Abstract
The last two decades had witnessed extensive investigation on bacterial degradation of carbazole, an N-heterocyclic aromatic hydrocarbon. Specifically, previous studies have reported the primary importance of angular dioxygenation, a novel type of oxygenation reaction, which facilitates mineralization of carbazole to intermediates of the TCA cycle. Proteobacteria and Actinobacteria are the predominant bacterial phyla implicated in this novel mode of dioxygenation, while anthranilic acid and catechol are the signature metabolites. Several studies have elucidated the degradative genes involved, the diversity of the car gene clusters and the unique organization of the car gene clusters in marine carbazole degraders. However, there is paucity of information regarding the environmental fate as well as industrial and medical importance of carbazole and its derivatives. In this review, attempt is made to harness this information to present a comprehensive outlook that not only focuses on carbazole biodegradation pathways, but also on its environmental fate as well as medical and industrial importance of carbazole and its derivatives.
Keywords: Carbazole, Angular dioxygenation, Environmental fate, Degradative pathways
Introduction
Carbazole: general description
Carbazole (C12H9N, dibenzopyrrole diphenylenimine, CAS No. 86-74-8) is a non-basic tricyclic aromatic N-heteroatomic compound (Fig. 1). It has a molecular weight of 167.21 g/mol, boiling and melting point of 355 and 246 °C (Lide 2003), water solubility of 1.2 mg/l (Johansen et al. 1997), vapor pressure of 1 × 10−4 Pa (Peddinghaus et al. 2012), and octanol/water partition coefficient (log K ow) of 3.72 (Blum et al. 2011). It is one of the π-excessive heterocycles (electron-rich rings) and is more recalcitrant than dibenzofuran but less than dibenzothiophene (Balaban et al. 2004). It is a white crystalline solid at ambient temperature. It sublimates, has a scent similar to creosote and exhibits strong fluorescence and long phosphorescence upon exposure to ultraviolet light (Collin and Höke 1986). It is one of the major N-heterocyclic aromatic hydrocarbons in fossil fuels (coal, crude oil, oil derived from oil shales pyrolysis) and is also found in cigarette smoke and emitted from coal and wood combustion (Odabasi et al. 2006).
Fig. 1.
Molecular structure of carbazole
Carbazole is used as a chemical feedstock for the production of dyes, reagents, explosives, insecticides, lubricants and it acts as a color inhibitor in detergents (Nojiri and Omori 2007). It is also widely used as a model compound for the study of biodegradation of aromatic N-heterocyclic hydrocarbons (Xu et al. 2006). However, its release into the environment from diverse anthropogenic sources is of serious health and environmental concern, as carbazole is both mutagenic and toxic and classified as “benign tumorigen” (Smith and Hansch 2000; Nojiri and Omori 2007).
Properties of carbazole
Solubility
Heterocyclic aromatic compounds are known to exhibit higher polarity and water solubility due to substitution of one carbon atom by nitrogen, sulfur or oxygen (Meyer and Steinhart 2000). These chemical properties lead to increase bioavailability and mobility as compared to the homologous polycyclic aromatic hydrocarbons resulting in various environmental influences of these compounds (Pearlman et al. 1984; Peddinghaus et al. 2012). Carbazole has an aqueous solubility at 25 °C of 1.2 mg/l. It is less soluble than dibenzothiophene (1.5 mg/l) and dibenzofuran (4.8 mg/l) but more soluble than xanthene (1.0 mg/l) in spite of its higher molecular weight (Table 1). It is readily soluble in acetone and dimethyl sulfoxide, slightly soluble in ether and ethanol, and barely soluble in chloroform, acetic acid, carbon tetrachloride, and carbon disulfide (Collin and Höke 1986).
Table 1.
Properties of some heterocyclic aromatic compounds
| Group | Compound | Molecular weight (g/mol) | Aqueous solubility at 25 °C (mg/l) | Log K ow |
|---|---|---|---|---|
| Nitrogen heteroaromatics | Pyrrole | 67.1 | 58,800 | 0.75 |
| Indole | 117.0 | 1875 | 2.00 | |
| Quinoline | 129.2 | 6718 | 2.03 | |
| Carbazole | 167.2 | 1.2 | 3.72 | |
| Acridine | 179.2 | 46.6 | 3.48 | |
| 6-Methylquinoline | 143.2 | 631 | 2.57 | |
| Sulphur heteroaromatics | Thiophene | 84.1 | 3600 | 1.81 |
| 1-Benzothiophene | 134.2 | 130 | 3.12 | |
| Dibenzothiophene | 184.3 | 1.5 | 4.38 | |
| Oxygen heteroaromatics | Benzofuran | 118.1 | 678 | 2.67 |
| Dibenzofuran | 168.2 | 4.8 | 4.12 | |
| 2-Methylbenzofuran | 132.2 | 160 | 3.22 | |
| Xanthene | 182.2 | 1.0 | 4.23 |
Aromaticity
Aromaticity is a property of planar, cyclic, conjugated molecules that act like unsaturated molecules and undergo substitution reaction rather than addition due to delocalization of electrons in the ring. It can also be considered a manifestation of cyclic delocalization and resonance (Balaban et al. 2004). The tendency to favor substitution rather than addition suggests that the parent unsaturated ring system has exceptional stability. Aromaticity cannot exist without conjugation (conjugation requires at least three overlapping p orbitals in the same plane). This is because aromatic molecules require planarity and overlapping p orbitals so that electron can be delocalized for better quality. In the same vein, resonance exists because of electron delocalization and emerges in different patterns based on the structure and arrangement within a molecule. Resonance gives extra stability due to electron delocalization and can be conferred sometimes on a molecule due to cycling double bonds.
Aromaticity in a molecule is premised on possession of four specific qualities (Katritzky et al. 2010). These are (1) Structure must be cyclic with conjugated Pi (π) bonds, (2) each atom in the ring must have an unhybridized p orbital, (3) all p orbitals must overlap continuously around the ring (planarity) and (4) 4n + 2 π electrons (n is an integer: 0,1,2,3…) in cyclic conjugation are associated with each ring.
Aromatic heterocyclic compounds electronic structure is in agreement with Huckel’s rule, which states that cyclic conjugated and planar systems having (4n + 2) π electrons are aromatic. The rings possess diamagnetic currents, react by substitution rather than addition, and bond orders and length tend to be intermediate between single and double (Balaban et al. 2004). Examples of these heterocycles are pyrrole, thiophene, and furan.
Carbazole (dibenzopyrrole) consists of two benzene rings fused together on either side of a pyrrole ring. Pyrrole is a five-membered ring in which the heteroatom has at least one pair of non-binding valence shell electrons. Hybridizing this heteroatom to an sp2 state creates a p orbital occupied by a pair of electrons and oriented parallel to the carbon p-orbitals resulting in a planar ring. Six electrons occupy the pi (π) system. Four of the electrons are from two double bonds and two from the heteroatom. Hence, these five sp2 hybridized atoms form planar six electrons delocalized π-cloud, which is responsible for the aromatic character of pyrrole.
The resonance energies of pyrrole, thiophene, and furan are 5.3, 6.5 and 4.3 kcal/mol, which gives the order of aromaticity as thiophene > pyrrole > furan. In essence, carbazole is less aromatic than dibenzothiophene but more aromatic than dibenzofuran (Balaban et al. 2004).
Toxicity
Heterocyclic aromatic compounds are highly ubiquitous in the environment and are known to exhibit diverse ecotoxic effects such as acute toxicity, developmental and reproductive toxicity, cytotoxicity, photo-induced toxicity, mutagenicity, and carcinogenicity (Bundy et al. 2001; Barron et al. 2004; Robbiano et al. 2004; Brack et al. 2007; Eisentraeger et al. 2008).
Human exposure to carbazole occurs through tobacco smoking and inhalation of polluted air (IARC 1983), while inhalation of vapors, dust, and dermal contact has been reported as possible routes of carbazole exposure to workers. There are no relevant epidemiological data to the carcinogenicity of carbazole to humans, though limited evidence in experimental animals for the carcinogenicity of carbazole has been reported (IARC 1999).
In a study conducted on groups of 50 male and 50 female, B6C3 F1 mice fed with different concentrations of technical grade carbazole (96% purity) for 96 weeks; neoplastic lesions were found in the livers and fore stomachs of the dead mice. The lesions were classified as neoplastic nodules and hepatocellular carcinomas. However, no such tumor was observed in the respective control groups (IARC 1983).
