Abstract
Extracellular electron transfer (EET) is intrinsically associated with the core phenomena of energy harvesting/energy conversion in natural ecosystems and biotechnology applications. However, the mechanisms associated with EET are complex and involve molecular interactions that take place at the “bionano interface” where biotic/abiotic interactions are usually explored. This work provides molecular perspective on the electron transfer mechanism(s) employed by Shewanella oneidensis MR-1. Molecular docking simulations were used to explain the interfacial relationships between two outer-membrane cytochromes (OMC) OmcA and MtrC and riboflavin (RF) and flavin mononucleotide (FMN), respectively. OMC-flavin interactions were analyzed by studying the electrostatic potential, the hydrophilic/hydrophobic surface properties, and the van der Waals surface of the OMC proteins. As a result, it was proposed that the interactions between flavins and OMCs are based on geometrical recognition event. The possible docking positions of RF and FMN to OmcA and MtrC were also shown.
I. INTRODUCTION
The discovery of extracellular electron transfer (EET) and its ubiquity and relevance to biogeochemical cycling and biotechnology has raised a number of important questions such as “How do bacteria transfer electrons to solid electron acceptors?,” “What is the role of excreted compounds such as flavins in the electron transfer process?,” “Do anode respiring bacteria share common EET mechanisms?.” Some of these questions found their answers over the years. For example, it has been determined that bacteria such as Shewanella spp. can interact with solid electron acceptors by three distinguished mechanisms of EET (Refs. 1–4)-mediated (MET) and direct (DET) electron transfer as well as long distance EET through nanowires. For Geobacter spp., which has been considered as the most active dissimilatory metal-reducing bacteria (DMRB), the only possible EET mechanism involves DET from outer membrane cytochromes (OMCs) and conductive pili.5 On the contrary to the nanowires, which are now thought to be outer-membrane extensions of Shewanella, it has been proposed that the electron transfer along Geobacter conductive pili is facilitated by aromatic amino acids such as tyrosine.6
The DET activity of Shewanella spp.7 has been associated with OMCs that are located in the outer cell membrane and periplasm.3 The electron transfer pathway of these species, more precisely Shewanella MR-1, involves a periplasmic decaheme cytochrome MtrA, an outer membrane β-barrel protein MtrB and outer membrane decaheme cytochromes OmcA and MtrC.8–11 This pathway is used to move electrons from the intracellular quinol pool to extracellular solid electron acceptors such as metal oxides or electrode surfaces,7,8,12 A synthetic electron conduit of mtrCAB has also been expressed in Escherichia coli demonstrating the importance of the cytochromes' electron “relay” in DET, and the ability of bacteria to reduce poised electrode surfaces.13 TerAvest et al. showed that the strain of E. coli having mtrCAB protein complex generated ∼eightfold higher current than a control strain.
All OMCs of Shewanella spp. share a common feature, which is the presence of heme groups. These hemes are the molecular conduits of electron transfer across the periplasm and the cell membrane as well as between the cytochromes. Specifically, OmcA and MtrC proteins have ten hemes arranged in a “staggered cross” configuration (Fig. 1).10,11 The distance between the hemes is in the range of 4–11 Å, small enough for rapid electron transfer.11,14 Each heme in the cytochrome molecule is covalently attached to the polypeptide chain via two cysteine residues in a CXXCH motif.11,12,14 When using electrochemical methods the multiple hemes in the OMCs have overlapping and experimentally undistinguishable redox potentials. They can be reduced over a broad potential window from 0.1 V to −0.4 vs SHE for MtrC (Ref. 15) and from −0.03 V to −0.325 vs SHE for OmcA.16 Therefore, computational approaches were used to calculate the redox potentials of the separate hemes and determine the free energy profile of EET.17,18
Fig. 1.
Crystal structure of MtrC (PDB ID: 4LM8). The polypeptide chains are shown in ribbon representation and colored from blue (N-terminus) to red (C-terminus). The Fe atoms of the hemes are represented as orange spheres and the porphyrin rings of the hemes are shown as red sticks.
While the DET pathways for Shewanella spp. have been explored for decades, the MET ability of these species has become a relatively recent topic of study. Shewanella MET has been correlated with the endogenous production and extracellular expression of flavins, which participate as electron shuttles and enhance EET rates.2,7,19–21 The two types of flavins excreted by Shewanella are flavin mononucleotide (FMN) and riboflavin (RF),20 where the amount of FMN secreted exceeds the amount of RF content by nearly five times in both anaerobic and aerobic conditions.2 The production and excretion of flavins in aerobic environment is higher, which is in contrast to what is expected based on the assumption that flavins are used in MET under anaerobic conditions2 and what has been shown for the flavins distribution within a microbial biofilm.22 The hypothesis of the shuttling role of flavins has been stated and confirmed many times using microbial fuel cell anodes as solid-phase electron acceptors.2,19 Monitoring the current generated by the bacterial flavin adenine (bfe dinucleotide exporter) mutant of Shewanella MR-1, unable to secrete flavins, has demonstrated their importance in EET.19 With the same concentration of artificially introduced flavins, Shewanella MR-1 wild type and the bfe mutant produced comparable currents. However, when flavins were not supplemented to the system, the bfe mutant generated 75% less current than the wild type. Also, strains lacking bfe could reduce Fe(III) oxide at only ∼25% of the rate of MR-1, demonstrating the importance of flavin electron shuttles under these conditions. It has also been shown that the Mtr pathway is important for the reduction of soluble flavins and that there is specificity of this activity with respect to OmcA and MtrC.7 Under electrode-reducing conditions, mutants of Shewanella MR-1 lacking OmcA reduced riboflavin at the same rate as the wild type, while ΔmtrC reduced RF at about half that rate.7 Coursolle et al. hypothesized that OmcA is required for attachment and/or biofilm formation on solid electron acceptors, while MtrC is the main electron conduit. The authors also suggested that under high flavin concentrations, OmcA may replace MtrC in flavin reduction and that the low current response from ΔmtrC is a result of the lack of viable cells. Although very important and timely, the manuscript of Coursolle et al. still did not provide the necessity insight into the role of flavins.
