.5°C for 1 minute, and 72°C for 1 minute. Parp1-specific primers used were: forward, 5′-CATGTTCGATGGGAAAGTCCC-3′; wild-type reverse, 5′-CCAGCGCAGCTCAGAGAAGCCA-3′; and mutant reverse, 5′-CATGTTCGATGGGAAAGTCCC-3′. The primers amplified a 112-bp fragment if wild type, a 350-bp fragment if Parp1 null, and both 112- and 350-bp fragments if heterozygous using amplification conditions consisting of 35 cycles at 94°C for 1 minute, 60°C for 1 minute, and 72°C for 3 minutes. PCR products were run in a 1.5% agarose gel containing ethidium bromide, and visualized using the Gel Doc XR+ Molecular Imager System (Bio-Rad). Mice were provided with drinking water supplemented with 0.5 g/l tetracycline hydrochloride (Sigma-Aldrich) and leukemia was induced by withdrawal of tetracycline. CML-CP–like leukemia was characterized by splenomegaly and leukocytosis associated with expansion of mature myeloid cells as described previously (18, 19).
Transfection of Parp1–/– and Parp1+/+ cells. mBMCs were infected with MSCV retroviruses containing AML1-ETO9a-IRES-GFP, HOXA9-IRES-MEIS1, BCR-ABL1-IRES-GFP, and IRES-GFP as described previously (49). GFP+ cells were sorted and used for experiments.
Microarray analysis. Gene expression data for 15 genes were used to select BRCA/DNA-PK–deficient and –proficient AMLs from a dataset of 172 samples arrayed on an Illumina HT-12 (supplemental data: AML gene expression levels). Peripheral blood or bone marrow samples were collected from subjects with AML after obtaining informed consent according to the procedures approved by the Research Ethics Board of the University Health Network (Toronto Princess Margaret Cancer Centre). The AML samples were cytogenetically normal. Bioinformatic analysis was performed in R. The log2 expression values for each gene were centered to a zero mean and Z scores were generated by dividing by the SD.
RT-qPCR
B-ALL.
Total RNA from human leukemia cells was extracted using an RNeasy Plus Micro Kit (QIAGEN) according to the manufacturer’s instructions. cDNA was generated using a poly(dT) oligonucleotide and SuperScript III Reverse Transcriptase according to the manufacturer’s protocol (Invitrogen). The levels of gene expression were measured using the SYBRGreenER mix (Invitrogen) with primers listed in Supplemental Table 5. The reactions were performed using the ABI 7900HT real-time PCR system (Applied Biosystems) and analyzed with the SigmaPlot version 12.0 software program.
t-MDS/AML.
Human CD34+ cells from t-MDS patients and normal controls were isolated by using magnetic bead selection (Miltenyi Biotec). Total RNA was extracted using the RNeasy Plus Micro Kit. First-strand cDNA was synthesized from 200 cells using a Superscript III First-Strand Synthesis System (Invitrogen) and preamplified for 18 cycles, and qPCR analysis performed by multiplex qPCR using the Fluidigm system and primers listed in Supplemental Table 6. Results are expressed as the ratio to β2 microglobulin.
Detection of DSB repair proteins
Flow cytometry.
The cells were fixed and permeabilized using a PerFix EXPOSE kit according to the manufacturer protocol (Beckman Coulter) followed by staining with specific antibodies: rabbit polyclonal anti-Ki67 (Abcam, ab15580), rabbit polyclonal anti-XRCC3 (Abcam, ab133736), rabbit polyclonal anti–ligase IV (Abcam, ab26039), mouse monoclonal IgG2b anti-BRCA1 (R&D Systems, MAB22101), mouse monoclonal IgG1 anti-BRCA2 (R&D Systems, MAB2476), rabbit polyclonal anti-PALB2 (Bethyl Laboratories, A301-246), rabbit polyclonal anti-KU80 (Bethyl Laboratories, A302-627A), rabbit polyclonal anti-KU70 (Bethyl Laboratories, A302-623A), rabbit polyclonal anti–DNA-PKcs (Bethyl Laboratories, A300-518A). For FACS analysis the antibodies were conjugated with fluorochromes (Alexa Fluor 405, Alexa Fluor 488, or allophycocyanin) using a Zenon labeling kit (Life Technologies). Isotype control antibodies (Abcam) were labeled and used as a background staining control. For DSB detection the rabbit polyclonal antibody against histone H2A.X phosphorylated at Ser193 (Biolegend, 613403) was used. Flow cytometric analysis was performed using the LSRFortessa (Becton Dickinson) equipped with lasers 355, 405, 488, and 635 nm.