Carbazole is mutagenic and toxic. Its toxicity to aquatic organisms is well documented (Eisentraeger et al. 2008; Peddinghaus et al. 2012). In a recent study on embryotoxic potential of NSO-heterocyclic compounds using groups of 3-month old zebrafish Danio rerio, carbazole displayed a very high embryotoxic potential with LC50 value of 1.07 mg/l, a value preceded only by acridine (0.7 mg/l) (Peddinghaus et al. 2012).
Although carbazole is not a human carcinogen, its hazardous derivatives such as N-methylcarbazole and 7-H-dibenzo (c,g) carbazole (and its derivatives) found in cigarette smoke and automobile emission are genotoxic and carcinogenic and have been categorized as “IARC Group 2B carcinogens” (Smith et al. 2000). 7H-dibenzo (c,g) carbazole is a potent multi-site and multi-species carcinogen (Szafarz et al. 1988; Warshawsky et al. 1996) that induces tumor at the site of application and at distant sites, specifically in the liver (Renault et al. 1998).
Synthetic methyl derivatives of 5,9-dimethyl dibenzo (c,g) carbazole, dimethyl-DBC and N-methyl-DBC exhibit specific attachment to liver and skin and together with the parent compound (DBC) induce significant levels of DNA strand-breaks, micronuclei, and DNA adducts in immortalized human keratinocytes HaCat cells (Valovicova et al. 2012).
Industrial and medical importance of carbazole
Carbazoles are dominant as structural motifs in various synthetic materials and naturally occurring alkaloids. It exhibits material properties as optoelectronic materials, conducting polymers and synthetic dyes (Roy et al. 2012). Several dyes such as Hydron Blue™, Naphthol™ dyes, anthraquinone vat dyes, styryl dyes, and dioxazine dyes are synthesized from carbazole. Similarly, 1,3,6,8-tetranitro-carbazole (Nitrosan™) is used as an insecticide while reaction of carbazole with phenol and formaldehyde in the presence of acidic catalysts forms Novalacs, which can be cured with hexamethylenetetramine to produce highly heat resistant polymers (Collin and Höke 1986). Carbazole is also used to synthesize the monomer, N-vinylcarbazole, which can be polymerized to form polyvinyl carbazole (PVK) (Pearson and Stolka 1981; Collin and Höke 1986).
Naturally occurring carbazoles manifest profound biological activities such as antitumor, psychotropic, anti-inflammatory, anti-histaminic, antibiotic and antioxidative activities (Fig. 2) (Lobastova et al. 2004; Roy et al. 2012). The structural features of such carbazole-based natural products are the presence of nuclear hydroxyl groups (major structural feature), quinine functionality and prenyl groups (Roy et al. 2012). In pharmaceutical industry, hydroxylated carbazole derivatives are value-added substances exhibiting strong antioxidant activity and widely used in the treatment of encephalopathy, cardiopathy, hepatopathy and arteriosclerosis (Seto 1991). Furthermore, carbazole moiety is considered as one of the pharmacophores in the cardiovascular pharmaceuticals carvedilol and carazolol, which are used in the treatment of hypertension, ischemic heart disease, and congestive heart failure (Roy et al. 2012).
Fig. 2.
Molecular structures of some natural carbazole alkaloids (1) mahanine (2) mahanimbicine and (3) mahanimbine
In the petroleum industry, the removal of nitrogen heteroaromatics is important for several reasons. First, their combustion directly causes the formation of nitrogen oxides (NOx), which contribute to acid rain and depletion of the ozone layer (Kirimura et al. 1999). Second, nitrogen-containing aromatic compounds presence can cause poisoning of refining catalysts, resulting in a decrease in yield (Girgis and Gates 1991; Williams and Chisti 2001). Carbazole directly affects the refining process by its conversion into basic derivatives during cracking, which allows it to adsorb to the active sites of the cracking catalyst. It also serves as a potent direct inhibitor of hydrodesulfurization, a property that enables it to be included in the refining process to meet sulfur content criteria (Benedik et al. 1998; Nojiri and Omori 2007). Finally, the presence of nitrogen heteroaromatics promotes corrosion of refining equipment, thereby increasing the refining costs (Benedik et al. 1998).
Environmental fate of carbazole
Atmospheric fate
Carbazole is a semi-volatile organic compound (SOC) found in ambient air in gas phase and sorbed to aerosol (Odabasi et al. 1999). The fate, transport and removal of carbazole from the atmosphere by dry and wet deposition processes are strongly influenced by its gas-particle partitioning (Bidleman 1988). The vapor pressure of carbazole (1 × 10−4 Pa) suggests that carbazole will exist in the vapor and particulate phases in the ambient atmosphere. Carbazole is released to the atmosphere in emissions from waste incineration, tobacco smoke, aluminum manufacturing, and rubber, petroleum, coal, and wood combustion (Smith et al. 1978; Jacobs and Billings 1985; Pereira et al. 1987). Upon its release into the atmosphere, photochemically produced hydroxyl radicals (estimated half-life of 3 h) rapidly degrade vapor-phase carbazole. In the particulate phase, photodegradation of carbazole is dependent on the adsorbing substrate as substrates containing more than 5% carbon can stabilize photodegradation and permit long-range global transport of the pollutant (Behymer and Hates 1988).
Terrestrial fate
Biodegradation by indigenous carbazole degraders in the soil is the dominant fate process for carbazole even though photolysis of carbazole in soil had been reported (Behymer and Hates 1988; Grosser et al. 1991). However, adsorption of carbazole to environmental substrates will limit or prevent photolysis. The average K oc (organic carbon normalized partition coefficient) value of carbazole is 637 (Ainsworth et al. 1989), which suggests low mobility of carbazole in soil. Sorption of carbazole to soil is non-linear and highly correlated with organic content of soils (Bi et al. 2007).
Aquatic fate
In aquatic environment, biodegradation and photolysis are the dominant fate processes for carbazole. Half-lives of carbazole in a river, pond, eutrophic lake, and oligotrophic lake have been estimated as 0.5, 10, 10, and 3 h, respectively (Smith et al. 1978). The absence of carbazole degraders in the microbial community will foreclose biodegradation as a fate process while adsorption of carbazole to sediment will make photolysis unattainable (Pereira et al. 1987; Grosser et al. 1991). Volatilization is not a fate process in aquatic environment since carbazole is non-volatile in water (Meylan and Howard 1991). Metabolism of carbazole to its N-methyl and N-acetyl derivatives by aquatic organisms has been reported. Furthermore, bioaccumulation and acute toxicity of NSO-heterocycles in aquatic organisms such as Daphnia, midge, and algae have been documented (Eisentraeger et al. 2008).
Bacterial degradation of carbazole
Diversity of carbazole-degrading bacteria
Various carbazole-degrading bacteria have been isolated from diverse niches by enrichment cultural technique using carbazole as the sole source of nitrogen, carbon and energy or carbon and energy. Majority of carbazole degraders reported in the literature are aerobic, Gram-negative bacteria with the exception of very few carbazole degraders such as Nocardioides aromaticivorans IC177 (Inoue et al. 2005), Gordonia sp. F.5.25.8 (Santos et al. 2006) and Microbacterium esteraromaticum strain SL6 (Salam et al. 2014) that are aerobic, Gram-positive bacteria (Table 2). About 23 and 39% of carbazole degraders isolated from activated sludge, soil, and freshwater samples belong to the genera Pseudomonas and Sphingomonas, respectively (Nojiri and Omori 2007). Recent research on carbazole degraders from marine environments using seawater-based enrichment culture has led to the isolation of novel carbazole degraders with unique carbazole degradative genes and enzymes different from those found in various carbazole degraders from soil, freshwater and activated sludge (Fuse et al. 2003; Maeda et al. 2009a, b, 2010).
Table 2.
