Abstract
Mature B-cell non-Hodgkin lymphoma is the most common subtype of non-Hodgkin lymphoma in childhood and adolescence. B-cell non-Hodgkin lymphomas are further classified into histological subtypes, with Burkitt lymphoma and Diffuse large B-cell lymphoma being the most common subgroups in pediatric patients. Translocations involving the MYC oncogene are known as relevant but not sufficient for Burkitt lymphoma pathogenesis. Recently published large-scale next-generation sequencing studies unveiled sets of additional recurrently mutated genes in samples of pediatric and adult B-cell non-Hodgkin lymphoma patients. ID3, TCF3 and CCND3 are potential drivers of Burkitt lymphomagenesis. In the study herein, frequency and clinical relevance of mutations in ID3, TCF3 and CCND3 were analyzed within a well-defined cohort of 84 uniformly diagnosed and treated pediatric B-cell non-Hodgkin lymphoma patients of the Berlin-Frankfurt-Münster group. Mutation frequency was 78% (ID3), 13% (TCF3) and 36% (CCND3) in Burkitt lymphoma (including Burkitt leukemia). ID3 and CCND3 mutations were associated with more advanced stages of the disease in MYC rearrangement positive Burkitt lymphoma. In conclusion, ID3-TCF3-CCND3 pathway genes are mutated in more than 88% of MYC-rearranged pediatric B-cell non-Hodgkin lymphoma and the pathway may represent a highly relevant second hit of Burkitt lymphoma pathogenesis, especially in children and adolescents.
Introduction
Non-Hodgkin lymphoma (NHL) belong to the most common hematological malignancies in childhood and adolescence.1 About two thirds of all pediatric NHL belong to mature B-cell lymphomas, with Burkitt lymphoma (BL) and Diffuse large B-cell Lymphoma (DLBCL) representing the most prevalent entities in this subgroup.2,3 During the past decades remarkable increases in patient survival were achieved by continuous advancement of treatment approaches worldwide.4 Nowadays, risk-stratified polychemotherapy treatment reaches up to a total of 90% probability of event-free survival (pEFS) in pediatric NHL.5 More than 95% of all pediatric patients diagnosed with mature B-cell lymphoma in Germany are registered in the clinical trials of the non-Hodgkin Lymphoma - Berlin-Frankfurt-Münster (NHL-BFM) study group and are treated according to standardized treatment plans.
Classical genetic and molecular pathological studies on the pathogenesis of B-cell lymphoma provided distinct pathogenetic features, like translocation of the tumor oncogene MYC in BL6 and specific molecular gene expression signatures in DLBCL.7 MYC translocation was shown to be involved in cell cycle regulation, cellular growth, metabolism and apoptosis.6,8,9 However, MYC translocation alone is not sufficient to initiate malignant transformation of B cells.10,11 In DLBCL multiple co-acting molecular alterations have been described. Differentiation between activated B-cell-like (ABC) and germinal center B-cell-like (GCB) by gene expression is well established, profiling revealed differences in prognosis, especially in adult patients.12
Recent next-generation sequencing (NGS) studies provided valuable insight into the landscape of genomic alterations in B-cell non-Hodgkin lymphoma (B-NHL) and independently introduced Inhibitor of DNA binding 3 (ID3) to be recurrently mutated in BL.13–15 ID3 encodes for a helix-loop-helix (HLH) protein that typically lacks a basic DNA-binding domain and therefore inhibits other HLH proteins from binding to their transcriptional target sites by heterodimerization.16–18 One such ID3-inhibited protein is Transcription Factor 3 (TCF3), which is consecutively expressed at high levels during B-cell development.19,20 TCF3 itself was also shown to be recurrently mutated in BL in the transcriptional study from Schmitz and colleagues, who additionally showed that both TCF3 and ID3 mutations resulted in increased expression of TCF3 targets,14 promoting growth and survival by activation of B-cell receptor signaling. A direct target of TCF3 is the cell cycle regulating Cyclin D3 (CCND3),21,22 which was also shown to harbor activating mutations in different subtypes of B-NHL cases.13,14 The above mentioned studies described ID3 mutations to accumulate in the HLH domain and functional analyses showed ID3 mutant proteins to be less effective or completely ineffective in inhibiting TCF3, thus forcing increased cell proliferation and survival via phosphoinositide 3-kinase (PI3K) and Cyclin D3.13–15 While TCF3 mutations also affected the basic HLH (bHLH) domain of its isoform E47, TCF3 mutant proteins did not lose their effect on downstream targets when compared to wild-type TCF3, but displayed ID3/TCF3 interaction, turning them immune to the inhibitory effect of ID3.14 CCND3 mutant proteins showed an increase in cell cycle stimulation when compared to unaffected CCND3, thereby indicating a gain-of-function.14 In summary, mutations in each of the candidate genes are thought to contribute to cellular growth, cell survival and proliferation.23,24
Within the index studies there was a large variation in the incidence of ID3 mutations in BL. The frequency of ID3 mutations varied between 34% (Love et al.), 58% (Schmitz et al.) and 68% (Richter et al.) (Online Supplementary Table S1). Schmitz et al. reported TCF3 mutations in 27% and additional CCND3 mutations in 38% of sporadic BL cases. CCND3 mutations were also analyzed in the study by Richter et al., who also reported 38% of the cases to display these aberrations.13–15
In this study we analyzed a well-defined cohort of 84 pediatric B-NHL patients, diagnosed and treated according to the NHL-BFM protocols for mutations in ID3, TCF3 and CCND3 to describe the incidence and relevance of such mutations in a uniformly diagnosed and treated representative pediatric cohort. Furthermore, we analyzed samples from 96 pediatric patients diagnosed with precursor B-cell acute lymphoblastic leukemia (pB-ALL), to examine whether disruption of this pathway also occurs in this precursor B-cell malignancy.
