Abstract
Human rhinoviruses are the most common respiratory viruses detected in patients after hematopoietic cell transplantation. Although rhinovirus appears to occasionally cause severe lower respiratory tract infection in immunocompromised patients, the clinical significance of rhinovirus detection in the lower respiratory tract remains unknown. We evaluated 697 recipients transplanted between 1993 and 2015 with rhinovirus in respiratory samples. As comparative cohorts, 273 recipients with lower respiratory tract infection caused by respiratory syncytial virus (N=117), parainfluenza virus (N=120), or influenza (N=36) were analyzed. Factors associated with mortality were analyzed using Cox proportional hazard models. Among 569 subjects with rhinovirus upper respiratory tract infection and 128 subjects with rhinovirus lower respiratory tract infection, probabilities of overall mortality at 90 days were 6% and 41%, respectively (P<0.001). The survival rate after lower respiratory tract infection was not affected by the presence of co-pathogens (55% in patients with co-pathogens, 64% in patients without, P=0.34). Low monocyte count (P=0.027), oxygen use (P=0.015), and steroid dose greater than 1 mg/kg/day (P=0.003) before diagnosis were significantly associated with mortality among patients with lower respiratory tract infection in multivariable analysis. Mortality after rhinovirus lower respiratory tract infection was similar to that after lower respiratory tract infection by respiratory syncytial virus, parainfluenza virus or influenza in an adjusted model. In summary, transplant recipients with rhinovirus detection in the lower respiratory tract had high mortality rates comparable to viral pneumonia associated with other well-established respiratory viruses. Our data suggest rhinovirus can contribute to severe pulmonary disease in immunocompromised hosts.
Introduction
Human rhinoviruses (HRVs) are the most common cause of respiratory virus infections in both immunocompetent and immunocompromised individuals.1–4 Although well documented cases suggested a possible role of HRV in severe disease,5 earlier cohort studies did not conclusively demonstrate the ability of HRV to cause lower respiratory tract disease, in part because of the presence of co-pathogens as well as a lack of sensitive molecular diagnostic techniques.6,7 With the widespread adoption of multiplex molecular detection platforms, HRV RNA is now detected frequently in the bronchoalveolar lavage (BAL) fluid of immunocompromised patients undergoing evaluation for pulmonary infiltrates.3,8,9 However, the significance of HRV RNA detection in the lower respiratory tract remains poorly defined.
In a recent large prospective study, detection of HRV in the upper respiratory tract of hematopoietic cell transplantation (HCT) candidates was associated with poor outcome after HCT.4 Another study also suggested poor outcomes in immunocompromised patients with HRV infection, comparable to those infected with 2009 H1N1 influenza.10 As for lower respiratory tract infection (LRI), recent studies of community-acquired pneumonia in immunocompetent patients identified HRV as the most common pathogen using molecular diagnostic techniques.1,11 HRV was also a key pathogen in immunocompromised patients originally diagnosed with idiopathic pneumonia syndrome, and its detection in the lower respiratory tract was associated with a particularly poor outcome.12 Overall, these data suggest that HRV may be a clinically significant pathogen with the potential to cause serious pulmonary disease. The purpose of this study was to determine the significance of detection of HRV RNA in the BAL fluid in HCT recipients.
Methods
Study design
This retrospective study includes patients who were transplanted between 1993 and 2015 at the Fred Hutchinson Cancer Research Center (FHCRC) and had virologically-confirmed HRV infection following HCT at the University of Washington Virology Laboratories.13 Only an individual’s first episode of HRV infection was analyzed. HRV upper respiratory tract infection (URI) was defined as HRV detection in a nasopharyngeal sample, and LRI was defined as HRV detection in a BAL or lung biopsy sample. Patients’ demographic data and transplant information closest to the HRV infection were retrieved from the FHCRC database, and other data related to the clinical course of HRV infections were collected by medical chart review. As comparative cohorts, patients with LRI caused by respiratory syncytial virus (RSV), parainfluenza virus (PIV), or influenza virus were included in the analyses.14–16 The first episode of LRI by any of the 4 viruses was selected and the cases with overlapped infections of the 4 viruses were excluded from the analyses. The study was approved by the Institutional Review Board at FHCRC.
Laboratory testing
Nasopharyngeal samples were collected when HCT recipients had URI symptoms and a BAL sample was obtained when patients had lower respiratory tract symptoms and a radiographic abnormality. HRV detection was performed by conventional culture and/or reverse transcription-polymerase chain reaction (RT-PCR) assay in respiratory samples. The culture was performed using 3 different culture systems (rhesus monkey kidney cells [RMK], adenocarcinomic human alveolar basal epithelial cells [A549], and human foreskin fibroblast [HFF]), and BAL samples were incubated for 10 days (an additional 11 days in HFFs culture) at 37°C. RT-PCR to detect HRV has been used routinely since 2007 in our center. RSV, PIV or influenza virus was detected by conventional culture, direct fluorescent antibody tests and/or PCR. The viral load of HRV was roughly estimated from the PCR cycle threshold (Ct) value of the sample that provided the initial diagnosis of HRV LRI.17
All biopsy or autopsy samples were inoculated in 3 different cell lines as above and also tested for HRV using RT-PCR. In autopsy samples, 2 curls from a frozen specimen of each side of lungs were separately tested for HRV using RT-PCR.
Statistical analysis
Patients’ demographic characteristics were summarized and compared between URI and LRI or among LRI with different viruses (HRV, RSV, PIV, and Influenza virus) using chi-square or Fisher’s exact test for categorical variables and Wilcoxon rank sum test for continuous variables (as appropriate). The probability of overall survival was estimated using the Kaplan-Meier method. The probability of mortality after HRV URI was estimated by cumulative incidence curves, treating progression to LRI as a competing risk. The probability of mortality from respiratory failure was estimated by cumulative incidence curves, treating death due to other causes as a competing risk. The log-rank test was used to compare hazards of time-to-event outcomes among patients’ characteristics. Cox proportional hazards models were used to evaluate unadjusted (uHR) and adjusted hazard ratios (aHR) of risk factors for overall mortality or respiratory mortality. Variables with P≤0.05 in the univariable models were candidates for multivariable models. Two-sided P values <0.05 were considered statistically significant. All statistical analyses were performed using SAS 9.3 for Windows.