Some carbazole-degrading bacteria
| Bacterial strain | Mediuma | Productsb | References |
|---|---|---|---|
| Ralstonia sp. RJGII.123 | Carbon | Anthranilic acid | Grosser et al. (1991); Schneider et al. (2000) |
| P. resinovorans CA10 | Carbon, nitrogen | Anthranilic acid, catechol | Ouchiyama et al. (1993); Nojiri et al. (1999) |
| P. resinovorans CA06 | Carbon, nitrogen | Anthranilic acid, catechol | Ouchiyama et al. (1993) |
| P. stutzeri ATCC31258 | Carbon | Anthranilic acid | Hisatsuka and Sato (1994) |
| Pseudomonas sp. LD2 | Carbon | Anthranilic acid | Gieg et al. (1996) |
| Burkholderia sp. CB1 | Carbon, nitrogen | Not detected | Shotbolt-Brown et al. 1996 |
| Xanthomonas sp. CB2 | Carbon, nitrogen | Not detected | Shotbolt-Brown et al. (1996) |
| Sphingomonas sp. CB3 | Carbon, nitrogen | Not detected | Shepherd and Lloyd-Jones (1998) |
| P. stutzeri OM1 | Carbon, nitrogen | Anthranilic acid | Ouchiyama et al. (1998) |
| Sphingomonas sp. CDH-7 | Carbon, nitrogen | Anthranilic acid | Kirimura et al. (1999) |
| Sphingomonas sp. GTIN11 | Nitrogen | Anthranilic acid | Kilbane II et al. (2002) |
| Sphingomonas sp. KA1 | Carbon | None | Habe et al. (2002) |
| Pseudomonas rhodesiae KK1 | Carbon | None | Yoon et al. (2002) |
| Neptunomonas naphthovorans CAR-SF | Carbon | None | Fuse et al. (2003) |
| Pseudomonas sp. XLDN4-9 | Nitrogen | None | Li et al. (2004) |
| Achromobacter sp. IC074 | Carbon, nitrogen | None | Inoue et al. (2005) |
| Stenotrophomonas sp. IC193 | Carbon, nitrogen | None | Inoue et al. (2005) |
| Janthinobacterium sp. J3 | Carbon, nitrogen | None | Inoue et al. (2004) |
| Pantoea sp. J14 | Carbon, nitrogen | None | Inoue et al. (2004) |
| Achromobacter sp. SL1 | Carbon | Anthranilic acid, catechol | Salam et al. (2014) |
| Pseudomonas sp. SL4 | Carbon | Anthranilic acid, catechol | Salam et al. (2014) |
| Microbacterium esteraromaticum SL6 | Carbon | Anthranilic acid, catechol | Salam et al. (2014) |
aCarbon and nitrogen: carbazole was added to the isolation medium as the carbon, nitrogen and energy sources; carbon carbazole was added to the medium as the carbon and energy source, nitrogen carbazole was added as the nitrogen source
bMajor metabolic intermediate produced when the bacterium is grown on carbazole
Interest in the study of bacterial degradation of carbazole is spurred partly because of the ubiquitous nature, mutagenic and toxic activities, and the fact that it is a structural analog of dioxins and carbazole-degrading enzymes can partly function as dioxin-degrading enzymes (Nojiri and Omori 2007).
Degradation pathways of carbazole
Three major degradation pathways have been reported for carbazole: Lateral dioxygenation at carbon positions 3 and 4, monohydroxylation at carbon positions 1, 2, and 3 and angular dioxygenation at carbon positions 1, and 9a (Grifoll et al. 1995; Lobastova et al. 2004; Nojiri 2012).
Lateral dioxygenation of carbazole
Grifoll and co-workers first suggested lateral dioxygenation of carbazole by fluorene-degrading bacteria Pseudomonas cepacia F297 at C3 and C4 carbons yielding 4-(3′-hydroxy-2′-indoyl)-2-oxo-3-butenoic acid as detected by GC-FID and GC–MS. However, strain F297 cannot utilize carbazole as source of carbon and energy (Fig. 3; Grifoll et al. 1995).
Fig. 3.
Lateral dioxygenation of carbazole at C3 and C4. The metabolites detected from the methylated acidic extract are 4-(3′-methoxy-2′-indolyl)-2-oxo-3-butenoic acid (Methylated, 19) and 4-(3′-oxo-2′-indolinyl)-2-oxo-3-butenoic acid (Methylated, 20)
Hydroxylation of carbazole
Transformation via hydroxylation appears to be a very common reaction in the metabolism of carbazole by bacteria. Lobastova et al. (2004) were able to identify 1-, 2- and 3-hydroxycarbazoles as the bioconversion products following monohydroxylation of carbazole at position 1, 2, and 3 by Aspergillus flavus VKM F-1024 using TLC, GC, MS and 1H NMR, respectively. 3-hydroxycarbazole was detected as the major product, while 1-hydroxy- and 2-hydroxycarbazoles were detected as minor products. Yamazoe et al. (2004), and Seo et al. (2006) also reported bioconversion of carbazole to hydroxycarbazoles.
Furthermore, bacterial dioxygenases such as naphthalene 1,2-dioxygenase from Pseudomonas sp. NCIB 9816-4 and biphenyl dioxygenase from Beijerinckia sp. B8/36 also catalyze the initial oxidation of carbazole to 3-hydroxycarbazole (Resnick et al. 1993). Hydroxylated carbazole derivatives have strong antioxidant activity and are value-added substances in pharmaceutical industry with diverse application in therapies for encepalopathy, cardiopathy, hepatopathy and arteriosclerosis (Lobastova et al. 2004).
Angular dioxygenation of carbazole
Some carbazole degraders reported in the literature degrade carbazole via angular dioxygenation, a novel type of oxidative attack that occurred at the ring-fused position and mediated by a multicomponent enzyme, carbazole 1,9a-dioxygenase (CARDO) with addictive preference for angular positions (Nojiri et al. 1999). In contrast to lateral dioxygenation and monohydroxylation, angular dioxygenation result in complete mineralization of carbazole with the resulting catechol converted to tricarboxylic acid (TCA) cycle intermediate (Nojiri and Omori 2002).
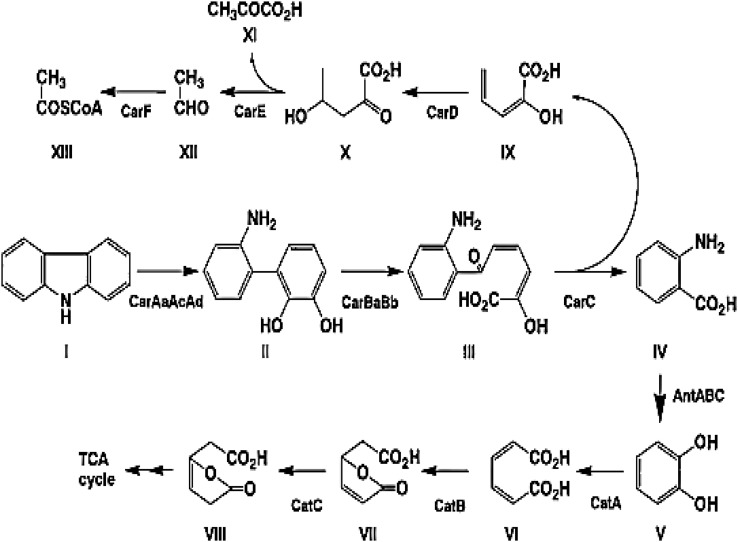
Ouchiyama and co-workers isolated a carbazole degrader, Pseudomonas resinovorans CA10, from activated sludge of a municipal wastewater treatment facility in Tokyo, Japan. The strain is capable of growth on carbazole as a sole source of carbon, nitrogen and energy and accumulates anthranilic acid and catechol as catabolic intermediates of carbazole. It also grows on anthranilic acid as carbon and nitrogen source and accumulates catechol suggesting carbazole conversion to catechol via anthranilic acid (Ouchiyama et al. 1993). Furthermore, production of 2′-aminobiphenyl-2,3-diol and its meta-cleavage product 2-hydroxy-6-oxo-6-(2′-aminopheny)-hexa-2,4-dienoate (HOADA) from the culture medium of CA10 grown on carbazole was suggested. Based on these findings and its similarity with dibenzofuran degradation pathway, a carbazole degradation pathway was proposed (Fig. 4). The pathway is divided into upper and lower pathway. The upper pathway encompasses the conversion of carbazole to catechol, while the lower pathway involves catechol mineralization (Nojiri 2012).
Fig. 4.
Carbazole degradation pathway in P. resinovorans CA10. Enzymes designations: CarAaAcAd, carbazole 1,9a-dioxygenase; CarBaBb, 2′-aminobiphenyl-2,3-diol 1,2-dioxygenase; CarC, 2-hydroxy-6-oxo-6-(2′-aminophenyl)-hexa-2,4-dienoate hydrolase; CarD, 2-hydroxypenta-2,4-dienoate hydratase; CarE, 4-hydroxy-2-oxovalerate aldolase; CarF, acetaldehyde dehydrogenase (acylating); AntABC, anthranilate 1,2-dioxygenase; CatA, catechol 1,2-dioxygenase; CatB, cis,cis-muconate lactonizing enzyme; CatC, muconolactone δ-isomerase. Compounds: I, CAR; II, 2′-aminobiphenyl-2,3-diol; III, 2-hydroxy-6-oxo-6-(2′-aminophenyl)-hexa-2,4-dienoate; IV, anthranilic acid; V, catechol; VI, cis,cis-muconate; VII, muconolactone; VIII, β-ketoadipic acid enol-lactone; IX, 2-hydroxy-penta-2,4-dienoate; X, 4-hydroxy-2-oxovalerate; XI, pyruvate; XII, acetaldehyde; XIII, acetyl coenzyme A (Nojiri et al. 2001)
Carbazole is dioxygenated at angular (C9a) and adjacent (C1) carbon atoms to produce an unstable hemiaminal (1-hydro-1,9a-dihydroxycarbazole) which is spontaneously cleaved to form 2′-aminobiphenyl-2,3-diol. This metabolic intermediate is converted to anthranilic acid via meta-cleavage and subsequent hydrolysis. Anthranilic acid is converted to catechol by dioxygenation at the C1 and C2 positions followed by spontaneous deamination and decarboxylation reactions (Kobayashi and Hiyaishi 1970). The resulting catechol is converted to a tricarboxylic acid (TCA)-cycle intermediate via ortho-cleavage (as in P. resinovorans CA10) or meta-cleavage (as in Pseudomonas stutzeri strain OM1) pathways (Ouchiyama et al. 1993; 1998; Fig. 4).
