Abstract
Autosomal recessive diseases (ARD) are typically caused by a limited number of mutations whose identification is challenged by their low prevalence. Our purpose was to develop a novel approach allowing an efficient search for mutations causing ARD and evaluation of their pathogenicity without a control group. We developed Iterative Sequencing and Variant Screening (ISVS) approach based on iterative cycles of gene sequencing and mutation screening, and ISVS Simulator software (http://zsibio.ii.pw.edu.pl/shiny/isvs/) for assessment of detected variants’ significance. As shown by simulations, ISVS efficiently identifies and correctly classifies pathogenic mutations except for cases where the gene of interest has extremely high number of low frequency nonpathogenic variants. By applying ISVS, we found 4 known and 9 novel (p.C73Y, p.S124L, p.C194Mfs*17, c.782 + 2 T > A, c.953-5 A > G, p.L325Q, p.D334Mfs*24, p.R436G, p.M448T) TMPRSS3 variants among deaf patients. For 3 known and 5 novel variants the disease association was supported by ISVS Simulator odds >90:1. Pathogenicity of 6 novel mutations has been supported by in-silico predictions of variants’ deleteriousness. By directly comparing variant prevalence in patients and controls, disease association was demonstrated only for two variants and it was relatively weak (P < 0.05). Summarizing, ISVS strategy and ISVS Simulator are useful for detection of genetic variants causing AR diseases.
Introduction
Autosomal recessive diseases (ARD) are often caused by a limited number of mutations. Accurate knowledge of pathogenic mutation profile in a given population is important as it helps to interpret results of diagnostic tests in the patients and may greatly facilitate design of efficient screening strategies. Whereas numerous in silico tools for mutation effect prediction are available they are not perfect and the assignment of pathogenicity remains a challenge, especially for variants with very low population frequency1. For such rare variants the association with disease defined as more frequent occurrence of a variant among the patients than controls is an important sine qua non criterion of pathogenicity2. However, the formal demonstration of an association may be difficult as it requires multiple patients with given mutation and a control group of appropriate size.
The purpose of our work was to develop and validate a novel approach allowing: (i) an efficient search for mutations causing ARD, (ii) evaluation of their pathogenicity by testing for disease association without using a control group.
The proposed mutation search strategy is based on iterative cycles of gene sequencing combined with focused mutation screening, which we termed Iterative Sequencing and Variant Screening (ISVS). Testing for association with disease relies on the analysis of statistical significance (using simulations) of co-occurrence among the patients of a given sequence variant with other rare potentially pathogenic variants in the same gene.
The developed approach was validated by searching for pathogenic variants in the TMPRSS3 gene in a cohort of 2247 subjects with sensorineural hearing loss (SNHL). We found 13 different rare TMPRSS3 variants, nine of which were novel. ISVS simulations showed for six of these variants (4 novel) strong (>1000:1) evidence for disease association, for two (1 novel) the evidence was moderate (90:1) and for five (4 novel) - weak.
Subjects and Methods
Principle of Iterative Sequencing and Variant Screening (ISVS)
ISVS is initiated by minimum one known pathogenic mutation (Mut. 1) which is screened for by a focused test (for example Real Time PCR – RT PCR) in the whole cohort of patients. Next, all the found samples heterozygous for the Mut. 1 are subjected to sequencing of all coding regions of gene (GENE) which is likely to reveal, in a number of cases, other variants either known as pathogenic or possibly pathogenic according to available criteria (database frequency, functional annotations, etc). Next, all these variants enter the second round of screening in the whole cohort of patients, similar as in the first round, and then GENE sequencing is similarly repeated. The procedure is iteratively continued until no potentially interesting variants are detected by sequencing.
In silico ISVS analysis
To assess the performance of ISVS method we first performed experiments in silico modelling search for pathogenic GENE mutations among patients with disease. We modelled the parameters used in this analysis according to the experiment performed later on using real samples and aimed at detecting TMPRSS3 mutations in approx. 2000 subjects with SNHL. In particular, we assumed that the prevalence of disease (f1) in the general population is 0.1%, as 1 to 2 in 1000 newborn children in developed countries are born with SNHL3. The frequency of cases affected by bi-allelic variants in GENE (f2) was assumed as 2% of all cases - an estimate based on the frequency of TMPRSS3 causative mutations among SNHL patients of different ethnicity4–10. Therefore, the frequency of individuals with TMPRSS3 homozygous or compound heterozygous variants in the general population f3 = f1*f2 = 0.002%.
