Abstract
Deriving the conformation of adsorbed proteins is important in the assessment of their functional activity when immobilized. This has particularly important bearings on the design of contemporary and new encapsulated enzyme-based drugs, biosensors, and other bioanalytical devices. Solid-state nuclear magnetic resonance (NMR) measurements can expand our molecular view of proteins in this state and of the molecular interactions governing protein immobilization on popular biocompatible surfaces such as silica. Here, the authors study the immobilization of ubiquitin on the mesoporous silica MCM41 by NMR and other techniques. Protein molecules are shown to bind efficiently at pH 5 through electrostatic interactions to individual MCM41 particles, causing their agglutination. The strong attraction of ubiquitin to MCM41 surface is given molecular context through evidence of proximity of basic, carbonyl and polar groups on the protein to groups on the silica surface using NMR measurements. The immobilized protein exhibits broad peaks in two-dimensional 13C dipolar-assisted rotational resonance spectra, an indication of structural multiplicity. At the same time, cross-peaks related to Tyr and Phe sidechains are missing due to motional averaging. Overall, the favorable adsorption of ubiquitin to MCM41 is accompanied by conformational heterogeneity and by a major loss of motional degrees of freedom as inferred from the marked entropy decrease. Nevertheless, local motions of the aromatic rings are retained in the immobilized state.
I. INTRODUCTION
An effective immobilization of enzymes and other functional biomolecules on submicron material surfaces that preserves functionality is key to the design of modern biosensors, biocompatible implantable devices, and bioanalytical tools.1–3 Numerous biotechnological constructs have been reported in recent years, which couple proteins as reporters of biological conditions to substrates that translate the biological information into electronic or spectroscopic analytical data.4 The success of future devices such as nanometric glucose sensors requires the development of better and higher-fidelity tools5 that (1) characterize the activity of the adsorbed enzyme and the mechanism of its operation, (2) determine the biomolecular structure in the finest possible detail, in situ, immobilized on the material, (3) elucidate the atomic details of the biomaterial interface that holds the construct together, and (4) track the kinetics and thermodynamics of immobilization faster and with higher sensitivity.
Atomic-force microscopy (AFM),6,7 attenuated total reflection Fourier transform infrared spectroscopy (ATR-FTIR),8 circular dichroism,9,10 surface-enhanced Raman spectroscopy,11 and transmission electron microscopy12 have proven very useful in studying the structure of immobilized enzymes and protein–material interfaces.13 Electrochemical techniques, quartz crystal microbalance,14 adsorption isotherms, Langmuir–Blodgett films,15 and isothermal titration calorimetry (ITC),16,17 as well as other techniques, have provided viable information both on the functionality of immobilized enzymes and on the energetics and rates of binding to the surface. In addition, techniques such as sum frequency generation,18–20 time-of-flight secondary ion mass spectrometry,21 and ATR-FTIR (Ref. 22) have successfully provided with important insights about the interfaces, i.e., the groups on the material surfaces and on the biomolecules involved in binding and the structure of these interfaces.
The effort to achieve a detailed atomic picture of these interfaces, including the structure of the protein and the material–surface adsorption site, is ongoing. To this end, tools for computational analysis that accommodate realistic views of the material surface for the prediction of protein structure are being developed rapidly.23–27 They are practically the only means for generating a successful detailed model of the biomaterial interface. Among the experimental techniques that can potentially address this challenge rapidly and is not yet considered is modern cryogenic transmission electron tomography.28 The other technique which can provide high-resolution insights into the interface formed between the residues on a protein and chemical groups on surfaces of materials is solid-state NMR spectroscopy.
In the last two decades, solid-state NMR has advanced from demonstrating protein–structure elucidation29–31 to tackling intractable structural biology challenges.32–35 However, a high-resolution structure of a protein bound to the surface of any material has not been reported yet, presumably due to the inherent heterogeneity of adsorption sites and biomolecule conformation adopted. Nevertheless, refined structural insights on proteins adsorbed onto solid surfaces23,36–41 or embedded inside materials42–46 were obtained using the technique.47 Two-dimensional (2D) magic-angle-spinning (MAS) NMR techniques were employed more recently in studying enamel-related proteins bound to hydroxyapatite crystallites,40 diatom frustule-related peptides,42,48–50 and entrapped enzymes43–45 in an effort to gather the necessary structural constraints to determine the structure of the immobilized protein.
Here, we combine macroscopic adsorption measurements with surface-analysis techniques and 2D MAS NMR measurements to study the immobilization of ubiquitin on the mesoporous silica material MCM41. This material with its network of nanopores which are ∼4 nm in diameter poses some interesting adsorbent characteristics owing to the commensurate pore diameter with ubiquitin's hydrodynamic diameter.51 The protein binds mainly through electrostatic forces to the porous surface of MCM41. However, from direct measurements of the biomaterial interface, it is apparent that hydrophobic and polar residues also participate in surface binding. The structural characteristics of adsorbed ubiquitin are reported. Preliminary evidence for dynamic sidechains in the immobilized state is also discussed. The NMR data are tied up to the thermodynamic measurements giving molecular details on the groups in charge of the protein's affinity to the silica surfaces.
II. RESULTS
Ubiquitin was adsorbed onto MCM41 at ambient temperature (25 °C) by mixing a solution of the protein in sodium-acetate buffer (100 mM, pH 5) with a suspension of MCM41 in the same buffer for 2 h, followed by centrifugation to collect the precipitate. The precipitate was washed with water twice. The supernatant and the content of washes were analyzed to infer the amount of protein in solutions and from that the amount adsorbed. For NMR measurements, the solid complex was dried by evaporation at 25 °C for 6 h.
A. Surface characteristics of ubiquitin-MCM41
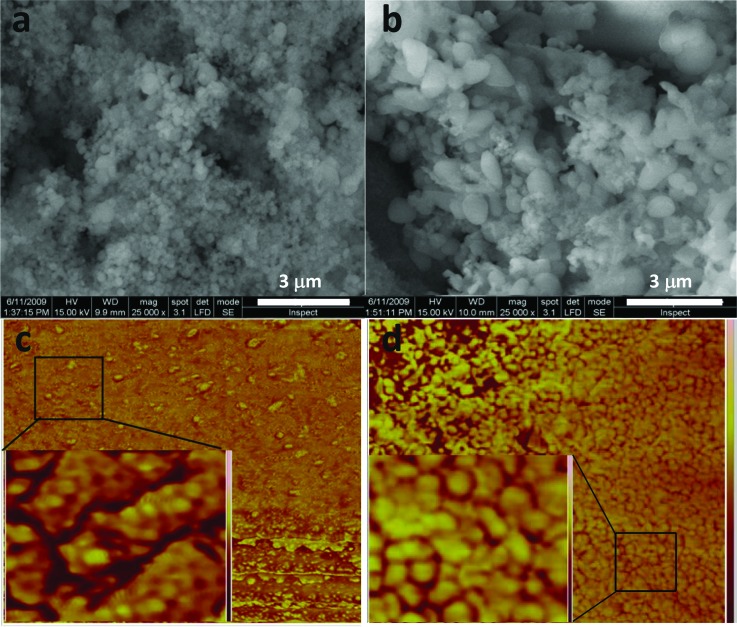
SEM and AFM micrographs of MCM41-immobilized ubiquitin were recorded in acetate buffer (100 mM, pH 5) and in double-distilled water at monolayer concentrations of the protein using values derived from the adsorption isotherm results shown below. The SEM measurements of MCM41 and ubiquitin-bound MCM41 are compared in Fig. 1. The values of the spherical particles in the micrograph of MCM41, with an average diameter of 300 (±50) nm, are in accordance with previous values reported for this material.52 In the micrograph of the ubiquitin-MCM41, the prolate particles are 2–3 times longer than those observed for MCM41. Only physical mixing of protein and silica in the buffer solution was performed, which suggests that ubiquitin molecules bind particles strongly and cause their agglutination. Energy dispersive x-ray spectroscopy (EDAX) analysis of the micrographs confirms the presence of the protein on the surface of the silica in ubiquitin-MCM41 through the detection of carbon and nitrogen at an atom% ratio of 5.5 (see Table S1),81 which is the same ratio of the corresponding atoms in ubiquitin. Tapping-mode AFM phase images, recorded with a high-resolution Si3N4 tip to probe the surface of the two samples, are also shown in Fig. 1. The topology images of these regions are shown in Fig. S1. The MCM41-phase image exhibits low contrast. The close-up image (inset) shows darker regions, which reflect surface roughness and are attributed to the porosity of the material. Ubiquitin-MCM41, on the other hand, exhibits a high-contrast phase image with bumps associated with the protein molecules located on the exposed surface. During ubiquitin-MCM41 scanning, damped tip oscillations were observed, with larger drag forces than those in MCM41 (data not shown), confirming that protein molecules cover the particle's outer surface.
