Abstract
Differences in chronic lymphocytic leukemia between the Asian and the Western population are widely known. To further clarify these ethnic differences, we profiled the molecular genetics in a cohort of 83 newly diagnosed patients from Taiwan. In detail, we assessed: (i) the usage and the mutational status of the clonotypic immunoglobulin heavy-chain variable region (IgHV) genes, (ii) the presence of VH CDR3 stereotypes, and (iii) TP53, NOTCH1, SF3B1, BIRC3, and MYD88 mutations. The IgHV gene repertoire was biased and distinct from that observed in the West with the most common IgHV genes being IgHV3-23, IgHV3-7, and IgHV3-48. In terms of IgHV gene mutational status, 63.8% of patients carried mutated rearrangements, whereas 22.4% of patients were assigned to stereotyped subsets (6.9% to major subsets and 15.5% to minor ones). The frequencies of NOTCH1, SF3B1, BIRC3 and MYD88 mutations were 9.6%, 7.2%, 1.2%, and 2.4%, respectively; however, the frequency of TP53 mutations was significantly higher (20.5%). Patients with TP53 mutations or del(17p), SF3B1 mutations and unmutated IgHV had a worse outcome compared to the other patients. In conclusion, the differences observed in IgHV properties suggest different pathogenetic factors implicated in the development of chronic lymphocytic leukemia, while the high frequency of TP53 mutations could in part explain the dismal outcome of these patients in Taiwan
Introduction
Chronic lymphocytic leukemia (CLL) is the most prevalent adult leukemia in the Western world, accounting for 5–11% of non-Hodgkin lymphomas (NHL).1 CLL belongs to indolent NHL, yet its clinical course varies widely.2 In the past 3 decades, many clinical and molecular features have been identified as predictors of outcome or response to therapy. Chromosome aberrations are important prognostic markers. With traditional cytotoxic agents and rituximab as the backbone of therapies, patients harboring 17p13 deletions have a very short survival with a median of less than 3 years, whereas those with 13q14 deletions, trisomy 12, or normal karyotype have much better outcomes, with a median survival up to 10 years or more, and those carrying 11q22-q23 deletions have a survival time in between the aforementioned.3 In addition, the mutational status of the immunoglobulin (IG) gene is one of the most robust prognostic factors; mutated IG is associated with better outcomes, compared to unmutated IG.4 With the advance of next-generation sequencing technology, some recurrent genetic mutations are found to possess prognostic significance, such as the negative survival impacts of TP53, NOTCH1, SF3B1 or BIRC3 mutations and the favorable survival impact of MYD88 mutations.5,6 These molecular and clinical markers of CLL greatly assist in our understanding of disease biology and, perhaps most importantly, in the optimization of individual patient’s management options.
CLL in Asia differs in terms not only of a lower prevalence7,8 but also of different treatment outcomes.9 Up to now, CLL has mostly been viewed as a “Western disease”, and the majority of knowledge is derived from studies in Western populations. This is especially true in terms of novel prognostic factors, which have not been thoroughly studied among Asian patients. Such a gap in knowledge becomes even more compelling since studies concerning the clinical course and the epidemiology of CLL have reported different outcomes in Taiwan and Japan compared to the West.9,10 Thus, any differences in the biological characteristics that may underlie the disparities in prognosis of CLL in Asian populations would be of great interest and value. The study herein was aimed at characterizing and validating the clinical implications of CLL immunogenetic and genetic features in a cohort of Taiwanese CLL patients in order to highlight any potential geographical differences that may underlie the outcome disparities. This knowledge could lead to the refinement of management strategies for Taiwanese CLL patients.
Methods
Patients, treatments, and survival
In total, 83 consecutive patients with newly diagnosed CLL at the National Taiwan University Hospital (NTUH) between February 1994 and December 2006 constituted the study cohort. The clinical characteristics and cytogenetic profiles in 80 of them have been reported previously.11 The diagnosis was made through the findings of blood cell counts, classification, morphology, and immunophenotying, all of which were carried out at the Specialized Hematology Laboratory in NTUH, according to the World Health Organization (WHO) classification.12 The patients’ demographics is summarized in the Online Supplementary Table S1. The study was approved by the Institutional Review Board of the National Taiwan University Hospital; written informed consent in accordance with the Declaration of Helsinki was obtained from all participants.