Methods
Patient samples
1117 Pediatric patients diagnosed with “Burkitt lymphoma”, “Burkitt leukemia”, “Diffuse large B-cell lymphoma” or “B-cell lymphoma, unclassifiable, with features intermediate between diffuse large B-cell lymphoma and Burkitt lymphoma” between January 2000 and December 2012 were eligible for the recruited population based study cohort. Pretreatment tumor samples from fresh frozen tissue, bone marrow or effusion samples were available for 84 patients (“study cohort”). For a more robust examination regarding the relevance of mutation status with respect to patient outcome, initial tumor samples from an additional 10 patients with a known history of relapse or progress were analyzed as the “extended cohort”.
All patients analyzed were registered in the NHL-BFM data center and treated according to the NHL-BFM protocols (NHL-BFM 95 and analogous protocol B-NHL BFM 04).3
Tumor DNA samples from 96 pediatric patients diagnosed with precursor B-ALL were kindly provided by the ALL-BFM study center, University of Kiel, Germany. All patients had previously been diagnosed between 2000 and 2006 and were treated according to the ALL-BFM 2000 protocol.25 More detailed clinical characteristics of the analyzed patients can be found in the online supplement (Online Supplementary Methods).
This study was approved by the Ethical Advisory Board of the University of Giessen, Germany (A89/11 Amendment 2013).
ID3, TCF3 and CCND3 mutation analysis
In the study cohort the full coding region of the ID3 gene, exon 17 of the TCF3 gene and the coding region of CCND3 exon 5 were sequenced. pB-ALL samples were analyzed for ID3 mutations only. Cases presenting with mutations were confirmed within a repetition experiment. More detailed descriptions of primer pairs, sequencing modalities, reference sequence annotation and exclusion of singular nuclear polymorphisms are given in the online supplement (Online Supplementary Methods).
Statistics
Statistical analysis was performed in order to identify differences in typical patient characteristics, such as sex, age, stage of disease, bone marrow (BM) involvement, central nervous system (CNS) involvement, lactate dehydrogenase (LDH) levels, diagnosis, pEFS and probability of overall survival (pOS) according to the mutational status of the analyzed candidate genes. MYC rearrangement status was available from the study database. Clinical data for each calculation referred to patients with successful investigation of the respective criteria. Differences in the distribution of individual parameters among patient subsets were analyzed using Pearson’s χ2 test26 or Fisher’s exact test27 where appropriate. pEFS was calculated according to Kaplan and Meier,28 taking into consideration the time between the date of diagnosis and either the date of event or date of last follow up. pOS was calculated according to Kaplan and Meier28 under consideration of the time between date of diagnosis and death from any cause. Survival estimates were compared by the log-rank test.29 Significant differences were assumed when the respective P value (P) was lower than 0.05. Calculations were conducted using the SAS statistical program (SAS-PC, Version 9.3, Cary, NC, USA: SAS Institute Inc.). Fisher’s exact tests were calculated using the software Prism 6 for Mac OS X (GraphPad Software, Version 6.0c, San Diego, CA, USA). The two-tailed option was used.
Results
Patient characteristics of the study cohort
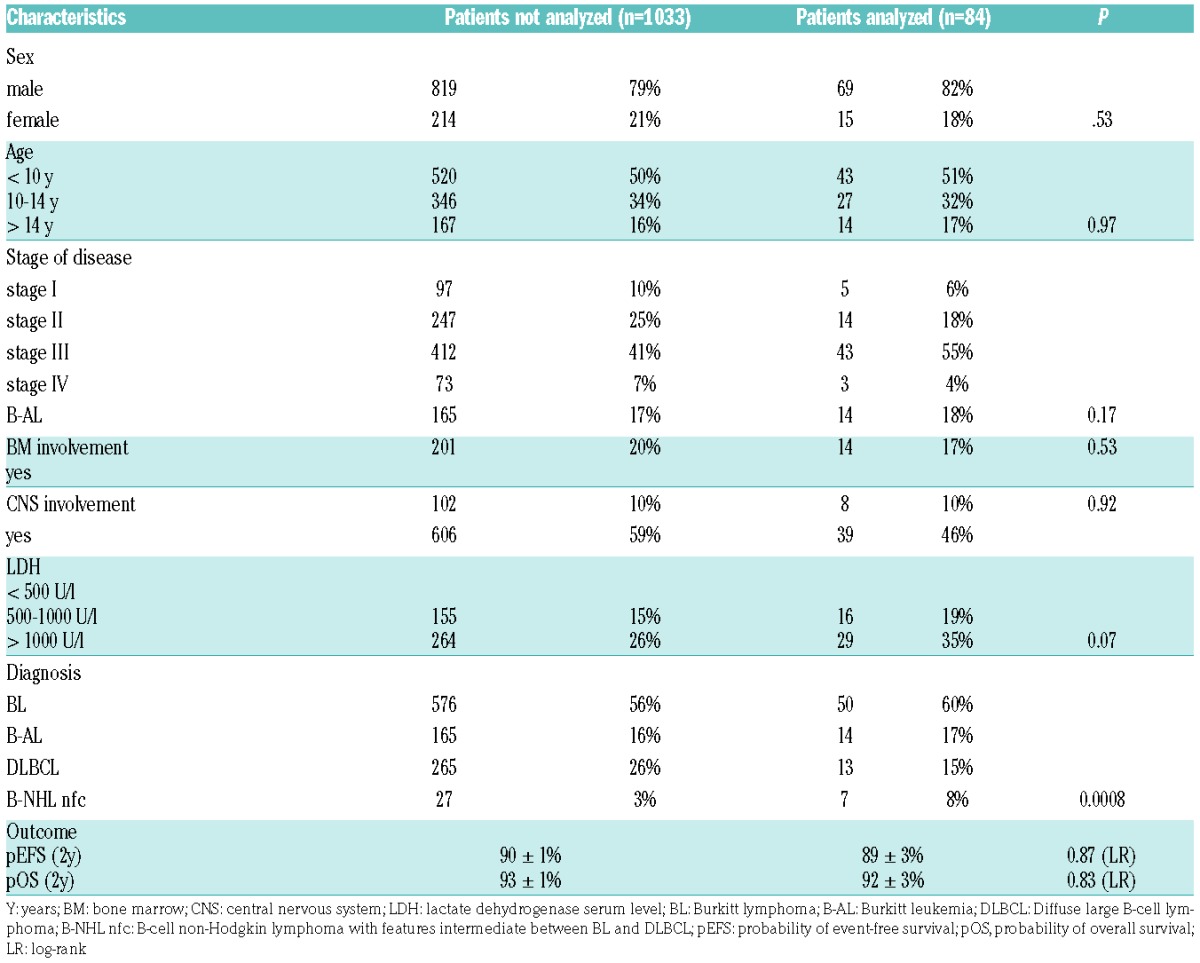