Fig. 1.
SEM images of MCM41 particles (a) before and (b) after ubiquitin immobilization. Noncontact tapping AFM phase 1×1 μm2 images of (c) MCM41 surface and (d) ubiquitin-MCM41 surface. Insets are magnified 175 × 175 nm2 images of the areas in the squares.
The preliminary evidence of the favorable interaction of ubiquitin with the MCM41 surface can be further expanded to explore which residues and groups, both on the protein and on the silica surface, contribute to it. Interactions of polar and charged groups on the protein with complementary groups, i.e., silanols (-Si-OH), protonated silanols (Si-OH2+) and anionic Si-O− groups immediately come to mind. However, other weaker interactions of hydrophobic and aromatic groups with these surface groups and with the less-polar siloxane groups may have non-negligible contributions as well. NMR experiments that can track atoms in the protein that are close to silica surface groups are very useful for breaking down the various contributions to the overall interaction of the biomolecule with surface sites.
B. Measurement of protein-side-chain proximity to MCM41 surface by NMR
The proximity of atoms in the protein to the MCM41 surface is inferred from the 13C{29Si} transferred-echo double resonance (TEDOR) magnetization transfer experiment.53 The experiment starts with a 1H-29Si cross-polarization (CP) step, which excites the silicon nuclear magnetization by adjacent hydrogen nuclei, then a transfer of magnetization between the silicon and carbon nuclei via magnetic dipole–dipole interactions followed by recording of the resultant carbon magnetization. The TEDOR spectrum of 15N-,13C-ubiquitin-MCM41, therefore, only shows carbon atoms in ubiquitin that are close to silicon-bearing groups on the MCM41 surface. It is typically compared to the standard 13C CP spectrum (recorded using 1H-13C CP) to indicate which carbon species out of the entire protein are involved in the interaction.
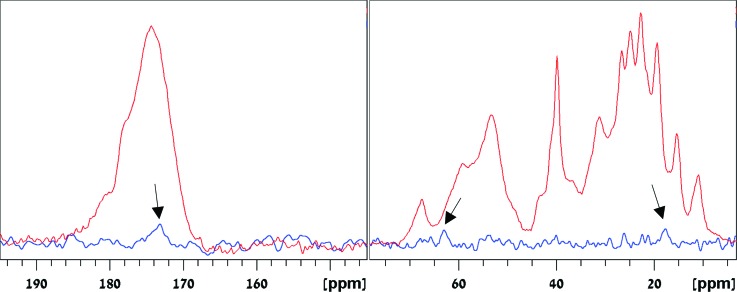
The 13C{29Si} TEDOR spectrum of the sample, recorded using a dipolar recoupling period of 4.6 ms whereby 29Si transverse magnetization is converted into antiphase magnetization, and a second dipolar recoupling period of 4.8 ms whereby antiphase magnetization is converted into in-phase 13C magnetization, is shown with the 13C CP spectrum in Fig. 2. Three peaks of carbon species coupled to silicon can be identified. An aliphatic sidechain carbon peak at 18 ppm, a Cα,β carbon peak at 64 ppm, and a carbonyl carbon peak at 173 ppm are observed. The 18-ppm peak suggests the proximity of Ile Cγ2, Met Cε, or Ala Cβ carbon to the silica surface. The 64-ppm peak is ascribed to carbons on the hydroxyl-bearing residues, that is, Ser Cβ or Thr Cα. The peak at 173 ppm can be assigned to any carbonyl group in the protein; however, carbonyls in the sidechain amide groups of Asn and Gln are more likely to be accessible for binding. Based on the signal intensity of the individual lines and the dephasing and recoupling times used, these carbons are estimated to be at distances greater than 4.9 Å from the surface silicon atoms. It cannot be overruled that acquiring more sensitive data would unravel additional carbons (e.g., in the region between 20 and 30 ppm) involved in interactions with the silica. The intensity of the lines in the TEDOR spectrum is less than 0.2%–0.5% of the signal in the CP spectrum. It is only 3–10 times smaller than the maximum signal expected, which is 2.35% of the signal in the CP experiment. The latter value 2.35% is derived by multiplying the 4.7% natural abundance of 29Si by 0.5 since only half of the 13C antiphase magnetization produced by the pair of 90° pulses is converted into observable 13C signal in the TEDOR experiment.
Fig. 2.
13C{29Si} TEDOR spectrum of 15N-,13C-ubiquitin-MCM41, recorded using dephasing and recoupling mixing times of 4.6 and 4.8 ms, respectively. The arrows mark the carbonyl peak (173.2 ppm) and some aliphatic carbons that are in proximity to the Si atoms on the MCM41 surface. Left and right shows respective carbonyl and aliphatic regions in the spectrum, respectively. For reference, the 13C cross polarization MAS spectrum of 15N-,13C-ubiquitin-MCM41 is also shown.
Notably, carbons from hydrophobic and polar residues are found to be in proximity to the MCM41 surface in the TEDOR measurements. To obtain further insight into the interactions involved in the adsorption of the protein to MCM41, we collected a complete thermodynamic profile of the process.