SF3B1, NOTCH1, MYD88, BIRC3 and TP53 mutations
Gene mutations were determined by polymerase chain reaction (PCR) of genomic DNA extracted from bone marrow samples, followed by direct Sanger sequencing in 83 patients. The PCR protocols of gene mutation analyses for MYD88 (exon 5),13 NOTCH1 (exon 34),14 SF3B1 (exons 14, 15, and 16),15 and TP53 (exons 3 to exon 9)16 were adapted from prior reports, and that for BIRC3 (exons 7, 8, and 9) was designed in-house. The primer sequences are summarized in the Online Supplementary Table S2. The results were compared with the data derived from Western cohorts in plots to delineate the differences and similarities between the populations.5,6
Amplification and analysis of Immunoglobulin (IG) gene rearrangements
Analysis of Immunoglobulin (IG) gene rearrangements was successfully performed in 58 patients. The methods for the amplification of IgHV-IgHD-IgHJ rearrangements sequences were adapted from Ghia et al.17 and Hamblin et al.4 Nucleotide sequences were aligned to the IMGT/V-QUEST database in order to determine the IG heavy-chain genes usage and somatic hypermutation status. The clusters of sequences with common heavy-chain complementarity determining region 3 (HCDR3) motifs, described as “stereotyped” B-cell receptors, were identified by the methods described by Bystry, Agathangelidis and Bikos et al.18 The results were compared with the data derived from Western cohorts in plots to delineate the differences and similarities between the populations.4,19,20
Conventional cytogenetic analysis and fluorescence in situ hybridization (FISH)
Chromosomal and FISH analyses were performed in 80 patients as previously described.21,22 The FISH panels, including the probes for the centromere of chromosome 12 (CEP12), 13q14.3 (LSI D13S319), 17p13 (p53), and 11q22.3 (ATM), were all from Vysis Inc. (Downers Grove, IL, USA).
Statistical analysis
χ2 or Fisher’s exact tests were used for the between-group comparison of discrete variables. Two-sample t-test was used for the between-group comparison of the means. Kaplan–Meier survival curves were used for the estimation of overall survival (OS). Log-rank test was used to test for the differences in the OS between groups. All the directional P-values were two-tailed and a P-value of <0.05 was considered as significant. All analyses were performed by using the PASW 18 software (SPSS, Inc., Chicago, IL, USA).
Results
CLL IGH gene rearrangement analysis
Using 2% difference from the closest germline gene as the cutoff, IG heavy-chain rearrangements were classified as mutated in 37 of the 58 subjects (63.8%). The median difference from the closest IgHV germline gene was 7.7% (range 2.1–11.9%). The IgHV gene repertoire in Taiwanese patients is summarized in Figure 1A: IgHV3-23, IgHV3-7, and IgHV3-48 genes were those most frequently expressed (17.2%, 15.5%, and 13.8%, respectively). All 10 IgHV3-23 gene rearrangements were mutated, whereas IgHV1-69 and IgHV3-15 rearrangements were unmutated (Figure 1A). IgHV3-23 and IgHV3-7 genes were more common in Taiwan as compared to the West (Figure 1B), whereas IgHV4 genes, especially IgHV4-34, one of the most overexpressed IgHV genes in Western CLL patients,4,19 and IgHV1-69, the most common IgHV gene in European patients,20 were underrepresented in the current study. The frequency of IgHV3-21 was 3.4%, similar to that reported in Ghia et al.20
Figure 1.
