Abstract
Glioma is the most common brain tumor. For the more aggressive form, glioblastoma, standard treatment includes surgical resection, irradiation with adjuvant temozolomide and, on recurrence, experimental chemotherapy. However, the survival of patients remains poor. There is a critical need for minimally invasive biomarkers for diagnosis and as measures of response to therapeutic interventions. Glioma shed extracellular vesicles (EVs), which invade the surrounding tissue and circulate within both the cerebrospinal fluid and the systemic circulation. These tumor-derived EVs and their content serve as an attractive source of biomarkers. In this review, we discuss the current state of the art of biomarkers for glioma with emphasis on their EV derivation.
Keywords: Cancer, CNS, glioma, exosomes, microvesicles, extracellular vesicles, biomarker, IDH1/2, EGFRvIII
1. Introduction
The consensus World Health Organization (WHO) classification of tumors of glial cells provides grading from lower to higher levels of aggression [astrocytoma (WHO grade II), anaplastic astrocytoma (WHO grade III), GBM (WHO grade IV)] with similar standards for those of oligodendrocytic origin [oligodendrogliomas (WHO grade II) and anaplastic oligodendroglioma (WHO grade III)]. GBM, the most aggressive, is characterized by cells with nuclear atypia and high mitotic rates with contiguous areas of new vessel formation and necrosis. The incidence of GBM is about 3.5 per 100.000 people per year with a mean overall survival of 1.5 years [1]. Patients with a less aggressive glioma have longer survival with concomitant morbidity but cure is uncommon. Neuro-oncologists and neurosurgeons understand primary glial tumors. However, their experience is not available to the molecular biologist, the developer of companion diagnostics, the reference pathologist or the healthcare administrator. Therefore, the aim of the present review is to offer a clinical perspective of glioma for the non-clinician.
The neuropathologic definition of “glioma” based on light microscopic morphology serves as a starting point. However, our review includes changes in classification that are based upon molecular studies of glioma-specific gene mutations and amplifications. Moreover, Extracellular vesicles (EVs), including exosomes, microvesicles, and oncosomes have entered the field of diagnosis. EVs are shed by a variety of cells into biofluids or surrounding tissues as lipid membrane structures with diameter 30–1000 nm [2]. Here, we review in particular the potential of EVs as diagnostic and prognostic biomarkers for glioma.
1.1. Challenges in Glioma Treatment. Epidemiology, Molecular Pathology and Survival
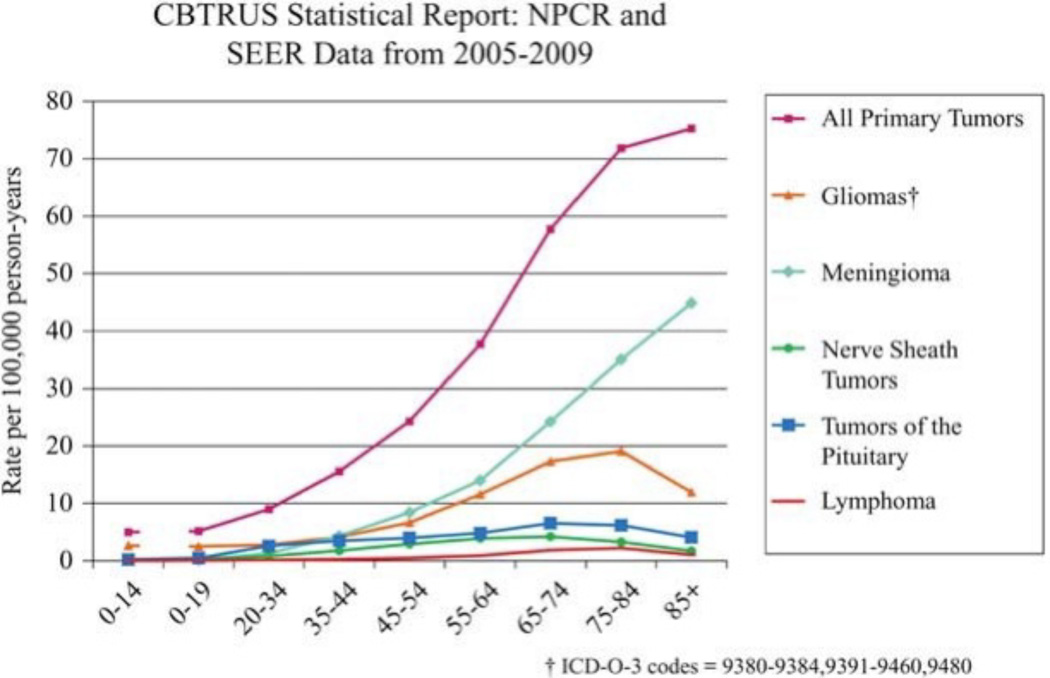
Amongst the 70,000 CNS tumors seen yearly in the United States, there appear those primary in brain and those metastatic to brain. All gliomas afflict 20.59 of 100,000 Americans [3] and represent 80% of tumors starting in the brain. Malignant glioma represent approximately one-third of all brain tumors; with glioblastoma (GBM) more common in males, Caucasians and individuals after the 5th decade (Fig.1). Childhood glioma account for one-quarter of pediatric tumors; second only to leukemia. Risk factors for gliomas include inherited neuro-cutaneous disorders (neurofibromatosis and tuberous sclerosis, basal cell nevus [Gorlin] syndromes); likely familial predisposition [4] and radiation exposure [5]. Malignant glioma in animals has been associated with JC papovapolyomavirus mutation of tumor suppressor genes [6]. Cytomegalovirus (CMV) genomic material has been found in GBM [7,8] and a recent study has proposed that CMV induces glioma by inhibition of tumor suppressor genes [9]. Thus, CMV genomic material has been used as the basis for anti-tumor immunization [10]. GBM are diffusely infiltrative of brain and grow in a microenvironment that favors angiogenesis and peritumoral growth of surrounding glial cells with suppression of the immune response [11–13]. There are 2 forms of GBM, primary and secondary GBM. Primary GBM, developing de novo, progress to death usually within less than 2 years. Secondary GBMs, evolving from tumors of lower grades in younger patients, progress over a number of years Although histologically indistinguishable from primary GBMs, these harbor different molecular alterations [14].
Figure 1.
All CNS tumors and types of brain tumors and their incidence in age categories represented as numbers of individuals versus age. Reprinted with permission from Oxford University Press.
As glial tumors are analyzed at increasing molecular resolution, their mutational heterogeneity is becoming clarified, even within the same histo-pathological subtype [15]. Molecular analysis of GBM has revealed alterations in signaling pathways for cellular proliferation, apoptosis, senescence, migration, and cell-to-cell communications. The Cancer Genome Atlas (TCGA) project has identified 3 critical pathways to be affected in most GBM including receptor tyrosine kinase/Ras/phosphatidylinositol-3 kinase, p53, and retinoblastoma signaling [16]. Several molecular subclassifications have been proposed for GBM such as (TCGA) division into proneural, neural, classical and mesenchymal subtypes based on expression of genes related to epidermal growth factor receptor (EGFR), neurofibromatosis type 1/2 (NF1), platelet-derived growth factor receptor (PDGFR), and isocitrate dehydrogenase 1/2 (IDH1) [16]. Noushmehrin et al [17] further subdivided the proneural subtype based on the expression of IDH1 mutation-associated CPG island methylator phenotype (G-CIMP) [17] occurring in younger patients showing prolonged survival. Multiple alterations in gene and protein expression patterns may be present in a single tumor and may change with time and treatment [15]. Amplification of wild type EGFR is the most common oncogenic alteration in GBM and is highly associated with the classical subtype [16]. Up to 50% of EGFR-amplified tumors also contain a unique EGFR mutation variant (EGFRvIII) resulting in ligandin-dependent constitutive activation of the EGFR pathway [18].
1.2 The Financial Consequence of Providing Care for Glioma Patients
Adult glioma is the most costly cancers in terms of treatment expense as well as the cost to society. Financial costs include those for provision of surgery, radiation therapy, chemotherapeutics, multiple hospitalizations, and supportive medications for brain edema and seizures. A cost utilization study of patients with malignant primary brain tumors in the United States from 1998–2000 (before the introduction of temozolomide) found that the total costs per patient were upwards of $50,000, the majority of which was driven by inpatient hospitalization and surgical fees [19]. To these costs must be added lost productivity, brain tumor-associated thromboembolic complications, seizure activity, infections and corticosteroid-induced muscle weakness, osteoporosis, obesity and diabetes.
2. The Current Care for Glioma
Here, we review diagnostic biomarkers of glioma including those expressed in EVs. We provide definitions of the subtypes of both low grade and high grade glioma. The nonclinician will benefit from an overview of the care of these patients, the paradigm of their care and their clinical outcome.
Patients afflicted by seizures, weakness of the extremities or changes in behavior are provided a neurologic examination to achieve a malignant glioma diagnosis within 3 months [20] rising up to 6 months for those with slowly evolving low grade tumors. As of 2007, there were almost 8000 MRI and over 10,000 CT scanning units in the United States; numbers exceeding the per capita availability of any country [21]. The U.S. MR scans costing $1200 (range, $500 to $3000) are commonly obtained by a general practitioner or by self-referral. MRI depicts and delineates intracerebral tumors, provides for surgical planning, and provides a metric of treatment response in longitudinal followup. GBM appears as a heterogeneous masses of low T1-signal intensity and high T2- signal intensity, with internal cysts and foci of blood products. Gadolinium contrast highlights an intense enhancement pattern, which often encompasses the non-enhancing necrotic central regions of the tumor. Unfortunately, MRI sensitivity for detection of masses is less than 90% and is limited further for the subtypes of glioma [22]. Low grade glioma, lymphoma, metastases, abscesses, and subacute infarcts may mimic the radiographic appearance of GBM. Even advanced MRI including spectroscopy and perfusion, provides diagnostic specificity ranging from 50 – 80% for distinction of GBM from the above mentioned conditions [23]. Spectroscopic MRI provides sensitivity for tumor diagnosis of 79% and specificity of 77%. The addition of MR perfusion imaging or diffusion studies increases the sensitivity of brain lesion detection to 81% but does not provide molecular information nor identification of tumor subtype [24]. Indeed over standard MRI, dynamic contrast enhanced MR and perfusion MR improved accuracy, sensitivity, and specificity of glioma grading from 64.9%. 78.6%, and 56.5% to 83.8%, 78.6% and 87.0%, respectively (Fig. 2) [25].
