Abstract
The heparin binding site (Hep II) of fibronectin plays a major role in tumor cell metastasis. Its interaction with heparan sulfate proteoglycans occurs in a variety of physiological processes including focal adhesion and migration. The melanoma inhibitory activity (MIA) is an important protein that is functionally involved in melanoma development, progression, and tumor cell invasion. After its secretion by malignant melanoma cells, MIA interacts with fibronectin and thereby actively facilitates focal cell detachment from surrounding structures and strongly promotes tumor cell invasion and the formation of metastases. In this report, the authors have determined the molecular basis of the interaction of MIA with the Hep II domain of fibronectin based on nuclear magnetic resonance spectroscopic binding assays. The authors have identified the type III modules 12 to 14 of fibronectin's Hep II as the major MIA binding sites. These results now provide a new target protein–protein binding interface for the discovery of novel antimetastatic agents against malignant melanoma in the future.
I. INTRODUCTION
The melanoma inhibitory activity (MIA) protein belongs to a family of non-cytosolic proteins that consists of MIA, OTOR/FDP, MIA-2, and TANGO [MIA-3, which bear four highly conserved cysteine-residues1–3 (Fig. 1)]. The 12 kDa MIA protein is secreted into the extracellular spaces and harbors an SH3 domain, an abundant cytoplasmic protein module known to interact with proline-rich motif found in SH2 domains (Fig. 1).4–8 MIA is highly expressed and secreted by melanoma cells after their malignant transformation.1 As a matter of fact, not only neoplastic tissues, such as malignant melanomas and chondrosarcomas, express MIA, but also non-neoplastic cartilage tissues.1 Both in vitro and in vivo data suggest an important functional role for MIA in melanoma metastasis and invasion.1 In contrast to other SH3 domains, MIA is not capable of binding to proline-rich peptide ligands4 but rather interacts with extracellular matrix molecules, such as laminin, tenascin, and fibronectin (FN). Through these interactions, MIA prevents cells from efficiently attaching to components of the extracellular matrix.4 Consequently, elevated levels of secreted MIA strongly boost tumor cell invasion that finally causes metastases in malignant melanoma.9
Fig. 1.
Sequence alignment of MIA, FDP, MIA-2, and TANGO. Conserved amino acids are highlighted according to Clustal Omega (www.ebi.ac.uk/Tools/msa/clustalo/): (*)/green—fully conserved residues; (:)/yellow—conservation between groups with strongly similar properties; (.)/orange—conservation between groups of weakly similar properties. Highly conserved cysteines are colored in red and their disulfide bridging pattern is shown by red connecting lines.
SH3 domains are abundant non-catalytic protein modules, which are structurally and functionally central to numerous intracellular signaling processes mediated by kinases, lipases, GTPases, adapter proteins, structural proteins, and viral regulatory proteins.10–15 SH3 domains consist of 55–70 amino acids and adopt a compact fold of antiparallel three-stranded β-sheets.16,17 Over the last decades, numerous studies revealed that SH3 domains participate in protein–protein interactions through binding to proline-rich peptide sequences located in SH2 domains (Ref. 13 and references therein). More extensive structural studies of the SH3 domain bound to proline-rich peptide ligands revealed that the hydrophobic, relatively shallow binding site consists of highly conserved aromatic amino side chains.10 Characteristic structural features of all SH3 domains are the RT-loop and n-Src loop (that confine the binding site), the distal hairpin (that can vary in its length), and sometimes a βI-turn.10,14,15,18,19 In addition to these structural elements, the extracellular MIA carries unique N- and C-terminal extensions and two conserved disulfide bonds.4–8
Fibronectin is a multimodular glycoprotein in extracellular matrix that is essential to many cellular processes such as adhesion and migration of cells, their proliferation and differentiation, as well as the organization of the cytoskeleton, and it is involved in fundamental and important processes, like embryogenesis, wound healing, and tumor metastasis.20 Fibronectin ubiquitously occurs in connective tissue, at cellular surfaces, in plasma, and in body fluids—either in an insoluble or a soluble form.21–24 In plasma, the soluble form of fibronectin is secreted from hepatocytes, which modulates blood clotting, wound healing, and phagocytosis. In the extracellular matrix, however, fibronectin adopts an insoluble state, after it has been secreted from fibroblasts, chondriocytes, and endo- as well as epithelial cells to create a network of fibrils that contributes to and even modulates tissue structure.25