Anthranilic acid has been detected in the culture extracts of several carbazole degraders and is regarded as the main metabolite of carbazole angular dioxygenation (Ouchiyama et al. 1993; Gieg et al. 1996; Ouchiyama et al. 1998; Kirimura et al. 1999; Schneider et al. 2000; Kilbane II et al. 2002; Inoue et al. 2005). Recently, carbazole degraders with addictive preference for angular dioxygenation were also isolated from hydrocarbon-contaminated tropical African soil in Lagos, Nigeria (Salam et al. 2014). The isolates, designated Achromobacter sp. strain SL1, Pseudomonas sp. strain SL4 and Microbacterium esteraromaticum strain SL6, produce anthranilic acid and catechol as the major metabolites of carbazole angular dioxygenation. Anthranilic acid is a biotic compound and is formed by the degradation of tryptophan in several living organisms (Hayaishi and Stanier 1951). It is an important intermediate in the metabolism of many N-heterocyclic compounds and plays an important role in Pseudomonas quinolone signal, which is involved in quorum sensing in Pseudomonas aeruginosa cells (Calfee et al. 2001).
It is worthy to note, however, that once angular dioxygenation and subsequent ring cleavage occur for carbazole, the resulting 2′-aminobiphenyl-2,3-diol is degraded via the analogous biphenyl degradation pathways (Furukawa et al. 2004).
The CARDO system in carbazole degraders and its substrate specificity
The extensively studied CARDO system in Pseudomonas resinovorans CA10 is a three-component dioxygenase system belonging to the Rieske nonheme iron oxygenase system (ROS) and consist of a terminal oxygenase and electron transport proteins (Sato et al. 1997a; Nam et al. 2002). The terminal oxygenase component of CARDO (CARDO-O) is a homotrimeric enzyme that contains one Rieske [2Fe-2S] cluster ([2Fe-2S]R and one active-site iron (Fe2+) in a single subunit (CarAa) (Nojiri and Omori 2007). The electron transport proteins of CARDO, which mediate electron transport from NAD(P)H to CARDO-O, comprise ferredoxin (CARDO-F; a monomer of CarAc), which contains one [2Fe-2S]R, and ferredoxin reductase (CARDO-R; a monomer of CarAd), which contains one FAD and one plant-type [2Fe-2S] cluster ([2Fe-2S]P) (Sato et al. 1997a; Nam et al. 2002).
Phylogenetic analysis revealed a very low homology (<19% overall length-wise identity) of the amino acid sequence of CARDO with almost all known catalytic subunits of ROS terminal oxygenases (Nojiri and Omori 2007). In addition, CARDO-O consists of only catalytic α subunit with the α3 configuration in contrast to typical class III ROSs whose terminal oxygenase components consist of both α and β subunits with the α3β3 (or α2β2) configuration (Nojiri and Omori 2007). This homotrimeric structure is typical of class IA ROSs, whose terminal oxygenases have α3 configurations (Ferraro et al. 2005).
CARDO catalyzes diverse oxygenation of aromatic compounds. Aside from angular dioxygenation, which is the most interesting feature of CARDO, biotransformation experiments with E. coli cells harboring carAa, carAc, and carAd revealed the ability of CARDO to catalyze lateral dioxygenation and monooxygenation of aromatic substrates exhibiting broad substrate specificity (Nojiri et al. 1999; Takagi et al. 2002). It was also observed that angular dioxygenation by CARDO occurs effectively at the angular position adjacent to an oxygen or nitrogen atom (due to high electronegativity of oxygen and nitrogen), but not a sulfur or carbon atom (Bressler and Fedorak 2000; Nojiri and Omori 2007).
Carbazole degradative genes
Pseudomonas-type car gene cluster
The CAR degradative genes of P. resinovorans CA10 have been extensively studied. Sato et al. (1997a, b) first succeeded in cloning the genes involved in upper pathway of carbazole degradation from P. resinovorans CA10 genome by shotgun cloning using meta-cleavage activity. The resultant gene fragment contains seven degradative genes, one open reading frame (ORF) that encoded a putative protein or unknown function, and two partial possible genes. Functional analysis of the degradative genes shows two identical copies of carAa, carAc, and carAd, which encode terminal oxygenase, ferredoxin, and ferredoxin reductase components of carbazole 1,9a-dioxygenase (CARDO); carBa and carBb, which encode structural and catalytic subunits of the meta-cleavage enzyme (2′-aminobiphenyl-2,3-diol 1,2-dioxygenase); and carC, which encodes the meta-cleavage compound (HOADA) hydrolase.
Gene walking around the car CA10 gene cluster revealed the entire gene structure. 2-hydroxypenta-2,4-dienoate (HPD degradative carDFE genes (meta-cleavage pathway genes) was found downstream of the carAd gene. In addition, antABC gene encoding anthranilate 1,2-dioxygenase was found in the 21-kb region upstream from carAa (Fig. 5a) (Nojiri et al. 2001). This anthranilate degradative gene cluster is a putative composite transposon flanked by two homologous insertion sequences ISPre1 and ISPre2. Furthermore, antR gene encoding a transcriptional regulator of the ant operon was found outside the putative composite transposon containing antABC, which regulates the inducible expression of the car gene cluster (Urata et al. 2004). Tn5 mutagenesis was used to isolate the β-ketoadipate pathway (ortho-cleavage pathway) genes involved in catechol mineralization from strain CA10 genome (Kimura et al. 1996).
Fig. 5.
Genetic structure of the gene clusters involved in carbazole biodegradation by a P. resinovorans CA10 and Janthinobacterium sp. J3, b Sphingomonas (Novosphingobium) sp. KA1 and Sphingomonas sp. GTIN11, c N. aromaticivorans IC177, and d Sphingomonas sp. CB3 (Nojiri and Omori 2007)
Carbazole-degrading bacteria from the genera Pseudomonas, Burkholderia, and Janthinobacterium have been reported that have nearly identical carbazole degradative genes with car CA10 and are designated Pseudomonas-type car gene cluster. Even though these carbazole degraders are isolated from different sources, comparison of the gene organization and flanking regions of their car gene clusters suggests evolutionary diversity as reflected in differences in copy number of car gene cluster among carbazole degraders (Inoue et al. 2004). This phenomenon may arise because car gene clusters are sometimes borne on plasmids or transposons and/or flanked by IS (insertion sequence) elements (Inoue et al. 2004).
Sphingomonas-type car gene cluster
The genus Sphingomonas was found to possess a car gene cluster homolog (though relatively low homology, <60% identity) showing similarity in gene organization and phylogeny with the car CA10 gene cluster. Isolation of car gene clusters in sphingomonads was first reported in Sphingomonas sp. GTIN11 (Kilbane II et al. 2002) and Sphingomonas sp. (reclassified as Novosphingobium sp) KA1 (Habe et al. 2002) and the car KA1/GTIN11 gene cluster homolog have been reported to occur in various carbazole-degrading Sphingomonas and related strains (Inoue et al. 2004, 2005).
The car gene clusters isolated from these two Sphingomonas strain are different from car CA10 gene cluster in two ways. First, unlike the car CA10 gene cluster, it does not contain the NAD(P)H:ferredoxin oxidoreductase gene involved in the initial dioxygenase, but contains the genes for terminal oxygenase (carAa) and ferredoxin (carAc), the meta-cleavage enzyme (carBaBb), and HOADA hydrolase (carC) (Fig. 5b). Second, though Sphingomonas CarAa exhibits significant homology with CA10 CarAa (>55% identity), its ferredoxin (CarAc) is neither related to CarAcCA10 nor with other Rieske ferredoxins but shows similarity to the putidaredoxin-type ferredoxins. Because the terminal oxygenase of strain KA1 (CarAaKA1) can receive electrons from strain KA1 ferredoxin (CarAcKA1) and catalyze angular dioxygenation of carbazole, it implies that ferredoxin selectivity differs between strain CarAaCA10 and CarAaKAI/GTIN11 (Inoue et al. 2004). Furthermore, two copies of car KA1 gene cluster (car-IKA1 and car-IIKA1) were found to be domiciled on a >250-kb circular plasmid pCAR3 in Novosphingobium sp. KA1 along with the presence of NAD(P)H:ferredoxin oxidoreductase genes (fdrI and fdrII) and a third putidaredoxin-type ferredoxin gene. These findings show clearly that the plasmid pCAR3 contains the complete set of genes responsible for carbazole mineralization in strain KA1 (Urata et al. 2006).