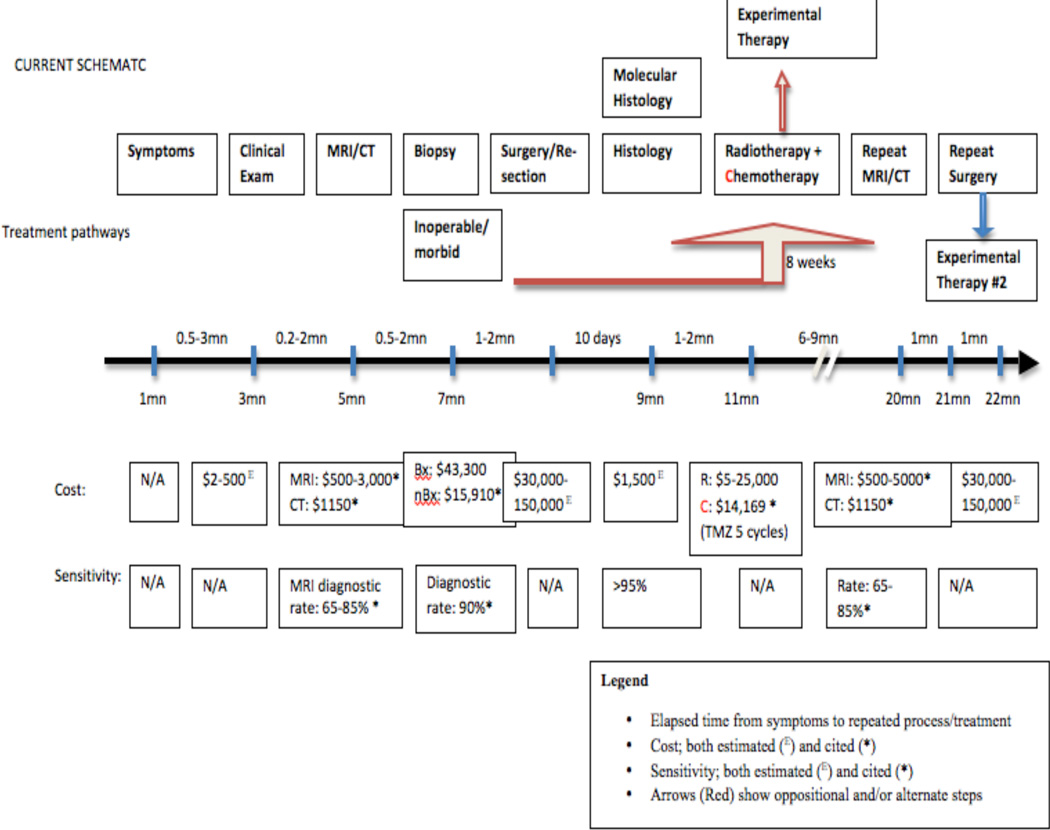
Figure 2.
The current paradigm of care for glioma patients.
Tumor diagnosis is made upon review of tissue obtained at biopsy or surgical resection. Molecular analyses include gene expression testing, DNA copy number, methylation profile, phospho-protein pathway profiling, genetic sequencing, and definition within the Cancer Genome Atlas [26]. Maximal safe resection, the treatment of choice for patients with GBM, enhances the effects of adjuvant therapies, reduces symptoms emerging from brain edema and epileptic seizures, and provides specimens for histologic and genomic studies [27]. In a preliminary review of the SEER database (BC) it was found that the surgical desire to obtain “gross-total resection” of tumor within MRI-delineated regions is achieved in fewer than 30% of patients. This goal is not reached for various reasons including difficulty distinguishing tumor cells from normal brain tissue and peri-tumoral reactive elements; the difference in goals and experience between surgeons in practice and in tertiary facilities and the availability of intraoperative MRI scanning. The Glioma Outcome Project reported a peri-operative complication rate of 24% in patients undergoing first craniotomy for glioma resection, with 8% displaying worsened neurologic status [28]. The incidence of peri-operative complications increased with subsequent operations (33% complications, 18% worsened neurologic status after second craniotomy) [28]. For patients with GBM, surgical therapeutic options include resection, implantation of a nitrosourea polymer wafer, the use of fluorescent guidance systems [29–31], irradiation of the tumor during operation via implanted “brachytherapy” isotopes [32,33] or post-operative radiation [34,35]. Approximately 30% of patients with GBM have tumors that permit only diagnostic biopsy. In an unpublished review (Noorbakhsh, in preparation) 22% of patients were found never provided with an operative diagnosis. The remainder receive biopsy or subtotal resection- a reflection of restrictions imposed by age, comorbidities, multi-focal masses, or tumor location. Pathologic diagnoses based upon biopsy carry with them issues of sampling errors due to tumor regional heterogeneity of architecture, vascularity, cellularity, and necrosis. Thus biopsies have limitations for tumor grading and diagnosis of GBM. Of 81 consecutive patient recipients of stereotactic biopsy [36], subsequent resection resulted in a changed diagnosis in 49%, of whom 26% experienced a change in clinical management. Similarly tumor heterogeneity imposes topographic limitations on mutational analyses [15].
Six weeks after surgery, patients are provided adjuvant therapy using temozolomide and fractionated 60 Gy radiation over 42 sessions, followed by 6 additional monthly cycles of temozolomide [37]. This therapy increased 2-years and 5-years survival rates to 27% and 11% from 11% and 2% respectively, with no significant adverse effects on quality of life [34]. The incremental cost of temozolomide is estimated to be $50,000 per life-year gained [38]. Resistance to temozolomide is described as a function of repair of damaged DNA by the enzyme O6–methylguanine-DNA methyltransferase (MGMT), by poly(ADP-ribose)polymerase (PARP) in the base excision repair (BER) pathway, or through tolerance of damaged DNA in mismatch repair-deficient cells [39]. However, other molecular alterations may cause resistance such as the MSH6 mutation [40,41]. Currently, methods for detection of temozolomide resistance other than de facto tumor progression do not exist. Thus, many patients undergo long and expensive therapies, which do not provide any benefit for their particular tumor. Biomarkers may provide a measure of response and progression.
At the present time, tumor response to therapy, whether reduction, recurrence or progression, is evaluated by longitudinal serial MRI, an approach with inherent limitations. Recurrence or tumor progression is measured by volumetric or cross-sectional changes in the tumor's T1 enhancement [42]. Treatment of recurrent GBM involves re-resection, [43], hypofractionated re-irradiation and anti-angiogenesis therapy with the use of bevacizumab and/or irinotecan [38, 44]. These therapies increase 6-month survival rates with attendant complications [38] and without curative intent. Salvage chemotherapy with experimental agents do not further improve survival and are provided late in the life of patients now afflicted with morbidities resulting from prior therapies. However, the MRI maximum spatial resolution of millimeters does not provide a measure of tumor cells, only tenths of micrometers in size, which increase in number before radiologic changes are apparent. Concomitant anti-angiogenic therapies make problematic the use of MRI for tumor monitoring [45]. The reduction of vascular permeability resulting from anti-angiogenic drugs reduces passage out of vessels of MRI contrast agents. As a result there is reduction of tumor contrast enhancement, even in the setting of paradoxical tumor growth [46]. Further limiting MRI specificity is the inability to distinguish radiation necrosis (‘pseudo-progression’) from progressive viable glioma [47]. These considerations stimulate the search for biomarkers of diagnosis and as metrics of therapeutic response.
3. EVs as Source of Biomarkers
EVs include exosomes, exosome-like vesicles, microvesicles, and oncosomes which are released by all cell types. The content of EVs and their functions vary with the cells of origin. For example, EVs released from tumor cells contain a wide variety of proteins and lipids, RNA and DNA which support tumor growth by altering multiple hallmarks of cancer. Thus EVs may explain features of oncogenesis including genetic instability, tumor growth, alterations of the microenvironment, cellular invasion, migration and metastasis and immune resistance. Please note that (A) the terminology of EVs is in flux and has yet to come to grips with the diagnostic implications of EV structural features and size differences, the effects of preparative techniques and the differing functional roles of EVs; (B) the number of novel biomarkers will, no doubt, increase in the coming years and (C) future correlative studies will validate the diagnostic value of EV biomarkers for subtypes of glial tumors [48]. EVs emerge from the endosomal compartment and are secreted into the extracellular space but can also detach from the plasma membrane of the cell. The preparative approaches for EV isolation include filtration, ultracentrifugation and column separation followed by electron microscopy and NanoSight analysis [2]. EVs have been isolated from multiple body fluids such as plasma and CSF. They participate in intercellular communication and modulate the microenvironment to alter the immune response. For example, EVs modulate expression of MHC class II molecules on the surface of dendritic cells as well as α-amino-3- hydroxy-5-methyl-4-isoxazole-propionic acid (AMPA) receptor on the surface of cortical neurons. EV-derived mRNA transfer to recipient cells is translated into protein with changes in the recipient cell genotype [2]. EVs from microglia are received by neurons with changes in synaptic function as are EVs from oligodendrocytes. Similar interactions exist between platelet-derived EVs and coagulation and by EVs and recipient cells to facilitate platelet adhesion. EVs contribute to cell invasion and migration by modulation of metalloproteases which alter the extracellular matrix. GBM-derived EVs facilitate angiogenesis. For example, EGFRvIII constitutively signals the initiation of a proangiogenic cascade in GBM. The EV cargo of mRNA, miRNA is transferred into GBM cells [49]. EV DNA and protein appears stable over years, and EV RNA is not likely to be degraded by RNAases and can be isolated at high levels from biofluids. The cargo has been extensively evaluated but the relationship between subsets of EVs and cargo-specificity remains obscure. The literature lacks speciation of EVs that carry specific types of RNA, ncRNA of retroviral origin and miRNA. Many of the latter (miRNA 1, 21, 181D) have been linked to GBM; but their specificity is lacking. For example, sera of GBM patients contain higher levels of miR-21 than sera from ‘normal’ patients. These single miRNAs may be able to target and regulate over 100 genes. Tumor EVs also contain retrotransposon elements, which can be transferred from tumor cells to normal cells [50]. Thus, genetic information encapsulated in specific fractions of tumor-derived EVs found in biofluids may serve as tumor markers.
4. Potential Glioma Biomarker Candidates
4.1.1. Epidermal growth factor receptor variant III mutation (EGFRvIII)
EGFRvIII mutation is highly specific for GBM. The exon 2-7 deletion from the EGFR gene is found in 20–25% of GBM cases; but reports of frequency vary. Its presence within a tumor is diagnostic and possibly prognostic, but only in patients that live beyond 2 years and in anaplastic astrocytomas [51,52]. Functionally, the presence of EGFRvIII presence is associated with amplification of EGFR wt, which subsequently upregulates the PI3K pathway and promotes abnormal cell proliferation and tumor progression [53].