In the extracellular matrix, fibronectin occurs as a heterodimeric protein, which consists of two similar 250 kDa subunits that are cross-linked via two disulfide bonds at their carboxy-termini. Each subunit consists of several independently folded homologous type I, II, and III modules, which are separated by linker sequences like pearls on a string.20 Human fibronectin harbors two type I, two type II, and 15–17 type III modules.26,27 These functional domains enable fibronectin to interact with various macromolecules in the extracellular matrix, such as collagen, fibrin, and cell surface proteins like integrins or syndecan.21–24 In addition, fibronectin can bind to proteoglycans and glycosaminoglycans such as heparin and heparansulfate that scaffold the extracellular matrix.21–24,28 Fibronectin is thus involved in regulating the adhesion and migration of cells.29 The heparin binding sites have been mapped onto fibronectin, referred to as heparin binding domains (Hep) I, II, and IIICS. Hep I is located at the N-terminus and Hep IIICS—an alternatively spliced type III connecting segment—is located at the C-terminus of fibronectin.30–32 Hep II is regarded as the most important binding site for heparin, which is also located at the C-terminus of fibronectin.29,32,33 Notably, the Hep II site is involved in a heparin-dependent adhesion of melanoma cells and it can influence the conformation of fibronectin itself.34 Furthermore, integrin α4β1 is able to bind to Hep II, which suggests that integrins might play a role in the metastasis of melanoma cells.35,36
The Hep II domain of fibronectin is composed of the type III modules 12–14, and it contributes almost exclusively to the overall affinity of fibronectin for heparin.37 These fibronectin modules consist of a sandwich of seven β strands that are packed to form two antiparallel β sheets.32 These type III modules 12–14 are separated by intermodule linkers and while the linker between modules 12 and 13 is rather flexible, the linker between modules 13 and 14 is rigid and causes these two type III modules to adopt a defined interdomain angle.32 The interaction between the Hep II domain and heparin is entirely electrostatic in nature with Arg and Lys side chains of fibronectin coordinating the sulfated sugar moieties.32,37 In fibronectin, heparin mainly contacts module 13 and is fine-tuned by modules 12 and 14.34 In particular, Arg98, Arg99, Arg101, Arg115, Lys117, and Arg146 located in module 13 are essential for the Hep II binding site as shown by mutational studies.32,37 These six basic residues form fibronectin's cationic fork. Next to the Hep II site located on module 13, a PRARI binding motif has been identified in module 14, which the cell adhesion receptor integrin α4β1 can bind to.29,32,38 Presumably, this PRARI region is stabilized by the rigid interdomain linker between type III modules 13 and 14 and it is located opposite to the Hep II region on the surface of fibronectin. This architecture could point toward a cooperative binding mechanism of heparin and integrin to fibronectin.32
Previously, a dodecapeptide FHWRYPLPLPGQ that was identified in a phage display experiment denoted as AR71 has been described as a MIA inhibitor, which led to reduced cell migration, reduced formation of metastases, and an increased immune response.1 Finally, a recent NMR-spectroscopic, fragment-based ligand screening (FBLS) study supported by an in silico screen of the ZINC database39 revealed N-(3-cyanophenyl)-3,5-dimethylbenzimidamide and (1-(1,2,3,4-tetrahydroisoquinoline-3-carbonyl)-N-(m-tolyl)pyrrolidine-2-carboxamide as low molecular weight antagonists of the interaction between MIA and fibronectin.39,40 These compounds antagonize the interaction between fibronectin and MIA. Based on these findings, this study aimed at characterizing the binding interface between MIA and fibronectin. Through optimizing the binding affinity of the previously identified peptide sequence AR71 into FRWRRRRR, we found that electrostatic charges determine and strengthen the interaction between MIA and its ligand. The fact that Arg-rich peptide sequences showed an improved binding to MIA enabled us to identify the binding site of MIA on fibronectin in NMR-based interaction studies between MIA and FN. Therefore, our study is the very first to structurally describe the MIA/FN complex at atomic resolution. It identifies the fibronectin FN12-14 Hep II domain as the binding site of MIA and characterizes the interaction between these two proteins on an atomic level.
II. EXPERIMENT
A. Protein expression and purification
Recombinant human MIA was expressed and purified as published previously.41 Fibronectin type III domains 12, 13, and 14 were cloned into pET-19b vectors (yielding a shift in sequence numbering of 72 in comparison to the PDB ID: 1FNH), expressed in BL21 (DE3), and purified as His-tagged proteins. The uniformly 15N-enriched protein samples were prepared by growing the bacteria in minimal media containing 15NH4Cl.41 The identity and purity of all isolated proteins were checked by sodium dodecyl sulfate polyacrylamide gel electrophoresis and matrix-assisted laser desorption/ionization time of flight. The isolated proteins were shown to be approx. 95% pure.
B. NMR spectroscopy
All protein NMR spectra were acquired at 298 K on Bruker DRX 600 and AVANCE 700 NMR spectrometers equipped with pulsed field gradients. Water suppression was achieved by incorporating a Watergate sequence into the various pulse sequences.42 All spectra were processed with NMRPipe and analyzed with ccpnmr Analysis and bruker topspin software packages.43,44 Complete 1H and 15N resonance assignments of human MIA have been published previously.5,41
Binding of peptides and proteins to MIA was characterized and validated by mapping chemical shift perturbations using two-dimensional (2D) 1H-15N HSQC NMR spectroscopy.15,19 The chemical shift pertubations were determined in units of ppm by multiplying the respective 15N and 1H Hz shifts by a correction factor of 1.44 for 1H and 0.23 for 15N.45 Binding was monitored by chemical shift perturbation analysis of backbone amide resonances of uniformly 15N-enriched MIA samples (0.1 mM, phosphate buffered saline buffer, pH 7, 298 K) through recording a series of 2D 1H-15N HSQC spectra with increasing ligand concentrations.46,47 The pH was maintained constant during the entire titration series.