Recently, Okamoto et al. dramatically changed the premise of EET mechanism in DMRB by providing an evidence that flavins not only serve as shuttling mediators but also as cofactors directly associated with the OMCs.23–26 This hypothesis is somewhat expected since flavins are cofactors in various proteins.27 Similarly, to Coursolle et al., Okamoto et al. demonstrated that flavins exhibit “preferences” toward different outer membrane cytochromes. For instance FMN specifically interacts with MtrC, while RF associates with OmcA.25 The reasoning behind such specificity, according to the authors, lies in the differences in flavins' redox potential (−0.11 V vs SHE for bound RF and −0.15 V vs SHE for bound FMN), which allows for the utilization of various terminal electron acceptors as a survival strategy.25 The findings of Okamoto et al. suggested relatively strong binding of flavins to OMCs. But when the apparent dissociation constants of binding FMN to OmcA and MtrC were experimentally determined through 31P NMR experiments,28 it was established that these proteins have weaker affinity for FMN. Based on these dissociation constants, FMN binds stronger to OmcA (Kd = 29 ± 11 μM) rather than MtrC (Kd = 255 ± 126 μM). It is also interesting to note that they found two binding sites of FMN in OmcA, while MtrC has only one binding site. The discrepancies between the findings of Okamoto et al. and Paquete et al. may be explained by the differences in the performed experiments. Okamoto et al. used whole Shewanella wild type and mutant cells, while Paquete used extracted and purified proteins for their measurements. We can assume that the interactions between OMCs and flavins will be strongly affected by the environment especially when proteins are membrane bound proteins in their natural environment. Therefore, the results obtained using whole cells can be considered as more reliable than the ones obtained with purified proteins.
The redox potential of bonded flavins appears to be at least 0.1 V more positive than the redox potential of free flavins in solution, which decreases the potential difference between flavins and OmcA-MtrCBA complex.24 Instead of the two electron reduction, originally proposed for the redox reaction of flavins, it has been discovered that bound flavins undergo one electron reduction by OMCs forming a semiquinone.24,25 It is believed that the formation of the OMC-flavin complexes leads to the stabilization of the semiquinone along with a positive shift of the redox potential. Okamoto et al. demonstrated that the EET enhancement at the Shewanella/electrode interface is due not only to the previously proposed shuttling mechanism but also to the redox activity of the oxidized flavin/semiquinone redox couple explicitly when the flavin is associated with a particular OMC.24 Recent reports have also proposed that Shewanella and Geobacter spp. share common EET mechanisms with the involvement of OMCs' associated flavins.26,29 Xu et al. confirmed Okamoto's hypothesis that flavins accelerate EET from Shewanella to the electrode surface primarily as cytochrome-bound cofactors, rather than free soluble shuttles.30 An important point in their study is the influence of electrode material on the obtained results and the conclusions drawn out, where MET is typically detected using glassy carbon electrodes that have a high adsorption affinity for flavins but lower surface area for bacteria attachment and biofilm formation promoting flavin-mediated EET. On the contrary, ITO electrodes have a low adsorption affinity for flavins and thus direct the EET through cytochromes and cytochrome-bound flavins. Flavins demonstrate an affinity to carbon materials20,31,32 and Fe(III) and Mn(IV) surfaces,20 which additionally supports the importance of associated flavins as opposed to freely diffusing mediators. It is hypothesized that adsorbed flavins not only enhance EET but also promote cell attachment and biofilm formation by increasing the redox gradient that is advantageous to microbial respiration.31,33
Molecular docking simulations were used in this work to explain the interfacial relationships between OmcA and MtrC, and RF and FMN, respectively. Molecular docking simulations have been previously explored in order to predict where electron shuttles will bind and interact with outer membrane cytochromes.28 Paquete et al. proposed that the interaction of electron shuttles and OmcA is predetermined by the charge of the docking molecule. Positively charged molecules such as phenazine attach near heme 10, while negatively charged AQDS and FMN dock near heme 2. The neutral RF binds close to hemes 9 and 10. In 2016, Hong et al. used AutoDock Vina to simulate the interactions of RF with OmcA and further refine the simulation by molecular dynamics (MD).34 The authors specifically modeled the interactions with hemes 2, 5, 7, and 10 and showed stronger RF affinity toward hemes 5 and 7 and weaker interactions with hemes 2 and 10. The weaker affinity of RF to hemes 2 and 10 is proposed to be a result of weaker van der Waals interactions since the isoalloxazine ring of RF is exposed to the solvent. Besides this work, AutoDock Vina has been successfully employed by our group to describe the specific interactions between proteins and small ligands.35,36 Here, we apply the same docking approach to predict binding sites of flavins (FMN and RF) to Shewanella oneidensis OMCs (OmcA, MtrC).35,37–39 The explored analysis is fast and provides information about the mechanism of the interactions between flavins and proteins and predicts the binding position of flavins in the cytochrome molecule. Although more advanced methods are required to fully describe EET,40 flavins binding position and the information about their interaction with MtrC and OmcA can be used to suggest which heme in the protein molecule is carrying out the EET to the flavin. Namely, the results of the simulations were used to determine the distance between N5 from the flavin molecule and the Fe in the nearest cytochrome heme, which can be used to give a qualitative picture of the EET rate if we assume DET from the reduced Fe of the heme to the flavin molecule. Additionally, in contrast to the work of Hong et al.,34 in this study, both flavins and the amino acids in the binding pocket were treated as flexible, which allows molecules/side chains to change conformation during the binding event. The latter assures more accurate prediction of the binding. AutoDock Vina, an improved version of Autodock 4, was used to perform the docking simulations. In contrast to other common docking simulation packages, Autodock Vina is based on a free energy force field that is obtained by “machine learning” rather than being restricted to the use of approximate mathematical terms, which are used to allow physical interpretation of different contributions to the force field.37 This machine learning approach extracts empirical information from the conformation preferences of the protein–ligand complex and the experimental affinity measurements. The use of AutoDock Vina is justified by the increase in speed and a better accuracy of the binding mode prediction. The scoring function estimates the standard chemical potential of the system, which in turn determines the preferred bound conformation and the free energy of binding, also known as binding affinity.