Skog et al. detected EGFRvIII from sera EVs from 30 GBM patients [49]. Fourteen of these patients had the EGFRvIII mutation present in matched tissue samples. Using nested RT-PCR, the EGFRvIII mutation was found in 7 of the 30 sera samples. Interestingly, 2 of 7 serum samples in which EGFRvIII was detected had matched tissue samples that had been negative for this mutation, supporting the idea that serum-based biomarkers may provide more sensitive tests of tumor characteristics than tissue specimens. The underlying heterogeneity of GBM cellular distribution may cause EGFRvIII to be undetectable in a small piece of tissue, but a serum sample presumably receives EVs from the entire tumor. After resection, GBM patient serum samples no longer contained EGFRvIII [49]. Analytic techniques have included immunohistochemistry [6], proteomic techniques and flow cytometry [54]. Analyses have been carried out on human tissue specimens [53,55] and non-human CSF [56] in addition to serum/plasma. Ongoing is a multi-center trial to validate the diagnostic utility of EV EGFrvIII mRNA in CSF and plasma. EGFR is the target of first generation inhibitors such as erlotinib and gefitinib, that have not resulted in survival benefits [57–59]. Forthcoming is a next generation of EGFR inhibitors, including afatinib, dacomitinib, and nimotuzumab [60].
4.1.2 Isocitrate dehydrogenase 1/2 mutations (IDH1/2)
IDH1/2 are NADP+-dependent dehydrogenases catalyzing the oxidative decarboxylation of isocitrate to α-ketoglutarate. Mutation in these genes alters the enzymatic activity of both IDH1 and IDH2. The mutations are found in 80% of low grade glioma and approximately 10% of GBM, but never in normal brain or bodily tissues [14]. IDH1/2 mutations are associated with young age, secondary GBM, and a longer overall survival [61]. Furthermore, the point mutation is found in cholangiocarcinoma, certain sarcomas, acute myelogenous leukemia and inborn accumulation of 2-hydroxyglutarate (see below). The IDH1R132H mutation represent 93% of glioma-associated IDH1 hotspot mutations [61]. The mutation in tissue correlates with survival approaching ten years in patients with low grade tumors and possibly improved response to temozolomide therapy in patients with secondary GBM. This response may reflect a correlation with methylation status in patients bearing this mutation. Analyses to detect the mutated versions of IDH1 have been performed on tissue using a monoclonal antibody for immunohistochemical detection followed by DNA sequencing and in EV-derived RNA from tissue and CSF using the highly-sensitive BEAMing qRT-PCR or digital PCR techniques and from DNA obtained from blood [62]. Peripheral blood samples have also been analyzed, and a murine model has been studied for clarification of disease mechanism [63,64]. Identification of the IDH1/2 mutations may serve not only as a “partial” diagnostic biomarker, but also a prognostic marker for improved survival in GBM. Large scale studies of IDH1/2 mutations are underway. The molecular mechanisms governing IDH1/2 mutations are not well understood. It has been stated that for both IDH1 and IDH2 mutations, mutant-mediated 2-hydroxyglutarate production inhibits enzymes involved in epigenetic regulation, collagen synthesis, or cell signaling [65]. Although 2-hydroxyglutarate accumulates in tumor tissue and serum, this accumulation alone does not support diagnostic biomarker utility of 2-hydroxyglutarate in addition to IDH1/2 mutations. The absence of documented cases in which IDH1/2 mutations co-exist may suggest that each mutation provides a sufficient independent growth advantage [66]. The discovery of mutated IDH1 within a body fluid sample would likely be tumor specific and diagnostic of glioma. Potentially, targeted agents under development might then be provided. Uncertain is whether these drugs, suppressive of the mutation, will amplify the expression of IDH1 wild type.
4.1.3. Drosophila capicua homolog (CIC)
CIC mutations are associated with 1p/19q deletion and with IDH1/2 mutations [67, 68]. The reported overall incidence of CIC mutations in oligodendrogliomas is reported at 46–69% [67,69]. The occurrence of the mutation is rare in astrocytomas (approximately 10%) [68,69]. The impact of CIC mutations on molecular pathways is not well understood. It is known to play a role in embryonal development, downstream of the RAS/MAPK pathway [65, 70]. In addition, sequence analysis in tissue has been performed [67–69].
4.1.4. Far upstream element binding protein 1 (FUBP1)
FUBP1 mutations are associated with 1p/19q deletion and with IDH1/2 mutations. FUBP1 mutations have been reported in 10–24% of oligodendroglioma [68,69] and in 10% of astrocytomas [69]. The mutated FUBP1 product exhibits loss of function and does not bind to its known target site in the Myc oncogene [65,71], resulting in loss of regulation and aberrant cell growth. Sequence analysis in tissue has been performed [68,69,71].
4.1.5. Alpha-thalassemia/mental retardation X-linked gene (ATRX)
ATRX mutations occur in 44% of pediatric GBM [72]; mutations have been reported on 33% of pediatric grade II glioma, 46% of pediatric grade III glioma, 57–80% of pediatric “secondary” GBM, 7% of pediatric “primary” GBM [69, 73], 71% of pediatric grade IIIII astrocytomas, and 68% of pediatric oligoastrocytomas [69]. Reportedly no ATRX mutations are found in pediatric oligodendroglioma [73]. The nonfunctional binding protein encoded by mutant ATRX permits inappropriate recombination, resulting in aberrant telomere lengthening [74]. ATRX mutations have been associated with the IDH1 mutation [73]. Sequencing studies have been carried out in tissue [69,72,73].
4.1.6. BRAF: BRAFV600E
BRAFV600E, is a point mutation that is frequently found in ganglioglioma and in about 65% of grade II xanthro-astrocytoma [75]. It is assumed that this alteration constitutively activates the RAS/RAK/MEK/ERK kinase pathway [75]. Incidence is reported to be 18% in brainstem ganglioglioma, 66% in pleomorphic xanthoastrocytoma with anaplasia, and 65% without anaplasia, 9% in pilocytic astrocytoma (PA); 3% in anaplastic astrocytoma [75], and 22.5% in pediatric grade II-IV tumors, but mutations are not found in pediatric grade I tumors [76]. The BRAFV600E mutation in tissues has been detected using in situ hybridization. The BRAFV600E mutation in melanoma has been targeted with a small-molecule BRAF kinase inhibitor vemurafenib (PLX4032), which therapy improves progression-free and overall survival [77,78]. When similar treatment effects are validated within low grade glioma, the drug could transform the BRAFV600E mutation from diagnostic marker to a marker which is predictive of response to therapy.
4.1.7. Telomerase reverse transcriptase-encoding gene (TERT)
TERT promoter mutations have been reported in 83% of 78 primary GBM, 10% of 40 astrocytomas, 78% of 45 oligodendroglioma, and 25% of 24 oligo-astrocytomas [74]. Overexpression results in 1.2–1.5 times increased risk of glioma occurrence [79]. TERT promoter mutations upregulate telomerase expression, enabling tumor cells to maintain sufficient telomere length in their genomes, thus removing an obstacle to prolonged cell proliferation. Analyses to detect TERT promoter mutations have been performed on tissue using RT-PCR and sequencing techniques [74].
4.1.8. Histone H3F3A gene
Unique to pediatric high grade glioma are mutations in the genes H3F3A and HIST1H3B which encode histone H3.3 [72]. Alterations in the H3F3A or HIST1H3B genes are present in approximately 80% of diffuse intrinsic pontine glioma and 20% of non-brain stem GBM in children [80]. There is a potential association of H3F3A mutations with ATRX mutations [72]. The effect of H3F3A mutations on gene expression is not well understood. H3F3A mutations have been proposed to affect epigenetic gene expression regulation, selective gene regulation, or telomere length/stability [80]. The various mutations of this gene all result in the same amino acid substitutions, suggesting a gain-of-function phenotype that could potentially be targeted by drug developers [80]. Like EGFRvIII deletion and IDH1 mutations, the specificity of the H3F3A mutation make it a promising sensitive and specific diagnostic biomarker, though it has yet to be investigated in biofluids.
4.2.Amplifications or mutations not unique to glioma
We have reviewed glioma-specific biomarkers (Table 1). In addition, there have been reports on amplification of receptors and overexpressed normal brain proteins. These are less compelling as diagnostic biomarkers as validation would involve identification of ‘cut-points’ or threshold values in accessible biofluids, which would separate glioma from both normal and carcinoma-afflicted individuals as well as those suffering nontumor neurologic syndromes.
Table 1.
Glioma-specific biomarkers and their relevance in the field of EVs.
| Biomarker | Reference Number | Glioma Subtype Correlate | Molecular Correlation | Analytic Technique | Tissue/Biofluid |
|---|---|---|---|---|---|
| EGFRvIII | [29, 40, 53, 63, 64, 98] | 24–67% of GBM; "primary" GBM; pediatric brainstem glioma | EGFRwt amplified; upregulate PI3K pathway | Immunohistochemical analysis, rtPCR, Western blot, flow cytometry | Tissue, CSF, plasma |
| IDH 1.132 | [5, 29, 69, 70, 74] | 50%–82% "secondary" GBM; 65–94% oligodendrogliomas/oligoastrocytomas | 2-hydroxyglutarate tissue; MGMT expression | Genomic analysis, gel electrophoresis, rtPCR, knock-in mouse tissue | FFPE tissue, peripheral blood samples |
| IDH 2 | [65, 70, 74] | 4.7% grade II oligodendrogliomas, 5.2% grade III anaplastic oligodendrogliomas, 6.2% grade III anaplastic oligoastrocytomas | 2 Hydroxyglutarate accumulation. No association with IDH1.132 | Knock-in mouse tissue, tissue sequencing | FFPE tissue |
| CIC (homolog of Drosophila Capicua) | [71, 72, 73, 74] | 46–69% oligodendrogliomas; app. 10% astrocytoma | FISH 1p/19q deletion; IDH 1/2 mutations. Downstream of RAS/MAPK pathway | Tissue sequencing | FFPE tissue |
| FUBP1 (Far Upstream Element [FUSE] Binding Protein 1) | [72, 73, 76] | 10–24% of oligos, 10% in astrocytomas | FISH tissue 1p/19q deletion; IDH 1/2 mutations; FUBP1 mutated gene does not bind to MYC oncogene | Tissue-based sequencing | FFPE tissue |
| ATRX (alpha thalassemia/mental retardation syndrome X-linked) | [69, 72, 73, 76] | 44% of pediatric GBM; 33%–71% grade II glioma, 68% oligoastroxytomas; 46% grade III glioma, 57–80% "secondary" GBM, 7% "primary" GBM; 0% in oligodendroglioma | Aberrant telomere lengthening. Associated with IDH1 mutation | Tissue sequencing | FFPE tissue |
| BRAF V600E | [75, 76] | 18% brainstem gangliogliomas; 66% pleomorphic xanthoastrocytoma; 9% pilocytic astrocytoma; 3% anaplastic astrocytoma; 22.5% pediatric grade II-IV tumors, 0% in grade I tumors | Activates RAS/RAF/MEK/ERK kinase pathway | DNA sequencing | Cell line |
| BRAF fusion | [75,80, 81, 84, 85] | Reported 100% in pediatric grade I tumors, 0% in grade II-IV tumors in 10 grade I, 31 grade II-IV gliomas respectively; reported up to 80% incidence in pilocytic astrocytomas; brainstem gangliogliomas; pleomorphic xanthoastrocytoma; pilocytic astrocytoma | KIAA1549-BRAF fusion-mediated upregulation of MAPK pathway | Clinical case report and sequencing | Tissue CSF |
| TERT | [48, 74] | 83% "primary" GBM, 10% astrocytomas, 78% oligodendrogliomas, 25% oligo-astrocytomas; increased glioma risk | Upregulation of telomerase expression | Tissue-based rtPCR and sequencing | Tissue |
| H3F3A/HIST1H3B (Histone H3.3) | [72, 73, 80] | 80% pediatric diffuse intrinsic pontine gliomas, 20% pediatric non-brainstem GBM; pediatric high grade glioma, 3.4% adult GBM | ATRX, selective gene regulation/ telomere length/stability | Whole-genome/targeted sequencing | FFPE tissue |
4.2.1. Epidermal growth factor receptor (EGFR)
Amplification of EGFR has been reported in tissues from 40–50% of all glioma [55,63], 45–60% of primary GBM [54, 81], 10% of secondary GBM [54], and in lung cancer [82]. It plays a fundamental role in normal tissue development. Overexpressed EGFR constitutively activates the PI3-K/AKT/mTOR pathway, resulting in cancer cell proliferation and invasive tumor progression [54, 55, 63, 82]. EGFR amplification in FFPE tissue is detected by genomics and proteomics [63, 82] and in peripheral blood [63].