C. Molecular docking
Docking studies were performed using haddock 2.1 and cns 1.3 software packages.48,49 The crystal structures of MIA and FN type III tandem domain 13–14 were obtained from the PDB databank (PDB ID: 1I1J and 1FNH).4,32 Within the HADDOCK process, active residues are forced to be part of the interface by applying ambiguous interaction restraints (AIR) while passive residue can also be part of the interface.48,50 Active interface residues of MIA and FN were defined based on chemical shift perturbations observed by 2D 1H-15N HSQC NMR titration of 15N-enriched MIA with FN. For both proteins, only residues that are part of the interface were treated as flexible. During the first docking iteration, 1000 structures were calculated. The 200 top-ranked models were then selected for further structural refinement. Figures were created using PyMOL.51
III. RESULTS AND DISCUSSION
A. NMR spectroscopy
Using NMR spectroscopy, we have tested and optimized previously published and new peptide sequences that bind to human MIA.1 Initially, the NMR-based binding experiments suggested that the previously structurally and functionally characterized peptide AR71, which was originally identified in a phage display experiment, exhibits a rather weak affinity for human MIA in solution.1 Therefore, we first optimized this peptide sequence in the current study and developed a new, Arg-enriched sequence FRWRRRRR that exhibits an increased affinity for MIA (Fig. 2). The binding site of both AR71 and the Arg-rich peptide is located the opposite to the canonical SH3 ligand binding site and is characterized by an acidic rim that harbors the basic side chains of the peptide ligands. Thus, through optimizing the binding affinity of the previously identified peptide sequence AR71 into the sequence FRWRRRRR we could show that electrostatic charges determine and strengthen the interaction between MIA and its ligand. The fact that Arg-rich peptide sequences showed an improved binding to MIA enabled us to identify the binding site of MIA on Fibronectin in NMR-based interaction studies between MIA and FN. Based on this result, we then identified the MIA-targeting amino acids located in fibronectin type III modules 12, 13, and 14. Ultimately, the Arg-rich peptide led to the identification of the FN Hep II domain as a target for MIA.
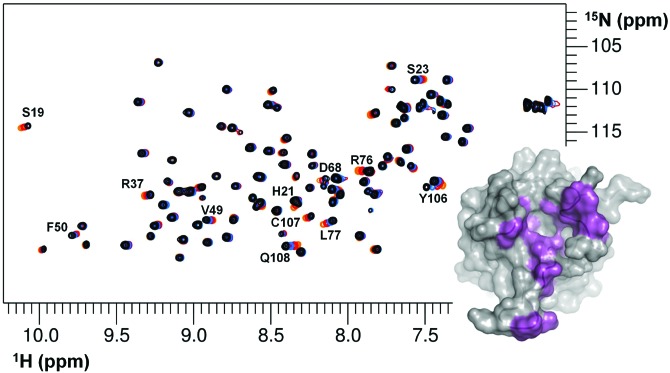
Fig. 2.
Superposition of 2D 1H-15N HSQC NMR spectra of 15N-enriched MIA in the absence (black) and presence of increasing amounts of the Arg-rich peptide FRWRRRRR (red to orange). Resonances originating from MIA that experience chemical shift perturbations upon peptide binding are labeled and projected onto the MIA surface in violet (lower right).
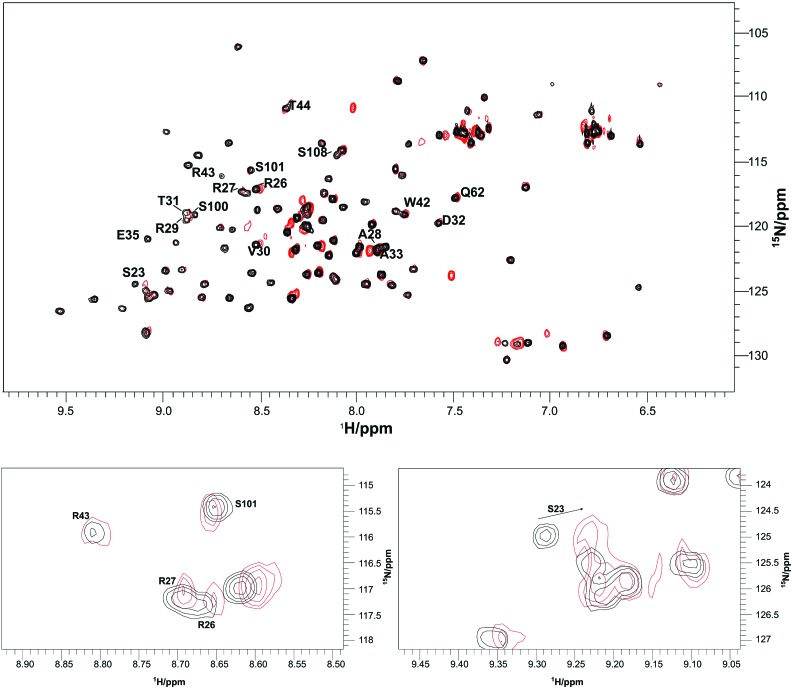
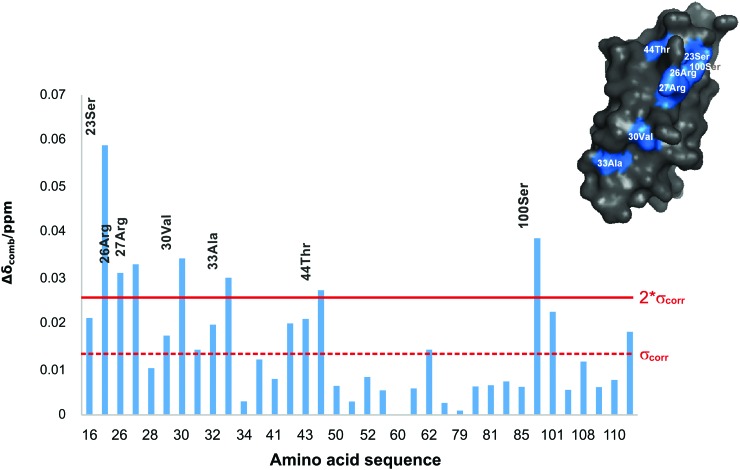
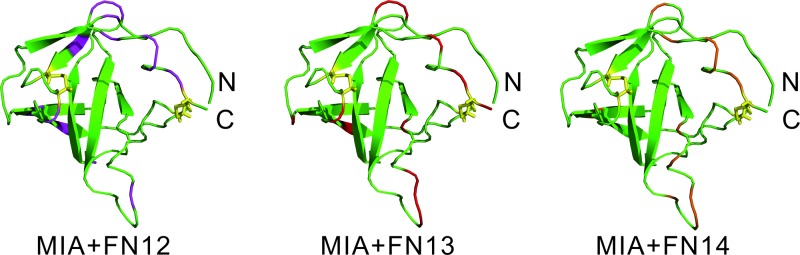