Results
Patient characteristics
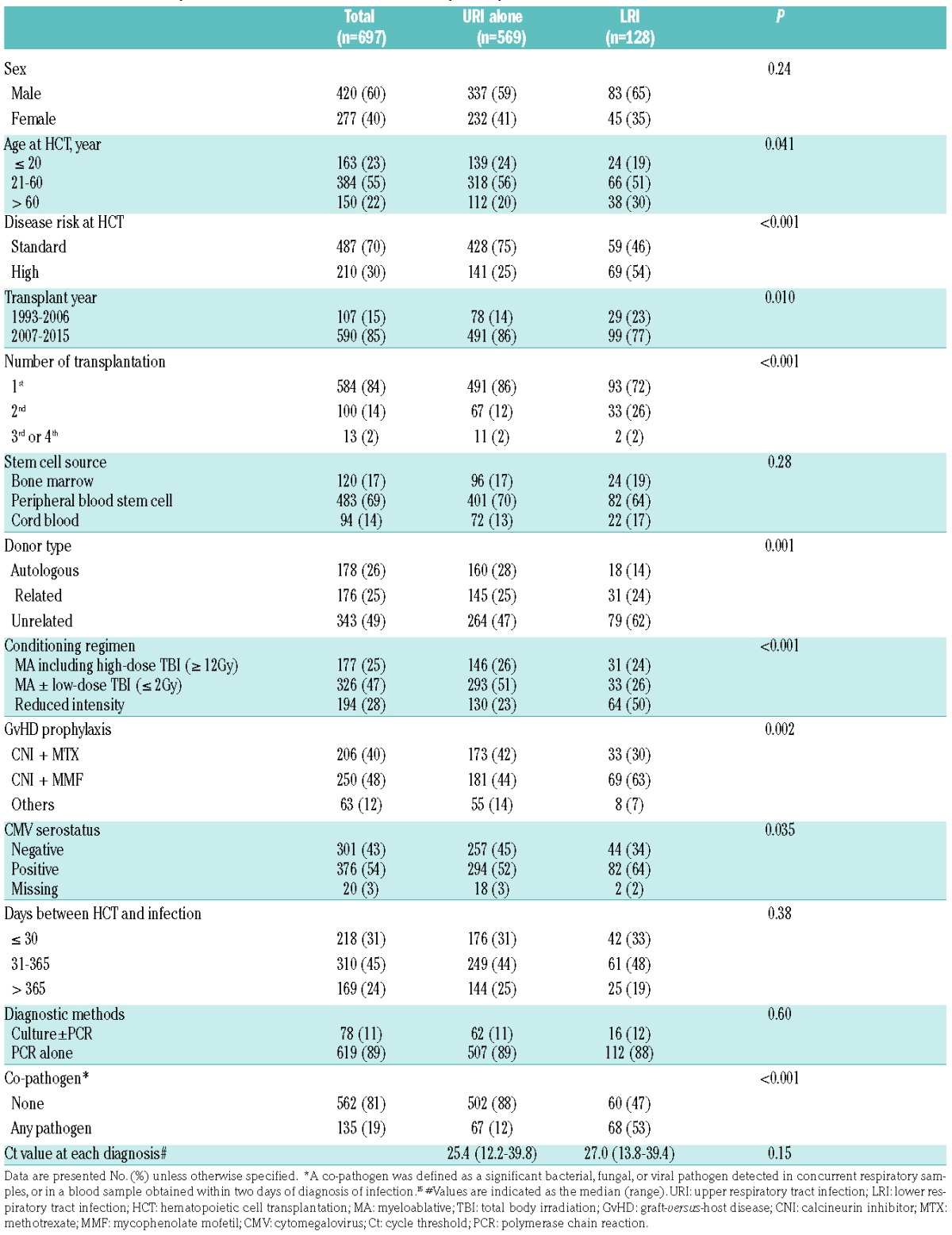
There were 697 patients diagnosed with HRV infection between 1993 and 2015; 569 (82%) and 128 (18%) had URI alone and LRI, respectively. Characteristics of each group are shown in Table 1. More than 80% of the HRV cases were diagnosed in and after 2007, reflecting the initiation of routine use of PCR to detect HRV. The median time to HRV URI and LRI following HCT was 74 days (range, 0 to 7063 days) and 87 days (range, 0 to 4309 days), respectively. Approximately half of patients with HRV LRI had co-pathogens at diagnosis, such as Aspergillus (N=11; including 5 cases positive by galactomannan alone) and Pseudomonas aeruginosa (N=5) considered to be non-viral co-pathogens, and RSV (N=3), PIV (N=3) and adenovirus (N=3) as viral co-pathogens.
Table 1.
Characteristics of patients with human rhinovirus infection (N=697),
Mortality after HRV infection and risk factors
There were 52 patients who died within 90 days after the onset of LRI. Forty-one patients (79%) died from pulmonary failure, and 5, 4 and 2 died from organ dysfunction, disease relapse and acute graft versus host disease (GvHD), respectively. The probabilities of overall mortality at 90 days following HRV diagnosis in patients with URI or LRI are shown in Figure 1A (6% in URI and 41% in LRI, P<0.001). The probabilities of 90-day survival after HRV LRI were 55% and 64% in patients with and without co-pathogens, respectively (P=0.34) (Figure 1B).
Figure 1.
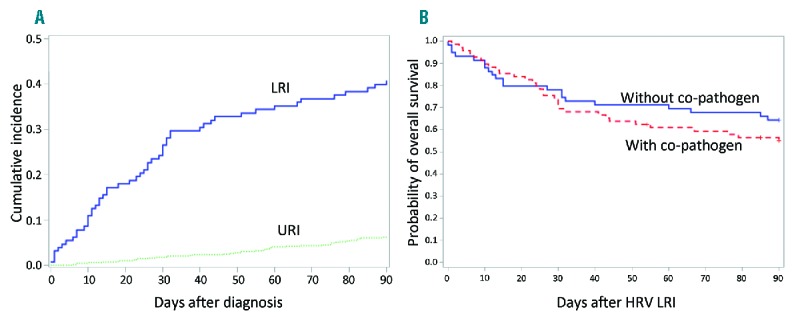
Probability of mortality after HRV infections. (A) Cumulative incidence of overall mortality after HRV URI or LRI (N=752, P=<0.001). (B) Kaplan-Meier estimate of overall survival after HRV LRI by presence of co-pathogens (N=128, P=0.34). HRV: human rhinovirus; LRI: lower respiratory tract infection; URI: upper respiratory tract infection.