4.2.2. BRAF: KIAA 1549-BRAF fusion gene
The KIAA 1549-BRAF fusion gene is present in up to 80% of PA [76,83–85]. PA, the most common brain tumor in childhood, is found in cerebellar and non-cerebellar locations [86]. The BRAF fusion gene has been shown to exert its pro-oncogenic activity by activation of the mitogen-activated protein kinase (MAPK) pathway. The prognosis is excellent for surgically resected lesions but this is accomplished in fewer than 50% of patients. Resected PAs contain the Sox-2 stem cell marker, and rarely synaptophysin. However these materials seldom immunostain for BRAF. It is likely that in PAs the RAF/MEK/ERK pathway is activated.
4.2.3. TP53 mutations
TP53 mutations are common in glioma, These mutations have been used as an astrocytic marker to differentiate types of glioma. Mutation of ATRX is found frequently in low grade astrocytomas and secondary GBM, but not in primary GBM. ATRX with TP53 and IDH mutations correlates with improved survival [65]. CIC and FUBP1 are frequently mutated in oligodendroglioma tumors, but needed are insights into their roles in tumor pathogenesis [65].
4.2.4. O-6-Methylguanine-DNA-methyltransferase (MGMT)promoter methylation status
Of marginal utility for diagnosis; but excellent utility for prognosis, are studies of the DNA repair enzyme O-6-methylguanine-DNA-methyltransferase (MGMT). When methylated, the MGMT promoter is silenced leading to improved response to alkylating agents [34, 87]. In a cohort of 301 patients, MGMT promoter methylation in 44% of the patients correlated with improved progression-free and overall survival [63]. The tumor response can be observed as MRI-delineated “pseudo-progression” masses, which are in fact focal, gadolinium-enhanced necrotic lesions. Thus, the MGMT status may serve as a biomarker of pseudo-progression otherwise only identifiable at the time of repeat surgery. MGMT promoter methylation occurs in GBM [55,87,88] including 40% of primary GBM, over 70% of secondary GBM, 50–80% of anaplastic glioma [62] and 50–93% of low-grade glioma [54,55]. Immunohistochemistry of the MGMT protein did not correlate with PCR analysis of methylation. Thus, the ‘gold standard for tissue analyses has yet to be defined and may include methylation-specific PCR pyrosequencing, and/or MPLA. GBM and grade 2–4 glioma tissue along with colon cancer tissue exhibit GCIMP, which correlates with presence of mutation IDH1R132H. This biomarker may be useful as a source of patient stratification for clinical trials. MGMT status can be identified in tissue and serum from GBM patients [89,90].
4.2.5. CHI3L1 (YKL-40)
CHI3L1, also known as YKL-40, has been shown to be highly overexpressed in GBM relative to normal brain and other CNS tumors. The overexpression favors the GBM mesenchymal subtype, and older age and is associated with poor prognosis [91]. The gene is not specific as expressed in conditions of extracellular matrix degradation and angiogenesis including severe arthritis, hepatic fibrosis, and other cancers. Elevated YKL-40 levels have been detected in the serum of glioma patients and have been shown to correlate with tumor grade and possibly tumor burden [92].
4.2.6. Phosphatase and tensin homolog gene (PTEN)
PTEN mutations occur in 28–60% of GBM, 7% of anaplastic astrocytomas, and no lower grade glioma [55,81]. Loss of PTEN function likely worsens survival for anaplastic glioma patients. Mutated PTEN gene products result in the loss of inhibition of the PI3K/AKT/mTOR pathway, leading to cell proliferation [55,81]. Analyses have been performed on tissue [55] and at least one GBM cell line [81] using genomics and proteomics [82].
4.2.7. c-Myc
Biofluids contain the c-Myc gene, characteristic of a subtype of childhood medulloblastoma. c-Myc amplification is characteristic of the group C medulloblastomas (Northcott et al.) which have significantly poorer progression-free and overall survival than the other three groups of childhood medulloblastomas [93]. Balaj et al. successfully measured c-Myc amplification in serum-derived EVs extracted from mice harboring human medulloblastoma xenografts [50].
5. Conclusion
A sensitive and glioma-specific biomarker diagnostic assay would benefit four underserved populations: 1) the 20% of Americans who currently never receive pathologic confirmation of their tumor; 2) the aged, infirm patient whose comorbidities preclude surgical evaluation; 3) patients whose masses are in ‘sensitive’ locations including the brainstem, the posterior fossa, speech and motor areas of cortex and subcortex, as well as those with non-discrete multifocal or diffusely infiltrative lesions; and 4) children for whom surgical morbidities may prove unacceptable. Although a plasma-based assay is preferable, there is consensus amongst neurosurgeons of the ABC2 Foundation Biomarker Consortium that a biomarker from CSF would be acceptable, less costly and safer than many delicate neurosurgical resections. Minimally invasive diagnostics would change the nature of stratification for clinical anti-cancer trials. The Chief Clinician at Cancer Research UK, and members of the Early Detection Research Network of the US National Cancer Institute recognize that molecular specification of tumors will create a novel clinical trial design by enabling personalized therapy based on a predominant driver mutation or amplification. This would be true for the glioma-specific mutations EGFRvIII and IDH1/2. The diagnostic biomarkers would then become the subject of quantitative, longitudinal studies as metrics of therapeutic success. In addition, multiplex assays likely will provide information about downstream pathways as well as resistance mechanisms in glial tumors. MGMT promoter methylation status has been observed to be a predictor of glioma resistance to chemotherapy [94]. The expression of IDH1/2 mutant proteins has also been shown to sensitize GBM cells to ionizing radiation-induced apoptosis improving overall survival of these patients [93, 94].
For many of the biomarkers identified (Table 1) novel therapeutics are in development. For targeting EGFRvIII a monoclonal antibody therapy has been offered as has induction of humoral reactivity against EGFRvIII [95]. Currently, EGFRvIII-targeted vaccination is undergoing evaluation in phase 3 clinical trials [96]. The immunotherapeutic agents which target EGFRvIII include rindopepimut which induces a humoral immune response [97]. Combination therapies also target wild-type EGFR and other growth factor receptors such as insulin-like growth factor receptor or PDGFRI [98]. Murine monoclonal antibodies have been synthesized against IDH1 mutations; in addition, drugs mimicking α-ketoglutarate have been proposed as a potential therapeutic option pertaining to IDH gene mutations [99]. Mutant IDH1 inhibition has been found to release restriction on glioma cell differentiation and deter tumoral growth, thus allowing cancer cells to differentiate within less invasive pathways [100]. Mutant IDH2 inhibition has been observed to have the same effects in leukemia cell lines [101]. The small-molecule agents used in these inhibitions are termed AG-221 and AG-110, respectively, from Agios Pharmaceuticals. An adenovirus vector-based combination PTEN/antisense hTERT therapy has shown benefit in a xenograft murine model [102]. Trastuzumab has been shown to upregulate PTEN activity and thus inhibit the PI3K pathway in metastatic breast cancer with intact PTEN [103]. Therefore, detection of PTEN within EVs could be of significant use to study the levels of PTEN elevated in this type of tumor. Several studies have provided proof-of-principle for BRAFV600E mutant inhibition in the context of malignant melanoma and have identified potent inhibitors, some of which are specific for BRAFV600E (e.g. PLX4720, sorafenib) and targeted inhibition of BRAFV600E in glioma has been proposed [104–109]. MGMT inhibitors are being investigated [110] and methylation-related resistance may correlate with methylation of HOXA7, 9, and 10 genes [111]. A telomerase inhibitor Imetelstat, is currently in phase 2 trials [112], but issues exist involving resistance via molecular pathways that lengthen telomeres [113]. In a broader sense, diagnostic biomarkers could improve the enrollment of glioma patients in phase 2 clinical trials. A 2006 evaluation of NIH-derived patient accrual reported a total of 6 patients accumulated for a phase 1 clinical study investigating glioma (Protocol #: 00-C-0173), and maximum monthly referrals of 2 individuals [114]. However, an estimated 80% of clinical trials fail to recruit subjects in their desired timeframe [115]. Phase 3 trials also carry concerns, as most accrue unselected patients instead of targeted populations [116]. Absent a targeted patient cohort, drug recipients may receive ineffective treatment which logically will bias trial outcomes.