C. Analysis of the energies involved in ubiquitin immobilization on MCM41
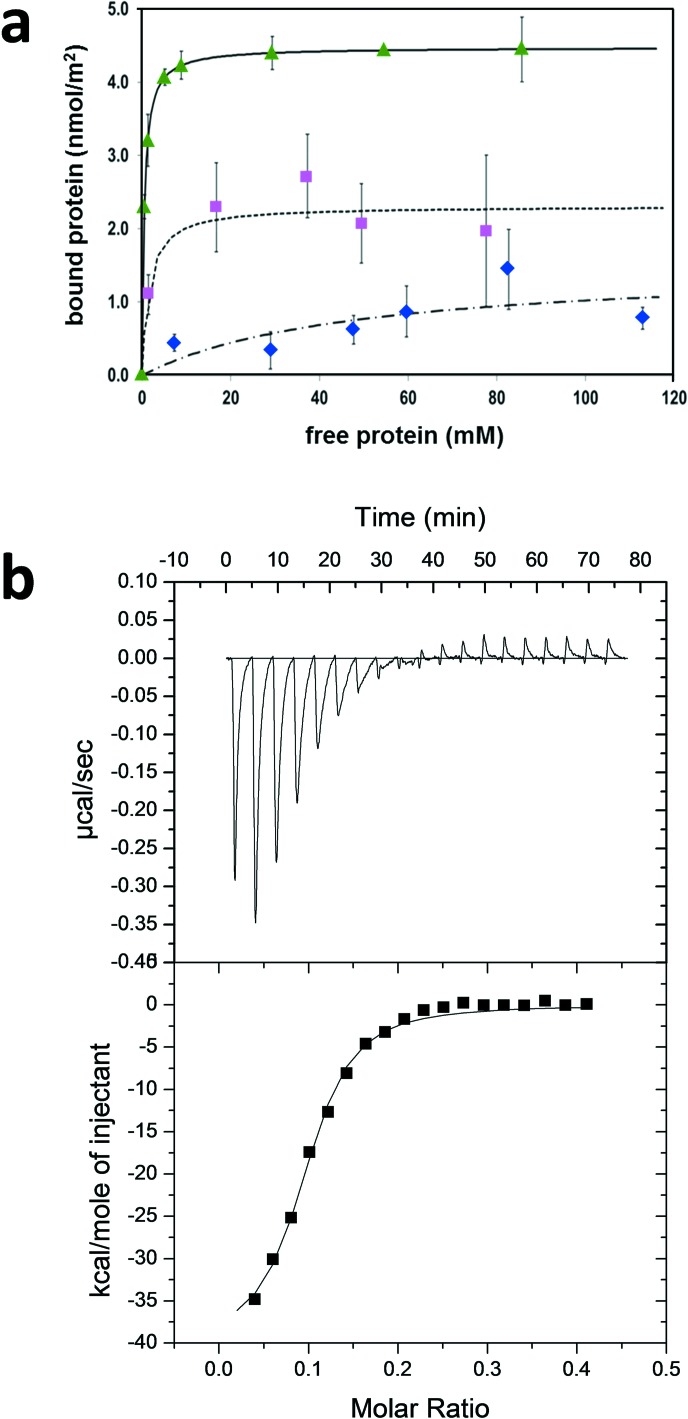
The apparent adsorption constant and free energy of ubiquitin binding to MCM41 were derived from adsorption isotherm measurements recorded at various nondenaturing pH values and under various ionic-strength conditions. The resulting isotherms, recorded at a fixed sodium acetate ionic strength of 100 mM, showing the bound protein amount normalized to the MCM41 surface area plotted versus the concentration of free protein at equilibrium at pH 5.0, 7.5, and 10.0, are shown in Fig. 3. Additional standard Langmuir-model fits to the experimental data are shown as solid, dotted, and dashed lines in the figure. As the data fit well to a single set of independent sites and adsorption saturation is reached, it is evident that the protein is adsorbed in a single layer.
Fig. 3.
(a) Adsorption isotherms showing ubiquitin adsorption to MCM41 at 25 °C. Data recorded at pH 5.0 (green triangle), pH 7.5 (pink square), and pH 10.0 (blue diamond) are shown. Parameters from Langmuir-based fits (lines) are given in Table I. (b) ITC measurement of ubiquitin binding to MCM41 suspended in acetate buffer (pH = 5.0). (Top) Time-dependent trace of ITC measurement showing the heat released upon ubiquitin binding to MCM41. (Bottom) Integrated heat produced in each injection (▪) and fit of the titration to a single set of equivalent adsorption sites (solid line).
The apparent equilibrium adsorption constants Kads and maximal coverages Nmax extracted from the isotherms are summarized in Table I. The binding affinity to the MCM41 surface is maximal under acidic conditions and drops by a factor of ∼3 at pH 7.5 and by a factor of ∼100 at pH 10.0. Similarly, ubiquitin coverage at pH 5.0 is maximal and decreases by a factor of 2 at pH 7.5 and a factor of 3 at pH 10.0. These trends can be qualitatively understood in terms of the net charge change on the protein (pI = 6.7) and on the MCM41 surface [point-of-zero-charge (PZC) = 3.2] at different pH values.
Table I.
pH dependency of equilibrium binding constants and surface coverage of ubiquitin to MCM41.
| pH | Kads (M−1) | Nmax (nmol/m2) |
|---|---|---|
| 5.0 | (1.94 ± 1.83) × 106 | 4.47 ± 0.05 |
| 7.5 | (6.78 ± 0.10) × 105 | 2.31 ± 0.03 |
| 10.0 | (2.08 ± 0.03) × 104 | 1.51 ± 0.49 |
At pH 5.0, the calculated charge on the protein is +1.90, and the charge on the silica surface is negative based on its PZC. Under electrostatic complementarity conditions, binding is very favorable, leading to the high Kads and coverage values recorded. At pH 7.5, the charges on the protein sum up to a small net negative charge (−0.01), lowering the binding affinity to the negatively charged surface to a third of its value at pH 5.0. Nevertheless, the local positive charge on the cationic four Arg and seven Lys residues is retained at this pH. Hence, their attraction to the anionic surface helps keep the coverage from dropping similarly. At pH 10.0, the Lys residues are mostly uncharged and the protein's net charge changes to −2.6. The repulsive interaction with the anionic MCM41 surface causes the Kads to drop by 2 orders of magnitude; yet, the coverage is only reduced by a factor of 3. This is attributed to the effective interaction of the hydrophobic residues with the silica surface, most probably with the siloxane groups which are less polar than silanols.
Additional adsorption experiments at constant pH 5.0, with decreasing ionic strength of the acetate buffer, result in the gradual increase in the affinity and surface coverage (data not shown). These results assert the electrostatic nature of the interactions that give rise to the strong binding observed at this pH value.
Separate enthalpic and entropic contributions to Gibbs free energy of adsorption of ubiquitin to MCM41 at pH 5.0 were deduced from ITC measurements. Ubiquitin concentration and MCM41 suspension used in the experiment are detailed in Sec. V. Starting from a dilute protein concentration in the ITC cell in the first injection, a strong signal of heat released is observed, producing a smooth curve with a high signal-to-noise ratio. The normalized heat change, calculated from integrating the heat signal in each injection, is plotted versus the molar ratio or stoichiometry of total protein concentration divided by the total surface site concentration.17 As more protein in acetate buffer was injected into the cell containing MCM41 suspension in the same buffer, the normalized heat signal decreased, as shown in Fig. 3 (middle). The integrated heat is plotted versus the molar ratio at the bottom of Fig. 3, after subtracting the heat produced per injection in a control experiment titrating the protein into the acetate-buffer solution. This subtraction eliminates the possibility that the enthalpy of binding observed is due to ubiquitin–ubiquitin interactions.
The heat change was fitted to a single set of binding sites, a model that was shown before to be equivalent to the Langmuir model of an independent set of adsorption sites.17 The excellent fit without divergent changes in the heat release confirms that ubiquitin adsorption to MCM41 follows a simple Langmuir behavior, reaching monolayer coverage at the last few injections, where heat changes become negligible. A major and discontinuous drop in the heat release is expected in the case of multilayer adsorption owing to the repulsive nature of the interaction between the positively charged protein molecules at pH 5.0. Repulsion between protein molecules impedes any accumulation of new protein layers.