Characteristics of the 84 analyzed patients in the study cohort are shown in Table 1. Clinical characteristics of patients analyzed and not analyzed were similar regarding age, sex, BM involvement, CNS involvement, stage of disease and outcome. Histological subtype was BL in 64 cases (including 14 Burkitt leukemias, B-AL), DLBCL in 13 cases and B-cell lymphoma, unclassifiable, with features intermediate between diffuse large B-cell lymphoma and Burkitt lymphoma (B-NHL not further classifiable [nfc]) in 7 cases. Comparing patient characteristics of the analyzed cohort with the not analyzed patients revealed a trend towards higher LDH levels in the study cohort and an overrepresentation of BL and B-NHL nfc over DLBCL cases. These mild differences, at least in part, might be related to the availability of tissue for molecular analysis. BM obtained in B-AL and tumor after ileocoecal resection of BL was more likely to be sent to the NHL-BFM study center for research than samples of small biopsies of e.g., cervical lymph nodes in DLBCL. These circumstances may also explain the trend towards higher LDH levels in the study cohort, as a high tumor burden is associated with higher LDH levels, which in turn is key to a larger availability of sample material in the study center. To compensate for this slight imbalance in the representation of histological subtypes, the analyses were run for the study cohort and for the subgroup of MYC rearrangement positive BL/B-AL separately.
Table 1.
Patient characteristics of the study cohort.
Incidence and relevance of ID3, TCF3 and CCND3 mutation status were analyzed in the study cohort. A detailed description of all genomic variants including the predicted change on protein level is presented in the Online Supplementary Table S2.
ID3, TCF3 and CCND3 sequencing results
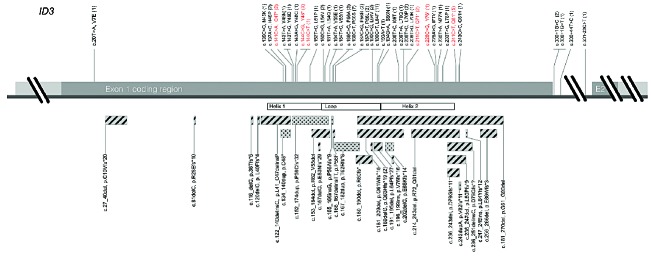
ID3 mutations were found and verified in 56 out of 84 B-NHL samples (Figure 1). Thirty-one of 56 cases showed multiple ID3 mutations, 26 cases with 2 mutations, 4 cases with 3 mutations and 1 case with 4 mutations. Ten of those cases were randomly selected for cloning, and biallelic involvement was shown in all cases. With respect to hotspots, single nucleotide substitutions affecting position C190 were the most frequent (13 cases), followed by C166 (10 cases) and C241 (5 cases). Four disambiguates were genomic variants that were not predicted to result in changes on amino acid levels: 144C>T, 193A>T, 300+44T>C, and 301-23C>T. Notably, each of the cases with one of those silent mutations also harbored at least a second ID3 mutation. On the genomic level, 77 of 93 (83%) mutations directly affected the functional HLH coding region. The remaining 18 mutations were allocated either close to the splice-site of exon 1 (4 mutations), upstream or downstream of the HLH domain (13 mutations) or in the intronic region between exon 1 and 2 (1 mutation). Again, all cases with mutations not directly affecting the HLH domain or the splice-site were associated with at least a second mutation in the HLH domain. The frequency of ID3 mutations according to diagnosis was 50/64 (78%) for BL/B-AL and 2/13 (15%) for DLBCL. In the subgroup of 7 analyzed B-NHL nfc, 4 showed mutations in ID3.
Figure 1.
ID3 gene plot with annotated mutations of the study cohort. ID3 coding region of exon 1 is illustrated with single base-pair substitutions on the upper and more complex alterations (insertions, deletions, InDels, duplications) on the lower site. Substitutions resulting in a nonsense mutation are depicted in red. Hatched bars delineate deletions and InDels, dotted bars characterize insertions and duplications. Each mutation is labeled with a correspondent description on the genomic and protein level, as well as the absolute number of occurrences in brackets. The functional helix-loop-helix domain is mapped according to UniProt (Q02535).
Mutations in TCF3 (8/84, 10%) were considerably lower compared to the high frequency of cases with ID3 and were only found in BL/B-AL cases (8/64, 13%). All 8 mutations occurred in the coding region of the bHLH binding domain of TCF3 (Online Supplementary Figure S1). Mutation 1675G>A was present in 2 cases.
Twenty-six cases harbored CCND3 mutations (Online Supplementary Figure S2). Mutations affected nucleotide C811 with a cytosine duplication in 9 cases, resulting in a protein elongating frameshift. Four cases presented with T869G substitution and 3 cases showed C580T mutations. Twenty-three out of 64 BL/B-AL cases (36%) presented with CCND3 mutations. In DLBCL there were 2 out of 14 cases affected. One mutation was present in a case with a B-NHL nfc diagnosis.