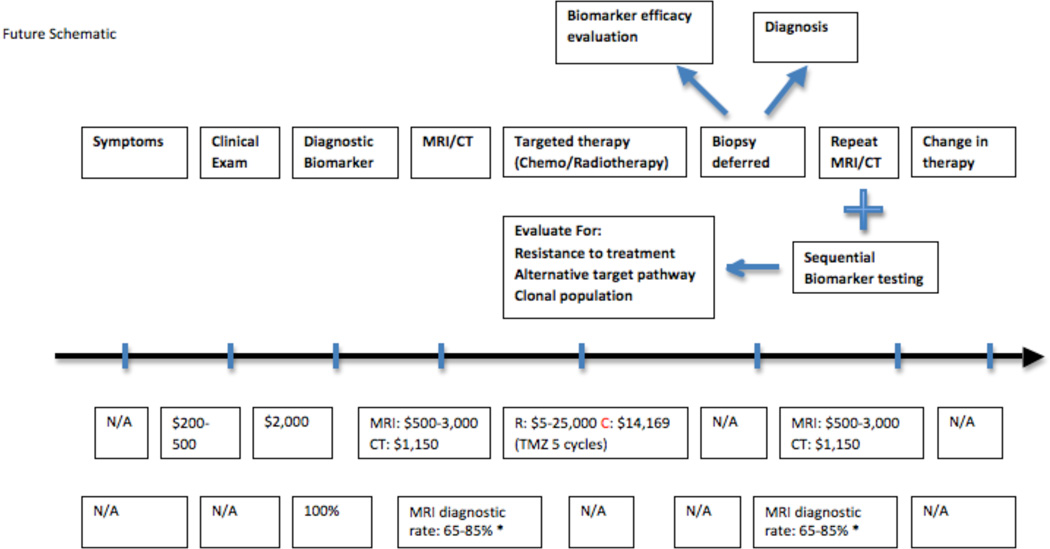
We propose a change in the paradigm of care. On the basis of symptoms, an MRI image will be obtained. A “suspicious” lesion is further evaluated using EV derived plasma or CSF biomarker analytics. From this will emerge the EV-associated molecular pathology of the tumor (Fig. 3). For inoperable patients there will be provided immediate nonsurgical therapy including radiation and/or mutation-specific chemotherapy. Operable candidates will receive pre-operative tumor-specific drug therapy prior to surgery or radiation treatment. For other patients, including children, identification of tumor-specific mutations in blood or CSF precludes biopsy of eloquent brain, or brainstem. This approach reduces cost and surgical morbidity. For example, the identification of an IDH mutant tumor should result in initiation of an anti-IDH1 chemotherapy without biopsy.
Figure 3.
The future perspective on the paradigm of care for glioma patients.
Identification of a molecular tumor signature, whose downstream pathways are known, opens the possibility of minimally invasive identification of drug-resistance pathways. These pathways may be modulated in real time, without the need for re-biopsy of tumor. The paradigm of diagnosis, resection, chemoradiation, imaging, re-biopsy would be replaced by clinical symptoms, diagnostic biomarker assay, targeted therapy, resection, chemoradiation, and response assessment by imaging, and biofluid biomarker assay.
This future paradigm relies on technologies within the grasp of the biomedical community. Readers of this review will likely see a number of these new strategies in place over the next 5–10 years.
6. Expert commentary
5.1. Short discussion of choices of an analysis with EGFRvIII and IDH1/2 mutations as examples
As a detection method, real time PCR assays provide sensitive detection of large gene deletions, mutations or rearrangements. For example, EGFRvIII with deletion of exon 2–7 can easily be detected using real time PCR assays. In the case of rare events and/or point mutations such as in the IDH1/2 genes, there is need for more sensitive assays (digital PCR or targeted sequencing) to increase sensitivity and produce more reliable results. This type of limitation in conventional techniques has stimulated studies to develop a system that can detect single nucleotide alterations in a gene and monitor tumor progression. Novel analytics are based upon evaluations performed of circulating nucleic acids using digital PCR and sequencing [117–119].
5.2. The spike-in cohort analysis and validation processes
It is important to define the detection limits for each assay. Digital PCR and sequencing can detect as low as 1 mutant copy in a background of 100,000 or even millions of wildtype copies. Before any assay can be adopted for clinical application to human samples, it is important to optimize the platform of the specific gene assay. This includes choice of optimal primers for the target sequence, amplification temperatures and times using spike-ins of ultramers or a plasmid containing the target sequence or synthetic or nonhuman mRNAs. Concerns exist regarding sources of contamination as well as endogenous and exogenous heparin-like molecules [120]. Currently, this paradigm moves from spike-ins in PBS to human normal pooled biofluids and subsequently glioma patient biofluids. These concerns are fundamental for determination of the sensitivity of assays of wild-type and mutant copies of the ‘diagnostic’ biomarker gene.
6.3. Diagnostics and stratification for trials
We have previously shown that blood-derived EVs contain nucleic acid from tumor cells [121]. EV-RNA analysis clearly distinguishes a tumor patient from controls. Upon further analysis of EVs, we can determine glioma-specific mRNAs, miRNAs, ncRNAs in comparison with those of normal controls. Identification of the EV gene expression signature at diagnosis would provide stratification criteria for patients in clinical trials. Thus, blood-derived EVs have been shown to contain a specific gene signature that can clearly distinguish GBM patients from controls [121]. Furthermore, EGFRvIII has been detected in circulating plasma and CSF EVs [49], and mutant IDH1G395A can be detected in CSF derived EVs [62]. These studies together with other biomarker studies [122] offer great hope for fast, specific, and “real-time” minimally-invasive molecular stratification and response evaluation for brain tumor patients.
Blood-based assays are more desirable compared to CSF-based assays as it is less invasive for patients. Ongoing studies have to address which biofluid offers the best detection rate for each molecular target. Detection of RNA or DNA within or removed from EVs may offer different answers. For example, we have been unable to detect mutant IDH1 mRNA in EVs from serum of patients whose tumor was positive for the IDH1 mutation [62], whereas this mutation has been readily detected in non-EV DNA from 60% of patients [123].
7. Five-year view
Treatment of patients with glioma evolved slowly in the last 3 decades. A “minimally invasive” diagnostic glioma biomarker will reshape the landscape by providing a rapid confirmation of the molecular subtype of benign and aggressive gliomas. Progression will be made in particular with isolation and characterization of brain tumor-specific EVs. Their mRNA, miRNA and ncRNA cargo will be sequenced to confirm the existence of diagnostic point mutations and amplifications and to identify novel mutations. These analyses, performed without surgical intervention, will create a nosology replacing that which is over 100 years old. New insights will emerge in neurooncology with respect to gliomagenesis, the roles of endogenous brain pleuripotential stem cells and genes that drive pathways of malignant change. These insights may inform whether these tumors stem from environmental, viral and/or genetic risks. The next 5 years can become productive in research neuro-oncological research in the following directions:
Classification of glioma will have a fundamental overhaul as a consequence of the mapping of EV-associated gene amplifications and mutations.
EV-related IDH1/2 mutations and associated changes in IDH wild type genes and their substrate (2-hydroxyglutarate) will become a scientific ‘hot bed’ as biofluid diagnostics become available, therapeutic decisions are made on the basis of these data and these become stratifiers for targeted drug trial entry.
International collaborations will provide rapid verification of novel biomarkers such as TERT.
EV-related studies of methylation status will lead to changes in dose/time schedules of radiation and temozolamide treatment.
The role of EV cargoes will be investigated in relation to tumor formation and attention will be focused on the role of peri-ependymal stem cells in both tumor formation and preservation of the ‘healthy brain’.
At least two clinically-useful FDA-approved EV-biomarkers of glioma will be available to clinicians, followed shortly thereafter by markers of metastases in brain. Neurosurgical study groups will coalesce to validate these markers.
With identification of the above, national priorities will be reset to consider costs of diagnostic approaches of brain tumors, prioritization in clinical trials and the provision of translational research funds.
Key Issues.
We provide a description of glioma-derived EVs, their detection as well as glioma-specific amplifications and mutations that can serve as diagnostic biomarkers.
The current sub-typing of glioma neither reflects our understanding of the mechanisms nor molecular pathways of gliomagenesis.
The treatment of these glioma has only marginally improved without curative intent in the last 30 years. The care of glioma patients remains among the mostly costly in the United States.
The current paradigm of care seldom takes into account the molecular features of the tumor with few trials driven by identified tumor-specific mutations or amplifications.
EVs of tumor origin can be isolated from plasma and CSF. Purification strategies are available along with sensitive analytics for amplified mRNA and mutations within glioma-specific genes. These analyses serve as the basis for detection of tumor-specific biomarkers in biofluids obtained with minimal risk for the patient. The most fruitful studies involve the creation of a diagnosis without surgical intervention. Two candidate glioma diagnostic biomarkers are EGFRvIII in plasma and CSF and IDH1/2 in CSF.
Validation of the sensitivity and specificity of glioma diagnostic biomarkers lead to a novel paradigm of targeted drug therapy.
Acknowledgements
The authors thank CJF van Noorden for his helpful review comments, corrections and criticisms (University of Amsterdam, the Netherlands). This work was supported by NCI: 2PO1CA069246-16A, Experimental Therapeutics and BioMonitoring of Brain Tumors (FH Hochberg) and NIH Office of the Director, Office of Strategic Coordination, NIH/NINDS Common Fund Award: 1 UH2 TR000931-01 (RS Carter, FH Hochberg) and The Richard Floor Biorepository Fund. NA Atai was recipient of 2009 AMC Scholarship, University of Amsterdam, the Netherlands.
Footnotes
Financial and competing interests disclosure
RS Carter serves on the Scientific Advisory Committee of Exosome Diagnostics, Inc., NY, USA. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties
No writing assistance was utilized in the production of this manuscript.