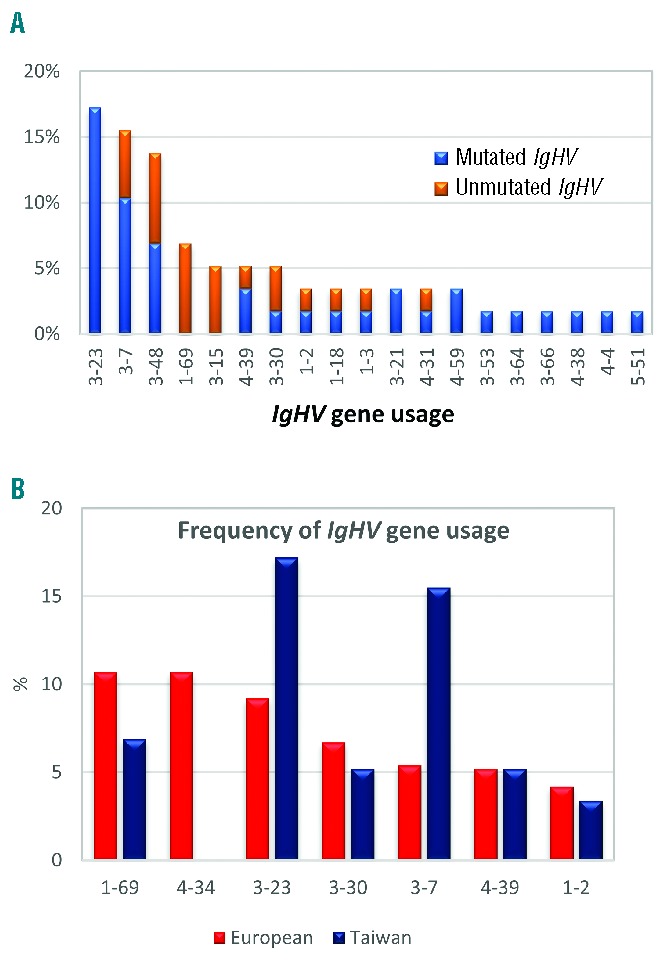
IgHV gene usage. (A) Frequency of the IgHV subgroups and somatic hypermutation status in Taiwanese CLL patients. (B) Comparison between Western and Taiwanese CLL patients regarding the frequency of IgHV gene usage. The data of these common IgHV genes, derived in the West for references, are from the study by Ghia et al.20
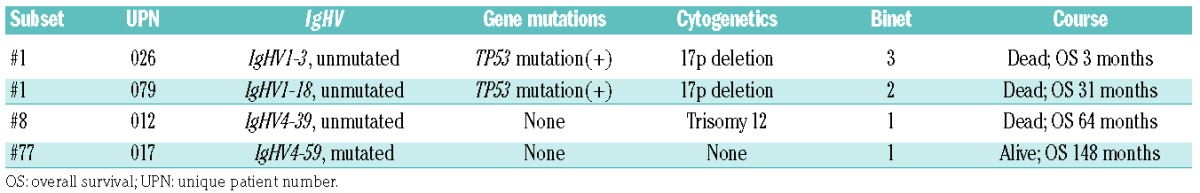
Among the analyzed IG gene rearrangements, 13 (22.4%) were assigned to stereotyped subsets based on a robust bioinformatics algorithm.23,24 Nine cases (15.5%) were assigned to minor subsets, and 4 (6.9%) to major subsets: 2 to subset #1, 1 to subset #8 and 1 to subset #77. The assignment to major subsets was further confirmed through the use of AssignSubsets, a novel major subset assignment tool.18 The clinical course and molecular profile of patients assigned to major subsets are summarized in Table 1. Similar to the report regarding prognostic impact,18 patients in aggressive subsets #1 and #8 had concurrent poor prognostic profiles and much shorter survival, whereas the patient in indolent subset #77 had a more favorable prognostic profile and longer survival.
Table 1.
Brief clinical profiles of patients in major CDR3 stereotype subsets.
Gene mutations in CLL patients
The profiles of gene mutations are summarized in Figure 2A and the Online Supplementary Table S3. Mutations in TP53, NOTCH1, MYD88, SF3B1 or BIRC3 were identified in 30 of the 83 patients (36.1%): 17 patients (20.5%) with TP53 mutations, 8 (9.6%) with NOTCH1 mutations, 6 (7.2%) with SF3B1 mutations, 2 (2.4%) with MYD88 mutations, and 1 (1.2%) with a BIRC3 mutation. Three patients had concurrent mutations in TP53 and NOTCH1, and 1 patient in TP53 and SF3B1. The summary of the gene mutations and cytogenetic changes is shown in Figure 2A. Compared with the data derived from Western populations,5 the frequency of TP53 mutation among newly diagnosed patients was dramatically higher in our cohort, whereas those of other mutations were quite similar (Figure 2B). In addition, a novel TP53 mutation, V31I, was noted in 3 cases in the current cohort. Cytogenetic changes detected by either conventional cytogenetics or FISH were available in 80 patients. Correlations of gene mutations with cytogenetic abnormalities demonstrated that TP53 mutations were closely associated with del(17p); 5 (71.4%) of the 7 patients with del(17p) had TP53 mutations while 12 (16.4%) of the 73 patients without del(17p) had TP53 mutations (P=0.004). On the other hand, SF3B1 mutations were correlated with del(11q); 5 (55.6%) of the 9 patients with del(11q) had SF3B1 mutations while only 1 (1.4%) of the 71 patients without del(11q) had SF3B1 mutations (P<0.001) (Online Supplementary Table S4).