Mutational pattern of ID3, TCF3 and CCND3 and correlation with MYC rearrangement status
In total, 63 out of 84 cases (75%) had at least 1 mutation in 1 of the investigated genes. Exclusive ID3 mutations were the most frequent (51%). This was followed by cases with concurrent ID3 and CCND3 mutation (31%). Cases 33 and 12 harbored mutations in all 3 genes. The pattern of mutations within the study cohort is depicted in Figure 2.
Figure 2.
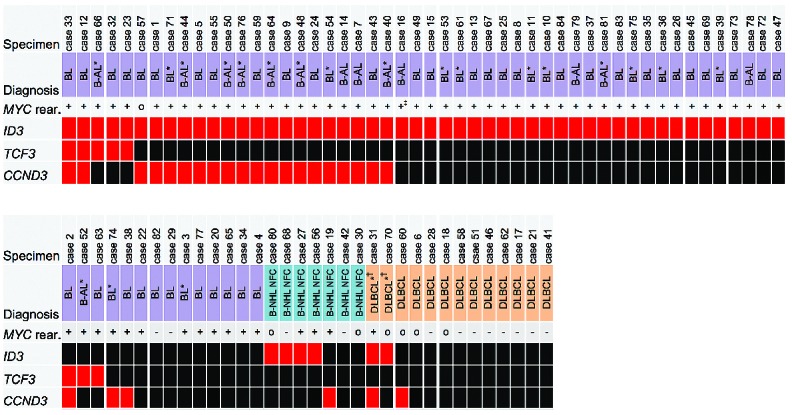
Overview of B-NHL patient sequencing results on ID3, TCF3 and CCND3. Reference diagnosis according to NHL-BFM study. Red block indicates a case with mutation, black block indicates wild-type. MYC rear.: MYC status as reported in the study database; +: MYC rearrangement positive; -: MYC rearrangement negative; o: MYC status unknown. ∗No reference pathology review available. Diagnosis according to study center review. †No reference diagnosis available. Diagnosis according to local pathology report. ‡MYC FISH analysis not available. However, MYC-Ig PCR report was positive for MYC rearrangement. BL: Burkitt lymphoma; B-AL: Burkitt leukemia; DLBCL: Diffuse large B-cell lymphoma; B-NHL nfc: B-NHL unclassifiable, with features intermediate between BL and DLBCL.
Results of fluorescence in situ hybridization (FISH) for detection of MYC rearrangements were available for 77 cases. Fifty-eight of 65 MYC rearrangement positive cases (89%) had at least one mutation in ID3 and/or TCF3 and/or CCND3. In contrast, within 12 MYC rearrangement negative patients only 1 case was affected by ID3 mutations (P<0.0001). In this patient (case 68) ID3 mutations 20T>A and 164T>A were present.
Clinical characteristics according to mutational status in ID3, TCF3 and CCND3
Clinical characteristics and outcome regarding ID3, TCF3 and CCND3 mutational status were first analyzed in the study cohort (Table 2). ID3 mutations were positively correlated with reference diagnosis of BL/B-AL (P=0.0003), higher LDH serum levels (P=0.0038) and higher stage of disease (P=0.03) (Table 2). ID3 mutations occurred at a higher frequency in BL/B-AL cases when compared to DLBCL (P=0.0001). However, these results are strongly biased by diagnosis, as BL/B-AL patients comprised higher LDH serum levels, higher stage of disease and a frequent discovery of ID3 mutations. To investigate the actual clinical relevance of mutations we further analyzed patient characteristics within BL/B-AL MYC rearrangement positive cases (n=61) (Online Supplementary Table S3). ID3 mutated cases were still associated with higher LDH serum levels (P=0.0431). Furthermore, CCND3 mutated cases were positively associated with advanced stage of disease (P=0.0482). Regarding pEFS and pOS, there were no significant differences between wild-type and mutated cases either in the study cohort or in subgroup analyses.
Table 2.
Clinical characteristics of study cohort regarding ID3, TCF3 and CCND3 mutation status.