REFERENCES
- 1.Atai NA, Renkema-Mills NA, Bosman J, et al. Differential activity of NADPH-producing dehydrogenases renders rodents unsuitable models to study IDH1R132 mutation effects in human glioblastoma. J. Histochem. Cytochem. 2011;59(5):489–503. doi: 10.1369/0022155411400606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Théry C, Zitvogel L, Amigorena S. Exosomes: composition, biogenesis and function. Nat. Rev. Immunol. 2002;2(8):569–579. doi: 10.1038/nri855. [DOI] [PubMed] [Google Scholar]
- 3.Dolecek TA, Propp JM, Stroup NE, Kruchko C. CBTRUS statistical report: primary brain and central nervous system tumors diagnosed in the United States in 2005–2009. Neuro-oncology. 2012;14(Suppl 5):v1–v49. doi: 10.1093/neuonc/nos218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grossman SA, Osman M, Hruban R, Piantadosi S. Central nervous system cancers in first-degree relatives and spouses. Cancer Invest. 1999;17(5):299–308. doi: 10.3109/07357909909032870. [DOI] [PubMed] [Google Scholar]
- 5.Neglia JP, Robison LL, Stovall M, et al. New primary neoplasms of the central nervous system in survivors of childhood cancer: a report from the childhood cancer survivor study. J. Nat. Cancer Inst. 2006;98(21):1528–1537. doi: 10.1093/jnci/djj411. [DOI] [PubMed] [Google Scholar]
- 6.Noch E, Khalili K. Oncogenic viruses and tumor glucose metabolism: like kids in a candy store. Molecular cancer therapeutics. 2012;11(1):14–23. doi: 10.1158/1535-7163.MCT-11-0517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mitchell DA, Xie W, Schmittling R, et al. Sensitive detection of human cytomegalovirus in tumors and peripheral blood of patients diagnosed with GBM. Neuro-oncology. 2008;10(1):10–18. doi: 10.1215/15228517-2007-035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lehrer S. Cytomegalovirus infection in early childhood may be protective against GBM multiforme, while later infection is a risk factor. Medical hypotheses. 2012;78(5):657–658. doi: 10.1016/j.mehy.2012.02.003. [DOI] [PubMed] [Google Scholar]
- 9.Price RL, Song J, Bingmer K, et al. Cytomegalovirus contributes to GBM in the context of tumor suppressor mutations. Cancer Res. 2013;73(11):3441–3450. doi: 10.1158/0008-5472.CAN-12-3846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sampson JH, Mitchell DA. Is cytomegalovirus a therapeutic target in GBM? Clin. Cancer Res. 2011;17(14):4619–4621. doi: 10.1158/1078-0432.CCR-11-0992. [DOI] [PubMed] [Google Scholar]
- 11.Ricci-Vitiani L, Pallini R, Biffoni M, et al. Tumour vascularization via endothelial differentiation of GBM stem-like cells. Nature. 2010;468(7325):824–828. doi: 10.1038/nature09557. [DOI] [PubMed] [Google Scholar]
- 12.Wang R, Chadalavada K, Wilshire J, et al. GBM stem-like cells give rise to tumour endothelium. Nature. 2010;468(7325):829–833. doi: 10.1038/nature09624. [DOI] [PubMed] [Google Scholar]
- 13.Bonavia R, Inda MM, Cavenee WK, Furnari FB. Heterogeneity maintenance in GBM: a social network. Cancer Res. 2011;71(12):4055–4060. doi: 10.1158/0008-5472.CAN-11-0153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yan H, Parsons DW, Jin G, et al. IDH1 and IDH2 mutations in gliomas. New Engl. J. Med. 2009;360(8):765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nickel GC, Barnholtz-Sloan J, Gould MP, et al. Characterizing mutational heterogeneity in a GBM patient with double recurrence. PloS one. 2012;7(4):e35262. doi: 10.1371/journal.pone.0035262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Verhaak RG, Hoadley KA, Purdom E, et al. Integrated genomic analysis identifies clinically relevant subtypes of GBM characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17(1):98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Noushmehr H, Weisenberger DJ, Diefes K, et al. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17(5):510–522. doi: 10.1016/j.ccr.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wong AJ, Ruppert JM, Bigner SH, et al. Structural alterations of the epidermal growth factor receptor gene in human gliomas. Proc. Nat. Acad. Sci. USA. 1992;89(7):2965–2969. doi: 10.1073/pnas.89.7.2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kutikova L, Bowman L, Chang S, Long SR, Thornton DE, Crown WH. Utilization and cost of health care services associated with primary malignant brain tumors in the United States. J. Neuro-oncol. 2007;81(1):61–65. doi: 10.1007/s11060-006-9197-y. [DOI] [PubMed] [Google Scholar]
- 20.Chittiboina P, Connor DE, Jr, Caldito G, Quillin JW, Wilson JD, Nanda A. Occult tumors presenting with negative imaging: analysis of the literature. J. Neurosurg. 2012;116(6):1195–1203. doi: 10.3171/2012.3.JNS112098. [DOI] [PubMed] [Google Scholar]
- 21. www.cdc.gov/nchs/data/hus/2011/123.pdf.
- 22.Khalid L, Carone M, Dumrongpisutikul N, et al. Imaging characteristics of oligodendrogliomas that predict grade. AJNR. American journal of neuroradiology. 2012;33(5):852–857. doi: 10.3174/ajnr.A2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weber MA, Zoubaa S, Schlieter M, et al. Diagnostic performance of spectroscopic and perfusion MRI for distinction of brain tumors. Neurology. 2006;66(12):1899–1906. doi: 10.1212/01.wnl.0000219767.49705.9c. [DOI] [PubMed] [Google Scholar]
- 24.Geer CP, Simonds J, Anvery A, et al. Does MR perfusion imaging impact management decisions for patients with brain tumors? A prospective study. AJNR. American journal of neuroradiology. 2012;33(3):556–562. doi: 10.3174/ajnr.A2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haegler K, Wiesmann M, Bohm C, et al. New similarity search based glioma grading. Neuroradiology. 2012;54(8):829–837. doi: 10.1007/s00234-011-0988-2. [DOI] [PubMed] [Google Scholar]
- 26.McLendon R, Friedman A, Bigner D, et al. Comprehensive genomic characterization defines human GBM genes and core pathways. Nature. 2008;455(7216):1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stummer W, Van Den Bent MJ, Westphal M. Cytoreductive surgery of GBM as the key to successful adjuvant therapies: new arguments in an old discussion. Acta neurochirurgica. 2011;153(6):1211–1218. doi: 10.1007/s00701-011-1001-x. [DOI] [PubMed] [Google Scholar]
- 28.Chang SM, Parney IF, Mcdermott M, et al. Perioperative complications and neurological outcomes of first and second craniotomies among patients enrolled in the Glioma Outcome Project. J. Neurosurg. 2003;98(6):1175–1181. doi: 10.3171/jns.2003.98.6.1175. [DOI] [PubMed] [Google Scholar]
- 29.Febbo PG, Ladanyi M, Aldape KD, et al. NCCN Task Force report: Evaluating the clinical utility of tumor markers in oncology. Journal of the National Comprehensive Cancer Network. JNCCN. 2011;9(Suppl 5):S1–S32. doi: 10.6004/jnccn.2011.0137. quiz S33. [DOI] [PubMed] [Google Scholar]
- 30.Westphal M, Hilt DC, Bortey E, et al. A phase 3 trial of local chemotherapy with biodegradable carmustine (BCNU) wafers (Gliadel wafers) in patients with primary malignant glioma. Neuro-oncology. 2003;5(2):79–88. doi: 10.1215/S1522-8517-02-00023-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stummer W, Kamp MA. The importance of surgical resection in malignant glioma. Current opinion in neurology. 2009;22(6):645–649. doi: 10.1097/WCO.0b013e3283320165. [DOI] [PubMed] [Google Scholar]
- 32.Souhami L, Seiferheld W, Brachman D, et al. Randomized comparison of stereotactic radiosurgery followed by conventional radiotherapy with carmustine to conventional radiotherapy with carmustine for patients with GBM multiforme: report of Radiation Therapy Oncology Group 93-05 protocol. Int. J. radiation oncology, biology, physics. 2004;60(3):853–860. doi: 10.1016/j.ijrobp.2004.04.011. [DOI] [PubMed] [Google Scholar]
- 33.Laperriere NJ, Leung PM, Mckenzie S, et al. Randomized study of brachytherapy in the initial management of patients with malignant astrocytoma. Int. J. radiation oncology, biology, physics. 1998;41(5):1005–1011. doi: 10.1016/s0360-3016(98)00159-x. [DOI] [PubMed] [Google Scholar]
- 34.Stupp R, Mason WP, Van Den Bent MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for GBM. New Engl J Med. 2005;352(10):987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 35.Stupp R, Hegi ME, Mason WP, et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in GBM in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. The Lancet Oncol. 2009;10(5):459–466. doi: 10.1016/S1470-2045(09)70025-7. [DOI] [PubMed] [Google Scholar]
- 36.Jackson RJ, Fuller GN, Abi-Said D, et al. Limitations of stereotactic biopsy in the initial management of gliomas. Neuro-oncology. 2001;3(3):193–200. doi: 10.1093/neuonc/3.3.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Preusser M, De Ribaupierre S, Wohrer A, et al. Current concepts and management of GBM. Annals of neurology. 2011;70(1):9–21. doi: 10.1002/ana.22425. [DOI] [PubMed] [Google Scholar]
- 38.Gutin PH, Iwamoto FM, Beal K, et al. Safety and efficacy of bevacizumab with hypofractionated stereotactic irradiation for recurrent malignant gliomas. Int. J. radiation oncol, biol, phys. 2009;75(1):156–163. doi: 10.1016/j.ijrobp.2008.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sarkaria JN, Kitange GJ, James CD, et al. Mechanisms of chemoresistance to alkylating agents in malignant glioma. Clin. Cancer. Res. 2008;14(10):2900–2908. doi: 10.1158/1078-0432.CCR-07-1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yip S, Miao J, Cahill DP, et al. MSH6 mutations arise in GBMs during temozolomide therapy and mediate temozolomide resistance. Clin. Cancer Res. 2009;15(14):4622–4629. doi: 10.1158/1078-0432.CCR-08-3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cahill DP, Levine KK, Betensky RA, et al. Loss of the mismatch repair protein MSH6 in human GBMs is associated with tumor progression during temozolomide treatment. Clin. Cancer Res. 2007;13(7):2038–2045. doi: 10.1158/1078-0432.CCR-06-2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Macdonald DR, Cascino TL, Schold SC, Jr, Cairncross JG. Response criteria for phase II studies of supratentorial malignant glioma. J. Clin Oncol. 1990;8(7):1277–1280. doi: 10.1200/JCO.1990.8.7.1277. [DOI] [PubMed] [Google Scholar]