II. COMPUTATIONAL DETAILS
Autodock Vina37 was used to perform the docking simulations of flavins on cytochromes OmcA and MtrC, which resulted in nine possible docking models in each run. The models are arranged in descending order of the protein–substrate binding affinity. To increase the reliability of the docking prediction, in each case, the simulations were performed at least three times with increasing exhaustiveness of the search. The biologically meaningful complex candidates were further selected using two criteria: (1) high frequency of occurrence based on a histogram of the frequencies of the output models acquired from different simulation runs, and (2) the minimum flavin–heme distance (N5 from the flavin molecule and the Fe in the nearest cytochrome heme) was set to be smaller than 11 Å, a criterion which enables rapid EET. For more details on the applied approach, see supplementary material and Fig. S1.46
For the docking simulations, the protein crystallographic structures were used [4LMH for OmcA (Ref. 11) and 4LM8 (Ref. 9) for MtrC as acquired from protein data bank]. The structures of riboflavin, FMN, and the semiquinone form of FMN in water were optimized using B3LYP/6-31 + G(d,p) level of theory and the polarizable continuum model as implemented in the Gaussian 09 quantum chemical package.41 AutoDock tools from MGL tools software42 were used for setting up the input files for Autodock Vina and for visualization of the docking models.
III. RESULTS AND DISCUSSION
Two sets of docking simulations were performed. The association of RF with the two S. oneidensis' MR-1 outer membrane cytochromes (OmcA and MtrC) was modeled first, followed by the docking of FMN to the same cytochromes.
A. Docking of riboflavin
Riboflavin was first docked to OmcA since Okamoto et al. demonstrated experimentally that this specific cytochrome interacts solely with RF.24,25 Figure 2 shows the two most probable docking positions for RF obtained using the modeling approach. The binding affinity of RF to OmcA in the two positions is similar (−7.4 kcal/mol for position 1 and −7.3 kcal/mol for position 2), indicating similar probability for the two models. In the first position, RF docks close to heme 5 (RF–heme 5 distance of 5.8 Å), and in the second position, RF is in the proximity of heme 7 (RF–heme 7 distance of 11.2 Å). The smaller distance between RF and heme 5 would suggest shorter distance for the electron transfer and thus faster and more efficient RF reduction by OmcA.
Fig. 2.
Docking of RF to OmcA, where in (a) the van der Waals surface of the protein is represented and in (b) the OmcA is represented with its secondary structure and the hemes colored in red. The RF is colored green.
It is known that binding of ligands to macromolecules occurs by intermolecular forces, such as ionic bonds, hydrogen bonds, van der Waals, or electrostatic forces. Electrostatic interactions were studied here by exploring the electrostatic potential of OmcA mapped on the van der Waals surface of the protein [Fig. 3(a)]. For clarity of the representation, the electrostatic potential of flavin molecules on their van der Waals surfaces are shown separate in Figs. 3(b) and S5.
Fig. 3.
(a) Docking of RF to OmcA, where the electrostatic potential was mapped on the van der Waals surface of the protein and (b) electrostatic potential of RF projected on the isodensity surface (0.05) as obtained using density functional theory and visualized using VMD. Blue surface is characterized with positive potential and red with negative. RF is colored according to the atom type.
The two binding pockets near hemes 5 and 7 of OmcA are positively charged. As it can be seen from Fig. 3(b), the isoalloxazine ring of RF is negatively charged near the quinone groups, but there is no clear charge separation in the flavin molecule. In addition, the RF binding pockets of MtrC (Fig. S2) are charged differently, i.e., not all of them are positively charged. Based on these, we can say that the electrostatic forces might play some role in OMCs-flavin binding, but no clear conclusion on the importance of the electrostatic interactions can be made based on these findings exclusively. At the same time, plotting the van der Waals surface for both RF and OmcA reveals that RF binds to the surface of the protein and does not penetrate the protein molecule (Fig. 4). In addition, based on the shape of the protein surface where the RF docks, it can be speculated that the interactions between RF and OmcA are based on stereospecificity for both docking positions. The same conclusion can be made for the interactions of RF with MtrC (Figs S2–S3), where in a similar manner docking simulations show that RF occupies the protein's surface pockets.
Fig. 4.
Docking of RF to OmcA, where the van der Waals surface of the protein is represented in white and the van der Waals surface of RF is in green. (a) The protein surface around heme 5 and (b) the protein surface around heme 5 after the docking of RF; (c) OmcA surface and the two docking positions of RF; (d) the protein surface around heme 7; and (e) the protein surface around heme 7 after the docking of RF.
Our finding disagrees with the findings of Paquete et al., who suggested electrostatic attraction as the main mechanism of flavins–OmcA interactions.28 Based on their docking simulations, RF binds close to hemes 9 and 10 while FMN docks near heme 2 of OmcA. The authors used AutoDock 4 for their simulations with all heme groups in their oxidized state. Both the protein and the docking ligands were considered rigid, i.e., they do not change conformation during the binding event. In the first set of our docking simulations, the flavin molecule was treated as flexible, and during the docking refinement, shown later in the paper, both the ligand and the amino acids in the binding pocket of the protein could change conformation during interaction. Breuer et al. also suggested that the flavin–OMCs binding is not based on electrostatic interactions but is mainly due to hydrogen binding.43 In addition, Hong et al. used molecular docking combined with MD simulation refinement and subsequent MD free energy calculations and found binding preference of RF at hemes 5 and 7 of OmcA, which agrees well with our findings. They also suggested that the interaction of flavins with cytochromes is based on aromatic stacking between isoalloxazine ring and hydrophobic residues in a similar fashion to flavoproteins. The discovery of Hong et al., which is based on the use of more sophisticated computational approach, overlaps with our findings and confirms the applicability of docking simulations as fast and simple screening method.