Figure 2.
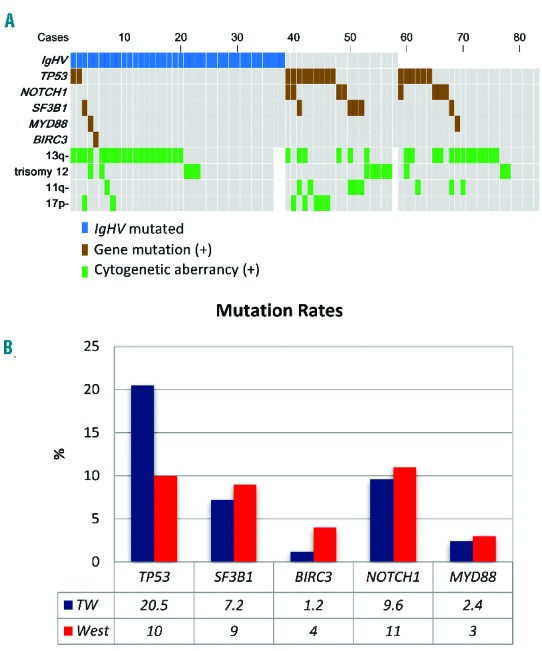
Gene mutation and cytogenetic status. (A) The mutation landscape including IgHV mutation, gene mutation, and cytogenetic aberrancy in the current study. Each column represents an untreated CLL case. (B) The frequencies of common gene mutations in Taiwanese CLL patients and Western CLL patients. The Western data for references are derived from the review by Foà et al.5 and Martinez-Trillos et al.6
Associations between IgHV mutational status and genetic alterations
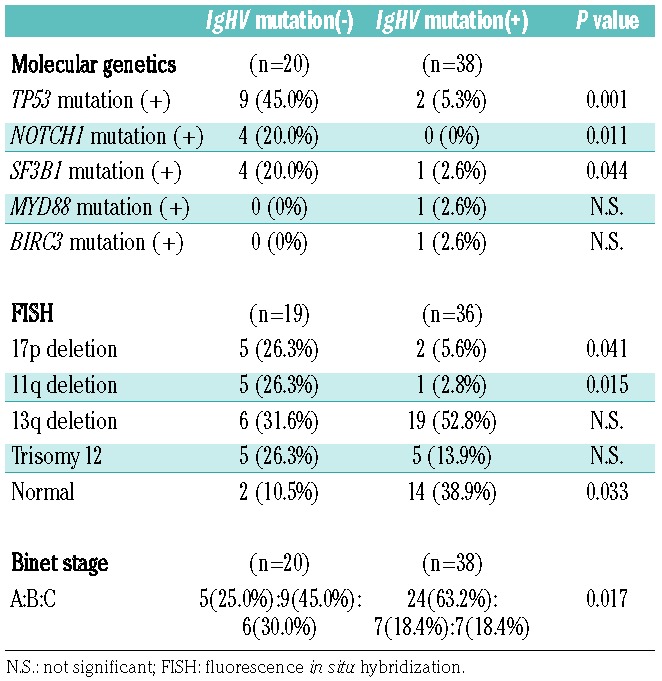
The mutational status of the rearranged IgHV gene correlated closely with the genetic profiles (Table 2). In more detail, patients with unmutated IgHV genes were more likely to have concurrent TP53, NOTCH1, or SF3B1 mutations, compared to those with mutated IgHV genes (45.0% vs. 5.3%, P=0.001; 20.0% vs. 0%, P=0.011; and 20.0% vs. 2.6%, P=0.044, respectively). Furthermore, the unmutated IgHV gene was correlated with del(17p) and del(11q) (26.3% vs. 5.6%, P=0.041; and 26.3% vs. 2.8%, P=0.015, respectively). Lastly, patients with mutated IgHV genes were diagnosed with early disease stage in comparison with unmutated cases (63.2% vs. 25.0% at Binet stage A, P=0.017).
Table 2.
The correlations of IgHV mutation status and other molecular genetics, cytogenetics, and clinical stages.
The prognostic impact of gene mutations
Novel gene mutations correlated with the patients OS: patients with either del(17p) or TP53 mutations had significantly shorter OS compared to those with neither aberrations (41.8 months vs. 86.8 months, P=0.048, Figure 3A). SF3B1 mutations were also associated with shorter OS (11.1 months vs. 86.8 months, P<0.001, Figure 3B), whereas patients with mutated IgHV genes had longer OS (89.2 months vs. 40.0 months, P=0.001, Figure 3C).