- 43.Brem H, Ewend MG, Piantadosi S, Greenhoot J, Burger PC, Sisti M. The safety of interstitial chemotherapy with BCNU-loaded polymer followed by radiation therapy in the treatment of newly diagnosed malignant gliomas: phase I trial. J. Neuro-oncol. 1995;26(2):111–123. doi: 10.1007/BF01060217. [DOI] [PubMed] [Google Scholar]
- 44.Vredenburgh JJ, Desjardins A, Herndon JE, 2nd, et al. Bevacizumab plus irinotecan in recurrent GBM multiforme. J. Clin. Oncol. 2007;25(30):4722–4729. doi: 10.1200/JCO.2007.12.2440. [DOI] [PubMed] [Google Scholar]
- 45.Iwamoto FM, Abrey LE, Beal K, et al. Patterns of relapse and prognosis after bevacizumab failure in recurrent GBM. Neurology. 2009;73(15):1200–1206. doi: 10.1212/WNL.0b013e3181bc0184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fischer I, Cunliffe CH, Bollo RJ, et al. High-grade glioma before and after treatment with radiation and Avastin: initial observations. Neuro-oncology. 2008;10(5):700–708. doi: 10.1215/15228517-2008-042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rock JP, Hearshen D, Scarpace L, et al. Correlations between magnetic resonance spectroscopy and image-guided histopathology, with special attention to radiation necrosis. Neurosurgery. 2002;51(4):912–919. doi: 10.1097/00006123-200210000-00010. discussion 919–920. [DOI] [PubMed] [Google Scholar]
- 48.Chen WW, Balaj L, Liau LM, et al. BEAMing and Droplet Digital PCR Analysis of Mutant IDH1 mRNA in Glioma Patient Serum and Cerebrospinal Fluid Extracellular Vesicles. Molecular therapy. Nucleic acids. 2013;2:e109. doi: 10.1038/mtna.2013.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Skog J, Wurdinger T, Van Rijn S, et al. GBM microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat. Cell Biol. 2008;10(12):1470–1476. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Balaj L, Lessard R, Dai L, et al. Tumour microvesicles contain retrotransposon elements and amplified oncogene sequences. Nat. Commun. 2011;2:180. doi: 10.1038/ncomms1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Heimberger AB, Hlatky R, Suki D, et al. Prognostic effect of epidermal growth factor receptor and EGFRvIII in GBM multiforme patients. Clin Cancer Res. 2005;11(4):1462–1466. doi: 10.1158/1078-0432.CCR-04-1737. [DOI] [PubMed] [Google Scholar]
- 52.Pelloski CE, Ballman KV, Furth AF, et al. Epidermal growth factor receptor variant III status defines clinically distinct subtypes of GBM. J. Clin Oncol. 2007;25(16):2288–2294. doi: 10.1200/JCO.2006.08.0705. [DOI] [PubMed] [Google Scholar]
- 53.Camara-Quintana JQ, Nitta RT, Li G. Pathology: commonly monitored GBM markers: EFGR, EGFRvIII, PTEN, and MGMT. Neurosurg. clin. of North America. 2012;23(2):237–246. viii. doi: 10.1016/j.nec.2012.01.011. [DOI] [PubMed] [Google Scholar]
- 54.Li G, Mitra SS, Monje M, et al. Expression of epidermal growth factor variant III (EGFRvIII) in pediatric diffuse intrinsic pontine gliomas. J. Neuro-oncol. 2012;108(3):395–402. doi: 10.1007/s11060-012-0842-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bruzzone MG, Eoli M, Cuccarini V, Grisoli M, Valletta L, Finocchiaro G. Genetic signature of adult gliomas and correlation with MRI features. Exp. Rev. Mol. Diagn. 2009;9(7):709–720. doi: 10.1586/erm.09.44. [DOI] [PubMed] [Google Scholar]
- 56.Ciesielski MJ, Kazim AL, Barth RF, Fenstermaker RA. Cellular antitumor immune response to a branched lysine multiple antigenic peptide containing epitopes of a common tumor-specific antigen in a rat glioma model. Cancer immunology, immunotherapy : CII. 2005;54(2):107–119. doi: 10.1007/s00262-004-0576-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lassman AB, Rossi MR, Raizer JJ, et al. Molecular study of malignant gliomas treated with epidermal growth factor receptor inhibitors: tissue analysis from North American Brain Tumor Consortium Trials 01-03 and 00-01. Clin. Cancer Res. 2005;11(21):7841–7850. doi: 10.1158/1078-0432.CCR-05-0421. [DOI] [PubMed] [Google Scholar]
- 58.Van Den Bent MJ, Brandes AA, Rampling R, et al. Randomized phase II trial of erlotinib versus temozolomide or carmustine in recurrent GBM: EORTC brain tumor group study 26034. J. Clin Oncol. 2009;27(8):1268–1274. doi: 10.1200/JCO.2008.17.5984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Brown PD, Krishnan S, Sarkaria JN, et al. Phase I/II trial of erlotinib and temozolomide with radiation therapy in the treatment of newly diagnosed GBMmultiforme: North Central Cancer Treatment Group Study N0177. J. Clin. Oncol. 2008;26(34):5603–5609. doi: 10.1200/JCO.2008.18.0612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tanaka S, Louis DN, Curry WT, Batchelor TT, Dietrich J. Diagnostic and therapeutic avenues for GBM: no longer a dead end? Nature Rev. Clin. Oncol. 2013;10(1):14–26. doi: 10.1038/nrclinonc.2012.204. [DOI] [PubMed] [Google Scholar]
- 61.Masui K, Cloughesy TF, Mischel PS. Review: molecular pathology in adult highgrade gliomas: from molecular diagnostics to target therapies. Neuropathology and applied neurobiology. 2012;38(3):271–291. doi: 10.1111/j.1365-2990.2011.01238.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen WW1, Balaj L, Liau LM, et al. BEAMing and Droplet Digital PCR Analysis of Mutant IDH1 mRNA in Glioma Patient Serum and Cerebrospinal Fluid Extracellular Vesicles. Mol. Therapy and Nucleic Acid. 2013;2:e109. doi: 10.1038/mtna.2013.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Weller M, Felsberg J, Hartmann C, et al. Molecular predictors of progression-free and overall survival in patients with newly diagnosed GBM: a prospective translational study of the German Glioma Network. J. Clin. Oncol. 2009;27(34):5743–5750. doi: 10.1200/JCO.2009.23.0805. [DOI] [PubMed] [Google Scholar]
- 64.Cairns RA, Mak TW. Oncogenic isocitrate dehydrogenase mutations: mechanisms, models, and clinical opportunities. Cancer discov. 2013 doi: 10.1158/2159-8290.CD-13-0083. [DOI] [PubMed] [Google Scholar]
- 65.Jones PS, Dunn GP, Barker FG, 2nd, Curry WT, Hochberg FH, Cahill DP. Molecular genetics of low-grade gliomas: genomic alterations guiding diagnosis and therapeutic intervention. 11th Annual Frye-Halloran Brain Tumor Symposium. Neurosurgical focus. 2013;34(2):E9. doi: 10.3171/2012.12.FOCUS12349. [DOI] [PubMed] [Google Scholar]
- 66.Hartmann C, Meyer J, Balss J, et al. Type and frequency of IDH1 and IDH2 mutations are related to astrocytic and oligodendroglial differentiation and age: a study of 1,010 diffuse gliomas. Acta neuropathol. 2009;118(4):469–474. doi: 10.1007/s00401-009-0561-9. [DOI] [PubMed] [Google Scholar]
- 67.Yip S, Butterfield YS, Morozova O, et al. Concurrent CIC mutations, IDH mutations, and 1p/19q loss distinguish oligodendrogliomas from other cancers. J. Pathol. 2012;226(1):7–16. doi: 10.1002/path.2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sahm F, Koelsche C, Meyer J, et al. CIC and FUBP1 mutations in oligodendrogliomas, oligoastrocytomas and astrocytomas. Acta neuropathol. 2012;123(6):853–860. doi: 10.1007/s00401-012-0993-5. [DOI] [PubMed] [Google Scholar]
- 69.Jiao Y, Killela PJ, Reitman ZJ, et al. Frequent ATRX, CIC, FUBP1 and IDH1 mutations refine the classification of malignant gliomas. Oncotarget. 2012;3(7):709–722. doi: 10.18632/oncotarget.588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lee CJ, Chan WI, Scotting PJ. CIC, a gene involved in cerebellar development and ErbB signaling, is significantly expressed in medulloblastomas. J. Neurooncol. 2005;73(2):101–108. doi: 10.1007/s11060-004-4598-2. [DOI] [PubMed] [Google Scholar]
- 71.Bettegowda C, Agrawal N, Jiao Y, et al. Mutations in CIC and FUBP1 contribute to human oligodendroglioma. Science. 2011;333(6048):1453–1455. doi: 10.1126/science.1210557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schwartzentruber J, Korshunov A, Liu XY, et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric GBM. Nature. 2012;482(7384):226–231. doi: 10.1038/nature10833. [DOI] [PubMed] [Google Scholar]
- 73.Liu XY, Gerges N, Korshunov A, et al. Frequent ATRX mutations and loss of expression in adult diffuse astrocytic tumors carrying IDH1/IDH2 and TP53 mutations. Acta neuropathol. 2012;124(5):615–625. doi: 10.1007/s00401-012-1031-3. [DOI] [PubMed] [Google Scholar]
- 74.Killela PJ, Reitman ZJ, Jiao Y, et al. TERT promoter mutations occur frequently in gliomas and a subset of tumors derived from cells with low rates of self-renewal. Proc. Nat. Acad. Sci. USA. 2013;110(15):6021–6026. doi: 10.1073/pnas.1303607110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Schindler G, Capper D, Meyer J, et al. Analysis of BRAF V600E mutation in 1,320 nervous system tumors reveals high mutation frequencies in pleomorphic xanthoastrocytoma, ganglioglioma and extra-cerebellar pilocytic astrocytoma. Acta neuropathol. 2011;121(3):397–405. doi: 10.1007/s00401-011-0802-6. [DOI] [PubMed] [Google Scholar]
- 76.Schiffman JD, Hodgson JG, Vandenberg SR, et al. Oncogenic BRAF mutation with CDKN2A inactivation is characteristic of a subset of pediatric malignant astrocytomas. Cancer Res. 2010;70(2):512–519. doi: 10.1158/0008-5472.CAN-09-1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chapman PB, Hauschild A, Robert C, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. New Engl. J. Med. 2011;364(26):2507–2516. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sosman JA, Kim KB, Schuchter L, et al. Survival in BRAF V600-mutant advanced melanoma treated with vemurafenib. New Engl. J Med. 2012;366(8):707–714. doi: 10.1056/NEJMoa1112302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chen P, Zou P, Yan Q, Xu H, Zhao P, Gu A. The TERT MNS16A polymorphism contributes to cancer susceptibility: meta-analysis of the current studies. Gene. 2013;519(2):266–270. doi: 10.1016/j.gene.2013.02.018. [DOI] [PubMed] [Google Scholar]
- 80.Wu G, Broniscer A, Mceachron TA, et al. Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem GBMs. Nat. Gen. 2012;44(3):251–253. doi: 10.1038/ng.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Xing WJ, Zou Y, Han QL, et al. Effects of epidermal growth factor receptor and phosphatase and tensin homologue gene expression on the inhibition of U87MG GBM cell proliferation induced by protein kinase inhibitors. Clin. Exp. pharmacol. physiol. 2013;40(1):13–21. doi: 10.1111/1440-1681.12026. [DOI] [PubMed] [Google Scholar]