Western analysis.
Cells were lysed at 95°C in buffer containing 50 mM Tris-HCl pH 6.8, 10% glycerol, and 2% SDS. Cell lysates were analyzed by Western blot using the antibodies detecting BRCA1 (R&D Systems, MAB22101), BRCA2 (R&D Systems, MAB2476), XRCC3 (Abcam, ab133736), RAD51 (Santa Cruz Biotechnology, sc6862), DNA-PKcs (Bethyl Laboratories, A300-518A), KU70 (Bethyl Laboratories, A302-623A), KU80 (Thermo Fisher Scientific, MA5-15873), cleaved caspase 3 (Cell Signaling Technology, 9661), PARylated proteins (Trevigen, 4335-MC-100), and β-actin (Sigma-Aldrich, A5316).
Immunocytofluorescence.
Nuclear expression of DNA-PKcs, KU70, BRCA1, and RAD51 was quantified as previously described with modifications (50). Briefly, Lin–CD34+ cells were cytospun followed by costaining with anti-Ki67 and anti–DNA-PKcs, -KU70, -BRCA1, or -RAD51 antibodies. Secondary antibodies conjugated with Alexa Fluor 488 or Alexa Fluor 568 were applied (Molecular Probes, A-11034 and A-11004). DNA was counterstained with DAPI (4′,6′diamedino-2-phenylindole). Specific nuclear staining was visualized using an inverted Olympus IX70 fluorescence microscope equipped with 100× UPlan Apo lens (numeric aperture 1.35), and a Cooke Sensicam QE camera (The Cooke Company). Images were acquired with Slidebook 3.0 (Intelligent Imaging Innovations). All graphic adjustments were performed using Adobe Photoshop.
HRR and NHEJ assays.
Xrcc6–/– and parental mESCs with EJ2-GFP and EJ5-GFP reporter cassettes and Brca2–/– V-C8 and Brca2+/+ V79 cells carrying DR-GFP reporter cassette were cotransfected with pCBASce1 (encoding I-Sce1) and pDsRed (transfection efficiency control) plasmids using Lipofectamine 2000 (Invitrogen) as described previously (10, 50). Olaparib (Selleckchem, AZD2281) or vehicle (DMSO) was added immediately after removing the transfection complexes. The percentage of GFP+DsRed+ cells in the DsRed+ population was detected after 72 hours by flow cytometry to assess DSB repair activity.
In vitro treatment.
TKIs such as imatinib and ponatinib, cytotoxic drugs (DNR, DA, cytarabine), and PARP1 inhibitors olaparib and BMN673 (all from Selleckchem) were added to the indicated cells for 3 to 5 days. Murine hematopoietic/leukemia cells were maintained in Iscove’s Modified Dulbecco’s Medium (IMDM) supplemented with 10% FBS, 100 ng/ml stem cell factor (SCF), and 20 ng/ml IL-3. Primary human hematopoietic cells were incubated in IMDM supplemented with 10% FBS and growth factors (CML, AML, and normal counterparts = 100 ng/ml SCF, 20 ng/ml IL-3, 100 ng/ml Flt-3 ligand, 20 ng/ml G-CSF, 20 ng/ml IL-6; B-ALL = 10 ng/ml IL-7) followed by plating in Methocult (StemCell Technologies) when indicated; colonies were scored after 5 to 7 days. For quiescent/proliferating cells, Lin– cells were stained with CFSE, cell trace violet (CTV), or Cell Proliferation Dye eFluor 670 (CPD) (eBioscience) and incubated for 5 days in StemSpan SFEM medium (Stem Cell Technologies) supplemented with the cocktail of growth factors (see above) and inhibitors where indicated. In addition, quiescent/proliferating cells were detected by immunostaining with anti-Ki67 antibody. Quiescent (CFSEmax, CPDmax, or Ki67–) and proliferating (CFSElo, CPDlo, or Ki67+) leukemia and normal hematopoietic stem and progenitor cells were detected by flow cytometry using fluorochrome-conjugated anti-Lin (catalog 340546), anti-CD34 (catalog 347203 and 555821), and anti-CD38 (catalog 355790 and 555460) antibodies (all from BD Pharmingen) as described previously (18). Cell viability was determined either by trypan blue exclusion or by annexin V staining.