The high apparent enthalpy of adsorption, ΔHads= −40.1 (±0.2) kcal mol−1, indicates strong attractive charge–charge or H-bond interactions between the protein and the silica surface. It is balanced by significant entropy decrease, ΔSads = −104 (±12) cal mol−1 K−1, due to loss of motional degrees of freedom upon protein immobilization. This net free energy change, ΔGads = 9.1 (±0.2) kcal mol−1, is, therefore, more moderate, with the associated equilibrium constant Kads = 3.3 (±0.7) × 106 M−1. This adsorption constant is consistent with the value derived from the adsorption isotherm measurements (see Table I).
D. Measurement of adsorbed ubiquitin structural characteristics
Proteins have been observed before to undergo conformational changes due to binding to silica.54–56 The strong binding to the MCM41 surface may affect the ubiquitin structure. We ran 2D NMR experiments on the complex of uniformly 13C-,15N-enriched protein and MCM41 (15N-,13C-ubiquitin-MCM41) to examine its structure. Experiments that indicate the proximity between unlike atoms such as protons and carbons are called heteronuclear-correlation (HETCOR) experiments. Experiments reporting the proximity between similar atoms (e.g., carbon–carbon) are called homonuclear-correlation (HOMCOR) experiments. These experiments give rise to 2D spectra which are basically 2D correlation maps where contours (peaks) represent the magnetization exchange between pairs of atoms that are close in space. The x and y coordinates of each peak are the chemical shifts of the two interacting atoms, and its intensity, that is, the number of contours, reflects, in general terms, how close the atoms are.
The 2D 1H-13C- and 1H-15N-HETCOR spectra of 15N-,13C-ubiquitin-MCM41, plotted on a common 1H axis, are shown in Fig. S2. They were recorded using homonuclear 1H decoupling in t1 to better resolve different proton lines. The spectra show predominant peaks/correlations between protons and carbons on the same residue in the protein. The broad peaks observed are typically implicated with the protein adopting multiple conformations when bound to MCM41. The 1H-15N- HETCOR spectrum shows peaks of sidechain nitrogen atoms which are unique. Apart from the Arg Nη (72 ppm) peak with directly bound Hη at 7.0 ppm and adjacent to the Hε proton at 7.7 ppm, another peak with a proton at 8.1 ppm appears. Similarly, Arg Nε (85 ppm) has a peak with a proton at the same chemical shift. The proton cannot be associated with the backbone amide proton of that residue, as it is too distant for efficient magnetization transfer, and is better ascribed to a protonated silanol group (Si-OH2+) on the surface of the MCM41.57,58 The Lys sidechain amine (32.9 ppm) shows a cross-peak with directly bound 1H (7.3 ppm) and with a proton at 8.2 ppm. The latter cross-peak, again, is unlikely to be the result of transfer from the intraresidue amide proton which is at a distance of 6.0–6.5 Å and is associated, as before, with a protonated silanol group on the surface.
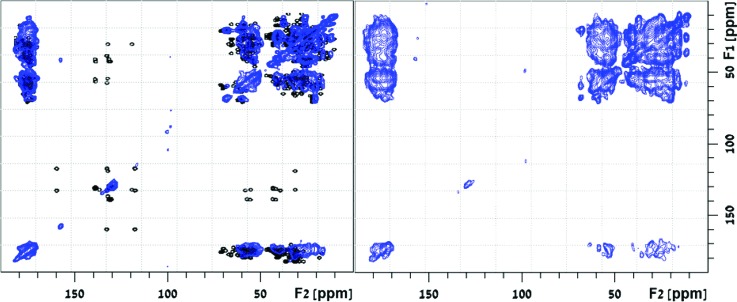
2D carbon–carbon HOMCOR experiments have been used earlier to obtain distance constraints between pairs of carbon atoms and incorporated in protein-folding programs to elucidate the structure of crystalline proteins.33–35,59 2D 13C DARR measurements60 were recorded here on 15N-,13C-ubiquitin-MCM41 at 298 K and 11 kHz spinning, using carbon–carbon mixing times of 90.9 and 272.7 ms. The correlation spectra shown in Fig. 4 exhibit cross-peaks with line widths ca. 1.5–3.0 ppm, owing to the dispersion of conformations that accompanies the adsorption.
Fig. 4.
2D 13C DARR spectra recorded at mixing times of 90.9 ms (left) and 272.7 ms (right) of [U-13C-,15N] ubiquitin bound to MCM41. Overlaid on the left spectrum is a simulated DARR spectrum (empty circles), computed based on two-bond intraresidue cross-peaks using chemical shift information deposited in the biological magnetic resonance bank (entry 7111) and 3D structure deposited in the protein databank (1ubq).
To aid in the analysis of the 2D correlation map, a simulated DARR spectrum was constructed using the “Peakr” program.61 The simulation uses structural data of crystalline ubiquitin deposited in the protein data bank (PDB code—1ubq),62 chemical shift predictions with the shiftX2 program63 and pH corrections to chemical shift values. It shows intraresidue peak correlations, by limiting the computation to through-space carbon–carbon magnetization transfers between nuclei that are up to 3.0 Å apart. The simulated spectrum (empty circles) is overlaid on the experimental DARR spectrum measured at a mixing time of 90.9 ms (Fig. 4, left). The broad cross peaks in the experimental spectrum and high ambiguity of peak assignment hamper the typical resonance assignment procedure. Nevertheless, the good overlap of the computed fingerprint spectrum without a substantial resonance shift in the adsorbed ubiquitin suggests that the backbone did not undergo drastic structural changes. A residual shift of 1.6 ppm is recorded for many of the cross peaks in the spectrum relative to the computed ones. For comparison, the use of methanol as solvent induces a conversion of the native form of ubiquitin to the A-form structure, manifested by replacement of the two c-terminal β-strands by a long α-helix.64,65 Such a massive transformation would result in a downfield shift of nearly half of the Cα carbons in the protein. The good overlap of experimental and computed data clearly shows that such structural transformation does not occur upon interaction with the silica surfaces. It does not, however, exclude the possibility of some more moderate conformational changes.
The cross-peaks of the Phe and Tyr ring carbons at 115–140 ppm with adjacent ring carbons and other sidechain carbons at 30–60 ppm seen in the simulated spectrum are missing in the experimental spectra. In comparison, in crystalline ubiquitin, these cross-peaks were observed, and the complete assignment of the carbons in the three aromatic residues in ubiquitin was obtained in certain cases.66 The absence of these peaks in the MCM41-adsorbed ubiquitin suggests that the aromatic rings have some mobility in the immobilized protein. Interestingly, one ring cross-peak is observed (at 157, 40 ppm) between Tyr59 Cζ and an adjacent aliphatic carbon, although no other Tyr-ring correlations can be seen. This Cζ is a hydroxyl-bearing carbon, noted before to have a major contribution to the stability of the loop in the Lys48-Gln62 region by forming an H-bond with the amide on Glu51.62 A plausible assignment of the coupled carbon at 40 ppm is, therefore, the Cγ on Glu51. This assignment suggests that the H-bond, which serves as an important tertiary constraint, is retained in the adsorbed protein. The exclusive appearance of this cross-peak in the whole ring can be explained based on the axial ring motion in the Cγ–Cζ direction. Such motion can, in principle, leave the Cζ at a fixed location with respect to the carbon to which it is coupled, that is, the Cγ on Glu51, while other ring carbons are in motion due to the ring rotation. Elaborate and more extensive NMR assessments of the type of motions that aromatic rings are experiencing in proteins were demonstrated earlier.67,68 They are valuable in showing that ring interaction in hydrophobic pockets retains some flexibility in the core of the protein.