A careful consideration of the RF docking positions on OmcA allowed us to identify the amino acids involved in the RF binding pocket. The docking simulations were then refined by allowing amino acids from the pocket to change their conformation during the interactions with RF, which has not been performed by other researchers. Figure 5(b) shows the docking of RF near heme 5 with the following amino acids assigned as flexible: Ser 356, Ala 357, His 358, His 359, Tyr 374, Gly 375, Gly 376, Tyr 517, and His 368. The searching space was centered on heme 5 with the size of 30 × 30 × 30 Å. After refining the computational model, the RF–heme 5 distance was determined to be 5.3 Å. It was also observed that in both, flexible and rigid simulations, the isoalloxazine ring of RF docks in the same binding pocket; however, in the flexible model, the quinone groups are positioned in the opposite direction than in the “rigid” model (Fig. 5). This observation suggests that stereospecificity is a possible mechanism for preferential RFOMC binding.
Fig. 5.
Docking of RF to OmcA in the proximity of heme 5, where the OmcA binding pocket for RF was assigned as (a) flexible and (b) as rigid. The amino acids in cyan color show the position of amino acids in the rigid simulations and blue-green their position after the flexible simulations. Heme 5 is colored red and RF is colored based on the element composition with N in blue, O in red, and C in gray.
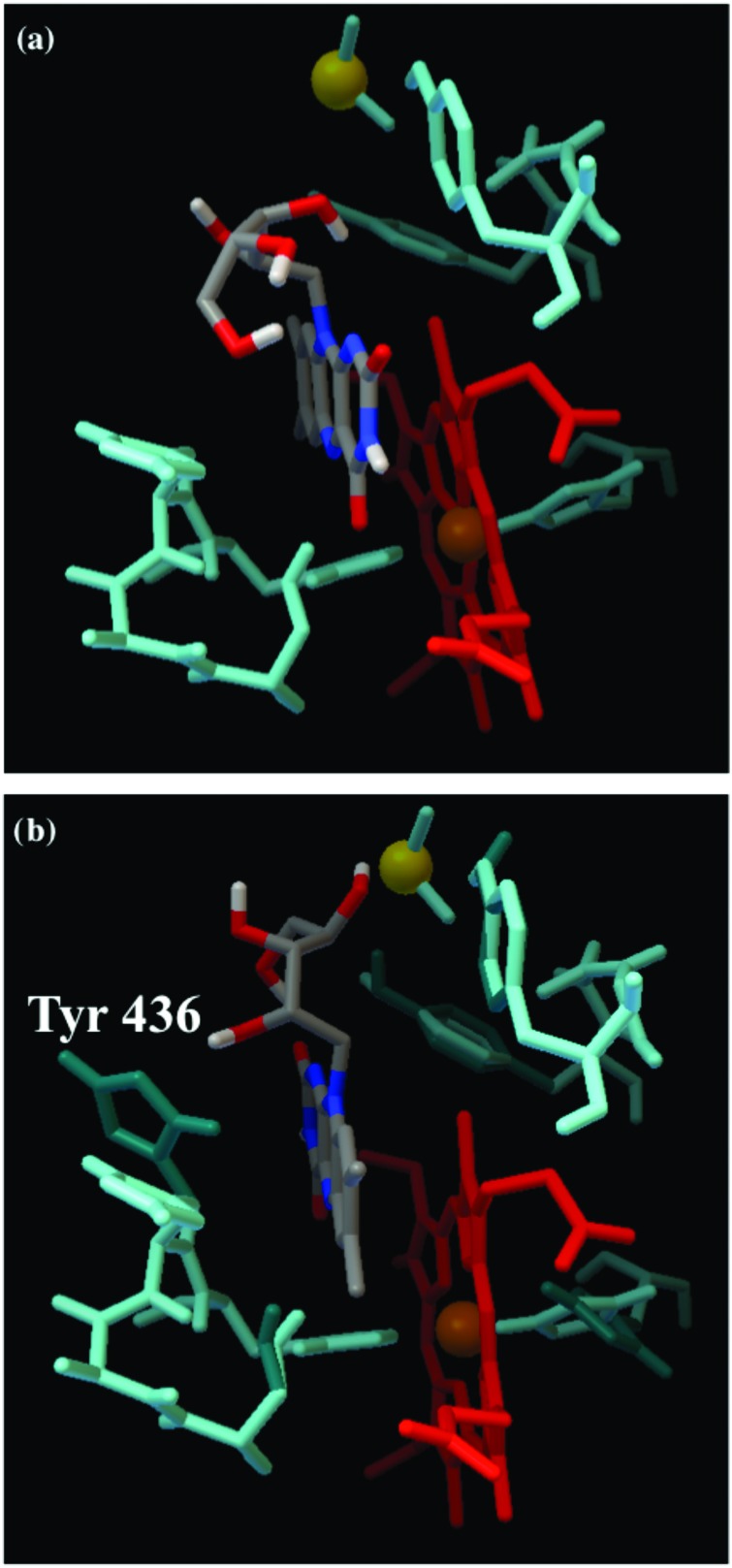
The same refinement in the docking simulation was performed for heme 7 docking position with a searching space of 30 × 30 × 30 Å centered on heme 7 and Ala 435, Tyr 436, Thr 437, Lys 438, Ser 440, Tyr 442, Tyr 460, Ser 461, Gly 464, Phe 465, Ala 466, Asn 469, Lys 471, Val 472, His 576, and Tyr 577 assigned as flexible. Figure 6 shows the docking of RF on OmcA in two tested scenarios: (1) OmcA being rigid [Fig. 6(a)] and (2) the amino acids from the RF binding pocket close to heme 7 assigned as flexible [Fig. 6(b)]. The flexibility of Tyr 436 and Tyr 460 allows RF to position closer to heme 7 (RF-Feheme 7 = 6.3 Å) and attach with improved binding affinity (−9.2 kcal/mol vs −7.3 kcal/mol). It is interesting that in this configuration, the isoalloxazine ring of RF is sandwiched between two tyrosine amino acids (Tyr 436 and Tyr 460) in a similar manner as the FMN in NADPH-cytochrome P450 oxidoreductase44 found in eukaryotic cells.