The interaction between MIA and the 15N-enriched fibronectin type III module FN13 was monitored by means of 2D 1H-15N HSQC NMR spectra. The MIA-dependent chemical shift perturbation (applying FN to MIA ratios of 1:1, 1:3, 1:8, 1:15, and 1:23) shows that a MIA-fibronectin complex is formed with weak micromolar affinity (Fig. 3). In detail, the following amino acids of the fibronectin type III module FN13 participate in binding to MIA: G16, S23, R26, R27, A28, R29, V30, T31, D32, A33, E35, W42, R43, T44, Q62, S100, S101, S108, and I111 (Fig. 4). Furthermore, we have also performed NMR-based binding studies with 15N-enriched human MIA and tested as to whether the individual fibronectin type III module FN12, FN13, and FN14 would bind. All of these analyzed fibronectin modules induce very similar chemical shift perturbations in 2D 1H-15N HSQC NMR spectra of MIA with similar affinities in the weak micromolar range. Projected onto the molecular surface of MIA, these chemical shift perturbations cluster to form the same binding site opposite to the canonical SH3 ligand binding groove (Fig. 5). Finally, these results allowed for mapping of the binding site of human MIA on the Hep II domain of fibronectin.
Fig. 3.
Superposition of 2D 1H-15N HSQC NMR spectra (full spectrum shown in upper panel, enlarged spectral regions depicted in lower panel) of 15N-enriched fibronectin type II module FN13 in the absence (black) and presence of an excess of human MIA (red). Resonances originating from fibronectin that experience chemical shift perturbations upon binding of MIA are labeled. For the sequence numbering scheme please refer to the experimental part.
Fig. 4.
Plot of the chemical shift perturbation vs the amino acid sequence of FN13 upon binding of MIA. Weighted chemical shift differences greater than two standard deviations were regarded as significant (2*σcorr) and projected in blue onto the molecular surface of FN13 (shown in the upper right corner). For the sequence numbering scheme please refer to the experimental part.
Fig. 5.
Chemical shift perturbations induced by FN12 (left), FN13 (middle), and FN14 (right) upon binding to 15N-enriched MIA projected onto its ribbon representation (highlighted in pink, red, and orange, respectively). The orientation of MIA matches the one shown in Fig. 2.
B. Molecular docking
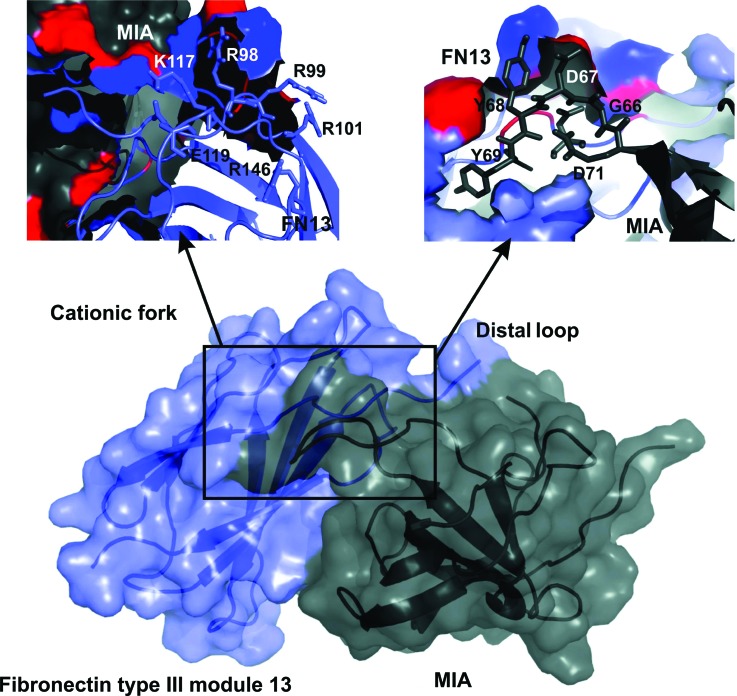
Using the software package haddock, a molecular dynamics-based docking approach was applied in order to create an atomic model of the MIA/FN13 based on the multidimensional NMR spectroscopic analysis (Fig. 6).48 The protein–protein interface was defined according to the chemical shift perturbations obtained from the NMR titrations of MIA to 15N-enriched FN13 and vice versa using 15N-enriched MIA.39 The affected amino acids (Figs. 3–5) were used as input for the HADDOCK protocol by classifying them as “active and passive residues” in order to generate AIR.48 For MIA, amino acids Q16, S19, D68, Y70, G71, A74, R76, L91, Y106 were defined as active and residues D15, E17, H20, G67, Y69, D72, L73, A75, L77, I90, K92, W103, and C107 as passive. For FN13, residues S95, R98, R99, T116, S172 were defined as active and residues P96, P97, A100, R115, K117, R171, and S173 as passive amino acids (according to the PDB ID: 1FNH). The HADDOCK-generated atomic model of the MIA/FN13 complex reveals that the cationic fork—one of the most important heparin binding sites on fibronectin—interacts with the acidic distal loop of MIA (Fig. 6). Furthermore, MIA binds to a region located to the right of the N-terminus of FN13.