The car gene cluster in Nocardioides aromaticivorans IC177
Quite distinct car gene cluster different from the Pseudomonas and Sphingomonas-types was found in a Gram-positive bacterium N. aromaticivorans IC177 (Inoue et al. 2005, 2006). The car gene was clustered in the carAaCBaBbAcAd and carDFE gene clusters encoding the enzymes responsible for degradation of carbazole to anthranilate and 2-hydroxypenta-2,4-dienoate (HPD) (upper pathway) and HPD to pyruvate and acetyl coenzyme A (lower pathway), respectively (Inoue et al. 2006).
However, the position of carC relative to carBaBb in strain IC177 is the opposite of that in car gene clusters of the Pseudomonas and Sphingomonas-types (Fig. 5c) (Inoue et al. 2006). In the car gene operons in strain IC177, the genes overlap each other by 1 or 4 bp with carDFE genes closely linked and located upstream of the carAaCBaBbAcAd gene cluster. In addition, organization of carbazole catabolic operon in strain IC177 occurred in a more orderly fashion as functional units than those in Gram-negative strains, such as strains CA10, J3, GTIN11, and KA1 (Nojiri and Omori 2007).
The car gene cluster in Sphingomonas sp. CB3
Interestingly, the car gene cluster of strain CB3 differs from those of the three above-mentioned types in terms of gene organization and phylogeny but showed marked similarity with naphthalene and biphenyl degradative bph gene cluster (Shepherd and Lloyd-Jones 1998). The car genes of strain CB3 are arranged in the order of carAaAbAcAdBCD, and the terminal oxygenase component of strain CB3, unlike those of other CAR degraders, which are composed of a single subunit, is composed of two subunits, CarAa and CarAb, respectively (Fig. 5d) (Shepherd and Lloyd-Jones 1998). Although carbazole metabolic activity of the enzymes encoded in carbazole catabolic operon in CB3 has not been confirmed, its transcription was detected when carbazole was used as source of carbon by strain CB3 (Nojiri and Omori 2007).
The car gene cluster in marine carbazole degraders
Carbazole-degrading bacteria from different genera such as Neptuniibacter, Erythrobacter, Marinobacter, Caulobacter, Hyphomonas, Lysobacter, Sphingosinicella, Kordiimonas, and Terrabacter have been isolated from marine environment (Fuse et al. 2003; Inoue et al. 2005; Maeda et al. 2009a, b). Southern hybridization analysis performed under strict conditions at 68 °C (hybridization conditions for similarity of >90%) and 55 °C (hybridization conditions for similarity >60%) using car CA10 and car KA1 gene cluster probes for 14 marine isolates showed that they lack car genes highly similar to car CA10 and car KA1. This suggests that marine isolates are evolutionarily different from their terrestrial counterpart, with unique car gene clusters and CARDO. Furthermore, hybridization analysis at 55 °C showed that eight of the 14 marine isolates have novel car gene cluster that are highly different from the car CA10 and car KA1 genes.
car gene cluster of Neptuniibacter sp. strain CAR-SF
The car gene cluster of strain CAR-SF is arranged in the order carAaBaBbC, resembling the order of arrangement of the Pseudomonas and Sphingomonas-type car gene clusters showing 48–77% similarity with car CA10 and carJ3 genes and thus designated as a Pseudomonas-type car gene cluster (Nagashima et al. 2010). However, in comparison with the car CA10 and carJ3 gene clusters, the car CAR-SF gene cluster lacks the ferredoxin carAc and ferredoxin reductase carAd genes, though a carAc CA10-like gene was revealed by Southern hybridization analysis. This shows that unlike in car CA10 and related Pseudomonas-type car gene clusters, ferredoxin gene of CARDO was in a different location in CAR-SF strain and not in the car CAR-SF gene cluster (Nagashima et al. 2010).
car gene cluster of Lysobacter sp. strain OC7
The car gene cluster in strain OC7 is arranged in the order carAaCBaBb, with the position of carC and carBaBb inverted when compared to their positions in Pseudomonas and Sphingomonas-type car gene clusters. However, the genes arrangement followed the same order as in the car gene cluster of strain IC177 (Maeda et al. 2009b). The open reading frames (ORFs) containing the car gene cluster of strain OC7 share 39-52% similarity with carAa, carC, carBa, and carBb genes of strains CA10 and KA1, and showed no similarity with car genes of strain CB3, making the car genes of strain OC7 phylogenetically distinct from previously reported car gene products. Furthermore, southern hybridization analysis shows that only Caulobacter sp. strain OC6, a phylogenetically different genus (α-proteobacteria), hybridized with the car OC7 gene cluster (from strain OC7 belonging to γ-proteobacteria group) probe with more than 90% similarity (Maeda et al. 2009b). This finding is interesting as it reveals the evolutionary diversity of car gene clusters and importance of genetic exchange in its distribution across different phylogenetic groups.
The product of carAa OC7 possessed consensus sequences of a Rieske-type [2Fe-2S] cluster and mononuclear heme iron (Maeda et al. 2009b). However, its ferredoxin and ferredoxin reductase genes are not located near the car gene cluster of strain OC7, as in strain CAR-SF. In addition, as in CAR-SF, E. coli harboring only carAa OC7 was unable to convert CAR but E. coli cells harboring pBOC77 (carAa OC7 AcAd CA10) converted CAR to 2′-aminobiphenyl-2,3-diol. However, the transformation ratio of CAR by pBOC77 (carAa OC7 AcAd CA10) was 32–36%, which is less than 99% recorded for E. coli cells harboring pUCARA (carAaAaAcORFcarAd) (Sato et al. 1997a) or pSF6 (carAa CAR-SF AcAd CA10) used as positive controls, thus revealing weak electron transfer efficiency of CarAaOC7AcAdCA10 and suggesting a different electron transfer components and RO class for CARDOOC7 (Maeda et al. 2009b).
car gene cluster of novel genus strain OC9
The CARDO system and the arrangement of car gene cluster in strain OC9 present a new question in relation to evolution and diversity of car genes in bacteria. First, the recovered ORFs of strain OC9 share 35–65% homology with previously reported car genes (carRAaCBaBb). However, a ferredoxin-like gene (carAc) found immediately downstream of carR does not show homology with any of the reported ferredoxin component of CARDO as it possesses a chloroplast-type ferredoxin (Maeda et al. 2010). This is a unique type of ferredoxin completely different from the Rieske and putidaredoxin-types reported for strains CA10, KA1, IC177, and CB3 CARDO systems (Sato et al. 1997a; Shepherd and Lloyd 1998; Inoue et al. 2006; Urata et al. 2006). Second, the car gene cluster of strain OC9 was arranged in the order carAcRAaCBaBb with carRAc and carAaBaBb having opposite orientation, thus suggesting that the carAc and carAa genes transcribed within different transcription units.
The product of carAa OC9 possessed consensus sequences of a Rieske-type [2Fe-2S] cluster and mononuclear heme iron (Maeda et al. 2010). However, unlike carAa CAR-SF and carAa OC7 that could not transform E. coli cells without CarAc, E. coli cells harboring only carAa OC9 in a resting cell reaction converted CAR to 2′-aminobiphenyl 2,3-diol, though the conversion ratio (12%) is low when compared to that of E. coli cells harboring genes for both carAa and carAc (100%), respectively (Maeda et al. 2010).
Conclusion
In summary, carbazole is a very important compound from the industrial, medical and environmental perspectives. Its use in the industry as chemical feedstock for the production of dyes, reagents, explosives, insecticides, lubricants and color inhibitor in detergent is well documented. The medical importance of naturally occurring carbazole and derivatives of hydroxylated carbazoles and their antitumor, psychotrophic, anti-inflammatory, anti-histaminic, antibiotic and antioxidative activities has also been reported extensively. However, environmentally, carbazole is of serious concern as it is recalcitrant, mutagenic and toxic with genotoxic and carcinogenic hazardous derivatives such as N-methylcarbazole and 7-H-dibenzo[c,g]carbazole found in cigarette smoke and automobile emissions categorized as “IARC Group 2B” carcinogens. Carbazole angular dioxygenation characterized with addictive preference for angular position results in complete mineralization to intermediates of the TCA cycle. The genes involved are evolutionarily diverse and have been detected in various microorganisms cutting across different bacteria phyla.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest in the publication.