Fig. 6.
Docking of RF to OmcA in the proximity of heme 7, where (a) the OmcA binding pocket for RF was assigned as flexible and (b) the OmcA binding for RF was assigned as rigid. The amino acids in cyan color show the position of amino acids in the rigid and blue-green their position after the flexible simulations. Heme 5 is colored red and RF is colored based on the element composition with N in blue, O in red, and C in gray.
The docking of RF to MtrC (Fig. 7) shows some similarities to the docking of RF on OmcA, such as comparable binding affinity and identical docking position; however, differences can also be observed (Table I). Based on the results of the docking simulations, three docking positions of RF to MtrC near hemes 1, 7, and 9 were considered as the possible sites for RF–MtrC interaction. RF–Feheme distance is the shortest between RF and heme 7, followed by heme 9 and heme 1. Heme 7 was experimentally proposed as a binding site for RF and FMN on MtrC.9 This binding position was suggested due to its location close to the disulfide bond of the protein, which appears to be important for the formation of flavin–cytochrome complex. Later, Breuer et al. used docking simulations to predict the interaction of MtrC and FMN, where the disulfide bond of MtrC was broken as suggested by Breuer et al. for the MtrC–flavin complex.43
Fig. 7.
Docking of RF to MtrC where the protein is represented with its secondary structure with the hemes colored in red. The RF is colored green.
Table I.
Parameters associated with the docking of RF and FMN to the outer membrane cytochromes OmcA and MtrC.
| RF | FMN | |||||
|---|---|---|---|---|---|---|
| OMC | Docking position | Heme | Affinity (kcal/mol) | RF–heme distance (Å) | Affinity (kcal/mol) | FMN–heme distance (Å) |
| OmcA | 1 | Heme 5 | −7.2 ± 0.1 | 5.8 ± 0.1 | −7.1 ± 0.3 | 6.6 ± 0.3 |
| 2 | Heme 7 | −7.2 ± 0.1 | 11.8 ± 0.5 | −7.7 ± 0.3 | 10.6 ± 0.3 | |
| MtrC | 1 | Heme 1 | −6.3 ± 0.8 | 8.9 ± 0.2 | −7.1 ± 0.4 | 8.6 ± 0.1 |
| 2 | Heme 4 | — | — | −7.0 ± 0.1 | 6.3 ± 0.1 | |
| 3 | Heme 7 | −6.2 ± 0.3 | 7.0 ± 0.7 | −7.3 ± 0.7 | 6.6 ± 0.3 | |
| 4 | Heme 9 | −6.6 ± 0.1 | 7.7 ± 0.2 | −7.6 ± 1.4 | 11.0 ± 0.3 | |
Breuer proposed heme 4 as most relevant docking position in this case, which agrees with our findings described later in the paper.
Edwards et al. demonstrated that one molecule of MtrC is capable of tightly binding a single riboflavin, which they attribute to the presence of a hydrophobic cleft on the surface of OmcA and MtrC implying hydrophobic OMC–flavin interactions. The results of the docking simulation of RF and FMN to MtrC show that the flavin most likely interacts with the hydrophilic amino acids not the hydrophobic ones [Figs. 8(b) and S4(b)]. Similar conclusions can be drawn for the OmcA–RF and OmcA–FMN complexes [Figs. 8(a) and S4(a)].
Fig. 8.
Hydrophilic (blue)/hydrophobic (red) amino acids distribution within the protein molecule of (a) OmcA and (b) MtrC aligned on the RF docking.
It should also be mentioned that in all docking models of RF to MtrC, the RF–Feheme distance is <10 Å, which is indicative of the ability of MtrC to reduce RF. Therefore, based on the parameters determined using the computational docking simulation of RF to MtrC, binding affinity, and RF–Feheme distance, there is no observable reason why MtrC could not reduce RF. Paquete et al. experimentally demonstrated that purified MtrC can reduce FMN and RF at comparable rates.28
It has also been proposed that the redox state of the hemes affects the structure of the flavin binding site and the affinity of the interaction.29 To test this hypothesis, we performed docking of RF to OmcA using AutoDock 4, which is, in contrast to AutoDock Vina, based on a different scoring function with an empirical free energy force field and assigns specific atomic charges for the atoms of the macromolecules and the ligands.39 When the docking of RF to OmcA was carried out with 0.00, 1.21,45 and 2.00 charge of the Fe in the hemes, no significant difference in the RF docking position and affinity was observed (data not shown). This also implies that the RF–OMCs interactions most likely are not based on electrostatics.
B. Docking of FMN
The docking simulations described in Sec. III A were repeated with FMN as the flavin of interest. FMN docking was explored with OmcA and MtrC, respectively (Fig. 9).
Fig. 9.
Docking of FMN to (a) OmcA and (b) MtrC, where the van der Waals surface of the protein is represented, FMN is colored green and some of the hemes are colored red.
The docking position close to heme 7 is of interest since Edwards et al. demonstrated the importance of the disulfide bond in FMN–MtrC complex.9 The authors suggested that three phenylalanine residues (Phe 438, Phe 504, and Phe 515) positioned close to heme 7 form the FMN binding pocket. As determined in this study, the amino acids in the FMN binding pocket of MtrC (Gln 250, Asn 251, Gln 254, Val 461, His 500, Thr 501, Glu 503, Phe 504, Glu 505, Ile 506, His 507, Lys 508, His 512, and Lys 563) include only one of the mentioned phenylalanine residues (Fig. 10).
Fig. 10.