Fig. 6.
Complex of MIA (gray) bound to FN13 (blue) based on HADDOCK docking calculations. The best scoring model is shown in surface representation. The amino acid interactions of the cationic fork from FN13 are shown in the upper left corner (according to the PDB ID: 1FNH), the contacts of the distal loop from MIA are shown in the upper right corner.
C. Discussion
Previously, several regions of human MIA were shown to be of functional relevance based on mutagenesis of certain amino acids.52 These regions not only contain parts of the canonical binding-site in homologous SH3 domains but also the highly flexible distal loop of this protein and regions on the opposite side of the MIA surface. It was concluded that these identified regions either constitute one continuous single ligand-binding site or point towards a bifunctional binding mode of MIA for two (different) ligands.52 This would indicate that the SH3-fold-based structure of MIA harbors an extended ligand binding site as shown for other SH3 proteins.14,32,40,48,53–56 The current study has now characterized the molecular interaction between MIA and fibronectin and found that, interestingly, the distal loop of human MIA is also involved in contacting fibronectin39,40 (Fig. 6). The chemical shift perturbation analysis clearly shows that the site of interaction for MIA on FN13 fully overlaps with the heparin binding site on the HEP II domain of fibronectin (Fig. 4) and is mainly mediated through electrostatic forces from the cationic fork. Obviously, the heparin binding site that is constituted by eight amino acids fully overlaps with the MIA binding site on the FN13 surface. In comparison, significant chemical shift perturbations could be detected for more than eight amino acids of FN13 upon binding to MIA (Figs. 3–5). This is in concordance with the bigger molecular interface between FN13 and MIA in comparison to the FN13/heparin complex.
The Hep II domain of fibronectin comprises the type III module 12–14 and is the most important fibronectin interaction site for heparin.32,37 The FN13 module exhibits the highest affinity for heparin as it contains a structural element known as the cationic fork.21–24 This structural feature is composed of six amino acids, R98, R99, R101, R115, K117, and R146. The basic side chains of these residues contact the sulfated sugar heparin.37 In addition to sugar binding, the Hep II domain of fibronectin can also bind to the integrin α4β1 receptor.29 This interaction is mediated by a PRARI motif located within fibronectin's type III module FN14 on the opposite side of the cationic fork. This relative orientation is stabilized through a rigid linker between FN13 and FN14.32 It is well established that integrin influences the adhesion of cells in the extracellular matrix, cell growth, and signal transduction.35,36 Previous studies were able to show that peptide sequences derived from fibronectin's Hep II domain that matched to the FN14 module could interact with MIA.1,6,39 Therefore, MIA could interfere with fibronectin/intregrin complex formation. The current study has now analyzed the interaction between MIA and fibronectin in more detail. Based on this study, all fibronectin type III modules of the Hep II domain—FN12, FN13, and FN14—can interact with human MIA with comparable affinities in the weak micromolar range (Fig. 5). Noticeably, MIA does not only bind to the main heparin contact module FN13, but also to module FN12 and FN14 that are known to fine-tune the FN/heparin interaction.32,34,37 Apparently, MIA does not only antagonize the interaction between fibronectin and integrin but could also interfere with fibronectin/heparin complex formation. This might be the molecular basis for MIA's capability to inhibit the attachment of melanoma cells to the extracellular matrix in vitro and its ability to promote metastasis in vivo.57,58 The non-canonical FN-binding interface on the MIA surface is located opposite to the (non-functional) canonical binding site for canonical SH3 polyproline ligands (Figs. 2 and 5). MIA mainly interacts through its rather extensive acidic surface patch that includes the distal loop with the basic patches found on each of the three type III modules of the fibronectin Hep II domain. This is in accordance with previous mutagenesis studies on MIA that identified residues of functional relevance within its acidic patch and recently identified low molecular weight antagonists of the MIA/fibronectin interaction.6,39,52
Interestingly, the distal loop is highly flexible on the pico- to nanosecond time scale in the apo-form of human MIA.59 In the HADDOCK-generated models of MIA bound to either FN13 or FN14,39 the distal loop is in contact with fibronectin (Fig. 6). It has been previously suggested that flexible loop regions might be of functional relevance in recognizing (different) ligands.60 Furthermore, through sampling numerous conformations in the flexible distal loop of MIA's apo-state, different ligands might select distinct conformations from the structural ensemble to provide selectivity in binding, thereby shifting the conformational equilibrium of the protein to its bound state. This mechanism has been described as conformational selection.61,62 Interestingly, it has also been shown that heparin itself exhibits intrinsic flexibility.63 Therefore, conformational selection might indeed play a crucial role for proper binding of MIA to the type III modules FN12–14 of the fibronectin Hep II domain.