References
- Ainsworth CC, Zachara JM, Smith SC. Carbazole sorption by surface and subsurface materials: influence of sorbent and solvent properties. Soil Sci Soc Am J. 1989;53:1391–1401. doi: 10.2136/sssaj1989.03615995005300050015x. [DOI] [Google Scholar]
- Balaban AT, Oniciu DC, Katritzky AR. Aromaticity as a cornerstone of heterocyclic chemistry. Chem Rev. 2004;104(5):2777–2812. doi: 10.1021/cr0306790. [DOI] [PubMed] [Google Scholar]
- Barron MG, Heintz R, Rice SD. Relative potency of PAHs and heterocycles as aryl hydrocarbon receptor agonists in fish. Mar Environ Res. 2004;58:95–100. doi: 10.1016/j.marenvres.2004.03.001. [DOI] [PubMed] [Google Scholar]
- Behymer TD, Hates RA. Photolysis of polycyclic aromatic hydrocarbons adsorbed on fly ash. Environ Sci Technol. 1988;22:1311–1319. doi: 10.1021/es00176a011. [DOI] [Google Scholar]
- Benedik MJ, Gibbs PR, Riddle RR, Willson RC. Microbial denitrogenation of fossil fuels. Trends Biotechnol. 1998;16:390–395. doi: 10.1016/S0167-7799(98)01237-2. [DOI] [PubMed] [Google Scholar]
- Bi E, Schmidt TC, Haderlain SB. Environmental factors influencing sorption of heterocyclic aromatic compounds to soil. Environ Sci Technol. 2007;41(9):3172–3178. doi: 10.1021/es0623764. [DOI] [PubMed] [Google Scholar]
- Bidleman TF. Atmospheric processes: wet and dry deposition of organic compounds are controlled by their vapour-particle partitioning. Environ Sci Technol. 1988;22:361–367. doi: 10.1021/es00169a002. [DOI] [Google Scholar]
- Blum P, Sagner A, Tiehm A, Martus P, Wendel T, Grathwohl P. Importance of heterocyclic aromatic compounds in monitored natural attenuation for coal tar contaminated aquifers: a review. Contam hydrol. 2011;126:181–194. doi: 10.1016/j.jconhyd.2011.08.004. [DOI] [PubMed] [Google Scholar]
- Brack W, Klamer HJ, Lopez de Alda M, Barcelo D. Effect-directed analysis of key toxicants in European river basins a review. Environ Sci Pollut Res. 2007;14:30–38. doi: 10.1065/espr2006.08.329. [DOI] [PubMed] [Google Scholar]
- Bressler DC, Fedorak PM. Bacterial metabolism of fluorene, dibenzofuran, dibenzothiophene, and carbazole. Can J Microbiol. 2000;46:397–409. doi: 10.1139/w00-017. [DOI] [PubMed] [Google Scholar]
- Bundy JG, Morriss AWJ, Durham DG, Campbell CD, Paton GI. Development of QSARs to investigate the bacterial toxicity and biotransformation potential of aromatic heterocyclic compounds. Chemosphere. 2001;42:885–892. doi: 10.1016/S0045-6535(00)00178-8. [DOI] [PubMed] [Google Scholar]
- Calfee MW, Coleman JP, Pesci EC. Interference with Pseudomonas quinolone signal synthesis inhibits virulence factor expression by Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 2001;98:11633–11637. doi: 10.1073/pnas.201328498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collin G, Hoke H. Carbazole. In: Elvers B, Hawkins S, Schultz G, editors. Ullman’s encyclopedia of industrial chemistry. 5. New York: VHS; 1986. [Google Scholar]
- Eisentraeger A, Brinkman C, Hollert H, Sagner A, Tiehm A, Neuwoehner J. Heterocyclic compounds: toxic effects using algae, daphnids, and the Salmonella/microsome test taking methodical quantitative aspects into account. Environ Toxicol Chem. 2008;27:1590–1596. doi: 10.1897/07-201.1. [DOI] [PubMed] [Google Scholar]
- Ferraro DJ, Gakhar L, Ramaswamy S. Rieske business: structure-function of Rieske non-heme oxygenases. Biochem Biophys Res Commun. 2005;338:175–190. doi: 10.1016/j.bbrc.2005.08.222. [DOI] [PubMed] [Google Scholar]
- Furukawa K, Suenaga H, Goto M. Biphenyl dioxygenases: functional versatilities and directed evolution. J Bacteriol. 2004;186:5189–5196. doi: 10.1128/JB.186.16.5189-5196.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuse H, Takimura O, Murakami K, Inoue H, Yamaoka Y. Degradation of chlorinated biphenyl, dibenzofuran, and dibenzo-p-dioxin by marine bacteria that degrade biphenyl, carbazole, or dibenzofuran. Biosci Biotechnol Biochem. 2003;67:1121–1125. doi: 10.1271/bbb.67.1121. [DOI] [PubMed] [Google Scholar]
- Gieg LM, Otter A, Fedorak PM. Carbazole degradation by Pseudomonas sp. LD2: metabolic characteristics and the identification of some metabolites. Environ Sci Technol. 1996;30:575–585. doi: 10.1021/es950345v. [DOI] [Google Scholar]
- Girgis MJ, Gates BC. Reactivities, reaction networks and kinetics in high pressure catalytic hydroprocessing. Ind Eng Chem Res. 1991;30:2021–2158. doi: 10.1021/ie00057a001. [DOI] [Google Scholar]
- Grifoll M, Selifonov SA, Chapman PJ. Transformation of substituted fluorenes and fluorine analogs by Pseudomonas sp. strain F274. Appl Environ Microbiol. 1995;57:3462–3469. doi: 10.1128/aem.61.9.3490-3493.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosser RJ, Warshawsky D, Vestal JR. Indigenous and enhanced mineralization of pyrene, benzo(a)pyrene and carbazole in soils. Appl Environ Microbiol. 1991;57:3462–3469. doi: 10.1128/aem.57.12.3462-3469.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habe H, Ashikawa Y, Saiki Y, Yoshida T, Nojiri H, Omori T. Sphingomonas sp. strain KA1, carrying a carbazole dioxygenase gene homologue, degrades chlorinated dibenzo-p-dioxins in soil. FEMS Microbiol Lett. 2002;211:43–49. doi: 10.1111/j.1574-6968.2002.tb11201.x. [DOI] [PubMed] [Google Scholar]
- Hayaishi O, Stanier RY. The bacterial oxidation of tryptophan III. Enzymatic activities of cell-free extracts from bacteria employing the aromatic pathway. J Bacteriol. 1951;62:691–709. doi: 10.1128/jb.62.6.691-709.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hisatsuka K, Sato M. Microbial transformation of carbazole to anthranilic acid by Pseudomonas stutzeriPseudomonas stutzeri. Biosci Biotech Biochem. 1994;58(1):213–214. doi: 10.1271/bbb.58.213. [DOI] [PubMed] [Google Scholar]
- IARC (1983) Monographs on the evaluation of the carcinogenic risks chemical of to humans. Vol.32. Polynuclear aromatic compounds, Part I, chemical, environmental and experimental data, pp. 37, 148, 156, 164, 278, 284, 316, 328, 338, 344, 385. International Agency for Research on Cancer [PubMed]
- IARC (1999) Monographs on the evaluation of the carcinogenic risks of chemical to humans. Re-evaluation of some organic chemicals, hydrazine and hydrogen peroxide. Vol.71, Part IIIb. International Agency for Research on Cancer, pp 1319–1323 [PMC free article] [PubMed]
- Inoue K, Widada J, Nakai S, Endoh T, et al. Divergent structures of carbazole degradative car operons isolated from Gram-negative bacteria. Biosci Biotechnol Biochem. 2004;68:1467–1480. doi: 10.1271/bbb.68.1467. [DOI] [PubMed] [Google Scholar]
- Inoue K, Habe H, Yamane H, Omori T, Nojiri H. Diversity of carbazole-degrading bacteria having the car gene cluster: isolation of a novel Gram-positive carbazole-degrading bacterium. FEMS Microbiol Lett. 2005;245:145–153. doi: 10.1016/j.femsle.2005.03.009. [DOI] [PubMed] [Google Scholar]