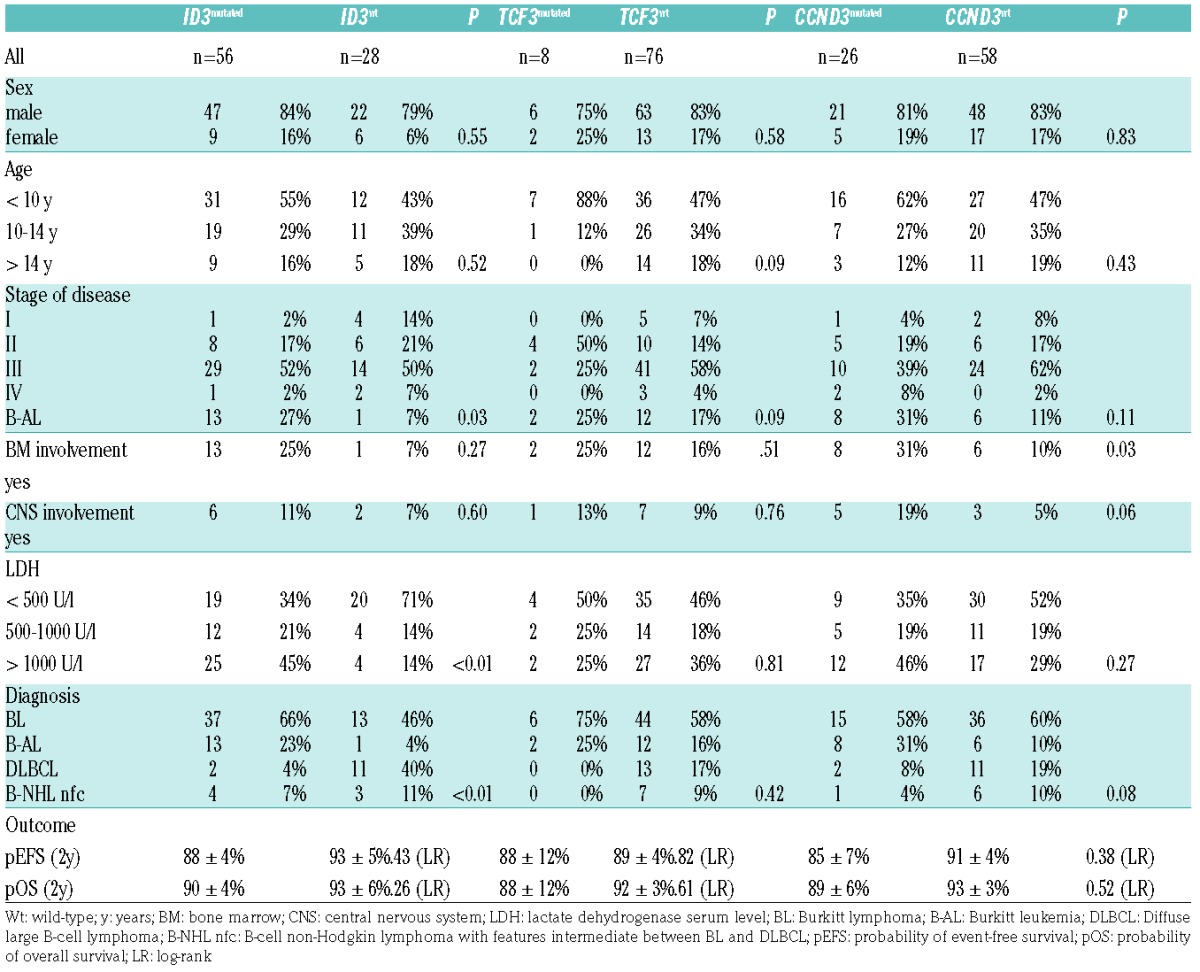
Next we analyzed for clinical relevance of certain mutational patterns. Of particular note was the combination of ID3 and/or TCF3 mutations, these cases were again associated with higher LDH levels (P=0.0023), which is also reflected in an increased frequency of these cases in higher risk groups. Also, in patients with simultaneous and exclusive ID3 and CCND3 mutations, the frequency of BM involvement (P=0.014) and as a consequence a diagnosis of B-AL (P=0.0175) was increased.
Regarding ID3 mutation hotspots it is of note that mutations affecting position 241C, resulting in Q81* nonsense mutation on the protein level, accumulated in cases with B-AL (P=0.0073) compared to BL. Further investigated ID3 hotspots at 190C and 166C did not show association to any clinical criteria.
Outcome and event-free survival with respect to mutational status
In the study cohort 9 out of 84 patients suffered disease progression or relapse. Detailed analyses of ID3, TCF3 and CCND3 mutation frequencies and mutational patterns were not significantly associated with pOS and pEFS (Table 2). This observation was confirmed within the analysis of an additional 10 initial BL samples from patients with a medical history of subsequent disease progression or relapse, adding up to a total of 19 cases with refractory or relapsed disease compared to 75 event-free cases.
ID3 sequencing results in pB-ALL
In the cohort of 96 pediatric pB-ALL patients DNA isolated from leukemic blasts was analyzed for ID3 mutations. There were no pathogenic ID3 mutations found.
Discussion
Burkitt Lymphoma is the most common subtype of NHL in children. With current polychemotherapy treatment regimen event-free survival rates of 90% can be achieved. However, the outcome of patients who suffer from relapse is often fatal. Most patients do not achieve second remission despite intensive salvage treatment. Therefore, new treatment concepts are urgently needed to salvage these patients. New drugs directly targeting pathogenetic pathways of lymphoma cells might represent one possible strategy. However, despite MYC-activating translocations, detectable in the majority of cases,30 little is known about Burkitt pathogenesis and molecular-based risk factors are lacking.
Therefore, the study herein aimed at identifying the frequency and clinical relevance of ID3-TCF3-CCND3 pathway mutations and presents the largest analysis of such mutations in pediatric B-NHL thus far. While the 3 NGS studies cited previously were the first to describe genomic alterations of ID3 and equally evaluated these finding as a new hallmark of BL, there were striking differences regarding the ID3 mutation frequency in the studied cohorts, ranging between 34% and 68%.13–15 Those variations point to relevant differences in inclusion criteria (i.e., histological-/morphological-/study-based vs. molecular-based definitions of BL) and clinical characteristics of the analyzed patients. This was also stressed by Havelange et al., who recently published a series of 13 pediatric and 11 adult BL patients with respect to age-related genetic differences.31 In their cohort they found 10 out of 10 evaluable ID3 mutated cases in the adult group compared to only 5 out of 13 pediatric ones and discussed a potential higher prevalence of ID3 mutations in adults. However, Richter et al. found an age-related correlation of ID3 mutations towards younger patients and another study of only adult BL patients reported a rather low mutation rate of 47%.32 With the finding of 78% ID3 mutations in BL in the current study and comparison of the age structure of the recent studies (Online Supplementary Table S1), we conclude that ID3 mutations occur at high frequency in pediatric BL patients. As patient age itself was not associated with mutation frequency within pediatric cases, these observations lead to the conclusion that ID3 mutation frequency might in fact be associated with more homogeneously presenting pediatric BL and occur less often in the more heterogeneous group of Burkitt-like adult B-cell lymphoma. This is also supported by previously found differences in molecular presentation of BL between pediatric and adult patients, with the general mutational load being significantly higher in older BL patients.33 Furthermore, BL in general is known for homogeneous gene expression profiles, especially in comparison to the related group of DLBCL.34,35 Results from whole genome sequencing of 13 pediatric BL cases of the NHL-BFM group supported these observations on the genomic level, showing a median of only 28 protein changing somatic mutations per tumor and a high frequency of recurrently affected genes, even in the small number of 13 cases.36 The frequency of TCF3 mutations in BL (13%) occurred less often in our cohort, while the incidence of CCND3 mutations in BL (36%) cases was consistent with the findings of other groups.13,14,31