At a mixing time of 272.7 ms, cross-peaks from distant carbons are also observed (Fig. 4, right). Most of the cross-peaks that are absent in the shorter mixing time, such as the N25, V26, and K27 intra/inter-residue Cα–Cβ peaks, appear at the longer mixing time. However, some are still missing, for instance, the Cα–C′ peaks on these same residues and the ring cross-peaks. The spectrum contains, overall, many fewer cross-peaks than those in the spectrum of the crystalline ubiquitin samples, indicating a less-efficient spin-diffusion process. This may be the result of motion or short 1H or 13C longitudinal relaxation in the adsorbed protein. The short 13C T1 measured for the sample at 0.63 (±0.04) s helps explain the limited spin diffusion observed. 2D rf-driven dipolar recoupling69 experiments using coherent shorter mixing times (data not shown) do not produce additional cross-peaks, indicating that motion is probably an important factor limiting the apparent magnetization exchange in the experiments.
As mentioned earlier, the wide peaks, reflecting a dispersion of the chemical environment on most of the carbons, are indicative of a distribution of ubiquitin conformers on adsorption sites in MCM41. These conformers may be static structures locked by the interaction with the surface or dynamic ones, representing some degree of motion that is slower than the NMR-measurement time scale. Since this conformational heterogeneity is linked to the strength of the binding, we can reduce the interaction with the surface sites and induce increased motions thermally or by adding a solvent to the dry complex. These environmental changes affect both the backbone and sidechains of the protein. Narrower 15N spectra are achieved by increasing the hydration level of the sample (Fig. S3). The Lys amine line at 32.9 ppm, for example, is significantly narrowed (FWHM reduced from 5.5 to 2.2 ppm) by solvation, which weakens its interaction with surface sites, rendering this sidechain a mobility in the fast exchange limit. Additionally, narrower 13C spectra are achieved by raising the temperature (Fig. S3), and the carbonyl line width is reduced by half due to the increase in motional exchange rates with the increase in thermal energy. Unfortunately, the accompanied decrease in cross-polarization and other magnetization-transfer processes compromises the sensitivity of the NMR measurements.
III. DISCUSSION
MAS NMR measurements provide evidence of the involvement of specific ubiquitin residues in the interaction with the surfaces of MCM41. These interactions contribute collectively to the apparent adsorption enthalpy measured through macroscopic ITC and adsorption isotherm measurements. For the polar Thr and Ser residues, the absence of cross-peaks at the 64- and 68-ppm carbon lines with either silanols at ∼2 ppm or protonated silanols at ∼8 ppm in the 1H–13C HETCOR spectrum leaves the only plausible site for binding as the siloxane site, despite the fact that it is the least hydrophilic site on the MCM41 surface. For the aliphatic carbons that bind to the surface according to the TEDOR data, the surface-binding sites can be either silanols or siloxane, due to the absence of cross-peaks with protonated silanols in the 1H–13C HETCOR spectrum. Preliminary data suggest that the protein does not unfold or undergo massive secondary structure changes, although subtle structural changes and disparity of conformations are observed. The existence of motion in the adsorbed state despite the strong affinity to MCM41 is unexpected and has far-reaching consequences to how we envision the biological activity of adsorbed proteins and the state of enzymes immobilized on rigid cellular constructs such as the cytoskeleton.
Surface topology has been shown before to have a large effect on the structure of adsorbed proteins.70 Nanoparticles have a concave down-curvature that can lower structural stability of the protein causing the globular fold to be less compact as particle size increases and surface curvature decreases. The porous MCM41 surface, in contrast, has a concave up-curvature of nanopores which may promote other changes to the adsorbed protein's globular fold.71 The conformation disparity observed cannot be associated for certain with either contraction or expansion, and further experiments are required to determine the tertiary fold of ubiquitin on the MCM41 surface. The existence of motion in the immobilized protein that can be further manipulated by simple perturbations implies that complete trapping of the protein molecules deep within the pores of the material probably does not take place but, rather, mainly adsorption to the outer surfaces of the porous particles, in accordance with the SEM measurements.
In previous studies, ubiquitin mixed in a suspension of citrate-capped gold nanoparticles (GNP) in phosphate buffer at pH 7.7 has shown transient binding to the nanoparticle surface. The low affinity of the protein to the GNP surface at this pH is readily understood since at this pH value both the citrate-covered particle surface and protein are negatively charged. The adsorption–desorption equilibrium induces chemical shift perturbations in the protein-solution NMR spectrum, in particular, in the Q2-I3 and L15-E18 segments of the flexible N-terminus β-turn-β motif.72,73 Minor conformational changes are in accordance with the weak binding and transient adsorption events. A later study has explained ubiquitin and other proteins adsorption to the gold particles in a three-step mechanism.74 On silver nanoparticles, on the other hand, the protein forms multilayers (coronas) by destabilizing the α-helix motif in favor of the β-motif.75,76 It is tempting to try and rationalize the conformational changes in terms of the surface site on the silica and the strength of local interaction of one residue with a surface site; however, for now, it is clear that rules to predict the effect of the adsorption event on the secondary and tertiary structure changes are not easy to predict and are certainly highly dependent on the surface chemistry and topology of the material surface. The preliminary structure analysis of ubiquitin immobilized on MCM41 will be further complemented by 3D heteronuclear spectral editing experiments to further investigate any conformational changes that the protein might have undergone by adsorbing to the size-commensurate pores of MCM41.
IV. CONCLUSIONS
Ubiquitin adsorbs readily to MCM41 under slightly acidic conditions and covers its external surface, efficiently promoting some silica particle agglutination. Adsorption isotherms and ITC measurements point to the formation of a single layer of ubiquitin molecules. The exothermic binding at acidic pH indicates that electrostatic interactions are dominant in ubiquitin adsorption to MCM41. NMR measurements showed that carbonyl carbons, polar sidechains of Ser or Thr, and hydrophobic sidechains in ubiquitin interact with MCM41 surface sites. Lys and Arg interactions with the surface sites were also implicated. These site-resolved proximity measurements give a detailed perspective on the overall favorable binding inferred from the macroscopic measurements. Although immobilization of the protein on MCM41 is quite efficient, the conformational heterogeneity observed indicates that different ubiquitin molecules accomodate slightly different binding sites which influence the local chemical environment on most of the nuclei observed.
In addition, some local motions were also observed in the NMR measurements in aromatic residues, although the complex was dried prior to the measurement. Such motions can be amplified by tuning solvent content in the sample to reduce the ensemble of structures adopted by the adsorbed protein and for facilitated structure examinations. However, this compromises the detection sensitivity. NMR characterization of biomolecular adsorption is promising for structural analysis and for identifying the molecular interactions that dictate the overall thermodynamic profile of the process.
V. MATERIALS AND METHODS
A. Materials
Commercial MCM41, sodium acetate and ubiquitin (8.5 kDa) were purchased from Sigma-Aldrich. In addition, the ubiquitin was expressed in the BL21 Escherichia coli strain using a pRSET plasmid. For the expression of uniform 13C and 15N labeled ubiquitin, standard protocols of bacterial growth in minimal media enriched with 15NH4Cl and [U-13C6] glucose were used. The protein was purified over GE health-sciences S-sepharose and monoS cation exchange columns in ammonium acetate buffer using an Akta explorer fast protein liquid chromatography system. A purity of more than 96% was verified using mass spectrometry analysis and sodium dodecyl sulfate polyacrylamide gel electrophoresis.