Docking of FMN to MtrC in the proximity of heme 7, where the MtrC binding pocket for FMN was assigned as flexible. The amino acids in cyan color show the position of amino acids before the simulations and blue-green—position after the flexible simulations. Heme 7 is colored red and FMN is colored based on the element composition with N in blue, O in red, and C in gray.
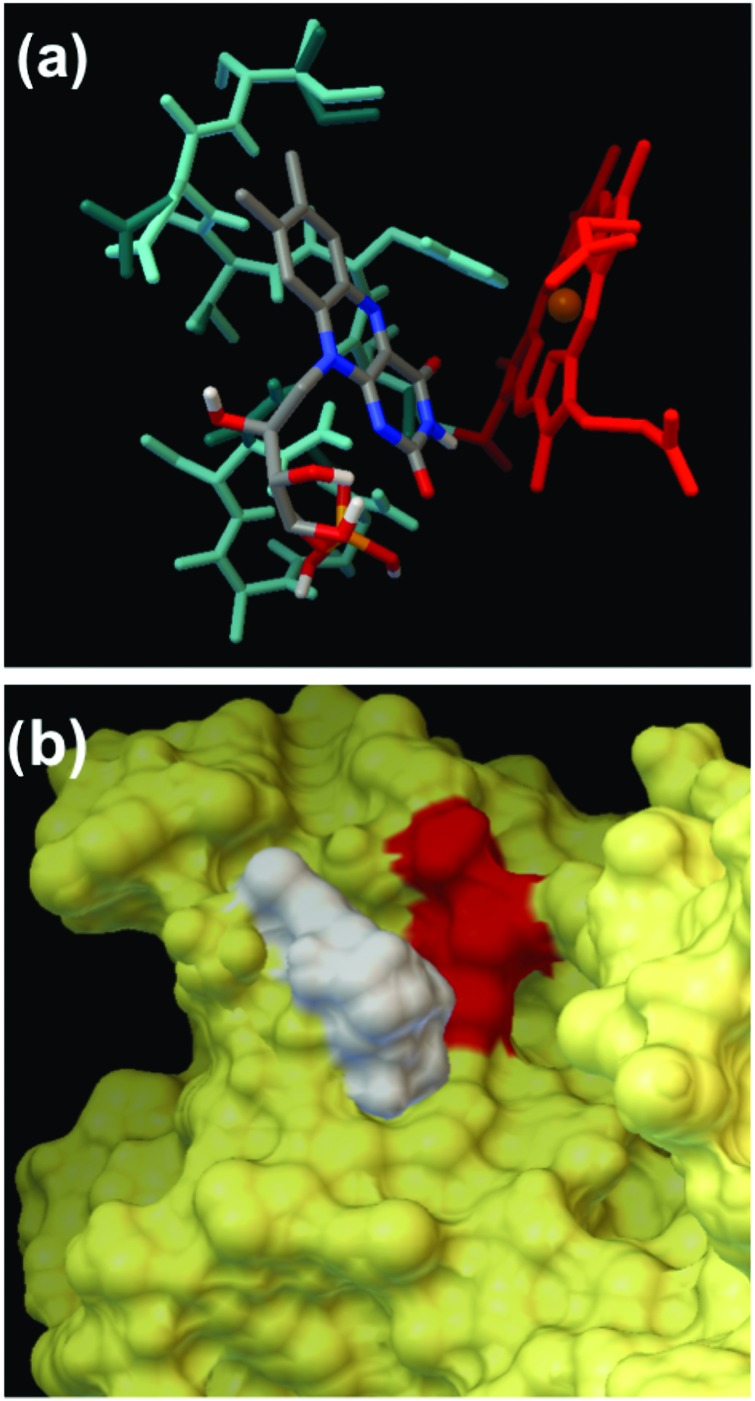
Table I summarizes the docking positions of FMN and RF, the FMN–Feheme and RF–Feheme distances, and the binding affinity of FMN and RF to OMCs at the given location of the protein molecule. The main difference between the docking of RF and FMN is in the docking position near heme 4 of FMN on MtrC, which was not observed with RF on MtrC. This docking position could be the key in the ability of MtrC to reduce FNM but not RF. The amino acids involved in the FMN binding pocket near heme 4 were determined to be Asn 234, Ala 235, Asn 236, Cys 257, Cys 285, His 286, Val 287, Asp 288, Ile 289, and Gly 296 (Fig. 11).
Fig. 11.
Docking of FMN to MtrC in the proximity of heme 4, where the MtrC binding pocket for FMN was assigned as flexible. (a) The rigid amino acids form the pocket are colored cyan, the flexible are blue-green, heme 4 is red and FMN is colored based on the element composition with N in blue, O in red and C in gray. (b) The van der Waals surface of MtrC is yellow, the FMN is colored white, and heme 4 is red.
Using 13P NMR, Paquete et al. demonstrated that FMN has two binding positions on OmcA and only one on MtrC.28 We can speculate that the two possible points of OmcA–FMN interactions are hemes 5 and 7 and the single binding site for FMN on MtrC is heme 4. The results shown here agree with the docking pose proposed by Breuer et al.,43 who suggested heme 4 as a “target” heme for FMN binding when the disulfide bond near heme 7 is broken. In presence of the disulfide bond though, Breuer et al. identify heme 2 as the docking site for FMN, which does not appear in our simulations.
Although the docking (association) of small ligands with proteins is usually reversible (dissociation) reaction, it can be expected that once the flavin docks on the cytochrome molecule, it is stably associated with it. This is the case with some flavoenzymes where the flavin remains tightly bound to the protein during reactions.27 To reveal the nature of the flavin binding to OMCs during the flavin reduction, the binding affinity of the fully oxidized FMN to MtrC was compared to the binding affinity of the semireduced form of FMN on the same cytochrome—near heme 4. The structure of the FMN semiquinone radical in water was first optimized using density functional theory and then used as an input for the docking simulations. The semiquinone of FMN docks in a similar manner as the completely oxidized form (Fig. 12). The binding affinity of MtrC and semiquinone FMN was found to be −6.4 ± 0.1 kcal/mol, which is identical to the binding affinity of the oxidized FMN in the proximity of heme 4. This result indicates that once the flavin is associated with the cytochrome, it is not released during or after the reduction process. The flavin attachment to the OMCs molecule agrees with the findings of Okamoto et al., suggesting that flavins can be electron relays in both directions: from the cytochrome toward the electron acceptor and vice versa.23 The strong flavin–OMC complex, though, disagrees with the experimentally determined Kd of flavin–OMC complexes, which indicate weak interactions.28
Fig. 12.