- 82.Maheswaran S, Sequist LV, Nagrath S, et al. Detection of mutations in EGFR in circulating lung-cancer cells. New Engl. J. Med. 2008;359(4):366–377. doi: 10.1056/NEJMoa0800668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Jeuken JW, Wesseling P. MAPK pathway activation through BRAF gene fusion in pilocytic astrocytomas; a novel oncogenic fusion gene with diagnostic, prognostic, and therapeutic potential. J. Pathol. 2010;222(4):324–328. doi: 10.1002/path.2780. [DOI] [PubMed] [Google Scholar]
- 84.Forshew T, Tatevossian RG, Lawson AR, et al. Activation of the ERK/MAPK pathway: a signature genetic defect in posterior fossa pilocytic astrocytomas. J. Pathol. 2009;218(2):172–181. doi: 10.1002/path.2558. [DOI] [PubMed] [Google Scholar]
- 85.Jones DT, Kocialkowski S, Liu L, et al. Tandem duplication producing a novel oncogenic BRAF fusion gene defines the majority of pilocytic astrocytomas. Cancer Res. 2008;68(21):8673–8677. doi: 10.1158/0008-5472.CAN-08-2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tihan T, Ersen A, Qaddoumi I, et al. Pathologic characteristics of pediatric intracranial pilocytic astrocytomas and their impact on outcome in 3 countries: a multi-institutional study. Am. J. Surg. Pathol. 2012;36(1):43–55. doi: 10.1097/PAS.0b013e3182329480. [DOI] [PubMed] [Google Scholar]
- 87.Hegi ME, Diserens AC, Gorlia T, et al. MGMT gene silencing and benefit from temozolomide in GBM. New Engl. J. Med. 2005;352(10):997–1003. doi: 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- 88.Dunn J, Baborie A, Alam F, et al. Extent of MGMT promoter methylation correlates with outcome in GBMs given temozolomide and radiotherapy. Br. J. Cancer. 2009;101(1):124–131. doi: 10.1038/sj.bjc.6605127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Balana C, Carrato C, Ramirez JL, et al. Tumour and serum MGMT promoter methylation and protein expression in GBM patients. Clin. Transl. Oncol. 2011;13(9):677–685. doi: 10.1007/s12094-011-0714-x. [DOI] [PubMed] [Google Scholar]
- 90.Weaver KD, Grossman SA, Herman JG. Methylated tumor-specific DNA as a plasma biomarker in patients with glioma. Cancer Invest. 2006;24(1):35–40. doi: 10.1080/07357900500449546. [DOI] [PubMed] [Google Scholar]
- 91.Pelloski CE, Mahajan A, Maor M, et al. YKL-40 expression is associated with poorer response to radiation and shorter overall survival in GBM. Clin. Cancer. Res. 2005;11(9):3326–3334. doi: 10.1158/1078-0432.CCR-04-1765. [DOI] [PubMed] [Google Scholar]
- 92.Tanwar MK, Gilbert MR, Holland EC. Gene expression microarray analysis reveals YKL-40 to be a potential serum marker for malignant character in human glioma. Cancer Res. 2002;62(15):4364–4368. [PubMed] [Google Scholar]
- 93.Li S, Chou AP, Chen W, et al. Overexpression of isocitrate dehydrogenase mutant proteins renders glioma cells more sensitive to radiation. Neuro-oncology. 2013;15(1):57–68. doi: 10.1093/neuonc/nos261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Baldewpersad Tewarie NM, Burgers IA, Dawood Y, et al. NADP+ -dependent IDH1 R132 mutation and its relevance for glioma patient survival. Med Hypotheses. 2013;80(6):728–731. doi: 10.1016/j.mehy.2013.02.022. [DOI] [PubMed] [Google Scholar]
- 95.Kuan CT, Wikstrand CJ, Bigner DD. EGF mutant receptor vIII as a molecular target in cancer therapy. Endocrine-related cancer. 2001;8(2):83–96. doi: 10.1677/erc.0.0080083. [DOI] [PubMed] [Google Scholar]
- 96.Choi BD, Archer GE, Mitchell DA, et al. EGFRvIII-targeted vaccination therapy of malignant glioma. Brain Pathol. 2009;19(4):713–723. doi: 10.1111/j.1750-3639.2009.00318.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Babu R, Adamson DC. Rindopepimut: an evidence-based review of its therapeutic potential in the treatment of EGFRvIII-positive GBM. Core evidence. 2012;7:93–103. doi: 10.2147/CE.S29001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Perry J, Okamoto M, Guiou M, Shirai K, Errett A, Chakravarti A. Novel therapies in GBM. Neurol. Res. Int. 2012;2012:428565. doi: 10.1155/2012/428565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Fu Y, Huang R, Du J, Yang R, An N, Liang A. Glioma-derived mutations in IDH: from mechanism to potential therapy. Biochem. Biophys. Res. Commun. 2010;397(2):127–130. doi: 10.1016/j.bbrc.2010.05.115. [DOI] [PubMed] [Google Scholar]
- 100.Rohle D, Popovici-Muller J, Palaskas N, et al. An inhibitor of mutant IDH1 delays growth and promotes differentiation of glioma cells. Science. 2013;340(6132):626–630. doi: 10.1126/science.1236062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wang F, Travins J, Delabarre B, et al. Targeted inhibition of mutant IDH2 in leukemia cells induces cellular differentiation. Science. 2013;340(6132):622–626. doi: 10.1126/science.1234769. [DOI] [PubMed] [Google Scholar]
- 102.You Y, Geng X, Zhao P, et al. Evaluation of combination gene therapy with PTEN and antisense hTERT for malignant glioma in vitro and xenografts. Cellular and molecular life sciences : CMLS. 2007;64(5):621–631. doi: 10.1007/s00018-007-6424-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Nagata Y, Lan KH, Zhou X, et al. PTEN activation contributes to tumor inhibition by trastuzumab, and loss of PTEN predicts trastuzumab resistance in patients. Cancer Cell. 2004;6(2):117–127. doi: 10.1016/j.ccr.2004.06.022. [DOI] [PubMed] [Google Scholar]
- 104.Dias-Santagata D, Lam Q, Vernovsky K, et al. BRAF V600E mutations are common in pleomorphic xanthoastrocytoma: diagnostic and therapeutic implications. PloS one. 2011;6(3):e17948. doi: 10.1371/journal.pone.0017948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Flaherty KT. Narrative review: BRAF opens the door for therapeutic advances in melanoma. Annals of internal medicine. 2010;153(9):587–591. doi: 10.7326/0003-4819-153-9-201011020-00008. [DOI] [PubMed] [Google Scholar]
- 106.Flaherty KT, Puzanov I, Kim KB, et al. Inhibition of mutated, activated BRAF in metastatic melanoma. New Engl. J. Med. 2010;363(9):809–819. doi: 10.1056/NEJMoa1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Joseph EW, Pratilas CA, Poulikakos PI, et al. The RAF inhibitor PLX4032 inhibits ERK signaling and tumor cell proliferation in a V600E BRAF-selective manner. Proc. Nat. Acad. Sci. USA. 2010;107(33):14903–14908. doi: 10.1073/pnas.1008990107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Tsai J, Lee JT, Wang W, et al. Discovery of a selective inhibitor of oncogenic BRaf kinase with potent antimelanoma activity. Proc. Nat. Acad. Sci. USA. 2008;105(8):3041–3046. doi: 10.1073/pnas.0711741105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.De Roock W, De Vriendt V, Normanno N, Ciardiello F, Tejpar S. KRAS, BRAF, PIK3CA, and PTEN mutations: implications for targeted therapies in metastatic colorectal cancer. The Lancet Oncol. 2011;12(6):594–603. doi: 10.1016/S1470-2045(10)70209-6. [DOI] [PubMed] [Google Scholar]
- 110.Bobustuc GC, Baker CH, Limaye A, et al. Levetiracetam enhances p53-mediated MGMT inhibition and sensitizes GBM cells to temozolomide. Neuro-oncology. 2010;12(9):917–927. doi: 10.1093/neuonc/noq044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Di Vinci A, Casciano I, Marasco E, et al. Quantitative methylation analysis of HOXA3, 7, 9, and 10 genes in glioma: association with tumor WHO grade and clinical outcome. J. Cancer. Res. Clin. Oncol. 2012;138(1):35–47. doi: 10.1007/s00432-011-1070-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Roth A, Harley CB, Baerlocher GM. Imetelstat (GRN163L)--telomerase-based cancer therapy. Recent results in cancer research. Fortschritte der Krebsforschung. Progres dans les recherches sur le cancer. 2010;184:221–234. doi: 10.1007/978-3-642-01222-8_16. [DOI] [PubMed] [Google Scholar]
- 113.Chen W, Xiao BK, Liu JP, Chen SM, Tao ZZ. Alternative lengthening of telomeres in hTERT-inhibited laryngeal cancer cells. Cancer Sci. 2010;101(8):1769–1776. doi: 10.1111/j.1349-7006.2010.01611.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.CSR, Incorporated. Evaluation of Patient Recruitment Strategies Phase I Feasibility Study. Final Report. 2006 [Google Scholar]
- 115.Marks RG, Conlon M, Ruberg SJ. Paradigm shifts in clinical trials enabled by information technology. Statistics in medicine. 2001;20(17–18):2683–2696. doi: 10.1002/sim.736. [DOI] [PubMed] [Google Scholar]
- 116.Tan DS, Thomas GV, Garrett MD, et al. Biomarker-driven early clinical trials in oncology: a paradigm shift in drug development. Cancer J. 2009;15(5):406–420. doi: 10.1097/PPO.0b013e3181bd0445. [DOI] [PubMed] [Google Scholar]
- 117.Diehl F, Li M, He Y, Kinzler KW, Vogelstein B, Dressman D. BEAMing: single-molecule PCR on microparticles in water-in-oil emulsions. Nat. Meth. 2006;3(7):551–559. doi: 10.1038/nmeth898. [DOI] [PubMed] [Google Scholar]
- 118.Li M, Chen WD, Papadopoulos N, et al. Sensitive digital quantification of DNA methylation in clinical samples. Nat. Biotechnol. 2009;27(9):858–863. doi: 10.1038/nbt.1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Li M, Diehl F, Dressman D, Vogelstein B, Kinzler KW. BEAMing up for detection and quantification of rare sequence variants. Nat. Meth. 2006;3(2):95–97. doi: 10.1038/nmeth850. [DOI] [PubMed] [Google Scholar]
- 120.Atai NA, Balaj L, Van Veen H, et al. Heparin blocks transfer of ectracellular vesicles between donor and recipient cells. J. Neurooncol. 2013;115(3):343–351. doi: 10.1007/s11060-013-1235-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Shao H, Chung J, Balaj L, et al. Protein typing of circulating microvesicles allows real-time monitoring of GBM therapy. Nat. Med. 2012;18(12):1835–1840. doi: 10.1038/nm.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Boisselier B, Gallego Perez-Larraya J, Rossetto M, et al. Detection of IDH1 mutation in the plasma of patients with glioma. Neurology. 2012;79(16):1693–1698. doi: 10.1212/WNL.0b013e31826e9b0a. [DOI] [PubMed] [Google Scholar]