B. Preparation of protein–silica complex
The physical characterization of ubiquitin-MCM41 and MCM41 by various techniques was performed in acetate buffer (100 mM, pH 5), unless otherwise noted. For NMR measurements, samples were prepared by mixing 10 mg of protein in sodium-acetate buffer (100 mM, pH 5) with 45 mg of MCM41 suspended in the buffer for 2 h, followed by centrifugation to collect the precipitate. The precipitate was washed with water and dried by evaporation at ambient temperature (25 °C) for 6 h. UV absorbance was used to assess the protein concentration in the solution before and after the adsorption. Further UV analysis of protein content in the washes with double-distilled water showed no evidence for desorption. Ubiquitin immobilized on MCM41 was packed into a 4-mm rotor for NMR measurements. The NMR sample in the rotor contained 45 mg of MCM41 and 5–6 mg of uniformly [13C (98%), 15N (99%)] labeled ubiquitin. The net charge on the ubiquitin at various pH values was calculated using the “LifeTein” algorithm.77
C. NMR experiments
Solid-state NMR measurements were carried out on an 11.74 T Bruker AvanceIII spectrometer equipped with a 4-mm MAS probe using a spinning rate of 11 kHz. Heteronuclear decoupling throughout the measurements employed a field of 85 kHz using the SPINAL decoupling scheme.78 The 1H spectra on all the samples were measured using direct excitation with a 90° pulse of 2.3 μs and a recycle delay of 1 s. The 1D 1H homonuclear decoupled spectra were acquired using a supercycled wPMLG (Ref. 79) [with a phase-modulated Lee–Goldburg (PMLG5) phase table] at a B1 field of 86 kHz to achieve 1H-1H decoupling during acquisition. The 15N Bloch decay spectrum was recorded using a 4 -μs 15N 90° pulse, with heteronuclear decoupling during acquisition, 8192 repetitions, and a 5-s recycle delay. The 1H-13C and 1H-15N CP experiments were performed at an 1H 90° pulse of 2.7 μs, followed by CP transfer using a ramped field between 35 and 75 kHz on 1H. The respective locking fields on 13C and 15N were 64 and 44 kHz, respectively, during 1H-13C CP and 1H-15N CP. The contact times were 2 ms for the 13C CP and 1 ms for the 15N CP. The 13C CP experiments were carried out using 512 scans and a 1 s recycle.
2D 1H-15N and 1H-13C HETCOR measurements were recorded using a homonuclear 1H decoupling during t1 with a PMLG5 scheme80 at an effective field of 105 kHz, using similar locking fields as those used in the CP experiments. 2D 13C DARR measurements were performed at 298 K, starting with 1H-13C- CP using the same conditions described above, an 11-kHz field on 1H during the mixing time and heteronuclear decoupling during t1 evolution and t2 detection periods. The DARR measurements were performed with mixing times of 90.9 and 272.7 ms. Each spectrum was acquired using 130 t1 points and 300 scans at a recycle delay of 1 s.
D. AFM measurements
All AFM measurements were performed on a dimension icon Bruker AXS microscope using a 2-nm high-resolution SiN (p-doped Si) tip with a force constant of 1.8–12.5 N/m operating at a frequency range of 110–220 kHz. Topography images (600 × 600 nm2) were collected using a tapping-mode scan at 1024 samples/line at a scan rate of 1 Hz. Samples of MCM41 suspended in acetate buffer and MCM41-adsorbed ubiquitin samples were measured as is and transferred to aqueous conditions. The samples were dried by vacuum before AFM scanning.
E. Adsorption isotherm and isothermal titration calorimetry
The adsorption measurements, recorded at 25 °C, were performed using the following initial ubiquitin solution concentrations: 80, 100, 150, 200, 250, 500, 750, and 1000 μg/ml. These protein solutions were mixed separately with 1.0 mg/ml MCM41, prewashed, and suspended in separate acetate-buffer solutions according to pH. A bicinchoninic acid assay was used to determine the concentration of the protein before and after adsorption, from which the adsorbed amount was deduced and plotted in the isotherms. The isothermal titration calorimetry measurements were recorded at 30 °C using 10–μl injections of ubiquitin solution (0.048 M) in acetate buffer (pH 5) from a 294 μl syringe into a stirred cell (250 rpm) containing 1.0 mg/ml of MCM41 suspended in 1.4 ml of 100 mM acetate buffer, with an adsorption-site concentration of 0.025 M, as derived from the adsorption isotherm measurements. The latter is determined from the maximal coverage back-converted from units of (nmol of proteins)/(m2 of MCM41) to molar concentration using the volume of solvent in which the complex is prepared.
ACKNOWLEDGMENTS
This research was supported by the Israel Science Foundation (Grant No. 1059/09). The authors thank Olga Girshevitz and Yael Goldfinger for their help with the AFM measurements and Ami Navon of the Weizmann Institute of Science for kindly providing us with the pRSET plasmid encoding ubiquitin expression in E. coli.