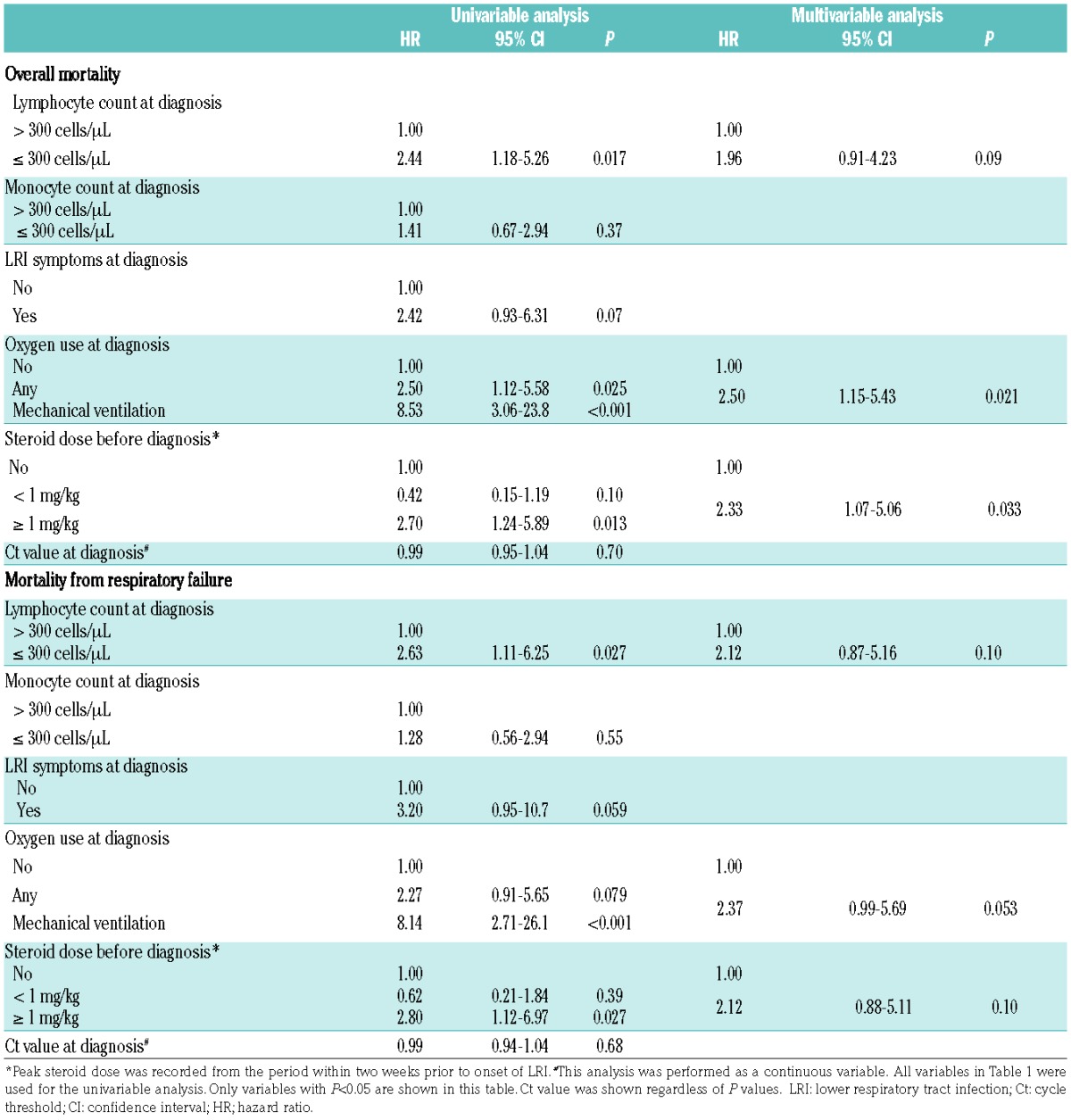
In a multivariable analysis of risk factors for overall mortality, low monocyte count, oxygen requirement at diagnosis, and steroid dose greater than 1 mg/kg/day before diagnosis were significantly associated with higher mortality. In the analysis of risk factors for mortality from respiratory failure, only low monocyte count at diagnosis and steroid use ≥1 mg/kg/day before diagnosis were significant factors in multivariable models (Table 2). Steroid use after diagnosis was also a significantly important factor (overall mortality: HR, 2.55; 95% confidence interval [CI], 1.39–4.69; P=0.003, mortality from respiratory failure: HR, 2.77; 95% CI, 1.39–5.52; P=0.004) (Online Supplementary Table S1), but allogeneic transplantation did not reach statistical significance (overall mortality: HR, 2.09; 95% CI, 0.76–5.81; P=0.16, mortality from respiratory failure: HR, 2.18; 95% CI, 0.67–7.06; P=0.19). To examine a more homogeneous cohort, we analyzed a cohort restricted to cases with allogeneic transplantation after 2007 and HRV infections within 2 years after HCT (Table 3). Oxygen requirement at diagnosis and steroid dose greater than 1 mg/kg/day before diagnosis were important factors for mortality (Table 4).
Table 2.
Risk factors for mortality from all causes or respiratory failure by day 90 after HRV LRI (N=128).