Figure 3.

The impact of molecular genetics on CLL patients overall survival in Taiwan. (A) The OS for patients with or without 17p deletion or TP53 mutation. (B) The OS for patients with or without SF3B1 mutation. (C) The OS for patients with or without IgHV gene mutation.
Discussion
It is well known that the incidence and prevalence of cases of CLL are much lower in Eastern countries.7,8 In addition to this disparity, quite a few studies have demonstrated that the cytogenetic profile and outcome of CLL in this area are different from that observed in the West.7,9,11 As for the molecular genetics of CLL, ethnical/geographical differences between populations have also been reported.8,25 In the study herein, we studied a series of immunogenetic and genetic features which are commonly evaluated at CLL diagnosis in the West, in a cohort of Taiwanese patients. The study results demonstrated that CLL in Taiwan is highly distinct compared to Western CLL in terms of molecular genetic profiles.
With regard to immunogenetics, 63.8% of patients in the current cohort had mutated IG rearrangements, a rate similar to that in the West where 50~60% of patients show somatic hypermutations.4,26 Associations between IgHV gene usage and somatic hypermutation status reported in the West were also evident in Taiwanese patients, with IgHV1-69 gene rearrangements belonging to the unmutated CLL fraction and IgHV3-23 to the mutated one (Figure 1A). On the contrary, the IgHV gene usage distribution in Taiwanese CLL patients was quite different from that in the West, characterized by the underrepresentation of the IgHV1-69 gene and the absence of IgHV4-34 gene rearrangements. A similar profile of IgHV subgroup gene usage was observed between our cohort and mainland Chinese patients, characterized by a higher frequency of the IgHV3 subgroup and a lower frequency of the IgHV4 subgroup compared to the West. However, the distribution of individual IgHV genes in Taiwanese patients was different from that in Western as well as Chinese patients, with the most characteristic example being that of the IgHV4-34 gene, which was the most common gene in Chinese patients,27 yet it was completely absent in our cohort. Since people from mainland China and Taiwan are expected to be similar in terms of genetic background, such differences in IgHV gene usage support the notion that environmental variables with population-specific antigenic selection may play a role in the development of CLL. Although IgHV3-7, IgHV3-26, and IgHV4-34 have been correlated with antibodies against influenza A, influenza B, and I antigen in cold agglutinin disease,28–32 respectively, no apparent racial differences in these diseases have been reported to support the possible associations between chronic stimulation resulting from these antigens or pathogens and CLL development in this region. Regarding stereotyped VH CDR3, although only a few patients were assigned to major subsets, the associations between stereotypes and clinical characteristics held true in the current cohort (Table 1) with subset #1 and #8 patients exhibiting aggressive disease behavior in contrast to the subset #77 patient who showed an indolent disease course.
Frequencies of SF3B1, NOTCH1, MYD88, and BIRC3 mutations in the current cohort were similar to those reported in the West. The only exception was the TP53 gene whose mutation rate at diagnosis was twice as high compared to the West (20.5% vs. 5~10%, Figure 2B),5 which is in line with previous studies in Korea.33 In this context, the fact that TP53 mutations frequently coincide with del(17p) together with the finding that del(17p) is also more common in Taiwan and other Asian cohorts11,34–36 corroborate our findings. Since TP53 mutations can be acquired during disease progression, one might question that this high frequency of TP53 mutations could result from diagnosis delay. However, the mutation analysis was performed on samples obtained at the time of diagnosis of all patients, and the mutation rate in the current Taiwanese cohort was even higher than that of patients after first treatment in the West (8~11%).5 In addition, SF3B1 mutations, which are also frequently acquired during disease progression,5 were not more common in Taiwan. With these observations, delayed diagnosis per se does not appear to be able to explain such a huge disparity of TP53 mutation rates. Since TP53 mutation/del(17p) is one of the strongest poor prognostic factors in CLL,5 the higher mutation rate might partially explain the dismal outcomes of CLL patients in Taiwan. A surprising observation was that the V31I mutation in exon 3 of TP53, which was recurrently identified in our cohort, has not been reported in Western CLL populations. According to the literature, this mutation could be of significance given the fact that it has been found in other hematological malignancies and solid cancers.16,37–42 In addition, findings from a functional study suggested that this mutation correlated with lower transcriptional activity and lower cell proliferation suppressing activity compared to wild-type TP53.43 The mechanisms and effects of this mutation in CLL pathogenesis need to be further clarified. Of note, it has been reported in the literature that around 3% to 12% of cases without del(17p) harbored TP53 mutations,5,44–46 but the mutation rate was higher, up to 16.4% (12 of 73 cases, Online Supplementary Table S4) in the current study. Such a high mutation rate in this population suggests that TP53 mutation analysis is of great importance in assessing the risk and prognosis in patients without del(17p) in Taiwan.