In a single experiment, the genotypes were generated for the cohort of N = 2000 individuals. For the given number m of distinct pathogenic mutations, the allele frequency spectrum was sampled from the beta distribution with parameters alpha = 0.5, beta = 10 and then scaled to ensure that cumulative allele frequency of all mutations equals to f3. Parameters of beta distribution were tuned based on the real allele frequency spectrum observed in the cohort described in this manuscript. Next, for each mutation the status of maternal and paternal alleles for all individuals has been randomly assigned to 0 or 1 using allele probabilities obtained in the previous step. Individuals with homozygous or compound heterozygous variants were labeled as affected due to mutations in GENE and (N*f1 - N*f3) subjects were marked as additional group of patients with deafness caused by other genetic or non-genetic factors.
In addition to pathogenic mutations, for every patient we sampled the zygosity status of k non-pathogenic variants assuming the cumulative frequency of these variants of 0.05. The rationale for choosing 0.05 was based on analysis of ExAC11 data which showed that the cumulative rare allele frequency (<0.01) of missense mutations in the TMPRSS3 gene was 0.027. It can be safely assumed that in majority of populations pathogenic variants with prevalence >0.01 are likely to be known and thus we can safely reject any variant with frequency >0.01 during ISVS process. Since some rare variants are not reported in ExAC during testing variant pathogenicity by in silico ISVS simulation we arbitrarily increased the value to 0.05. As discussed further on, assuming higher cumulative frequency of non-pathogenic variants is conservative as the performance of ISVS decreases with increase of this parameter.
For such a cohort of all affected individuals (with or without GENE mutations) we performed an ISVS simulation, which works as follows. First, we randomly select a single individual with homozygous or compound heterozygous pathogenic GENE mutation. Next, remaining affected subjects are screened for the mutations found in the first individual. All carriers of these mutations are then checked for additional variants across the whole gene (which corresponds to sequencing part in the real experiment). New individuals with homozygous and compound heterozygous variants are added to the list of patients in whom disease is potentially caused by mutations in GENE. Then the screening is performed again using the list of novel variants and it is followed by the sequencing. This procedure is repeated until there are no novel variants to be screened in the next step. Finally, we summarize the results of experiments by calculating the fraction of identified variants out of all m mutations considered in the experiment.
Testing variant pathogenicity by in silico ISVS simulation
To cause a recessive condition a pathogenic variant on one allele has to co-occur with another pathogenic variant on the other allele. Thus, in patients with recessive diseases causative variants are likely to co-occur (in-trans configuration) with other pathogenic variants but this is not the case for rare variants with no phenotypic effect. We tested if this information can be used to discriminate pathogenic and non-pathogenic variants identified in ISVS experiment using the following procedure.
First, the set of ISVS simulations (i.e. in silico repetitions of the ISVS experiment) is randomly split into the training (80% of simulations) and testing part (20% of simulations). Next, using the data from training simulations we build a classification model. For each variant identified in the ISVS experiment we calculate two numbers: (i) the number of individuals for which this variant occurs in-trans with any other variant identified by ISVS procedure, and (ii) the total number of occurrences of this variant in the analyzed cohort. These two features along with the predefined class label for the variant (i.e. pathogenic/non-pathogenic) are then used to train Support Vector Machine (SVM) classifier12. Performance of other classification methods and details of SVM model tuning are described in supplementary material. Subsequently, the classification model is used to predict the class of variants from the set of test simulations. Finally, to calculate the algorithm performance we repeat above steps under 5-fold cross validation. In addition, to provide a single “pathogenicity” score for an individual variant we also calculate the likelihood ratio, i.e. the probability of variant being pathogenic vs non-pathogenic.
Development of a web application to in silico simulate ISVS experiments
We have developed a software which performs a series of in-silico simulations of ISVS experiment (publicly available at http://zsibio.ii.pw.edu.pl/shiny/isvs/). Application is implemented in R programming language (https://cran.r-project.org/) using shiny framework and rCharts library (https://ramnathv.github.io/rCharts/) to create user interface. To speed up the computations these simulations are run on the server in parallel using 30 cores on our local cluster.