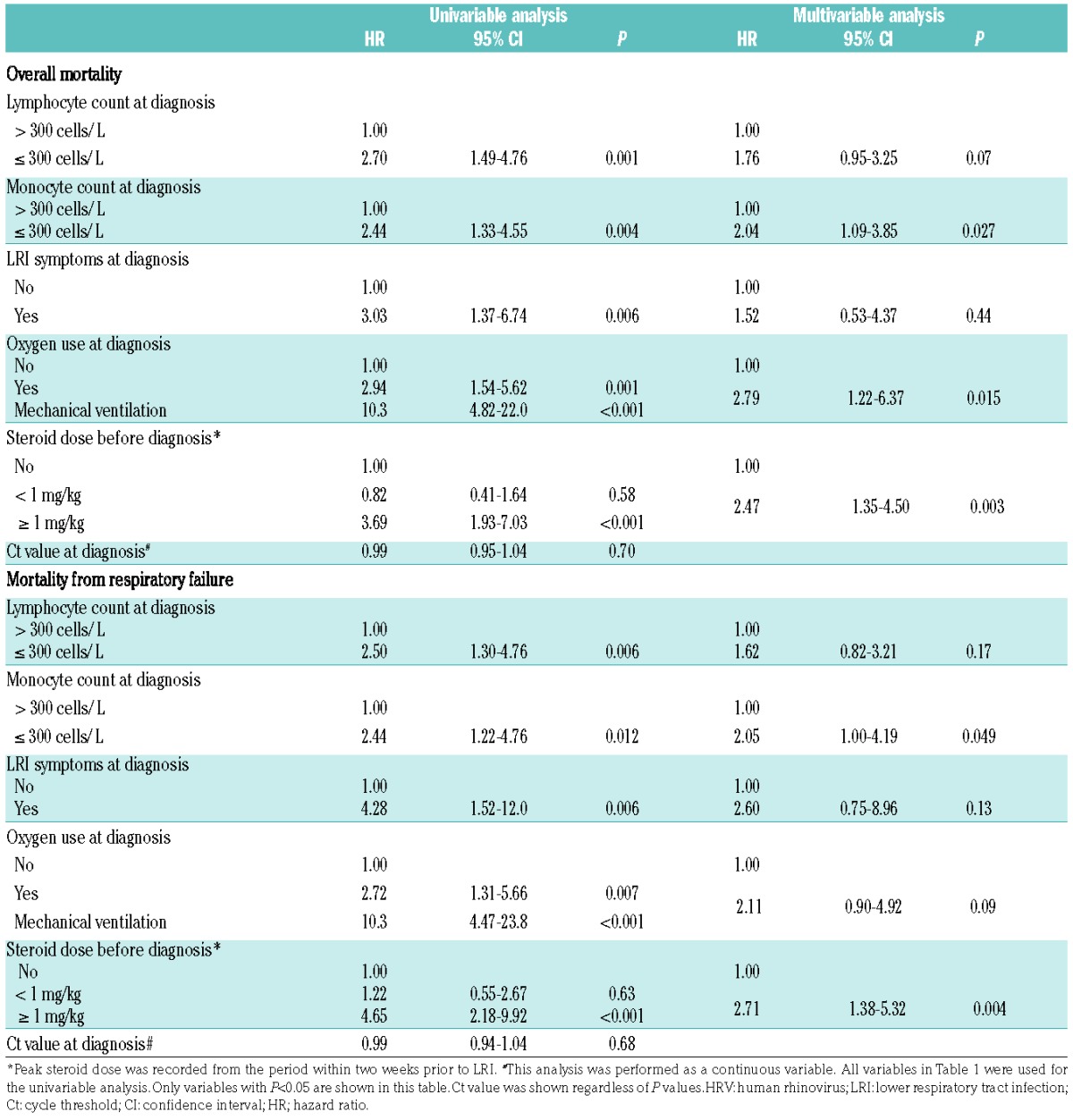
Table 3.
Characteristics of patients with human rhinovirus infection in a restrictive cohort consisting of allogeneic transplant recipients after 2007 with LRI within 2 years after HCT (N=434).
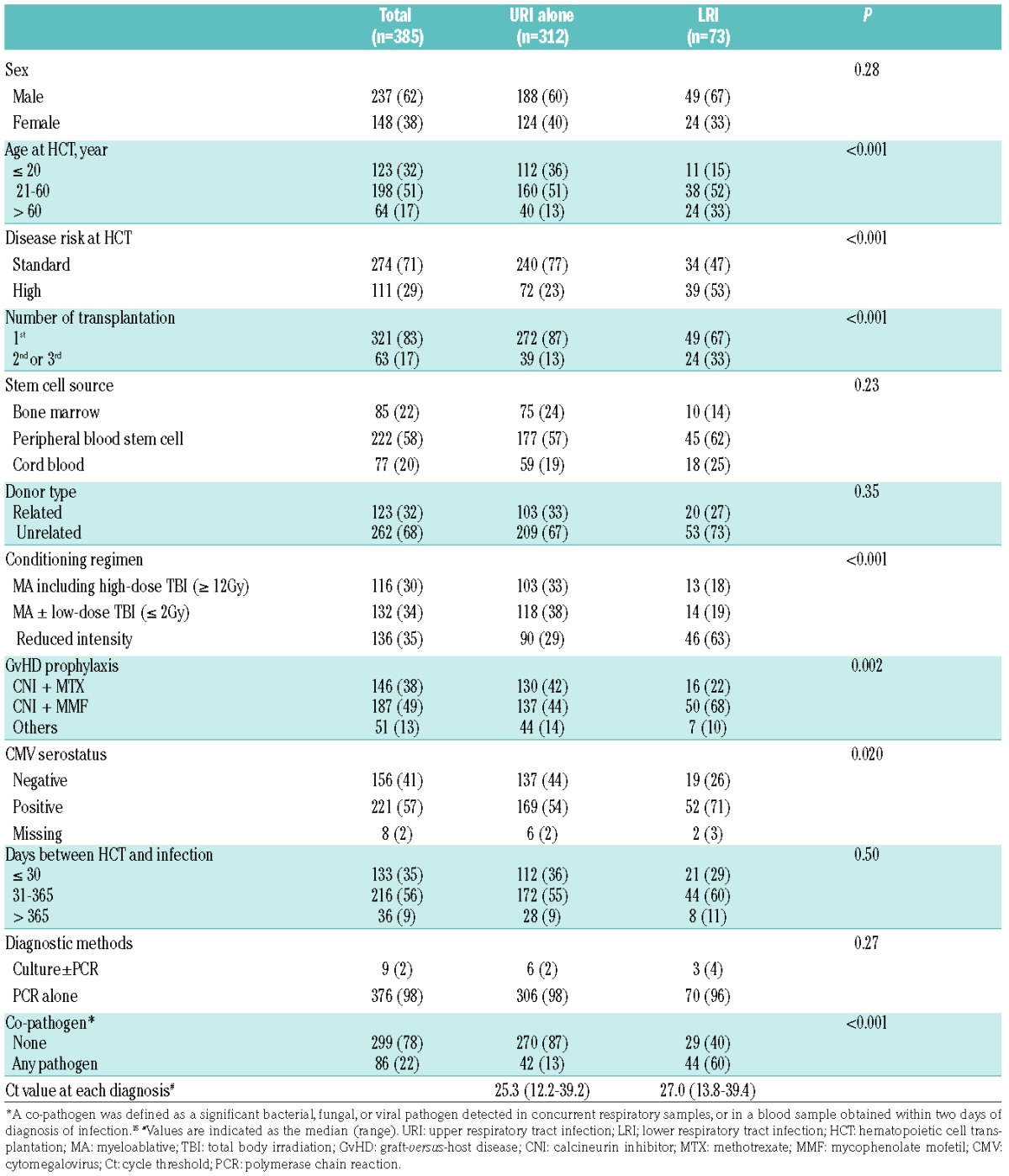
Table 4.
Risk factors for mortality from all causes or respiratory failure by day 90 after HRV LRI onset in a restricted cohort consisting of allogeneic transplant recipients after 2007 with LRI within 2 years after HCT (N=73).