In summary, the distinct usage of IgHV genes in Taiwanese CLL patients suggests different pathogenetic mechanisms with distinct antigenic elements being implicated in the development of the disease. In addition, we demonstrated herein a significantly higher TP53 mutation frequency, a finding compatible with the overall dismal outcome of CLL patients in Taiwan. Further studies are needed to better characterize and understand the actual impact of these differences in the development and progression of CLL among different ethnic and geographical groups.
Supplementary Material
Acknowledgements
This work was supported by a grant from the Ministry of Science and Technology (104WFA0150879), Taiwan, ROC, and a grant from the Hematology Society of Taiwan, Taiwan, ROC. We thank the staff of the Third Core Lab, Department of Medical Research, National Taiwan University Hospital for technical support.
Footnotes
Check the online version for the most updated information on this article, online supplements, and information on authorship & disclosures: www.haematologica.org/content/102/6/1085
References
- 1.Rozman C, Montserrat E. Chronic lymphocytic leukemia. N Engl J Med. 1995; 333(16):1052–1057. [DOI] [PubMed] [Google Scholar]
- 2.Nabhan C, Rosen ST. Chronic lymphocytic leukemia: a clinical review. JAMA. 2014; 312(21):2265–2276. [DOI] [PubMed] [Google Scholar]
- 3.Dohner H, Stilgenbauer S, Benner A, et al. Genomic aberrations and survival in chronic lymphocytic leukemia. N Engl J Med. 2000; 343(26):1910–1916. [DOI] [PubMed] [Google Scholar]
- 4.Hamblin TJ, Davis Z, Gardiner A, Oscier DG, Stevenson FK. Unmutated Ig V(H) genes are associated with a more aggressive form of chronic lymphocytic leukemia. Blood. 1999;94(6):1848–1854. [PubMed] [Google Scholar]
- 5.Foa R, Del Giudice I, Guarini A, Rossi D, Gaidano G. Clinical implications of the molecular genetics of chronic lymphocytic leukemia. Haematologica. 2013;98(5):675–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martinez-Trillos A, Pinyol M, Navarro A, et al. Mutations in TLR/MYD88 pathway identify a subset of young chronic lymphocytic leukemia patients with favorable outcome. Blood. 2014;123(24):3790–3796. [DOI] [PubMed] [Google Scholar]
- 7.Wu SJ, Huang SY, Lin CT, Lin YJ, Chang CJ, Tien HF. The incidence of chronic lymphocytic leukemia in Taiwan, 1986–2005: a distinct increasing trend with birth-cohort effect. Blood. 2010;116(22):4430–4435. [DOI] [PubMed] [Google Scholar]
- 8.Wu SJ, Chiang CJ, Lin CT, Tien HF, Lai MS. A nationwide population-based cross-sectional comparison of hematological malignancies incidences between Taiwan and the United States of America. Ann Hematol. 2016;95(1):165–167. [DOI] [PubMed] [Google Scholar]
- 9.Wu SJ, Chiang CJ, Lin CT, Tien HF, Lai MS. Improving but inferior survival in patients with chronic lymphocytic leukemia in taiwan: a population-based study, 1990–2004. PloS One. 2013;8(4):e62930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tomomatsu J, Isobe Y, Oshimi K, et al. Chronic lymphocytic leukemia in a Japanese population: varied immunophenotypic profile, distinctive usage of frequently mutated IGH gene, and indolent clinical behavior. Leuk Lymphoma. 2010;51(12):2230–2239. [DOI] [PubMed] [Google Scholar]
- 11.Wu SJ, Lin CT, Huang SY, et al. Chromosomal abnormalities by conventional cytogenetics and interphase fluorescence in situ hybridization in chronic lymphocytic leukemia in Taiwan, an area with low incidence–clinical implication and comparison between the West and the East. Ann Hematol. 2013;92(6):799–806. [DOI] [PubMed] [Google Scholar]
- 12.Jaffe ES, H N, Stein H, Vardiman JW. Pathology and genetics of tumours of haematopoietic and lymphoid tissues: WHO/IARC classification of tumours, 3rd edition, 2001. [Google Scholar]