Within the subgroup of 61 MYC rearrangement positive BL, ID3 mutations were significantly associated with a more disseminated presentation of disease and CCND3 showed positive correlation to an advanced stage of disease, supporting their pro-proliferative and cell cycle driving role. These effects became even more evident when evaluating cases with ID3 and/or TCF3 mutations, which can be regarded as equal with respect to the resulting functional disruption of the pathway. The frequency of simultaneous ID3 and CCND3 mutations was significantly higher in patients with BM involvement, hinting at a potential relevance in terms of blast migration. Havelange et al. found a poorer outcome for patients affected by such simultaneous mutations in ID3 and CCND3.31 In our cohort we could not confirm this finding for pediatric patients, and it is, however, difficult to compare pediatric and adult patients with respect to clinical characteristics and prognosis as treatment regimens are generally different and outcome is inferior in adult patients.37 In our study there was no association between clinical outcome and mutational status. These findings represent a contrast to previous data of Richter et al., where superior outcome for patients with ID3 mutations was reported. Again, one possible explanation are the aforementioned general differences between BL in pediatric and adult patients. The high number of ID3 mutations and recurrent involvement of its partners suggest a role of these alterations in Burkitt-lymphomagenesis, rather than a role for disease recurrence in a small subgroup of patients. In the current study, 89% of BL with positive MYC translocation had mutations in at least 1 of the 3 investigated candidate genes, representing affection of the ID3-TCF3-CCND3 pathway in the vast majority of pediatric BL cases (Figure 3).
Figure 3.
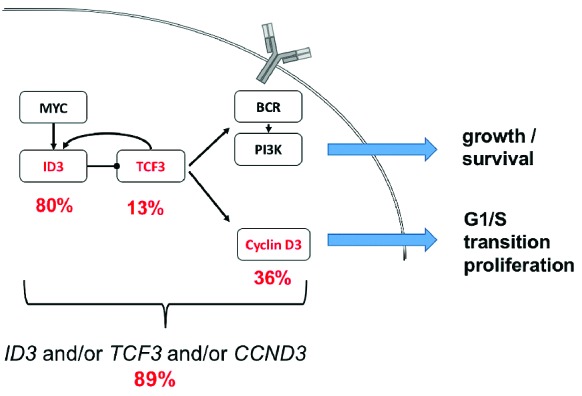
ID3, TCF3, Cyclin D3 pathway with frequencies of respective mutations in MYC rearrangement positive BL. BL with positive MYC translocation had mutations in at least one of the three investigated candidate genes in 89%, representing affection of the ID3-TCF3-CCND3 pathway in the vast majority of pediatric BL cases.
The study cohort included 7 cases of B-NHL nfc and 13 cases of DLBCL. Among them we detected 5 and 3 cases with ID3-TCF3-CCND3 pathway mutations, respectively. Information on the MYC rearrangement status was limited in some of these patients, however, the overall strong association of ID3-TCF3-CCND3 pathway mutations with MYC rearrangements in BL could similarly be observed in B-NHL nfc and DLBCL. Despite the defining histological diagnosis some of those cases show Burkitt-like features with respect to the genetic findings. In many malignancies certain discrepancies between histological and molecular diagnosis have been observed after the establishment of molecular profiling. In this context, proof of ID3-TCF3-CCND3 pathway impairment might be helpful to better discriminate such borderline cases as BL in the future. This is supported by the recently updated World Health Organization (WHO) classification of lymphoid neoplasms, wherein ID3 and TCF3 mutations were added to the molecular characteristics of BL.38
The analysis of 96 samples of pB-ALL patients representing an immature B-cell malignancy did not show any pathogenic ID3 mutations, supposing their exclusive occurrence in mature B-cell lymphoma. Regarding the process of malignant transformation in mature B cells, initial studies attributed the occurrence of the pathognomonic MYC translocations in BL to altered recombination-activating gene (RAG)-mediated recombination, however, more recently it has been widely accepted that aberrant somatic hypermutation processes involving activation-induced cytidine deaminase (AID) lead to these changes.39,40 Regarding ID3, it is of note that mutations were shown to recurrently occur in the RGYW-motif that is favorably affected by AID as well.13 As MYC translocation alone seems not to be sufficient to induce lymphomagenesis,10 one might speculate that subsequent impairment of the investigated pathway serves as a relevant second hit for BL development.
This hypothesis is furthermore supported by the detection of ID3 mutations in mature B-cell malignancies exclusively. In addition, the lack of associations with clinical characteristics or prognosis may even imply an essential function of ID3-TCF3-CCND3 pathway disruption. Cases presenting without mutations might still be affected by focal loss of ID3 or mutations in other functional partners that are involved up- or downstream within the same pathway, likely in the B-cell receptor (BCR), PI3K and cyclin-dependent kinase (CDK)4/6 pathways and their regulators. However, additional candidates will less likely present at similar high frequencies, as NGS studies thus far should have covered most of the highly recurrent genomic events in BL.13–15,41
The overall high number of affected cases asks for therapeutic targeting of this pathway. There is initial promising evidence for successful application and efficacy of the orally available CDK4/6 inhibitor, palbociclib (PD) 0332991, as demonstrated by tumor mass reduction in a BL mouse model by Schmitz et al.14 CDK4/6 inhibitors have also recently been shown to be effective in renal cell carcinoma cell lines and breast cancer cell lines42,43 and are in preparation for clinical phase I and II studies in breast cancer patients (clinicaltrials.gov Identifier: 02297438). Further functional investigation of this pathway will shed more light on molecular processes in BL and hopefully reveal more specific therapeutic options.