The HRV PCR Ct value of the BAL sample obtained at diagnosis was not significantly associated with mortality after HRV LRI (Tables 2 and 4). There were 20 cases with sequential BALs within 90 days following diagnosis of LRI. Fourteen of those 20 patients had decreased viral load, of whom 6 (43%) died; the remaining 6 patients had stable or increased viral load, and all six died. There were 13 patients with sequential nasopharyngeal samples and 7 and 6 had decreased and increased viral loads, respectively. Each group had 1 patient who died.
Detection of HRV in tissue samples
To investigate whether HRV could be potentially responsible for lower respiratory tract disease or subsequent death, we examined the biopsy and/or autopsy samples for HRV using conventional culture and the RT-PCR assay. Of 15 biopsies performed at LRI diagnosis and 7 after diagnosis, 15 (100%) and 6 (86%), respectively, were HRV positive by RT-PCR; 2 (13%) and 4 (57%), respectively, were HRV positive by culture.
Among 128 patients with HRV LRI, 52 died within 90 days after onset of HRV LRI. Among them, 12 patients underwent autopsy (median days between LRI diagnosis and autopsy, 10 days; range, 2 to 67 days) and HRV was detected in 6 samples by RT-PCR. Both samples obtained from right and left lungs were positive in 5 of 6 positive cases. No samples were positive by culture.
Mortality comparison with LRI due to other respiratory viruses
We compared mortality after HRV LRI with that after LRI caused by RSV (N=117), PIV (N=120), or influenza virus (N=36). Patients’ characteristics are shown in Table 3. We observed differences among the various virus cohorts with regard to time of onset after transplantation, cell source, and oxygen requirements at diagnosis.
Overall survival by 90 days among groups without co-pathogens were similar (P=0.62 in Figure 2A). Since oxygen requirement at diagnosis is an important risk factor for mortality and is a reflection of the degree of lung injury, we analyzed survival by oxygen requirement. As shown in Figure 2B and 2C, the 4 groups of patients with respiratory viral LRI were comparable in overall survival (P=0.76 in Figure 2B, P=0.95 in Figure 2C).
Figure 2.
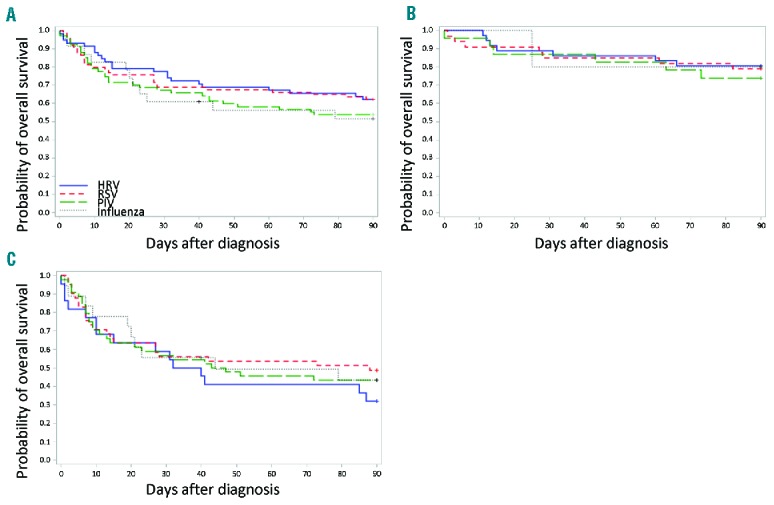
Probability of overall survival in patients without co-pathogens by viral type. (A) Kaplan-Meier estimate of overall survival by viral type (N=222, P=0.62). (B) Kaplan-Meier estimate of overall survival in patients without oxygen requirement at diagnosis by viral type (N=97, P=0.76). (C) Kaplan-Meier estimate of overall survival in patients with oxygen requirements at diagnosis by viral type (N=125, P=0.95). HRV: human rhinovirus; RSV: respiratory syncytial virus; PIV: parainfluenza virus.
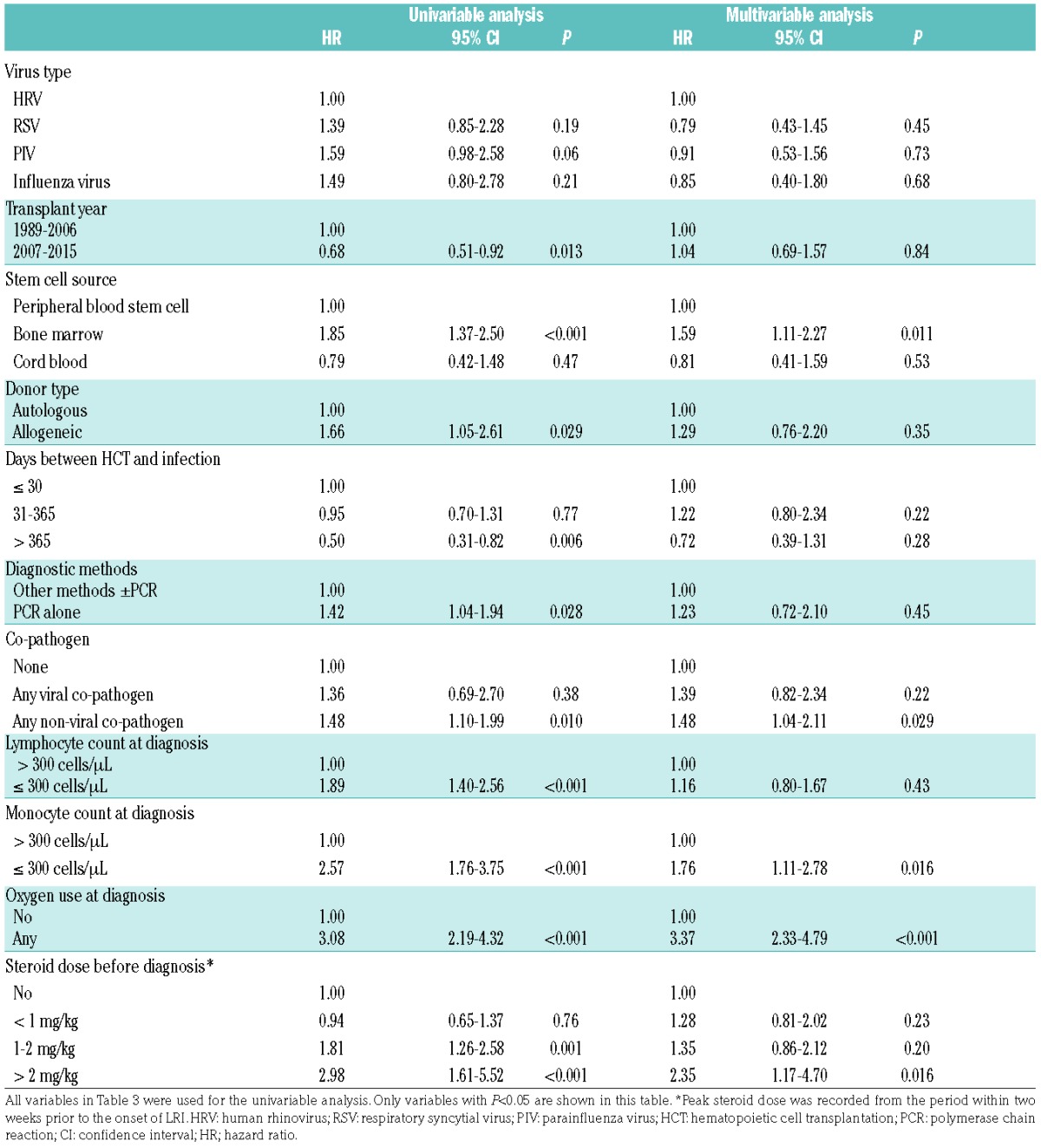
To confirm that mortality after HRV LRI is similar to that after RSV, PIV or influenza virus, we performed a multivariable analysis among the 388 total patients (Table 5). In an adjusted model, mortality in HRV LRI remained similar to that after LRI by other respiratory viruses (Table 6).