The current study characterizes the molecular basis for the interaction between MIA and the Hep II domain of fibronectin, which differs from the canonical ligand interaction of classical SH3 domains. The identified acidic fibronectin binding patch of MIA surrounds the small ligand binding pocket, which has recently been identified and shown to be a valuable target for MIA/fibronectin antagonists.39 Therefore, the current study provides a more detailed view on the molecular basis of the MIA/fibronectin interaction that might pave the way for future therapeutic strategies against malignant melanoma disease.
IV. SUMMARY AND CONCLUSIONS
The MIA protein plays a crucial role in the tumor progression of malignant melanoma. Unlike melanocytes, melanoma cells specifically express and secrete this protein in ample amounts. In melanoma patients, the serum levels of MIA correlate with metastasizing disease states III and IV and, thus, MIA can serve—among lactate dehydrogenase and the S100 calcium-binding protein B—as a relevant clinical marker protein. Previously, numerous studies reported that MIA interacts with proteins from the extracellular matrix, such as fibronectin for instance. This interaction competitively interferes with melanoma cell adhesion and, ultimately, contributes to or even promotes metastasis in malignant melanoma. Based on previous investigations that reported fibronectin-derived peptide ligands for MIA, this study focused on the characterization of the MIA/fibronectin complex on an atomic level. After having optimized known peptide ligands for MIA, we analyzed the interaction between MIA and the Hep II domain of fibronectin using multidimensional heteronuclear NMR spectroscopy. Here, we could show that MIA binds to the type III modules 12, 13, and 14 of fibronectin that constitute the Hep II domain. In addition, we have mapped the binding interface between MIA and FN13 at atomic resolution. These results now provide a molecular basis for a detailed understanding of the MIA/FN interaction, which is of physiological importance for metastasis in malignant melanoma. Finally, the current study provides a detailed description of the target MIA/FN protein–protein binding interface at atomic resolution for the discovery of novel antimetastatic agents that might serve as a starting point for future therapeutic strategies against malignant melanoma.
ACKNOWLEDGMENTS
K.T.Y. has been supported by fellowships from the Schäfersnolte-Gedächtnis-Stiftung, the SFB 642, and the Jürgen Manchot Foundation. The authors gratefully acknowledge generous support from the RUB Research SchoolPlus and the Else Kröner-Fresenius-Stiftung.
References
- 1. Schmidt J. et al. , PLoS One 7, 1 (2012). 10.1371/journal.pone.0037941 [DOI] [Google Scholar]
- 2. Bosserhoff Anja-Katrin, Kaufmann Martin, Kaluza Brigitte, Bartke Ilse, Zirngibl Hubert, Hein Rüdiger, Stolz Wilhelm, and Buettner Reinhard, Cancer Res. 57, 3149 (1997). [PubMed] [Google Scholar]
- 3. Sanmamed M. F. et al. , Clin. Chim. Acta 429, 168 (2014). 10.1016/j.cca.2013.11.034 [DOI] [PubMed] [Google Scholar]
- 4. Lougheed J. C., Holton J. M., Alber T., Bazan J. F., and Handel T. M., Proc. Natl. Acad. Sci. U.S.A. 98, 5515 (2001). 10.1073/pnas.091601698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lougheed J. C., Domaille P. J., and Handel T. M., J. Biomol. NMR 22, 211 (2002). 10.1023/A:1014961408029 [DOI] [PubMed] [Google Scholar]
- 6. Stoll R. et al. , EMBO J. 20, 340 (2001). 10.1093/emboj/20.3.340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Stoll R. and Bosserhoff A., Curr. Protein Pept. Sci. 9, 221 (2008). 10.2174/138920308784534014 [DOI] [PubMed] [Google Scholar]
- 8. Bosserhoff A.-K., Stoll R., Sleeman J. P., Bataille F., Buettner R., and Holak T. A., Lab. Invest. 83, 1583 (2003). 10.1097/01.LAB.0000097191.12477.5D [DOI] [PubMed] [Google Scholar]
- 9. Scott D. E., Coyne A. G., Hudson S. A., and Abell C., Biochemistry 51, 4990 (2012). 10.1021/bi3005126 [DOI] [PubMed] [Google Scholar]
- 10. Dummer R. et al. , J. Clin. Oncol. 31, 9028 (2013). [Google Scholar]