In vivo treatment.
ICR SCID mice (Taconic Biosciences) were total-body irradiated (250 cGy) and injected i.v. with 1 × 105 BCR-ABL1–transformed GFP+ mBMCs. NSG mice (The Jackson Laboratory) were total-body irradiated (250 cGy) and inoculated i.v. with 1 × 106 AML and ALL PLX cells, 2.5 × 106 CD34+ CML-CP cells, or 1 × 107 CD34+ CML-AP cells. Seven or 10 days later mice were treated with vehicle (control), BMN673 (0.33 mg/kg/day by oral gavage for 7 or 14 days; see ref. 51), olaparib (50 mg/kg daily i.p. for 14 days; see ref. 52), DA (1.5 mg/kg/day i.v. [days 1–3]) + ara-C (50 mg/kg i.v. [days 1–5]; see ref. 53), imatinib (100 mg/kg twice daily by oral gavage; see ref. 54), and a combination of imatinib + BMN673/olaparib or DA + BMN673. Leukemia burden was analyzed by flow cytometry 7 days after the end of treatment. Murine leukemia cells were GFP+. Human leukemia cells were detected by anti–human CD45 antibody and leukemia stem and progenitor cells were detected by anti–human CD34 and CD38 antibodies as described previously (18). Median survival time was determined.
Statistics.
Data are expressed as mean ± SD. When conducting subgroup comparisons between 2 groups, a 2-tailed t test was used. When multiple comparisons were conducted using a t test, Bonferroni’s method was used for adjusting P values for multiple testing. The synergistic effect was evaluated by 2-way ANOVA. MST of the mice ± SEM was calculated by Kaplan-Meier log-rank survival analysis. P values less than 0.05 were considered significant.
Study approval.
Human studies were approved by the appropriate IRBs and met all requirements of the Declaration of Helsinki. Animal studies were approved by the Temple University IACUC.
Author contributions
MNS performed in vitro experiments with primary cells and assisted with experiments in mice. KS performed experiments with AML/ALL PLX and BCR-ABL1 leukemia in mice. YD tested dual cellular synthetic lethality in adherent cell lines and examined HR and NHEJ activities in primary cells. PPB analyzed protein expression by immunofluorescence. GH performed in vivo experiments in NSG mice injected with CML cells. SM generated data in NALM-6 cell lines. EM and DDM generated in vitro data with AML1-ETO in Parp1–/– and wild-type BMCs. EBG assisted in analyzing the effect of PARP1i on quiescent and proliferating cells. KCM performed experiments using B-ALL PLX cells. JL analyzed expression of HR and NHEJ genes in B-ALLs by RT-qPCR. KE and LB performed meta-analyses of gene expression patterns in the cohorts of AMLs. AS assessed the expression of DNA repair proteins in CML cells. ML, SB, and RB obtained t-MDS/AML cells and analyzed them by RT-qPCR. DG performed TCGA analyses. SCR, IS, and PV provided primary CML cells. T. Stoklosa contributed to the in vivo studies with CML cells. KP supervised PPB. CIC contributed AML/ALL PLX cells. MM supervised JL. JED, JCYW, MDM, and KE shared data from transcriptome microarrays on AML samples. MDM provided AML samples. HZ performed statistical analyses. VG provided materials and expertise in measuring HR and NHEJ in primary cells. SMS supervised EM and DDM. T. Skorski designed the studies, supervised the experiments, and wrote the manuscript.