In the context of relatively homogeneous genomic alterations in pediatric Burkitt lymphoma, the high number of ID3 mutations found in this study of pediatric B-NHL patients suggests an essential role for this pathway with respect to lymphomagenesis and the phenotype of Burkitt lymphoma.
Supplementary Material
Acknowledgments
We thank the patients, their parents and participating clinics of the NHL-BFM studies.
Footnotes
Check the online version for the most updated information on this article, online supplements, and information on authorship & disclosures: www.haematologica.org/content/102/6/1091
Funding
The research on MYC-positive lymphomas is supported by the BMBF in the framework of E:bio Network Molecular Mechanisms in Malignant Lymphoma with MYC-Deregulation (MMML-MYC-SYS) Grant 0316166. The ICGC MMML-Seq Network is supported by the grants of the BMBF (01KU1002A-J and 01KU1505G). Support of research infrastructure by the Forschungshilfe-Peiper, Giessen, is acknowledged. WK, RS, MS and IO are supported by the Kinderkrebsinitiative Buchholz, Holm-Seppensen (KKI). The research of JL is supported by the Clinician-Scientist Programme of the Cells-in-Motion Cluster of Excellence, Muenster, Germany.
References
- 1.Swerdlow SH, Campo E, Harris NL, et al. (eds.). WHO classification of tumours of hematopoietic and lymphoid tissue. IARC press, Lyon: 2008 [Google Scholar]
- 2.Burkhardt B, Zimmermann M, Oschlies I, et al. The impact of age and gender on biology, clinical features and treatment outcome of non-Hodgkin lymphoma in childhood and adolescence. Br J Haematol. 2005; 131(1):39–49. [DOI] [PubMed] [Google Scholar]
- 3.Woessmann W, Seidemann K, Mann G, et al. The impact of the methotrexate administration schedule and dose in the treatment of children and adolescents with B-cell neoplasms: a report of the BFM Group Study NHL-BFM95. Blood. 2005;105(3):948–958. [DOI] [PubMed] [Google Scholar]
- 4.Rossig C, Juergens H, Schrappe M, et al. Effective childhood cancer treatment: The impact of large scale clinical trials in Germany and Austria. Pediatr Blood Cancer. 2013;60(10):1574–81. [DOI] [PubMed] [Google Scholar]
- 5.Worch J, Rohde M, Burkhardt B. Mature B-Cell lymphoma and leukemia in children and adolescents-review of standard chemotherapy regimen and perspectives. Pediatr Hematol Oncol. 2013;30(6):465–83. [DOI] [PubMed] [Google Scholar]
- 6.Boxer LM, Dang CV. Translocations involving c-myc and c-myc function. Oncogene. 2001;20(40):5595–5610. [DOI] [PubMed] [Google Scholar]
- 7.Miles RR, Raphael M, McCarthy K, et al. Pediatric diffuse large B-cell lymphoma demonstrates a high proliferation index, frequent c-Myc protein expression, and a high incidence of germinal center subtype: Report of the French-American-British (FAB) international study group. Pediatr Blood Cancer. 2008;51(3):369–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sanchez-Beato M, Sanchez-Aguilera A, Piris MA. Cell cycle deregulation in B-cell lymphomas. Blood. 2003;101(4):1220–1235. [DOI] [PubMed] [Google Scholar]
- 9.Dang CV. c-Myc target genes involved in cell growth, apoptosis, and metabolism. Mol Cell Biol. 1999;19(1):1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roschke V, Kopantzev E, Dertzbaugh M, Rudikoff S. Chromosomal translocations deregulating c-myc are associated with normal immune responses. Oncogene. 1997; 14(25):3011–3016. [DOI] [PubMed] [Google Scholar]
- 11.Nepal RM, Zaheen A, Basit W, Li L, Berger SA, Martin A. AID and RAG1 do not contribute to lymphomagenesis in Emu c-myc transgenic mice. Oncogene. 2008;27(34):4752–4756. [DOI] [PubMed] [Google Scholar]
- 12.Rosenwald A, Wright G, Chan WC, et al. The use of molecular profiling to predict survival after chemotherapy for diffuse large-B-cell lymphoma. N Engl J Med. 2002; 346(25):1937–1947. [DOI] [PubMed] [Google Scholar]
- 13.Richter J, Schlesner M, Hoffmann S, et al. Recurrent mutation of the ID3 gene in Burkitt lymphoma identified by integrated genome, exome and transcriptome sequencing. Nat Genet. 2012;44(12):1316–1320. [DOI] [PubMed] [Google Scholar]
- 14.Schmitz R, Young RM, Ceribelli M, et al. Burkitt lymphoma pathogenesis and therapeutic targets from structural and functional genomics. Nature. 2012;490(7418):116–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Love C, Sun Z, Jima D, et al. The genetic landscape of mutations in Burkitt lymphoma. Nat Genet. 2012;44(12):1321–1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Benezra R, Davis RL, Lassar A, et al. Id: a negative regulator of helix-loop-helix DNA binding proteins. Control of terminal myogenic differentiation. Ann N Y Acad Sci. 1990;599:1–11. [DOI] [PubMed] [Google Scholar]
- 17.Benezra R, Davis RL, Lockshon D, Turner DL, Weintraub H. The protein Id: a negative regulator of helix-loop-helix DNA binding proteins. Cell. 1990;61(1):49–59. [DOI] [PubMed] [Google Scholar]
- 18.Murre C. Helix-loop-helix proteins and lymphocyte development. Nat Immunol. 2005;6(11):1079–1086. [DOI] [PubMed] [Google Scholar]
- 19.Hsu LY, Lauring J, Liang HE, et al. A conserved transcriptional enhancer regulates RAG gene expression in developing B cells. Immunity. 2003;19(1):105–117. [DOI] [PubMed] [Google Scholar]