Docking of semiquinone FMN to MtrC in the proximity of heme 4, where the MtrC binding pocket for FMN was assigned as flexible. FMN is colored based on the element composition with N in blue, O in red, and C in gray, heme 4 is red and hemes 1, 2, 3, and 5 are colored light red.
IV. CONCLUSIONS
Molecular docking simulations have been used as a fast and simple tool for exploring the OMC–flavin interactions and predicting various possible docking positions of RF and FMN to OmcA and MtrC. Both RF and FMN bind to the surface of the proteins and do not penetrate the protein molecule. In addition, based on the shape of the protein surface where the flavins dock, it can be speculated that the interactions in between flavins and OMCs are based on stereospecificity for all docking positions.
The results of the simulations further suggest that OmcA could interact with both RF and FMN through hemes 5 and 7, with the binding affinity of RF–OmcA complex slightly higher than that of FMN-OmcA along with a shorter flavin-Feheme distance in the RF–OmcA complex. Both parameters indicate a stronger interaction of OmcA with RF than FMN. Three identical docking positions were identified for RF and FMN on MtrC close to hemes 1, 9, and 7 and one additional binding site in the proximity of heme 4 was found for FMN on MtrC. This additional binding site near heme 4 is a good candidate for FMN–MtrC interactions. Further experimental investigation is needed to confirm the importance of heme 4 in the FMN–MtrC associated electron transfer mechanism.
ACKNOWLEDGMENTS
Roddenberry Foundation Award NNX13AI31A and Bill and Melinda Gates Grant No. OPP1139954 financially supported the work. Part of the computational work was performed using the computational resources of EMSL, a national scientific user facility sponsored by the Department of Energy's Office of Biological and Environmental Research and located at Pacific Northwest National Laboratory, NERSC, supported by the Office of Science of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231, and CNMS, sponsored at Oak Ridge National Laboratory by the Scientific User Facilities Division, Office of Basic Energy Sciences, U.S. Department of Energy.
References
- 1. Yang Y., Xua M., Guo J., and Sun G., Process Biochem. 47, 1707 (2012). 10.1016/j.procbio.2012.07.032 [DOI] [Google Scholar]
- 2. von Canstein H., Ogawa J., Shimizu S., and Lloyd J. R., Appl. Environ. Microbiol. 74, 615 (2008). 10.1128/AEM.01387-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Pirbadian S. et al. , Proc. Natl. Acad. Sci. 111, 12883 (2014). 10.1073/pnas.1410551111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Nakano C. M., Byun H. S., Ma H., Wei T., and El-Naggar M. Y., Comput. Phys. Commun. 193, 1 (2015). 10.1016/j.cpc.2015.03.009 [DOI] [Google Scholar]
- 5. Bond D. R., Strycharz-Glaven S. M., Tender L. M., and Torres C. I., ChemSusChem 5, 1099 (2012). 10.1002/cssc.201100748 [DOI] [PubMed] [Google Scholar]
- 6. Feliciano G. T., Steidl R. J., and Reguera G., Phys. Chem. Chem. Phys. 17, 22217 (2015). 10.1039/C5CP03432A [DOI] [PubMed] [Google Scholar]
- 7. Coursolle D., Baron D. B., Bond D. R., and Gralnick J. A., J. Bacteriol. 192, 467 (2010). 10.1128/JB.00925-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ross D. E., Ruebush S. S., Brantley S. L., Hartshorne R. S., Clarke T. A., Richardson D. J., and Tien M., Appl. Environ. Microbiol. 73, 5797 (2007). 10.1128/AEM.00146-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Edwards M. J. et al. , Sci. Rep. 5, 11677 (2015). 10.1038/srep11677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Edwards M. J., Fredrickson J. K., Zachara J. M., Richardson D. J., and Clarke T. A., Biochem. Soc. Trans. 40, 1181 (2012). 10.1042/BST20120132 [DOI] [PubMed] [Google Scholar]
- 11. Edwards M. J. et al. , FEBS Lett. 588, 1886 (2014). 10.1016/j.febslet.2014.04.013 [DOI] [PubMed] [Google Scholar]
- 12. Richardson D. J. et al. , Mol. Microbiol. 85, 201 (2012). 10.1111/j.1365-2958.2012.08088.x [DOI] [PubMed] [Google Scholar]
- 13. TerAvest M. A., Zajdel T. J., and Ajo-Franklin C. M., ChemElectroChem 1, 1874 (2014). 10.1002/celc.201402194 [DOI] [Google Scholar]
- 14. Clarke T. A. and Edwards M. J., “Crystal structure of the outer membrane decaheme cytochrome OmcA,” Protein Data Bank (2014).