- 13.Varettoni M, Arcaini L, Zibellini S, et al. Prevalence and clinical significance of the MYD88 (L265P) somatic mutation in Waldenstrom’s macroglobulinemia and related lymphoid neoplasms. Blood. 2013; 121(13):2522–2528. [DOI] [PubMed] [Google Scholar]
- 14.Weng AP, Ferrando AA, Lee W, et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306(5694):269–271. [DOI] [PubMed] [Google Scholar]
- 15.Hou HA, Liu CY, Kuo YY, et al. Splicing factor mutations predict poor prognosis in patients with de novo acute myeloid leukemia. Oncotarget. 2016;7(8):9084–9101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hou HA, Chou WC, Kuo YY, et al. TP53 mutations in de novo acute myeloid leukemia patients: longitudinal follow-ups show the mutation is stable during disease evolution. Blood Cancer J. 2015;5:e331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ghia P, Stamatopoulos K, Belessi C, et al. ERIC recommendations on IGHV gene mutational status analysis in chronic lymphocytic leukemia. Leukemia. 2007;21(1):1–3. [DOI] [PubMed] [Google Scholar]
- 18.Bystry V, Agathangelidis A, Bikos V, et al. ARResT/AssignSubsets: a novel application for robust subclassification of chronic lymphocytic leukemia based on B cell receptor IG stereotypy. Bioinformatics. 2015;31(23):3844–3846. [DOI] [PubMed] [Google Scholar]
- 19.Fais F, Ghiotto F, Hashimoto S, et al. Chronic lymphocytic leukemia B cells express restricted sets of mutated and unmutated antigen receptors. J Clin Invest. 1998;102(8):1515–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ghia P, Stamatopoulos K, Belessi C, et al. Geographic patterns and pathogenetic implications of IGHV gene usage in chronic lymphocytic leukemia: the lesson of the IGHV3-21 gene. Blood. 2005;105(4):1678–1685. [DOI] [PubMed] [Google Scholar]
- 21.Ko BS, Tang JL, Lee FY, et al. Additional chromosomal abnormalities and variability of BCR breakpoints in Philadelphia chromosome/BCR-ABL-positive acute lymphoblastic leukemia in Taiwan. Am J Hematol. 2002;71(4):291–299. [DOI] [PubMed] [Google Scholar]
- 22.Huang SY, Yao M, Tang JL, et al. Clinical significance of cytogenetics and interphase fluorescence in situ hybridization analysis in newly diagnosed multiple myeloma in Taiwan. Ann Oncol. 2005;16(9):1530–1538. [DOI] [PubMed] [Google Scholar]
- 23.Carstensen B, P M, Laara E, Hills M. Epi: a package for statistical analysis in epidemiology. R package version 1.1. 40 ed. 2012. [Google Scholar]
- 24.Darzentas N, Hadzidimitriou A, Murray F, et al. A different ontogenesis for chronic lymphocytic leukemia cases carrying stereotyped antigen receptors: molecular and computational evidence. Leukemia. 2010; 24(1):125–132. [DOI] [PubMed] [Google Scholar]
- 25.Marinelli M, Ilari C, Xia Y, et al. Immunoglobulin gene rearrangements in Chinese and Italian patients with chronic lymphocytic leukemia. Oncotarget. 2016; 7(15):20520–20531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Damle RN, Wasil T, Fais F, et al. Ig V gene mutation status and CD38 expression as novel prognostic indicators in chronic lymphocytic leukemia. Blood. 1999;94(6):1840–1847. [PubMed] [Google Scholar]
- 27.Xia Y, Fan L, Wang L, et al. Frequencies of SF3B1, NOTCH1, MYD88, BIRC3 and IGHV mutations and TP53 disruptions in Chinese with chronic lymphocytic leukemia: disparities with Europeans. Oncotarget. 2015;6(7):5426–5434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pascual V, Victor K, Spellerberg M, Hamblin TJ, Stevenson FK, Capra JD. VH restriction among human cold agglutinins. The VH4-21 gene segment is required to encode anti-I and anti-i specificities. J Immunol. 1992;149(7):2337–2344. [PubMed] [Google Scholar]
- 29.Schutte ME, van Es JH, Silberstein LE, Logtenberg T. VH4.21-encoded natural autoantibodies with anti-i specificity mirror those associated with cold hemagglutinin disease. J Immunol. 1993;151(11):6569–6576. [PubMed] [Google Scholar]