Table 5.
Characteristics of patients with LRI due to respiratory virus (N=388).
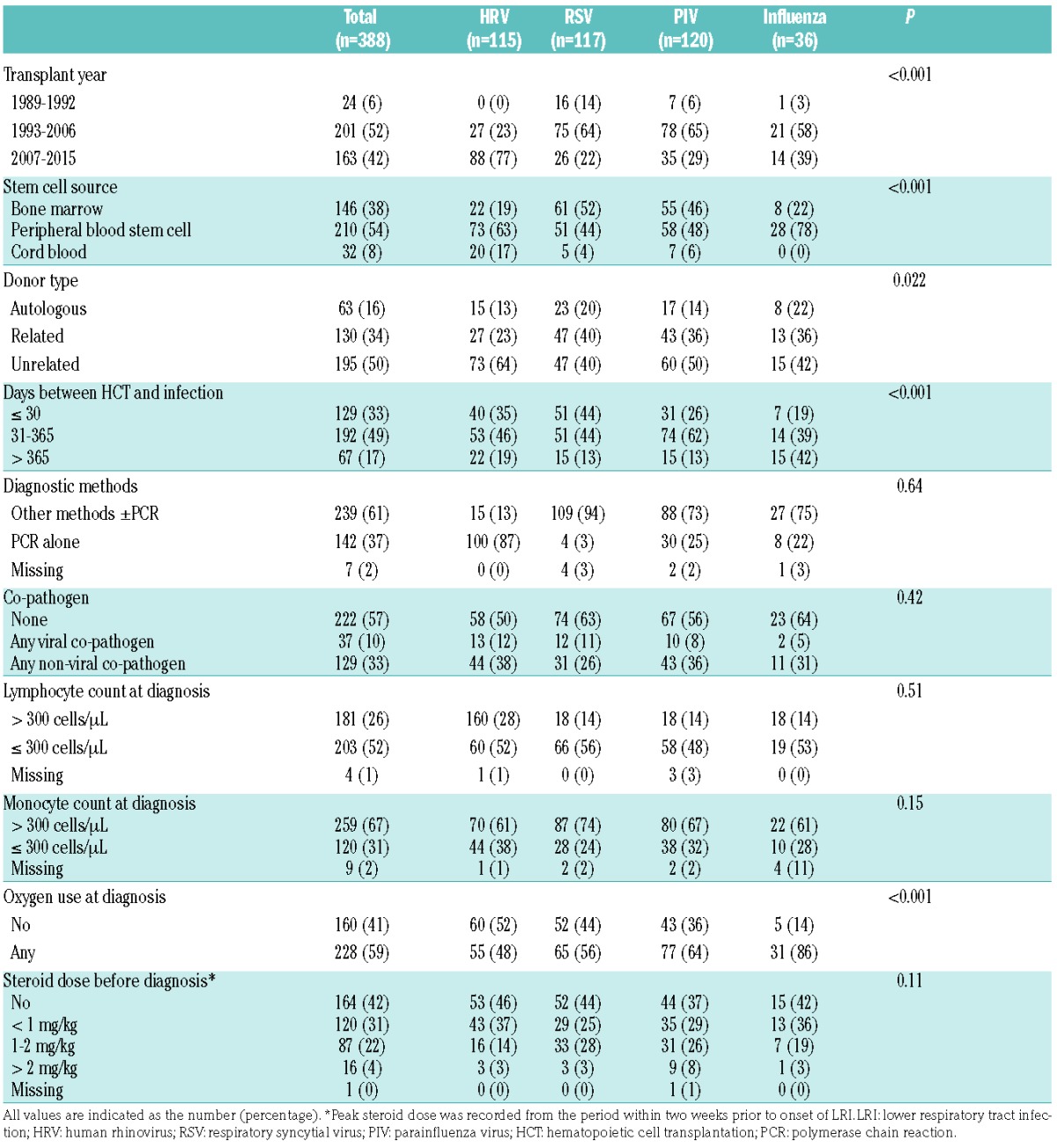
Table 6.
Risk factors for overall mortality comparing each respiratory virus (N=388).
Discussion
The study herein showed that patients who have HRV detected in the lower respiratory tract have similar outcomes to patients with LRI due to known pathogenic viruses such as RSV, PIV and influenza virus, even after excluding other potential bacterial, fungal and viral co-pathogens. Although the detection of HRV RNA by RT-PCR is not proof of ongoing viral replication, we were also able to detect replicating virus from the lung tissue of a subset of patients with a fatal outcome, thus demonstrating that these viruses were viable and potentially responsible for the pulmonary disease in these patients.
To prove a pathogen-disease association for a pathogen that is difficult to cultivate,18 especially in the molecular era, is difficult, as Koch’s postulates or other assessments of direct viral injury cannot be easily applied despite the detection of viral nucleic acid. We employed multiple strategies to examine the plausibility of HRV as a significant pathogen, including: (i) a comparison of mortality in patients with HRV detection in the upper and lower respiratory tract, (ii) a comparison between patients with and without co-pathogens in outcome after HRV LRI, (iii) an evaluation of factors associated with death following HRV detection in BAL samples, (iv) a comparison of mortality with respiratory viruses of well-established pathogenicity, and (v) tissue detection of the virus.