- Inoue K, Habe H, Yamane H, Nojiri H. Characterization of novel carbazole catabolism genes from the Gram-positive carbazole degrader Nocardioides aromaticivorans ICI77. Appl Environ Microbiol. 2006;72:3321–3329. doi: 10.1128/AEM.72.5.3321-3329.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs BW, Billings CE. Characterization and temperature dependence of PAH emissions from a simulated rubber combustion operation. Am Ind Hyg Assoc J. 1985;46:547–554. doi: 10.1080/15298668591395337. [DOI] [PubMed] [Google Scholar]
- Johansen SS, Hansen AB, Mosbaek H, Arvin E. Identification of heteroaromatic and other organic compounds in ground water at creosote-contaminated sites in Denmark. Groundw Monit Rem. 1997;17:106–115. doi: 10.1111/j.1745-6592.1997.tb01283.x. [DOI] [Google Scholar]
- Katritzky AR, Ramsden CA, Joule JA, Zhdankin VV. Handbook of heterocyclic chemistry. 3. Amsterdam: Elsevier; 2010. [Google Scholar]
- Kilbane JJ, II, Daram A, Abbasian J, Kayser KJ. Isolation and characterization of Sphingomonas sp. GTIN11 capable of carbazole mineralization in petroleum. Biochem Biophys Res Commun. 2002;297:242–248. doi: 10.1016/S0006-291X(02)02183-6. [DOI] [PubMed] [Google Scholar]
- Kimura T, Zhang Y, Kodama T, Omori T. Isolation and characterization of Tn5-induced mutants deficient in carbazole catabolism. FEMS Microbiol Lett. 1996;135:65–70. doi: 10.1111/j.1574-6968.1996.tb07967.x. [DOI] [Google Scholar]
- Kirimura K, Nakagawa H, Tsuji K, Matsuda K, Kurane R, Usami S. Selective and continuous degradation of carbazole contained in petroleum oil by resting cells of Sphingomonas sp. CDH-7. Biosci Biotechnol Biochem. 1999;63:1563–1568. doi: 10.1271/bbb.63.1563. [DOI] [PubMed] [Google Scholar]
- Kobayashi S, Hayaishi O. Anthranilic acid conversion to catechol (Pseudomonas) Methods Enzymol. 1970;17A:505–510. doi: 10.1016/0076-6879(71)17231-X. [DOI] [Google Scholar]
- Li L, Xu P, Blankespor HD. Degradation of carbazole in the presence of non-aqueous phase liquids by Pseudomonas sp. Biotechnol Lett. 2004;26:581–584. doi: 10.1023/B:BILE.0000021959.00819.d5. [DOI] [PubMed] [Google Scholar]
- Lide DR. Handbook of chemistry and physics. 84. Boca Raton: CRC Press; 2003. [Google Scholar]
- Lobastova TG, Sukhodolokaya GV, Nikolayeva VM, Baskunov BP, Turchin KF, Donova MV. Hydroxylation of carbazoles by Aspergillus flavus VKM F-1024. FEMS Microbiol Lett. 2004;235:51–56. doi: 10.1111/j.1574-6968.2004.tb09566.x. [DOI] [PubMed] [Google Scholar]
- Maeda R, Nagashima H, Widada J, Iwata K, Omori T. Novel marine carbazole-degrading bacteria. FEMS Microbiol Lett. 2009;292:203–209. doi: 10.1111/j.1574-6968.2009.01497.x. [DOI] [PubMed] [Google Scholar]
- Maeda R, Nagashima H, Zulkharnain AB, Iwata K, Omori T. Isolation and characterization of a car gene cluster from the naphthalene, phenanthrene, and carbazole-degrading marine isolate Lysobacter sp. strain OC7. Curr Microbiol. 2009;59:154–159. doi: 10.1007/s00284-009-9414-y. [DOI] [PubMed] [Google Scholar]
- Maeda R, Ito Y, Iwata K, Omori T. Comparison of marine and terrestrial carbazole-degrading bacteria. Curr Res Technol Educ Top Appl Microbiol Microb Biotechnol. 2010;2:1311–1321. [Google Scholar]
- Meyer S, Steinhart H. Effects of heterocyclic PAHs (N, S, O) on the biodegradation of typical tar oil PAHs in a soil/compost mixture. Chemosphere. 2000;40:359–367. doi: 10.1016/S0045-6535(99)00237-4. [DOI] [PubMed] [Google Scholar]
- Meylan WM, Howard PH. Bond contribution method for estimating Henry’s law constants. Environ Toxicol Chem. 1991;10:1283–1293. doi: 10.1002/etc.5620101007. [DOI] [Google Scholar]
- Nagashima H, Zulkharnain AB, Maeda R, Fuse H, Iwata K, Omori T. Cloning and nucleotide sequences of carbazole degradation genes from marine bacterium Neptuniibacter sp. strain CAR-SF. Curr Microbiol. 2010;61(1):50–56. doi: 10.1007/s00284-009-9575-8. [DOI] [PubMed] [Google Scholar]
- Nam J, Nojiri H, Noguchi H, Uchimura H, Yoshida T, Habe H, Yamane H, Omori T. Purification and characterization of carbazole 1,9a-dioxygenase, a three-component dioxygenase system of Pseudomonas resinovorans strain CA10. Appl Environ Microbiol. 2002;68:5882–5890. doi: 10.1128/AEM.68.12.5882-5890.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nojiri H. Structural and molecular genetic analyses of the bacterial carbazole degradation system. Biosci Biotechnol Biochem. 2012;76(1):1–18. doi: 10.1271/bbb.110620. [DOI] [PubMed] [Google Scholar]
- Nojiri H, Omori T. Molecular bases of aerobic bacterial degradation of dioxins: involvement of angular dioxygenation. Biosci Biotechnol Biochem. 2002;66:2001–2016. doi: 10.1271/bbb.66.2001. [DOI] [PubMed] [Google Scholar]
- Nojiri H, Omori T. Carbazole metabolism by Pseudomonads. In: Ramos J-L, Filloux A, editors. Pseudomonas. New York: Springer; 2007. pp. 107–145. [Google Scholar]
- Nojiri H, Nam JW, Kosaka M, Morii K-I, Takemura T, et al. Diverse oxygenations catalyzed by carbazole 1,9a-dioxygenase from Pseudomonas sp. strain CA10. J Bacteriol. 1999;181(10):3105–3113. doi: 10.1128/jb.181.10.3105-3113.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nojiri H, Sekiguchi H, Maeda K, Urata M, Nakai S, Yoshida T, Habe H, Omori T. Genetic characterization and evolutionary implications of a car gene cluster in the carbazole degrader Pseudomonas sp. strain CA10. J Bacteriol. 2001;183:3663–3679. doi: 10.1128/JB.183.12.3663-3679.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odabasi M, Vardar N, Tasdemir Y, Sofuoglu A, Holsen TM. Polycyclic aromatic hydrocarbons in Chicago air. Sci Total Environ. 1999;227:57–67. doi: 10.1016/S0048-9697(99)00004-2. [DOI] [Google Scholar]
- Odabasi M, Cetin B, Sofuoglu A. Henry’s law constant, octanol-air partition coefficient and super cooled liquid vapour pressure of carbazole as a function of temperature: application to gas/particle partitioning in the atmosphere. Chemosphere. 2006;62:1087–1096. doi: 10.1016/j.chemosphere.2005.05.035. [DOI] [PubMed] [Google Scholar]
- Ouchiyama N, Zhang Y, Omori T, Kodama T. Biodegradation of carbazole by Pseudomonas spp. CA06 and CA10. Biosci Biotechnol Biochem. 1993;57:455–460. doi: 10.1271/bbb.57.455. [DOI] [Google Scholar]
- Ouchiyama N, Miyachi S, Omori T. Cloning and nucleotide sequence of carbazole catabolic genes from Pseudomonas stutzeri strain OM1, isolated from activated sludge. J Gen Appl Microbiol. 1998;44:57–63. doi: 10.2323/jgam.44.57. [DOI] [PubMed] [Google Scholar]
- Pearlman RS, Yalkowsky SH, Banerjee S. Water solubilities of polynuclear aromatic and heteroaromatic compounds. J Phys Chem Ref Data. 1984;13:555–562. doi: 10.1063/1.555712. [DOI] [Google Scholar]
- Pearson JM, Stolka M (1981) Poly(N-vinylcarbazole) In: Polymer Monographs No. 6. Gordon & Breach, New York
- Peddinghaus S, Brinkmann M, Bluhm K, Sagner A, et al. Quantitative assessment of the embryotoxic potential of NSO-heterocyclic compounds using zebrafish (Danio rerio) Reproduc Toxicol. 2012;33:224–232. doi: 10.1016/j.reprotox.2011.12.005. [DOI] [PubMed] [Google Scholar]