Supplementary Material
Acknowledgments
This work was funded by NIH/NCI grants R01 CA134458 and CA186238, Leukemia and Lymphoma Society New Idea Award NIA-8999-14, and Leukemia SPORE Developmental Research Award from MD Anderson Cancer Center (all to T. Skorski); NIH/NCI R00 CA158461, the W.W. Smith Foundation, and the American Society of Hematology Junior Scholar Award (all to S.M.S); the European Union Innovative Economy Operational Program: Innovative methods of stem cell applications in medicine (grant 01.01.02-00-109/09-00) (to G.H.); the Canadian Institutes of Health Research and Cole Foundation (to K.E.); EU program: FP7-REGPOT-2012-CT2012-316254-BASTION (to T. Stoklosa); Poland National Science Centre grants UMO-2014/15/D/NZ3/05187 (to P.P.B.) and 2011/01/B/NZ3/02145 (to K.P.); the Rotary Foundation Global Grant Scholarship in the field of “Disease Prevention and Treatment” (to D.D.M.); and the Austrian Science Funds (FWF) SFB grant F4704-B20 (to P.V.). We thank Jaroslav Jelinek (Fels Institute for Cancer Research & Molecular Biology, Temple University School of Medicine) for help with analysis of TCGA database.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information:J Clin Invest. 2017;127(6):2392–2406.https://doi.org/10.1172/JCI90825.
Contributor Information
Margaret Nieborowska-Skorska, Email: mnieboro@temple.edu.
Katherine Sullivan, Email: tue83264@temple.edu.
Yashodhara Dasgupta, Email: y.dasgupta@temple.edu.
Paulina Podszywalow-Bartnicka, Email: p.podszywalow@nencki.gov.pl.
Grazyna Hoser, Email: graho@cmkp.edu.pl.
Silvia Maifrede, Email: silviam@temple.edu.
Esteban Martinez, Email: esteban.martinez@fccc.edu.
Daniela Di Marcantonio, Email: daniela.dimarcantonio@fccc.edu.
Artur Slupianek, Email: aslupian@temple.edu.
Daniel Gritsyuk, Email: dgritsyuk@gmail.com.
Sabine Cerny-Reiterer, Email: sabine.cerny-reiterer@meduniwien.ac.at.
Ilona Seferynska, Email: iseferynska@wp.pl.
Tomasz Stoklosa, Email: tomasz.stoklosa@wum.edu.pl.
Lars Bullinger, Email: lars.bullinger@uniklinik-ulm.de.
Huaqing Zhao, Email: tuf08292@temple.edu.
Vera Gorbunova, Email: vera.gorbunova@rochester.edu.
Katarzyna Piwocka, Email: k.piwocka@nencki.gov.pl.
Peter Valent, Email: peter.valent@meduniwien.ac.at.
John E. Dick, Email: jdick@uhnresearch.ca.
Jean C.Y. Wang, Email: jwang@uhnresearch.ca.
Smita Bhatia, Email: sbhatia@peds.uab.edu.
Ravi Bhatia, Email: rbhatia@uabmc.edu.
Mark D. Minden, Email: mark.minden@uhn.ca.
Tomasz Skorski, Email: tskorski@temple.edu.