Our primary comparison was between patients with HRV URI and those with HRV detection in the BAL (Figure 1A). While the comparison of mortality after URI and LRI alone is not conclusive evidence for a causative effect of HRV for LRI, the two curves are significantly different, and in addition, look remarkably similar to curves previously documented for other respiratory viruses, such as PIV, in HCT recipients.15 Second, the fact that mortality in patients with or without co-pathogens is similar could suggest that HRV by itself is a cause of LRI, because otherwise one would expect the outcome to be improved in patients without co-pathogens. Next, factors associated with mortality following HRV BAL positivity were similar to those for other respiratory viruses (RSV, PIV, HMPV, influenza viruses) reported by our group and others,14–16,19–23 and included oxygen use, cytopenias, and high-dose corti costeroid use (Tables 2 and 4). An important analysis was the comparison of the outcome of HRV LRI with other well-established viral pneumonias. We used 3 previously described cohorts14–16 and analyzed overall mortality in a multivariable Cox model that adjusted for key factors that are associated with poor outcome. There was no difference in mortality after LRI between HRV and the other 3 viruses, even after adjusting for several important factors, such as the presence of co-pathogens and oxygen use at the time of diagnosis (Table 6, Figure 2), providing strong evidence that HRV detection in the BAL is indeed a clinically significant finding. Although the Cox model adjusted for oxygen use, we performed a subset analysis of patients who presented without oxygen use (Figure 2B). This group likely had minimal or no acute lung injury at the time of diagnosis and the subsequent outcome in these patients was more likely to be associated with the viral insult rather than the inflammatory changes associated with acute lung injury. Although mortality was, as expected, lower in this group, mortality rates approaching 20% by 3 months after diagnosis were documented, indicating an important negative impact on survival. Importantly, there were no apparent differences in LRI outcomes between HRV and RSV, PIV or influenza virus.
We also examined virologic factors to evaluate the role of HRV in lung disease. Tissue documentation of a pathogen is considered a key factor supporting a pathogenic role of an infectious agent.24,25 Although we were limited by the small number of tissue samples from lung biopsies and autopsies in our cohort, we were able to culture HRV from lung biopsies in 6 of 22 patients who had cultures performed. This is a remarkable result because our culture system is not optimized for HRV recovery (which in general is improved by incubation at lower temperatures and other methods, such as roller flasks, as used in previous papers),7 and suggests a high viral load may have been present. Since viral culture positivity from biopsy material is generally considered to indicate invasive viral disease for other respiratory viruses and cytomegalovirus (CMV),24,26,27 we consider this finding to be highly significant. We also detected HRV RNA by PCR in archived frozen tissue from lung biopsies and autopsy samples. The HRV PCR Ct values of the BAL samples, which provide a rough estimate of the HRV viral load, were not associated with mortality in our cohort (Tables 2 and 4). The lack of association does not rule out a pathogenic role of HRV, since studies with other well-established pathogens such as CMV, RSV, and PIV also failed to demonstrate an association of viral load in the BAL with mortality.12,15,28,29 Methodologic issues may be responsible for these results, including the inability to account for BAL dilution and the potentially differential amplification efficiency of different HRV serotypes.30 Additional studies are needed to conclusively study the role of the viral load in the BAL for HRV and other viruses.
The study herein has both strengths and limitations. We examined a large number of cases of HRV RNA positive BALs and comparative cases with other viral lower respiratory diseases which allowed us to perform appropriate statistical analyses that carefully accounted for co-pathogens and stages of acute lung injury. Our study was not restricted to specific transplant types or time periods after HCT, however, a subgroup analysis of recent allogeneic transplant recipients showed similar results. Due to a lack of specific information about the severity of GvHD in some cases, the association between mortality after HRV LRI and GvHD remains unclear, although we used steroid dose as a surrogate marker for GvHD severity. Our largely unbiased approach of evaluating patients with pulmonary infiltrates by bronchoscopy and BAL and the availability of lung tissue are additional strengths. According to previous reports, approximately 30% of HRV URI cases were asymptomatic4 and 5% of BAL samples from asymptomatic HCT recipients included HRV LRI;12 we therefore think that asymptomatic lower respiratory tract infection is only a minor factor in the present study. Other limitations include the lack of tissue immunohistochemical studies and the lack of direct viral quantitation, as well as our inability to adequately culture for HRV in autopsy or biopsy specimens. Currently available antibodies for HRV are serotype-specific.31 Because we did not identify the HRV serotypes in our tissue samples, we could not perform immunohistochemistry to confirm tissue detection of HRV. Furthermore, we acknowledge that certain HRV strains (particularly HRV-C viruses) may not have been identified earlier in this study and would be unlikely to be successfully cultivated. Thus, infections with HRV-C might be underrepresented in the patients prior to 2008.
In conclusion, we provide further evidence that HRV is a serious pathogen in the lower respiratory tract of HCT recipients. This conclusion is supported by similar outcomes of patients with HRV in the BAL compared to those with well-established respiratory pathogens such as RSV and influenza virus and by the presence of HRV in lung tissue. The data are also consistent with the recent literature on HRV in other clinical settings, which supports a potential causal role in serious clinical disease,1 and agree with earlier studies in HCT recipients carried out prior to the use of molecular diagnostic techniques.6,7 However, while these data are supportive of a pathogenic role of HRV, ultimate proof could best be provided by randomized placebo-controlled trials. These data provide the rationale for the rapid development and clinical evaluation of new therapeutics.
Supplementary Material
Acknowledgments
The authors would like to thank Zachary Stednick for database services, Terry Stevens-Ayers for laboratory assistance, and Amanda C Moklebust for preparation of pathology samples.
Footnotes
Check the online version for the most updated information on this article, online supplements, and information on authorship & disclosures: www.haematologica.org/content/102/6/1120
Funding
This work was partially supported by grants [K24HL093294, K23AI114844, and CA15704] from the National Institutes of Health. SS is a recipient of a fellowship from the Joel Meyers Endowment Scholarship.