- Perreira WE, Rostard CE, Updegraft DL, Bennett JM. Fate and movement of azaarenes and their anaerobic biotransformation products in an aquifer contaminated by wood-treatment chemicals. Environ Toxicol Chem. 1987;6:163–176. doi: 10.1002/etc.5620060302. [DOI] [Google Scholar]
- Renault D, Tombalan F, Brault D, Perin F, Thybaud V. Comparative mutagenicity of 7H-dibenzo (c, g) carbazole and two derivatives in MutaMouse liver and skin. Mutat Res. 1998;417:129–140. doi: 10.1016/S1383-5718(98)00101-6. [DOI] [PubMed] [Google Scholar]
- Resnick SM, Torok DS, Gibson DT. Oxidation of carbazole to 3-hydroxycarbazole by naphthalene 1,2-dioxygenase and biphenyl 2,3-dioxygenase. FEMS Microbiol Lett. 1993;113:297–302. doi: 10.1111/j.1574-6968.1993.tb06530.x. [DOI] [PubMed] [Google Scholar]
- Robbiano L, Baroni D, Carrozzino R, Mereto E, Brambilla G. DNA damage and micronuclei induced in rat and human kidney cells by six chemicals carcinogenic to the rat kidney. Toxicology. 2004;204:187–195. doi: 10.1016/j.tox.2004.06.057. [DOI] [PubMed] [Google Scholar]
- Roy J, Jana AK, Mal D. Recent trends in the synthesis of carbazoles: an update. Tetrahedron. 2012;68:6099–6121. doi: 10.1016/j.tet.2012.05.007. [DOI] [Google Scholar]
- Salam LB, Ilori MO, Amund OO, et al. Carbazole angular dioxygenation and mineralization by bacteria isolated from hydrocarbon-contaminated tropical African soil. Environ Sci Pollut Res. 2014;21:9311–9324. doi: 10.1007/s11356-014-2855-2. [DOI] [PubMed] [Google Scholar]
- Santos SC, Alviano DS, Alviano CS, Padula M, et al. Characterization of Gordonia sp. strain F.5.25.8 capable of dibenzothiophene desulfurization and carbazole utilization. Appl Microbiol Biotechnol. 2006;71:355–362. doi: 10.1007/s00253-005-0154-z. [DOI] [PubMed] [Google Scholar]
- Sato S, Nam JW, Kasuga K, Nojiri H, Yamane H, Omori T. Identification and characterization of genes encoding carbazole 1,9a-dioxygenase in Pseudomonas sp. strain CA 10. J Bacteriol. 1997;179:4850–4858. doi: 10.1128/jb.179.15.4850-4858.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato S, Ouchiyama N, Kimura T, Nojiri H, Yamane H, Omori T. Cloning of genes involved in carbazole degradation of Pseudomonas sp, strain CA10: nucleotide sequences of genes and characterization of meta-cleavage enzymes and hydrolase. J Bacteriol. 1997;179:4841–4849. doi: 10.1128/jb.179.15.4841-4849.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider J, Grosser RJ, Jayasimhulu K, Xue W, Kinkle B, Warshawsky D. Biodegradation of carbazole by Ralstonia sp. RJGII.123 isolated from a hydrocarbon-contaminated soil. Can J Microbiol. 2000;46:269–277. doi: 10.1139/w99-131. [DOI] [PubMed] [Google Scholar]
- Seo JS, Keum Y-S, Cho IK, Li QX. Degradation of dibenzothiophene and carbazole by Arthrobacter sp. P1-1. Int Biodeterior Biodegrad. 2006;58:36–43. doi: 10.1016/j.ibiod.2006.04.005. [DOI] [Google Scholar]
- Seto H (1991) New 3-hydroxy-4-aminocarbazole compounds. JP, Patent 03227971
- Shepherd JM, Lloyd-Jones G. Novel carbazole degradation genes of Sphingomonas CB3: sequence analysis, transcription, and molecular ecology. Biochem Biophys Res Commun. 1998;247:129–135. doi: 10.1006/bbrc.1998.8750. [DOI] [PubMed] [Google Scholar]
- Shotbolt-Brown J, Hunter DWF, Aislabie J. Isolation and description of carbazole-degrading bacteria. Can J Microbiol. 1996;42:79–82. doi: 10.1139/m96-012. [DOI] [PubMed] [Google Scholar]
- Smith CJ, Hansch C. The relative toxicity of compounds in mainstream cigarette smoke condensate. Food Chem Toxicol. 2000;38:637–646. doi: 10.1016/S0278-6915(00)00051-X. [DOI] [PubMed] [Google Scholar]
- Smith JH, Mabey WR., Bahonos N, Holt BR, Lee SS, Chou TW, Venberger DC, Mill T (1978) Environmental pathways of selected chemicals in freshwater systems: Part II Laboratory studies. Interagency Energy-Environment Research and Development Program Report. EPA-600/7-78-074. U.S. Environmental Protection Agency, Athens GA 30605
- Smith CJ, Perfetti TA, Mullens MA, Rodgman A, Doolittle DJ. “IARC Group 2B Carcinogens” reported in cigarette mainstream smoke. Food Chem Toxicol. 2000;38:825–848. doi: 10.1016/S0278-6915(00)00066-1. [DOI] [PubMed] [Google Scholar]
- Szafarz D, Perin F, Valero D, Zajdela F. Structure and carcinogenicity of dibenzo (c, g) carbazole derivatives. Biosci Rep. 1988;8:633–643. doi: 10.1007/BF01117342. [DOI] [PubMed] [Google Scholar]
- Takagi T, Nojiri H, Yoshida T, Habe H, Omori T. Detailed comparison between the substrate specificities of two angular dioxygenases, dibenzofuran 4,4a-dioxygenase from Terrabacter sp. and carbazole 1,9a-dioxygenase from Pseudomonas resinovorans. Biotechnol Lett. 2002;24:2099–2106. doi: 10.1023/A:1021325816766. [DOI] [Google Scholar]
- Urata M, Miyakoshi M, Kai S, Maeda K, Habe H, Omori T, Yamane H, Nojiri H. Transcriptional regulation of the ant operon, encoding two-component anthranilate 1,2-dioxygenase, on the carbazole-degradative plasmid pCARI of Pseudomonas resinovorans strain CA10. J Bacteriol. 2004;186:6815–6823. doi: 10.1128/JB.186.20.6815-6823.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urata M, Uchimura H, Noguchi H, Sakaguchi T, et al. Plasmid pCAR3 contains multiple gene sets involved in the conversion of carbazole to anthranilate. Appl Environ Microbiol. 2006;72:3198–3205. doi: 10.1128/AEM.72.5.3198-3205.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valovicova Z, Mesarosova M, Trilecova L, Hruba E, et al. Genotoxicity of 7H-dibenzo (c, g)carbazole and its methyl derivatives in human keratinocytes. Mutat Res. 2012;743:91–98. doi: 10.1016/j.mrgentox.2011.12.030. [DOI] [PubMed] [Google Scholar]
- Warshawsky D, Talaska G, Xue W, Schneider J. Comparative carcinogenicity, metabolism, mutagenicity, and DNA binding of 7H-dibenzo (c, g) carbazole and dibenzo(a, j) acridine. Crit Rev Toxicol. 1996;26:213–249. doi: 10.3109/10408449609017932. [DOI] [PubMed] [Google Scholar]
- Williams PT, Chisti HM. Reaction of nitrogen and sulphur compounds during catalytic hydrotreatment of shale oil. Fuel. 2001;80:957–963. doi: 10.1016/S0016-2361(00)00189-7. [DOI] [Google Scholar]
- Xu P, Yu B, Li F, Cai X, Ma C. Microbial degradation of sulfur, nitrogen, and oxygen heterocycles. Trends Microbiol. 2006;14:398–405. doi: 10.1016/j.tim.2006.07.002. [DOI] [PubMed] [Google Scholar]
- Yamazoe A, Yagi O, Oyaizu H. Biotransformation of fluorene, diphenyl ether, dibenzo-p-dioxin and carbazole by Janibacter sp. Biotechnol Lett. 2004;26:479–486. doi: 10.1023/B:BILE.0000019554.49484.40. [DOI] [PubMed] [Google Scholar]
- Yoon BJ, Lee D-H, Kang Y-S, Oh D-C, Kim S-I, Oh K-H, Kahng H-Y. Evaluation of carbazole degradation by Pseudomonas rhodesiae strain KK1 isolated from soil contaminated with coal tar. J Basic Microbiol. 2002;42:434–443. doi: 10.1002/1521-4028(200212)42:6<434::AID-JOBM434>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]