References
- 1.Skorski T. Oncogenic tyrosine kinases and the DNA-damage response. Nat Rev Cancer. 2002;2(5):351–360. doi: 10.1038/nrc799. [DOI] [PubMed] [Google Scholar]
- 2.Curtin NJ. DNA repair dysregulation from cancer driver to therapeutic target. Nat Rev Cancer. 2012;12(12):801–817. doi: 10.1038/nrc3399. [DOI] [PubMed] [Google Scholar]
- 3.Karanam K, Kafri R, Loewer A, Lahav G. Quantitative live cell imaging reveals a gradual shift between DNA repair mechanisms and a maximal use of HR in mid S phase. Mol Cell. 2012;47(2):320–329. doi: 10.1016/j.molcel.2012.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Metzger MJ, Stoddard BL, Monnat RJ. PARP-mediated repair, homologous recombination, and back-up non-homologous end joining-like repair of single-strand nicks. DNA Repair (Amst) 2013;12(7):529–534. doi: 10.1016/j.dnarep.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ying S, Hamdy FC, Helleday T. Mre11-dependent degradation of stalled DNA replication forks is prevented by BRCA2 and PARP1. Cancer Res. 2012;72(11):2814–2821. doi: 10.1158/0008-5472.CAN-11-3417. [DOI] [PubMed] [Google Scholar]
- 6.Farmer H, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434(7035):917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 7.Bryant HE, et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434(7035):913–917. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- 8.Lutz C, et al. Quiescent leukaemic cells account for minimal residual disease in childhood lymphoblastic leukaemia. Leukemia. 2013;27(5):1204–1207. doi: 10.1038/leu.2012.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cancer Genome Atlas Research Network Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368(22):2059–2074. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Howard SM, Yanez DA, Stark JM. DNA damage response factors from diverse pathways, including DNA crosslink repair, mediate alternative end joining. PLoS Genet. 2015;11(1):e1004943. doi: 10.1371/journal.pgen.1004943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fujita M, et al. Interference in DNA replication can cause mitotic chromosomal breakage unassociated with double-strand breaks. PLoS ONE. 2013;8(4):e60043. doi: 10.1371/journal.pone.0060043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dedes KJ, Wilkerson PM, Wetterskog D, Weigelt B, Ashworth A, Reis-Filho JS. Synthetic lethality of PARP inhibition in cancers lacking BRCA1 and BRCA2 mutations. Cell Cycle. 2011;10(8):1192–1199. doi: 10.4161/cc.10.8.15273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang M, et al. PARP-1 and Ku compete for repair of DNA double strand breaks by distinct NHEJ pathways. Nucleic Acids Res. 2006;34(21):6170–6182. doi: 10.1093/nar/gkl840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Seluanov A, Mao Z, Gorbunova V. Analysis of DNA double-strand break (DSB) repair in mammalian cells. J Vis Exp. 2010;43(43):e2002. doi: 10.3791/2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sarry JE, et al. Human acute myelogenous leukemia stem cells are rare and heterogeneous when assayed in NOD/SCID/IL2Rγc-deficient mice. J Clin Invest. 2011;121(1):384–395. doi: 10.1172/JCI41495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Podszywalow-Bartnicka P, et al. Downregulation of BRCA1 protein in BCR-ABL1 leukemia cells depends on stress-triggered TIAR-mediated suppression of translation. Cell Cycle. 2014;13(23):3727–3741. doi: 10.4161/15384101.2014.965013. [DOI] [PubMed] [Google Scholar]
- 17.Deutsch E, et al. BCR-ABL down-regulates the DNA repair protein DNA-PKcs. Blood. 2001;97(7):2084–2090. doi: 10.1182/blood.V97.7.2084. [DOI] [PubMed] [Google Scholar]
- 18.Bolton-Gillespie E, et al. Genomic instability may originate from imatinib-refractory chronic myeloid leukemia stem cells. Blood. 2013;121(20):4175–4183. doi: 10.1182/blood-2012-11-466938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koschmieder S, et al. Inducible chronic phase of myeloid leukemia with expansion of hematopoietic stem cells in a transgenic model of BCR-ABL leukemogenesis. Blood. 2005;105(1):324–334. doi: 10.1182/blood-2003-12-4369. [DOI] [PubMed] [Google Scholar]