References
- 1. Adeloju S. B. and Wallace G. G., Analyst 121, 699 (1996). 10.1039/an9962100699 [DOI] [PubMed] [Google Scholar]
- 2. Knopp D., Tang D. P., and Niessner R., Anal. Chim. Acta 647, 14 (2009). 10.1016/j.aca.2009.05.037 [DOI] [PubMed] [Google Scholar]
- 3. Ispas C. R., Crivat G., and Andreescu S., Anal. Lett. 45, 168 (2012). 10.1080/00032719.2011.633188 [DOI] [Google Scholar]
- 4. Barone P. W. and Strano M. S., J. Diabetes Sci. Technol. 3, 242 (2009). 10.1177/193229680900300204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Marchenko S. V., Soldatkin O. O., Kasap B. O., Kurc B. A., Soldatkin A. P., and Dzyadevych S. V., Nanoscale Res. Lett. 11, 173 (2016). 10.1186/s11671-016-1386-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Prajapati S., Tao J. H., Ruan Q. C., De Yoreo J. J., and Moradian-Oldak J., Biomaterials 75, 260 (2016). 10.1016/j.biomaterials.2015.10.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tao J., Buchko G., Shaw W., Yoreo De J., and Tarasevich B., Langmuir 31, 10451 (2015). 10.1021/acs.langmuir.5b02549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chittur K. K., Biomaterials 19, 357 (1998). 10.1016/S0142-9612(97)00223-8 [DOI] [PubMed] [Google Scholar]
- 9. Wu C. S. and Chen G. C., Anal. Biochem. 177, 178 (1989). 10.1016/0003-2697(89)90036-5 [DOI] [PubMed] [Google Scholar]
- 10. Santos O., Kosoric J., Hector M. P., Anderson P., and Lindh L., J. Colloid Interface Sci. 318, 175 (2008). 10.1016/j.jcis.2007.11.015 [DOI] [PubMed] [Google Scholar]
- 11. Yonzon C. R., Haynes C. L., Zhang X. Y., Walsh J. T., and Van Duyne R. P., Anal. Chem. 76, 78 (2004). 10.1021/ac035134k [DOI] [PubMed] [Google Scholar]
- 12. Reymond- Laruinaz S., Saviot L., Potin V., and de Lucas M. D. M., Appl. Surf. Sci. 389, 17 (2016). 10.1016/j.apsusc.2016.07.082 [DOI] [Google Scholar]
- 13. Martins M. C. L., Sousa S. R., Antunes J. C., and Barbosa M. A., “ Protein adsorption characterization,” in Nanotechnology in Regenerative Medicine: Methods and Protocols, edited by Navarro M. and Planell J. A. ( Humana, Totowa, NJ, 2012), pp. 141–161. [DOI] [PubMed] [Google Scholar]
- 14. Carlsson N., Gustafsson H., Thorn C., Olsson L., Holmberg K., and Akerman B., Adv. Colloid Interface Sci. 205, 339 (2014). 10.1016/j.cis.2013.08.010 [DOI] [PubMed] [Google Scholar]
- 15. Boussaad S., Dziri L., Arechabaleta R., Tao N. J., and Leblanc R. M., Langmuir 14, 6215 (1998). 10.1021/la980319l [DOI] [Google Scholar]
- 16. Matlahov I., Geiger Y., and Goobes G., Phys. Chem. Chem. Phys. 16, 9031 (2014). 10.1039/c3cp55520h [DOI] [PubMed] [Google Scholar]
- 17. Goobes R., Goobes G., Campbell C. T., and Stayton P. S., Biochemistry 45, 5576 (2006). 10.1021/bi052321z [DOI] [PubMed] [Google Scholar]
- 18. Bellucci L., Ardevol A., Parrinello M., Lutz H., Lu H., Weidner T., and Corni S., Nanoscale 8, 8737 (2016). 10.1039/C6NR01539E [DOI] [PubMed] [Google Scholar]
- 19. Baio J., Zane A., Jaeger V., Roehrich A., Lutz H., Pfaendtner J., Drobny G., and Weidner T., J. Am. Chem. Soc. 136, 15134 (2014). 10.1021/ja5078238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mermut O., Phillips D., York R., McCrea K., Ward R., and Somorjai G., J. Am. Chem. Soc. 128, 3598 (2006). 10.1021/ja056031h [DOI] [PubMed] [Google Scholar]
- 21. Wang Y.-C. and Castner D. G., Microsc. Microanal. 22, 346 (2016). 10.1017/S1431927616002580 [DOI] [Google Scholar]
- 22. Mangiagalli M. et al. , FEBS J. 284, 163 (2017). 10.1111/febs.13965 [DOI] [PubMed] [Google Scholar]
- 23. Masica D. L., Ash J. T., Ndao M., Drobny G. P., and Gray J. J. Structure 18, 1678 (2010). 10.1016/j.str.2010.09.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Addison W. N., Masica D. L., Gray J. J., and McKee M. D., J. Bone Miner. Res. 25, 695 (2010). 10.1359/jbmr.090832 [DOI] [PubMed] [Google Scholar]
- 25. Wright L. B., Rodger P. M., Corni S., and Walsh T. R., J. Chem. Theory Comput. 9, 1616 (2013). 10.1021/ct301018m [DOI] [PubMed] [Google Scholar]
- 26. Rimola A., Costa D., Sodupe M., Lambert J. F., and Ugliengo P., Chem. Rev. 113, 4216 (2013). 10.1021/cr3003054 [DOI] [PubMed] [Google Scholar]
- 27. Latour R. A., Colloids Surf., B 124, 25 (2014). 10.1016/j.colsurfb.2014.06.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nogales E., Nat. Methods 13, 24 (2016). 10.1038/nmeth.3694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Castellani F., van Rossum B., Diehl A., Schubert M., Rehbein K., and Oschkinat H., Nature 420, 98 (2002). 10.1038/nature01070 [DOI] [PubMed] [Google Scholar]
- 30. Zech S. G., Wand A. J., and McDermott A. E., J. Am. Chem. Soc. 127, 8618 (2005). 10.1021/ja0503128 [DOI] [PubMed] [Google Scholar]
- 31. Franks W. T., Wylie B. J., Schmidt H. L. F., Nieuwkoop A. J., Mayrhofer R. M., Shah G. J., Graesser D. T., and Rienstra C. M., Proc. Natl. Acad. Sci. U.S.A. 105, 4621 (2008). 10.1073/pnas.0712393105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Loquet A. et al. , Nature 486, 276 (2012). 10.1038/nature11079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Morag O., Sgourakis N. G., Baker D., and Goldbourt A., Proc. Natl. Acad. Sci. U.S.A. 112, 971 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Liu C. et al. , Nat. Commun. 7, 10714 (2016). 10.1038/ncomms10714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Barbet-Massin E. et al. , Biophys. J. 107, 941 (2014). 10.1016/j.bpj.2014.05.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Shaw W., Long J., Dindot J., Campbell A., Stayton P., and Drobny G., J. Am. Chem. Soc. 122, 1709 (2000). 10.1021/ja9842823 [DOI] [Google Scholar]
- 37. Goobes G., Goobes R., Schueler-Furman O., Baker D., Stayton P., and Drobny G., Proc. Natl. Acad. Sci. U.S.A. 103, 16083 (2006). 10.1073/pnas.0607193103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ndao M., Ash J., Breen N., Goobes G., Stayton P., and Drobny G., Langmuir 25, 12136 (2009). 10.1021/la901647n [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Masica D., Gray J., and Shaw W., J. Phys. Chem. C 115, 13775 (2011). 10.1021/jp202965h [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chen P., Tseng Y., Mou Y., Tsai Y., Guo S., Huang S., Yu S., and Chan J., J. Am. Chem. Soc. 130, 2862 (2008). 10.1021/ja076607y [DOI] [PubMed] [Google Scholar]
- 41. Faure N., Halling P., and Wimperis S., J. Phys. Chem. C 118, 1042 (2014). 10.1021/jp4098414 [DOI] [Google Scholar]
- 42. Roehrich A. and Drobny G., Acc. Chem. Res. 46, 2136 (2013). 10.1021/ar300321e [DOI] [PubMed] [Google Scholar]