- 20.Beck K, Peak MM, Ota T, Nemazee D, Murre C. Distinct roles for E12 and E47 in B cell specification and the sequential rearrangement of immunoglobulin light chain loci. J Exp Med. 2009;206(10):2271–2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Song S, Cooperman J, Letting DL, Blobel GA, Choi JK. Identification of cyclin D3 as a direct target of E2A using DamID. Mol Cell Biol. 2004;24(19):8790–8802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cato MH, Chintalapati SK, Yau IW, Omori SA, Rickert RC. Cyclin D3 is selectively required for proliferative expansion of germinal center B cells. Mol Cell Biol. 2011;31(1):127–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Campo E. New pathogenic mechanisms in Burkitt lymphoma. Nat Genet. 2012;44(12):1288–1289. [DOI] [PubMed] [Google Scholar]
- 24.Schmitz R, Ceribelli M, Pittaluga S, Wright G, Staudt LM. Oncogenic mechanisms in Burkitt Lymphoma. Cold Spring Harb Perspect Med. 2014;4(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moricke A, Zimmermann M, Valsecchi MG, et al. Dexamethasone vs prednisone in induction treatment of pediatric ALL: results of the randomized trial AIEOP-BFM ALL 2000. Blood. 2016;127(17):2101–2112. [DOI] [PubMed] [Google Scholar]
- 26.Pearson K. X. On the criterion that a given system of deviations from the probable in the case of a correlated system of variables is such that it can be reasonably supposed to have arisen from random sampling. Philos Mag. 1900;50(302):157–175. [Google Scholar]
- 27.Fisher RA. On the Interpretation of 2 from Contingency Tables, and the Calculation of P. J R Stat Soc. 1922;85(1):87–94. [Google Scholar]
- 28.Kaplan EL, Meier P. Nonparametric estimation from incomplete observations. J Am Stat Assoc. 1958;53(282):457–481. [Google Scholar]
- 29.Mantel N. Evaluation of survival data and two new rank order statistics arising in its consideration. Cancer Chemother Rep. 1966;50(3):163–170. [PubMed] [Google Scholar]
- 30.Salaverria I, Martin-Guerrero I, Wagener R, et al. A recurrent 11q aberration pattern characterizes a subset of MYC-negative high-grade B-cell lymphomas resembling Burkitt lymphoma. Blood. 2014; 123(8):1187–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Havelange V, Pepermans X, Ameye G, et al. Genetic differences between paediatric and adult Burkitt lymphomas. British journal of haematology. Br J Haematol. 2016; 173(1):137–144. [DOI] [PubMed] [Google Scholar]
- 32.Forero-Castro M, Robledo C, Lumbreras E, et al. The presence of genomic imbalances is associated with poor outcome in patients with burkitt lymphoma treated with dose-intensive chemotherapy including rituximab. Br J Haematol. 2016;172(3):428–438. [DOI] [PubMed] [Google Scholar]
- 33.Trautmann H, Hadzidimitriou A, Darzentas N, et al. Evidence for antigen-driven development of molecularly classified Burkitt Lymphomas. Blood (ASH Annual Meeting Abstracts). 2009; 114(22):Abstract Number 317. [Google Scholar]
- 34.Lenze D, Leoncini L, Hummel M, et al. The different epidemiologic subtypes of Burkitt lymphoma share a homogenous micro RNA profile distinct from diffuse large B-cell lymphoma. Leukemia. 2011; 25(12):1869–1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hummel M, Bentink S, Berger H, et al. A biologic definition of Burkitt’s lymphoma from transcriptional and genomic profiling. N Engl J Med. 2006;354(23):2419–2430. [DOI] [PubMed] [Google Scholar]
- 36.Rohde M, Schlesner M, Richter J, et al. Pediatric Burkitt Lymphoma of the NHL-BFM group analyzed within the ICGC-MMML-SEQ: Whole genome sequencing data from 12 cases. Hematol Oncol. 2013; 31 Suppl 1(Abstract number 081):96–150.22961993 [Google Scholar]
- 37.Perkins AS, Friedberg JW. Burkitt lymphoma in adults. Hematology Am Soc Hematol Educ Program. 2008;341–348. [DOI] [PubMed] [Google Scholar]
- 38.Swerdlow SH, Campo E, Pileri SA, et al. The 2016 revision of the World Health Organization classification of lymphoid neoplasms. Blood. 2016;127(20):2375–2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Goossens T, Klein U, Kuppers R. Frequent occurrence of deletions and duplications during somatic hypermutation: implications for oncogene translocations and heavy chain disease. Proc Natl Acad Sci USA. 1998;95(5):2463–2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Casellas R, Yamane A, Kovalchuk AL, Potter M. Restricting activation-induced cytidine deaminase tumorigenic activity in B lymphocytes. Immunology. 2009; 126(3):316–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kretzmer H, Bernhart SH, Wang W, et al. DNA methylome analysis in Burkitt and follicular lymphomas identifies differentially methylated regions linked to somatic mutation and transcriptional control. Nat Genet. 2015;47(11):1316–1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Logan JE, Mostofizadeh N, Desai AJ, et al. PD-0332991, a potent and selective inhibitor of cyclin-dependent kinase 4/6, demonstrates inhibition of proliferation in renal cell carcinoma at nanomolar concentrations and molecular markers predict for sensitivity. Anticancer Res. 2013; 33(8):2997–3004. [PubMed] [Google Scholar]
- 43.Finn RS, Dering J, Conklin D, et al. PD 0332991, a selective cyclin D kinase 4/6 inhibitor, preferentially inhibits proliferation of luminal estrogen receptor-positive human breast cancer cell lines in vitro. Breast Cancer Res. 2009;11(5):R77. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.