- 20.Alcalay M, et al. Acute myeloid leukemia fusion proteins deregulate genes involved in stem cell maintenance and DNA repair. J Clin Invest. 2003;112(11):1751–1761. doi: 10.1172/JCI17595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fan J, Li L, Small D, Rassool F. Cells expressing FLT3/ITD mutations exhibit elevated repair errors generated through alternative NHEJ pathways: implications for genomic instability and therapy. Blood. 2010;116(24):5298–5305. doi: 10.1182/blood-2010-03-272591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Esposito MT, et al. Synthetic lethal targeting of oncogenic transcription factors in acute leukemia by PARP inhibitors. Nat Med. 2015;21(12):1481–1490. doi: 10.1038/nm.3993. [DOI] [PubMed] [Google Scholar]
- 23.Jørgensen HG, Holyoake TL. Characterization of cancer stem cells in chronic myeloid leukaemia. Biochem Soc Trans. 2007;35(Pt 5):1347–1351. doi: 10.1042/BST0351347. [DOI] [PubMed] [Google Scholar]
- 24.Viale A, et al. Cell-cycle restriction limits DNA damage and maintains self-renewal of leukaemia stem cells. Nature. 2009;457(7225):51–56. doi: 10.1038/nature07618. [DOI] [PubMed] [Google Scholar]
- 25.Nabatiyan A, Szüts D, Krude T. Induction of CAF-1 expression in response to DNA strand breaks in quiescent human cells. Mol Cell Biol. 2006;26(5):1839–1849. doi: 10.1128/MCB.26.5.1839-1849.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Abbotts R, Thompson N, Madhusudan S. DNA repair in cancer: emerging targets for personalized therapy. Cancer Manag Res. 2014;6:77–92. doi: 10.2147/CMAR.S50497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Helleday T. The underlying mechanism for the PARP and BRCA synthetic lethality: clearing up the misunderstandings. Mol Oncol. 2011;5(4):387–393. doi: 10.1016/j.molonc.2011.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu W, Wang M, Wu W, Singh SK, Mussfeldt T, Iliakis G. Repair of radiation induced DNA double strand breaks by backup NHEJ is enhanced in G2. DNA Repair (Amst) 2008;7(2):329–338. doi: 10.1016/j.dnarep.2007.11.008. [DOI] [PubMed] [Google Scholar]
- 29.Veuger SJ, Curtin NJ, Richardson CJ, Smith GC, Durkacz BW. Radiosensitization and DNA repair inhibition by the combined use of novel inhibitors of DNA-dependent protein kinase and poly(ADP-ribose) polymerase-1. Cancer Res. 2003;63(18):6008–6015. [PubMed] [Google Scholar]
- 30.Löser DA, Shibata A, Shibata AK, Woodbine LJ, Jeggo PA, Chalmers AJ. Sensitization to radiation and alkylating agents by inhibitors of poly(ADP-ribose) polymerase is enhanced in cells deficient in DNA double-strand break repair. Mol Cancer Ther. 2010;9(6):1775–1787. doi: 10.1158/1535-7163.MCT-09-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mohrin M, et al. Hematopoietic stem cell quiescence promotes error-prone DNA repair and mutagenesis. Cell Stem Cell. 2010;7(2):174–185. doi: 10.1016/j.stem.2010.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bryant HE, et al. PARP is activated at stalled forks to mediate Mre11-dependent replication restart and recombination. EMBO J. 2009;28(17):2601–2615. doi: 10.1038/emboj.2009.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yarden RI, Pardo-Reoyo S, Sgagias M, Cowan KH, Brody LC. BRCA1 regulates the G2/M checkpoint by activating Chk1 kinase upon DNA damage. Nat Genet. 2002;30(3):285–289. doi: 10.1038/ng837. [DOI] [PubMed] [Google Scholar]
- 34.Choi E, et al. BRCA2 fine-tunes the spindle assembly checkpoint through reinforcement of BubR1 acetylation. Dev Cell. 2012;22(2):295–308. doi: 10.1016/j.devcel.2012.01.009. [DOI] [PubMed] [Google Scholar]
- 35.Patel AG, Sarkaria JN, Kaufmann SH. Nonhomologous end joining drives poly(ADP-ribose) polymerase (PARP) inhibitor lethality in homologous recombination-deficient cells. Proc Natl Acad Sci USA. 2011;108(8):3406–3411. doi: 10.1073/pnas.1013715108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Du W, Amarachintha S, Wilson AF, Pang Q. Hyper-active non-homologous end joining selects for synthetic lethality resistant and pathological Fanconi anemia hematopoietic stem and progenitor cells. Sci Rep. 2016;6:22167. doi: 10.1038/srep22167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chao MP, Seita J, Weissman IL. Establishment of a normal hematopoietic and leukemia stem cell hierarchy. Cold Spring Harb Symp Quant Biol. 2008;73:439–449. doi: 10.1101/sqb.2008.73.031. [DOI] [PubMed] [Google Scholar]