- 43. Fragai M., Luchinat C., Martelli T., Ravera E., Sagi I., Solomonov I., and Udi Y., Chem. Commun. 50, 421 (2014). 10.1039/C3CC46896H [DOI] [PubMed] [Google Scholar]
- 44. Ravera E., Michaelis V., Ong T., Keeler E., Martelli T., Fragai M., Griffin R., and Luchinat C., Chemphyschem 16, 2751 (2015). 10.1002/cphc.201500549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ravera E., Schubeis T., Martelli T., Fragai M., Parigi G., and Luchinat C., J. Magn. Reson. 253, 60 (2015). 10.1016/j.jmr.2014.12.019 [DOI] [PubMed] [Google Scholar]
- 46. Matlahov I., Iline-Vul T., Abayev M., Lee E. M. Y., Nadav-Tsubery M., Keinan-Adamsky K., Gray J. J., and Goobes G., Chem. Mater. 27, 5562 (2015). 10.1021/acs.chemmater.5b01696 [DOI] [Google Scholar]
- 47. Goobes G., Israel J. Chem. 54, 113 (2014). 10.1002/ijch.201300113 [DOI] [Google Scholar]
- 48. Geiger Y., Gottlieb H., Akbey U., Oschkinat H., and Goobes G., J. Am. Chem. Soc. 138, 5561 (2016). 10.1021/jacs.5b07809 [DOI] [PubMed] [Google Scholar]
- 49. Zane A., Michelet C., Roehrich A., Emani P., and Drobny G., Langmuir 30, 7152 (2014). 10.1021/la501444t [DOI] [PubMed] [Google Scholar]
- 50. Jantschke A., Koers E., Mance D., Weingarth M., Brunner E., and Baldus M., Angew. Chem. Int. Ed. 54, 15069 (2015). 10.1002/anie.201507327 [DOI] [PubMed] [Google Scholar]
- 51. Yiu H. and Wright P., J. Mater. Chem. 15, 3690 (2005). 10.1039/b506090g [DOI] [Google Scholar]
- 52. Raj D., Biju S., and Reddy M., J. Mater. Chem. 19, 7976 (2009). 10.1039/b913786f [DOI] [Google Scholar]
- 53. Hing A. W., Vega S., and Schaefer J., J. Magn. Reson., Ser. A 103, 151 (1993). 10.1006/jmra.1993.1146 [DOI] [Google Scholar]
- 54. Lundqvist M., Sethson I., and Jonsson B., Langmuir 20, 10639 (2004). 10.1021/la0484725 [DOI] [PubMed] [Google Scholar]
- 55. Meissner J., Prause A., Bharti B., and Findenegg G., Colloid Polym. Sci. 293, 3381 (2015). 10.1007/s00396-015-3754-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Tian Y., Li Z., Zhang W., Li Y., Yuan Y., and Liu C., J. Nanosci. Nanotechnol. 16, 5528 (2016). 10.1166/jnn.2016.11743 [DOI] [PubMed] [Google Scholar]
- 57. Amitay-Rosen T., Kababya S., and Vega S., J. Phys. Chem. B 113, 6267 (2009). 10.1021/jp810572r [DOI] [PubMed] [Google Scholar]
- 58. Baccile N., Laurent G., Bonhomme C., Innocenzi P., and Babonneau F., Chem. Mater. 9, 1343 (2007). 10.1021/cm062545j [DOI] [Google Scholar]
- 59. Manolikas T., Herrmann T., and Meier B. H., J. Am. Chem. Soc. 130, 3959 (2008). 10.1021/ja078039s [DOI] [PubMed] [Google Scholar]
- 60. Takegoshi K., Nakamura S., and Terao T., Chem. Phys. Lett. 344, 631 (2001). 10.1016/S0009-2614(01)00791-6 [DOI] [Google Scholar]
- 61. Schneider R., Odronitz F., Hammesfahr B., Hellkamp M., and Kollmar M., Bioinformatics 29, 1134 (2013). 10.1093/bioinformatics/btt125 [DOI] [PubMed] [Google Scholar]
- 62. Vijaykumar S., Bugg C. E., and Cook W. J. J. Mol. Biol. 194, 531 (1987). 10.1016/0022-2836(87)90679-6 [DOI] [PubMed] [Google Scholar]
- 63. Han B., Liu Y. F., Ginzinger S. W., and Wishart D. S., J. Biomol. NMR 50, 43 (2011). 10.1007/s10858-011-9478-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Wilkinson K. and Mayer A., Arch. Biochem. Biophys. 250, 390 (1986). 10.1016/0003-9861(86)90741-1 [DOI] [PubMed] [Google Scholar]
- 65. Brutscher B., Bruschweiler R., and Ernst R. R., Biochemistry 36, 13043 (1997). 10.1021/bi971538t [DOI] [PubMed] [Google Scholar]
- 66. Schubert M., Manolikas T., Rogowski M., and Meier B. H., J. Biomol. NMR 35, 167 (2006). 10.1007/s10858-006-9025-x [DOI] [PubMed] [Google Scholar]
- 67. Gall C., Cross T., Diverdi J., and Opella S., Proc. Natl. Acad. Sci. U.S.A. 79, 101 (1982). 10.1073/pnas.79.1.101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Vugmeyster L., Ostrovsky D., Villafranca T., Sharp J., Xu W., Lipton A., Hoatson G., and Vold R., J. Phys. Chem. B 119, 14892 (2015). 10.1021/acs.jpcb.5b09299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Bennett A. E., Ok J. H., Griffin R. G., and Vega S., J. Chem. Phys. 96, 8624 (1992). 10.1063/1.462267 [DOI] [Google Scholar]
- 70. Shang W., Nuffer J. H., Dordick J. S., and Siegel R. W., Nano Lett. 7, 1991 (2002). 10.1021/nl070777r [DOI] [PubMed] [Google Scholar]
- 71. Ravindra R., Shuang Z., Gies H., and Winter R., J. Am. Chem. Soc. 126, 12224 (2004). 10.1021/ja046900n [DOI] [PubMed] [Google Scholar]
- 72. Calzolai L., Franchini F., Gilliland D., and Rossi F., Nano Lett. 10, 3101 (2010). 10.1021/nl101746v [DOI] [PubMed] [Google Scholar]
- 73. Brancolini G., Kokh D. B., Calzolai L., Wade R. C., and Corni S., ACS Nano 6, 9863 (2012). 10.1021/nn303444b [DOI] [PubMed] [Google Scholar]
- 74. Wang A. L., Vangala K., Vo T., Zhang D. M., and Fitzkee N. C., J. Phys. Chem. C 118, 8134 (2014). 10.1021/jp411543y [DOI] [Google Scholar]
- 75. Ding F., Radic S., Chen R., Chen P. Y., Geitner N. K., Brown J. M., and Ke P. C., Nanoscale 5, 9162 (2013). 10.1039/c3nr02147e [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Brahmkhatri V. P., Chandra K., Dubey A., and Atreya H. S., Nanoscale 7, 12921 (2015). 10.1039/C5NR03047A [DOI] [PubMed] [Google Scholar]
- 77.“ Protein/peptide property calculator,” www.lifetein.com/peptide-analysis-tool.html.
- 78. Brauniger T., Wormald P., and Hodgkinson P., Monatsh. Chem. 133, 1549 (2002). 10.1007/s00706-002-0501-z [DOI] [Google Scholar]
- 79. Vinogradov E., Madhu P. K., and Vega S., Chem. Phys. Lett. 354, 193 (2002). 10.1016/S0009-2614(02)00060-X [DOI] [Google Scholar]
- 80. Vinogradov E., Madhu P. K., and Vega S., Chem. Phys. Lett. 314, 443 (1999). 10.1016/S0009-2614(99)01174-4 [DOI] [Google Scholar]
- 81.See supplementary material http://dx.doi.org/10.1116/1.4983273 E-BJIOBN-12-327702 for a table with the EDAX analysis of the MCM41 and ubiquitin-bound MCM41, AFM surface topology images of MCM41 and ubiquitin-bound MCM41, 2D 1H-13C and 1H-15N HETCOR spectra of U-13C, 15N-ubiquitin-bound MCM41, 1D 15N, and 13C spectra of U-13C,15N-ubiquitin-bound MCM41 at various hydration levels and various temperatures, respectively.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- See supplementary material http://dx.doi.org/10.1116/1.4983273 E-BJIOBN-12-327702 for a table with the EDAX analysis of the MCM41 and ubiquitin-bound MCM41, AFM surface topology images of MCM41 and ubiquitin-bound MCM41, 2D 1H-13C and 1H-15N HETCOR spectra of U-13C, 15N-ubiquitin-bound MCM41, 1D 15N, and 13C spectra of U-13C,15N-ubiquitin-bound MCM41 at various hydration levels and various temperatures, respectively.