- 15. Hartshorne R. S., Jepson B. N., Clarke T. A., Field S. J., Fredrickson J., Zachara J., Shi L., Butt J. N., and Richardson D. J., J. Biol. Inorg. Chem. 12, 1083 (2007). 10.1007/s00775-007-0278-y [DOI] [PubMed] [Google Scholar]
- 16. Firer-Sherwood M., Pulcu G. S., and Elliott S. J., J. Biol. Inorg. Chem. 13, 849 (2008). 10.1007/s00775-008-0398-z [DOI] [PubMed] [Google Scholar]
- 17. Breuer M., Zarzycki P., Blumberger J., and Rosso K. M., J. Am. Chem. Soc. 134, 9868 (2012). 10.1021/ja3027696 [DOI] [PubMed] [Google Scholar]
- 18. Breuer M., Rosso K. M., and Blumberger J., Proc. Natl. Acad. Sci. 111, 611 (2014). 10.1073/pnas.1316156111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kotloski N. J. and Gralnick J. A., mBio 4, e00553-12 (2013). 10.1128/mBio.00553-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Marsili E., Baron D. B., Shikhare I. D., Coursolle D., Gralnick J. A., and Bond D. R., Proc. Natl. Acad. Sci. 105, 3968 (2008). 10.1073/pnas.0710525105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wang V. B. et al. , Electrochem. Commun. 41, 55 (2014). 10.1016/j.elecom.2014.01.025 [DOI] [Google Scholar]
- 22. Nguyen H. D., Renslow R., Babauta J. T., Ahmed B., and Beyenal H., Sens. Actuators, B 161, 929 (2012). 10.1016/j.snb.2011.11.066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Okamoto A., Hashimoto K., and Nealson K. H., Angew. Chem. 53, 10988 (2014). 10.1002/amie.201407004 [DOI] [PubMed] [Google Scholar]
- 24. Okamoto A., Hashimoto K., Nealson K. H., and Nakamura R., Proc. Natl. Acad. Sci. 110, 7856 (2013). 10.1073/pnas.1220823110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Okamoto A., Kalathil S., Deng X., Hashimoto K., Nakamura R., and Nealson K. H., Sci. Rep. 4, 5628 (2014). 10.1038/srep05628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Okamoto A., Saito K., Inoue K., Nealson K. H., Hashimoto K., and Nakamura R., Energy Environ. Sci. 7, 1357 (2014). 10.1039/C3EE43674H [DOI] [Google Scholar]
- 27. Joosten V. and van Berkel W. J., Opin. Chem. Biol. 11, 195 (2007). 10.1016/j.cbpa.2007.01.010 [DOI] [PubMed] [Google Scholar]
- 28. Paquete C. M., Fonseca B. M., Cruz D. R., Pereira T. M., Pacheco I., Soares C. M., and Louro R. O., Front. Microbiol. 5, 318 (2014). 10.3389/fmicb.2014.00318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Okamoto A., Nakamura R., Nealson K. H., and Hashimoto K., ChemElectroChem 1, 1808 (2014). 10.1002/celc.201402151 [DOI] [Google Scholar]
- 30. Xu S., Jangir Y., and El-Naggar M. Y., Electrochim. Acta 198, 49 (2016). 10.1016/j.electacta.2016.03.074 [DOI] [Google Scholar]
- 31. Cornejo J. A., Lopez C., Babanova S., Santoro C., Artyushkova K., Ista K. L., Schuler A. J., and Atanassov P., J. Electrochem. Soc. 162, H597 (2015). 10.1149/2.0271509jes [DOI] [Google Scholar]
- 32. Malinauskas A., Chemija 19, 1 (2008). [Google Scholar]
- 33. Roy J. N., Garcia K., Luckarift H., Falase A., Cornejo J., Babanova S., Schuler A. J., Johnson G., and Atanassov P., J. Electrochem. Soc. 160, H866 (2013). 10.1149/2.001401jes [DOI] [Google Scholar]
- 34. Hong G. and Pachter R., J. Phys. Chem., B 120, 5617 (2016). 10.1021/acs.jpcb.6b03851 [DOI] [PubMed] [Google Scholar]
- 35. Babanova S., Matanovic I., Chavez M. S., and Atanassov P., J. Am. Chem. Soc 137, 7754 (2015). 10.1021/jacs.5b03053 [DOI] [PubMed] [Google Scholar]
- 36. Matanovic I., Babanova S., Chavez M. S., and Atanassov P., J. Phys. Chem. B 120, 3634 (2016). 10.1021/acs.jpcb.6b01616 [DOI] [PubMed] [Google Scholar]
- 37. Trott O. and Olson A. J., J. Comput. Chem. 31, 455 (2010). 10.1002/jcc.21334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Morris G. M., Huey R., Lindstrom W., Sanner M. F., Belew R. K., Goodsell D. S., and Olson A. J., J. Comput. Chem. 30, 2785 (2009). 10.1002/jcc.21256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Huey R., Morris G. M., Olson A. J., and Goodsell D. S., J. Comput. Chem. 28, 1145 (2007). 10.1002/jcc.20634 [DOI] [PubMed] [Google Scholar]
- 40. Sjulstok E., Olsen J. M. H., and Solovyov I. A., Sci. Rep. 5, 18446 (2015). 10.1038/srep18446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Frisch M. J. et al. , Gaussian 09, Revision B.01, Gaussian, Inc., Wallingford, CT, 2009. [Google Scholar]
- 42. Sanner M. F., J. Mol. Graphics Mod. 17, 57 (1999). [PubMed] [Google Scholar]
- 43. Breuer M., Rosso K. M., and Blumberger J., Biophys. J. 109, 2614 (2015). 10.1016/j.bpj.2015.10.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zhao Q. et al. , Protein Sci. 8, 298 (1999). 10.1110/ps.8.2.298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Autenrieth F., Tajkhorshid E., Baudry J., and Luthey-Schulten Z., J. Comput. Chem. 25, 1613 (2004). 10.1002/jcc.20079 [DOI] [PubMed] [Google Scholar]
- 46.See supplementary material at http://dx.doi.org/10.1116/1.4984007 E-BJIOBN-12-333702 for a detailed description of the docking simulations procedure. It contains results from the modeling of the interactions between RF and MtrC as well as hydrophilic/hydrophobic amino acids distribution within the protein molecule of OmcA and MtrC aligned in the FMN docking.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- See supplementary material at http://dx.doi.org/10.1116/1.4984007 E-BJIOBN-12-333702 for a detailed description of the docking simulations procedure. It contains results from the modeling of the interactions between RF and MtrC as well as hydrophilic/hydrophobic amino acids distribution within the protein molecule of OmcA and MtrC aligned in the FMN docking.