- 30.Krause JC, Tsibane T, Tumpey TM, et al. Epitope-specific human influenza antibody repertoires diversify by B cell intraclonal sequence divergence and interclonal convergence. J Immunol. 2011;187(7):3704–3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lucas AH, Reason DC. Polysaccharide vaccines as probes of antibody repertoires in man. Immunol Rev. 1999;171(1):89–104. [DOI] [PubMed] [Google Scholar]
- 32.Xu GJ, Kula T, Xu Q, et al. Viral immunology. Comprehensive serological profiling of human populations using a synthetic human virome. Science. 2015;348(6239):aaa0698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim J-A, Hwang B, Park SN, et al. Ethnic difference in genomic profiles of chronic lymphocytic leukemia in Korea: targeted exome sequencing and molecular cytogenetics. Blood. 2015;126(23):1726–1726. [Google Scholar]
- 34.Irons RD, Le A, Bao L, et al. Characterization of chronic lymphocytic leukemia/small lymphocytic lymphoma (CLL/SLL) in Shanghai, China: molecular and cytogenetic characteristics, IgV gene restriction and hypermutation patterns. Leuk Res. 2009;33(12):1599–1603. [DOI] [PubMed] [Google Scholar]
- 35.Qiu HX, Xu W, Cao XS, et al. Cytogenetic characterisation in Chinese patients with chronic lymphocytic leukemia: a prospective, multicenter study on 143 cases analysed with interphase fluorescence in situ hybridisation. Leuk Lymphoma. 2008;49(10):1887–1892. [DOI] [PubMed] [Google Scholar]
- 36.Xu W, Li JY, Pan JL, et al. Interphase fluorescence in situ hybridization detection of cytogenetic abnormalities in B-cell chronic lymphocytic leukemia. Int J Hematol. 2007;85(5):430–436. [DOI] [PubMed] [Google Scholar]
- 37.Kishimoto Y, Murakami Y, Shiraishi M, Hayashi K, Sekiya T. Aberrations of the p53 tumor suppressor gene in human non-small cell carcinomas of the lung. Cancer Res. 1992;52(17):4799–4804. [PubMed] [Google Scholar]
- 38.Mori S, Ito G, Usami N, et al. p53 apoptotic pathway molecules are frequently and simultaneously altered in nonsmall cell lung carcinoma. Cancer. 2004;100(8):1673–1682. [DOI] [PubMed] [Google Scholar]
- 39.Jeck WR, Parker J, Carson CC, et al. Targeted next generation sequencing identifies clinically actionable mutations in patients with melanoma. Pigment Cell Melanoma Res. 2014;27(4):653–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen CH, Dickman KG, Moriya M, et al. Aristolochic acid-associated urothelial cancer in Taiwan. Proc Natl Acad Sci USA. 2012;109(21):8241–8246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang L, Yamaguchi S, Burstein MD, et al. Novel somatic and germline mutations in intracranial germ cell tumours. Nature. 2014; 511(7508):241–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu L, Gu ZH, Li Y, et al. Genomic landscape of CD34+ hematopoietic cells in myelodysplastic syndrome and gene mutation profiles as prognostic markers. Proc Natl Acad Sci USA. 2014;111(23):8589–8594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yamada H, Shinmura K, Okudela K, et al. Identification and characterization of a novel germ line p53 mutation in familial gastric cancer in the Japanese population. Carcinogenesis. 2007;28(9):2013–2018. [DOI] [PubMed] [Google Scholar]
- 44.Zenz T, Vollmer D, Trbusek M, et al. TP53 mutation profile in chronic lymphocytic leukemia: evidence for a disease specific profile from a comprehensive analysis of 268 mutations. Leukemia. 2010;24(12):2072–2079. [DOI] [PubMed] [Google Scholar]
- 45.Zenz T, Eichhorst B, Busch R, et al. TP53 mutation and survival in chronic lymphocytic leukemia. J Clin Oncol. 2010;28(29):4473–4479. [DOI] [PubMed] [Google Scholar]
- 46.Yu L, Kim HT, Kasar SN, et al. Survival of del17p CLL depends on genomic complexity and somatic mutation. Clin Cancer Res. 2017;23(3):735–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