- 38.Slupianek A, et al. BCR/ABL stimulates WRN to promote survival and genomic instability. Cancer Res. 2011;71(3):842–851. doi: 10.1158/0008-5472.CAN-10-1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Choudhury A, et al. Targeting homologous recombination using imatinib results in enhanced tumor cell chemosensitivity and radiosensitivity. Mol Cancer Ther. 2009;8(1):203–213. doi: 10.1158/1535-7163.MCT-08-0959. [DOI] [PubMed] [Google Scholar]
- 40.Brenner JC, et al. Mechanistic rationale for inhibition of poly(ADP-ribose) polymerase in ETS gene fusion-positive prostate cancer. Cancer Cell. 2011;19(5):664–678. doi: 10.1016/j.ccr.2011.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pfäffle HN, et al. EGFR-activating mutations correlate with a Fanconi anemia-like cellular phenotype that includes PARP inhibitor sensitivity. Cancer Res. 2013;73(20):6254–6263. doi: 10.1158/0008-5472.CAN-13-0044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Garnett MJ, et al. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature. 2012;483(7391):570–575. doi: 10.1038/nature11005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hähnel PS, et al. Targeting components of the alternative NHEJ pathway sensitizes KRAS mutant leukemic cells to chemotherapy. Blood. 2014;123(15):2355–2366. doi: 10.1182/blood-2013-01-477620. [DOI] [PubMed] [Google Scholar]
- 44.Figueroa ME, et al. DNA methylation signatures identify biologically distinct subtypes in acute myeloid leukemia. Cancer Cell. 2010;17(1):13–27. doi: 10.1016/j.ccr.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moskwa P, et al. miR-182-mediated downregulation of BRCA1 impacts DNA repair and sensitivity to PARP inhibitors. Mol Cell. 2011;41(2):210–220. doi: 10.1016/j.molcel.2010.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Anantha RW, et al. Requirement of heterogeneous nuclear ribonucleoprotein C for BRCA gene expression and homologous recombination. PLoS One. 2013;8(4):e61368. doi: 10.1371/journal.pone.0061368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yan D, et al. Targeting DNA-PKcs and ATM with miR-101 sensitizes tumors to radiation. PLoS One. 2010;5(7):e11397. doi: 10.1371/journal.pone.0011397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Saeki H, et al. Suppression of the DNA repair defects of BRCA2-deficient cells with heterologous protein fusions. Proc Natl Acad Sci USA. 2006;103(23):8768–8773. doi: 10.1073/pnas.0600298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sykes SM, et al. AKT/FOXO signaling enforces reversible differentiation blockade in myeloid leukemias. Cell. 2011;146(5):697–708. doi: 10.1016/j.cell.2011.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Slupianek A, et al. BCR/ABL regulates mammalian RecA homologs, resulting in drug resistance. Mol Cell. 2001;8(4):795–806. doi: 10.1016/S1097-2765(01)00357-4. [DOI] [PubMed] [Google Scholar]
- 51.Shen Y, et al. BMN 673, a novel and highly potent PARP1/2 inhibitor for the treatment of human cancers with DNA repair deficiency. Clin Cancer Res. 2013;19(18):5003–5015. doi: 10.1158/1078-0432.CCR-13-1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weston VJ, et al. The PARP inhibitor olaparib induces significant killing of ATM-deficient lymphoid tumor cells in vitro and in vivo. Blood. 2010;116(22):4578–4587. doi: 10.1182/blood-2010-01-265769. [DOI] [PubMed] [Google Scholar]
- 53.Wunderlich M, et al. AML cells are differentially sensitive to chemotherapy treatment in a human xenograft model. Blood. 2013;121(12):e90–e97. doi: 10.1182/blood-2012-10-464677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schemionek M, et al. BCR-ABL enhances differentiation of long-term repopulating hematopoietic stem cells. Blood. 2010;115(16):3185–3195. doi: 10.1182/blood-2009-04-215376. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.