- 11. Irwin J. J., Sterling T., Mysinger M. M., Bolstad E. S., and Coleman R. G., J. Chem. Inf. Model. 52, 1757 (2012). 10.1021/ci3001277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Weisel M., Proschak E., and Schneider G., Chem. Central J. 17, 1 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dalgarno D. C., Botfield M. C., and Rickles R. J., Biopolymers 43, 383 (1997). 10.1002/(SICI)1097-0282(1997)43:5%3C383::AID-BIP4%3E3.0.CO;2-R [DOI] [PubMed] [Google Scholar]
- 14. Congreve M., Carr R., Murray C., and Jhoti H., Drug Discovery Today 8, 876 (2003). 10.1016/S1359-6446(03)02831-9 [DOI] [PubMed] [Google Scholar]
- 15. Clarkson J. and Campbell I. D., Biochem. Soc. Trans. 31, 1006 (2003). 10.1016/S1359-6446(03)02831-9 [DOI] [PubMed] [Google Scholar]
- 16. Kim K. B., Chesney J., Robinson D., Gardner H., Shi M. M., and Kirkwood J. M., Clin. Cancer Res. 17, 7451 (2011). 10.1158/1078-0432.CCR-11-1747 [DOI] [PubMed] [Google Scholar]
- 17. Grimaldi A. M., Cassidy P. B., Leachmann S., and Ascierto P. A., Cancer Treat. Res. 159, 443 (2014). 10.1007/978-3-642-38007-5_25 [DOI] [PubMed] [Google Scholar]
- 18. Pellecchia Maurizio, Meininger David, Dong Qing, Chang Edcon, Jack Rick, and Sem Daniel S., J. Biomolecular NMR 21, 165 (2002). [DOI] [PubMed] [Google Scholar]
- 19. Zuiderweg E. R. P., Biochemistry 41, 1 (2002). 10.1021/bi011870b [DOI] [PubMed] [Google Scholar]
- 20. Humphries M. J., Obara M., Olden K., and Kenneth M., Cancer Investigation 7, 373 (1989). 10.3109/07357908909039866 [DOI] [PubMed] [Google Scholar]
- 21. Mosher D. F. and Furcht L. T., J. Invest. Dermatol. 77, 175 (1981). 10.1111/1523-1747.ep12479791 [DOI] [PubMed] [Google Scholar]
- 22. Pankov R. and Yamada K. M., J. Cell Sci. 115, 3861 (2002). 10.1242/jcs.00059 [DOI] [PubMed] [Google Scholar]
- 23. Singh P., Carraher C., and Schwarzbauer J. E., Annu. Rev. Cell Dev. Biol. 26, 397 (2010). 10.1146/annurev-cellbio-100109-104020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. To W. S. and Midwood K. S., Fibrog. Tissue Repair 4, 1 (2011). 10.1186/1755-1536-4-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ayad S., Boot-Handford R. P., Humphries M. J., Kadler K. E., and Shuttleworth C. A., The Extracellular Matrix ( Harcourt Brace and Company, San Diego, CA, 1998). [Google Scholar]
- 26. Gao M., Craig D., Lequin O., Campbell I. D., Vogel V., and Schulten K., Proc. Natl. Acad. Sci. U.S.A. 100, 14784 (2003). 10.1073/pnas.2334390100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sachchidanand, Lequin Olivier, Staunton David, Mulloy Barbara, Forster Mark J., Yoshida Keiichi, and Campbell Iain D., J. Biol. Chem. 277, 50629 (2002). 10.1074/jbc.M208956200 [DOI] [PubMed] [Google Scholar]
- 28. Nikitovic D., Mytilinaiou M., Berdiaki A., Karamanos N. K., and Tzanakakis G. N., Biochim. Biophys. Acta 1840, 2471 (2014). 10.1016/j.bbagen.2014.01.031 [DOI] [PubMed] [Google Scholar]
- 29. Peterson J. A., Sheibani N., David G., Garcia-Pardo A., and Peters D. M., J. Biol. Chem. 280, 6915 (2005). 10.1074/jbc.M406625200 [DOI] [PubMed] [Google Scholar]
- 30. Dalton B. A., Mcfarland C. D., Underwood P. A., and Steele J. G., J. Cell Sci. 2092, 2083 (1995). [DOI] [PubMed] [Google Scholar]
- 31. Mostafavi-Pour Z., Askari J. A., Whittard J. D., and Humphries M. J. U., Matrix Biol. 20, 63 (2001). 10.1016/S0945-053X(00)00131-1 [DOI] [PubMed] [Google Scholar]
- 32. Sharma A., Askari J. A., Humphries M. J., Jones E. Y., and Stuart D. I., EMBO J. 18, 1468 (1999). 10.1093/emboj/18.6.1468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pechik I., Nachman J., Ingham K., and Gilliland G. L., Proteins Struct. Funct. Genet. 16, 43 (1993). 10.1002/prot.340160105 [DOI] [PubMed] [Google Scholar]
- 34. Benecky M. J., Kolvenbach C. G., Amrani D. L., and Mosesson M. W., Biochemistry 27, 7565 (1988). 10.1021/bi00419a058 [DOI] [PubMed] [Google Scholar]
- 35. Akiyama S. K., Olden K., and Yamada K. M., Cancer Metastasis Rev. 14, 173 (1995). 10.1007/BF00690290 [DOI] [PubMed] [Google Scholar]
- 36. Johansson S., Svineng G., Wennerberg K., Armulik A., and Lohikangas L., Front. Biosci. 2, 126 (1997). [DOI] [PubMed] [Google Scholar]