References
- 1.Jain S, Self WH, Wunderink RG, Team CES. Community-acquired pneumonia requiring hospitalization among U.S. adults. N Engl J Med. 2015;373(5):415–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Milano F, Campbell AP, Guthrie KA, et al. Human rhinovirus and coronavirus detection among allogeneic hematopoietic stem cell transplantation recipients. Blood. 2010;115(10):2088–2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jacobs SE, Lamson DM, Soave R, et al. Clinical and molecular epidemiology of human rhinovirus infections in patients with hematologic malignancy. J Clin Virol. 2015;71:51–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Campbell AP, Guthrie KA, Englund JA, et al. Clinical outcomes associated with respiratory virus detection before allogeneic hematopoietic stem cell transplant. Clin Infect Dis. 2015;61(2):192–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gutman JA, Peck AJ, Kuypers J, Boeckh M. Rhinovirus as a cause of fatal lower respiratory tract infection in adult stem cell transplantation patients: a report of two cases. Bone Marrow Transplant. 2007;40(8):809–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ison MG, Hayden FG, Kaiser L, Corey L, Boeckh M. Rhinovirus infections in hematopoietic stem cell transplant recipients with pneumonia. Clin Infect Dis. 2003;36(9):1139–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ghosh S, Champlin R, Couch R, et al. Rhinovirus infections in myelosuppressed adult blood and marrow transplant recipients. Clin Infect Dis. 1999;29(3):528–532. [DOI] [PubMed] [Google Scholar]
- 8.Jacobs SE, Soave R, Shore TB, et al. Human rhinovirus infections of the lower respiratory tract in hematopoietic stem cell transplant recipients. Transpl Infect Dis. 2013;15(5):474–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mikulska M, Del Bono V, Gandolfo N, et al. Epidemiology of viral respiratory tract infections in an outpatient haematology facility. Ann Hematol. 2014;93(4):669–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kraft CS, Jacob JT, Sears MH, et al. Severity of human rhinovirus infection in immunocompromised adults is similar to that of 2009 H1N1 influenza. J Clin Microbiol. 2012;50(3):1061–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gadsby NJ, Russell CD, McHugh MP, et al. Comprehensive molecular testing for respiratory pathogens in community-acquired pneumonia. Clin Infect Dis. 2016; 62(7):817–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seo S, Renaud C, Kuypers JM, et al. Idiopathic pneumonia syndrome after hematopoietic cell transplantation: evidence of occult infectious etiologies. Blood. 2015;125(24):3789–3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu X, Holloway B, Dare RK, et al. Real-time reverse transcription-PCR assay for comprehensive detection of human rhinoviruses. J Clin Microbiol. 2008;46(2):533–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Waghmare A, Campbell AP, Xie H, et al. Respiratory syncytial virus lower respiratory disease in hematopoietic cell transplant recipients: viral RNA detection in blood, antiviral treatment, and clinical outcomes. Clin Infect Dis. 2013;57(12):1731–1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seo S, Xie H, Campbell AP, et al. Parainfluenza virus lower respiratory tract disease after hematopoietic cell transplantation: viral detection in the lung predicts outcome. Clin Infect Dis. 2014;58(10):1357–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Choi SM, Boudreault AA, Xie H, et al. Differences in clinical outcomes after 2009 influenza A/H1N1 and seasonal influenza among hematopoietic cell transplant recipients. Blood. 2011;117(19):5050–5056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martin EK, Kuypers J, Chu HY, et al. Molecular epidemiology of human rhinovirus infections in the pediatric emergency department. J Clin Virol. 2015;62:25–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fredricks DN, Relman DA. Infectious agents and the etiology of chronic idiopathic diseases. Curr Clin Top Infect Dis. 1998;18:180–200. [PubMed] [Google Scholar]
- 19.Seo S, Campbell AP, Xie H, et al. Outcome of respiratory syncytial virus lower respiratory tract disease in hematopoietic cell transplant recipients receiving aerosolized ribavirin: significance of stem cell source and oxygen requirement. Biol Blood Marrow Transplant. 2013;19(4):589–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shah DP, Ghantoji SS, Shah JN, et al. Impact of aerosolized ribavirin on mortality in 280 allogeneic haematopoietic stem cell transplant recipients with respiratory syncytial virus infections. J Antimicrob Chemother. 2013;68(8):1872–1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chemaly RF, Hanmod SS, Rathod DB, et al. The characteristics and outcomes of parainfluenza virus infections in 200 patients with leukemia or recipients of hematopoietic stem cell transplantation. Blood. 2012;119(12):2738–2745. [DOI] [PubMed] [Google Scholar]
- 22.Renaud C, Xie H, Seo S, et al. Mortality rates of human metapneumovirus and respiratory syncytial virus lower respiratory tract infections in hematopoietic cell transplantation recipients. Biol Blood Marrow Transplant. 2013;19(8):1220–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ljungman P, de la Camara R, Perez-Bercoff L, et al. Outcome of pandemic H1N1 infections in hematopoietic stem cell transplant recipients. Haematologica. 2011; 96(8):1231–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ljungman P, Griffiths P, Paya C. Definitions of cytomegalovirus infection and disease in transplant recipients. Clin Infect Dis. 2002;34(8):1094–1097. [DOI] [PubMed] [Google Scholar]
- 25.Harrington RD, Hooton TM, Hackman RC, et al. An outbreak of respiratory syncytial virus in a bone marrow transplant center. J Infect Dis. 1992;165(6):987–993. [DOI] [PubMed] [Google Scholar]
- 26.Boeckh M, Berrey MM, Bowden RA, et al. Phase 1 evaluation of the respiratory syncytial virus-specific monoclonal antibody palivizumab in recipients of hematopoietic stem cell transplants. J Infect Dis. 2001; 184(3):350–354. [DOI] [PubMed] [Google Scholar]
- 27.Erard V, Huang ML, Ferrenberg J, et al. Quantitative real-time polymerase chain reaction for detection of adenovirus after T cell-replete hematopoietic cell transplantation: viral load as a marker for invasive disease. Clin Infect Dis. 2007;45(8):958–965. [DOI] [PubMed] [Google Scholar]
- 28.Erard V, Guthrie KA, Seo S, et al. Reduced mortality of cytomegalovirus pneumonia after hematopoietic cell transplantation due to antiviral therapy and changes in transplantation practices. Clin Infect Dis. 2015; 61(1):31–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Campbell AP, Chien JW, Kuypers J, et al. Respiratory virus pneumonia after hematopoietic cell transplantation (HCT): associations between viral load in bronchoalveolar lavage samples, viral RNA detection in serum samples, and clinical outcomes of HCT. J Infect Dis. 2010; 201(9):1404–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jacobs SE, Lamson DM, St George K, Walsh TJ. Human rhinoviruses. Clin Microbiol Rev. 2013;26(1):135–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mosser AG, Brockman-Schneider R, Amineva S, et al. Similar frequency of rhinovirus-infectible cells in upper and lower airway epithelium. J Infect Dis. 2002; 185(6):734–743. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