- 37. Busby T. F., Argraves W. S., Brew S. A., Pechik I., Gilliland G. L., and Ingham K. C., J. Biol. Chem. 270, 18558 (1995). 10.1074/jbc.270.31.18558 [DOI] [PubMed] [Google Scholar]
- 38. Humphries J. D., Byron A., and Humphries M. J., J. Cell Sci. 119, 3901 (2006). 10.1242/jcs.03098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yip King Tuo, Zhong Xue Yin, Seibel Nadia, Pütz Stefanie, Autzen Jasmin, Gasper Raphael, Hofmann Eckhard, Scherkenbeck Jürgen, and Stoll Raphael, Sci. Rep. 6, 25119 (2016). 10.1038/srep25119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Shuker S. B., Hajduk P. J., Meadows R. P., and Fesik S. W., Sci. Rep. 274, 1531 (1996). 10.1126/science.274.5292.1531 [DOI] [PubMed] [Google Scholar]
- 41. Stoll R., Renner C., Ambrosius D., Golob M., Voelter W., Buettner R., Bosserhoff A.-K., and Holak T. A., J. Biomol. NMR 17, 87 (2000). 10.1023/A:1008306918293 [DOI] [PubMed] [Google Scholar]
- 42. Liu Maili, Mao Xi-an, Ye Chaohui, Huang He, Nicholson Jeremy K., and Lindon John C., J. Mag. Res. 129, 125 (1998). 10.1006/jmre.1998.1405 [DOI] [PubMed] [Google Scholar]
- 43. Delaglio Frank, Grzesiek Stephan, Vuister Geerten W., Zhu Guang, Pfeifer John, and Bax Ad, J. Biol. Chem. 6, 277 (1995). 10.1007/BF00197809 [DOI] [PubMed] [Google Scholar]
- 44. Vranken W. F. et al. , Proteins: Structure, Function, Bioinformatics 696, 687 (2005). 10.1002/prot.20449 [DOI] [Google Scholar]
- 45. Maurer T. et al. , Proc. Natl. Acad. Sci. U.S.A. 109, 5299 (2012). 10.1073/pnas.1116510109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Schumann F. H., Riepl H., Maurer T., Gronwald W., Neidig K.-P., and Kalbitzer H. R., J. Biomol. NMR 39, 275 (2007). 10.1007/s10858-007-9197-z [DOI] [PubMed] [Google Scholar]
- 47. Fielding L., Progress NMR Spect. 51, 219 (2007). 10.1016/j.pnmrs.2007.04.001 [DOI] [Google Scholar]
- 48. Dominguez C., Boelens R., and Bonvin A. M. J. J., J. Am. Chem. Soc. 125,1731 (2003). 10.1021/ja026939x [DOI] [PubMed] [Google Scholar]
- 49. Brünger A. T. et al. , Acta Crystallogr., Sect. D 54, 905 (1998). 10.1107/S0907444998003254 [DOI] [PubMed] [Google Scholar]
- 50. Trellet M., Melquiond A. S. J., and Bonvin A. M. J. J., PLoS One 8, e58769 (2013). 10.1371/journal.pone.0058769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. DeLano W. L., “ The PyMOL Molecular Graphics System,” De-Lano Scientific, San Carlos, CA, 2002, http://www.pymol.org.
- 52. Stoll R., Lodermeyer S., and Bosserhoff A.-K., Biol. Chem. 387, 1601 (2006). 10.1515/BC.2006.199 [DOI] [PubMed] [Google Scholar]
- 53. Seeliger D. and De Groot B. L., J. Comput. Aided Mol. Des. 24, 417 (2010). 10.1007/s10822-010-9352-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Mayer M. and Meyer B., J. Am. Chem. Soc. 123, 6108 (2001). 10.1021/ja0100120 [DOI] [PubMed] [Google Scholar]
- 55. D'Silva L., Ozdowy P., Krajewski M., Rothweiler U., and Holak T. A., J. Am. Chem. Soc. 127, 13220 (2005). 10.1021/ja052143x [DOI] [PubMed] [Google Scholar]
- 56. Shamsara J., Int. J. Comput. Biol. Drug Des. 7, 80 (2014). 10.1504/IJCBDD.2014.058594 [DOI] [PubMed] [Google Scholar]
- 57. Bosserhoff A. K., Hein R., Bogdahn U., and Buettner R., J. Biol. Chem. 271, 490 (1996). 10.1074/jbc.271.1.490 [DOI] [PubMed] [Google Scholar]
- 58. Bosserhoff A.-K., Golob M., Buettner R., Landthaler M., and Hein R., Hautarzt 49, 762 (1998). 10.1007/s001050050822 [DOI] [PubMed] [Google Scholar]
- 59. Stoll R., Renner C., Buettner R., Voelter W., Bosserhoff A.-K., and Holak T. A., Protein Sci. 12, 510 (2003). 10.1110/ps.0222603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Renner C. and Holak T. A., Eur. J. Biochem. 268, 1058 (2001). 10.1046/j.1432-1327.2001.01965.x [DOI] [PubMed] [Google Scholar]
- 61. Vogt A. D. and Cera E., Biochemistry 52, 5723 (2014). 10.1021/bi400929b [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Karassek S. et al. , J. Biolog. Chem. 285, 33979 (2010). 10.1074/jbc.M109.095968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Mulloy B. and Forster M. J., Glycobiology 10, 1147 (2000). 10.1093/glycob/10.11.1147 [DOI] [PubMed] [Google Scholar]