Application of ISVS for screening for TMPRSS3 mutations among patients with hearing loss (HI)
The patients were selected from the Polish SNHL subjects consulted in the Genetic Department of the Institute of Physiology and Pathology of Hearing (IPPH) between 2000 and 2014. Genomic DNA (gDNA) was extracted from the peripheral blood of all patients using salting out method. The hearing loss was determined by pure-tone audiometry at 500 Hz, 1 kHz, 2 kHz, 4 kHz and 8 kHz and was at least 20 dBHL. The group included 2247 (1204 females and 1043 males) unrelated SNHL patients from IPPH. The mean level of HI was moderate (i.e. 41~70 dBHL) at 500 kHz, 1 kHz, 2 kHz and severe (i.e. 71~95 dBHL) at 4 kHz, 8 kHz. The mean age of SNHL onset was 12 years. We excluded from this study patients who already had established cause of HI, that is those with syndromic SNHL of known genetic origin, and also carriers of two mutations in connexin genes GJB2 and/or GJB6 (recessive inheritance). GJB2/GJB6 linked deafness is relatively frequent in Caucasians accounting for up to 20% of nonsydromic cases13, 14. Whereas different pathogenic mutations may co-occur by chance it remains a rare event3, 15, thus the exclusion of patients with already defined cause of SNHL can be expected to increase the yield of the search for mutations in any gene(s) not tested previously.
Written informed consent was obtained from all the subjects or parents/guardians of participating children and all procedures were approved by the bioethical commission at Institute of Physiology and Pathology of Hearing, Warsaw, Poland.
Screening of the background Polish population was performed using DNA samples from anonymous unrelated individuals who underwent paternity testing at Department of Forensic Medicine, Medical University of Warsaw, all these subjects gave written informed consent for anonymous use of their DNA for research.
Naming of all TMPRSS3 mutations is in accordance with the recommendations of Human Genome Variation Society (HGVS). NM_024022.2 was applied as the reference TMPRSS3 sequence for cDNA, while at the protein level NP_076927.1 sequence was used.
Whole Exome Sequencing (WES)
One female proband with congenital, profound SNHL from a family suggestive of recessive type of inheritance (healthy parents and a sister with SNHL) with excluded common GJB2 mutations and NC_012920.1:m.1555 A > G, NC_012920.1:m.3243A>G was selected for WES. WES was performed at the ICMB in Kiel on HiSeq2000 platform with 2 × 100 bp reads using TruSeq Exome Enrichment Kit (Illumina). The data were analyzed as previously described16.
Laboratory steps of ISVS based TMPRSS3 screening among SNHL patients
ISVS was initiated by WES in a single proband which revealed two frameshift variants NM_024022.2:c.[208delC(;)579dupA]. These two variants were screened for by RT PCR in the whole cohort of patients with SNHL. Next, all samples heterozygous for any of the two variants underwent Sanger sequencing of the TMPRSS3 gene which revealed, in 20 cases, variants either known as pathogenic or tentatively classified by us as such. All these variants were then screened for by RT PCR among the cohort of SNHL patients similar as in the former round and then Sanger sequencing was similarly repeated. The procedure was continued until no potentially interesting variants were detected by sequencing.
The tentative assignment of pathogenicity (and therefore inclusion in RT PCR screening) was based on previous reports and/or prevalence <1% in 1000Genomes or CG6917 ESPSP650018 and ExAC11 databases. Furthermore, the prevalence of each variant was determined in a cohort of 597 samples from background population of Poland using RT PCR. Detected variants with prevalence in the abovementioned cohorts <1% were regarded as ‘potentially pathogenic’.
Screening with RT PCR was performed using Assay on Demand reagents (Life Technologies, Carlsbad, CA, USA). Sanger sequencing was performed with ABI PRISM 3500XL capillary sequencer (Life Technologies) and covered all coding exons and boundary intronic sequences of TMPRSS3 (sequences of primers used are shown in Supplementary Table 1 (Supp. Table 1).
Testing of potentially pathogenic TMPRSS3 variants for association with HI
All potentially pathogenic TMPRSS3 variants were tested for association with SNHL using two approaches: (i) prevalence of each variant was compared among SNHL patients and controls by Fisher’s exact test using SPSS 11.5 K (SPSS Inc., Chicago, USA), and p values of less than 0.05 were considered statistically significant, (ii) among SNHL patients in whom given variant was found in at least one chromosome, the prevalence of additional potentially pathogenic rare variants (present presumably in the other chromosome) was compared with distribution obtained from ISVS simulations.
RNA expression study
For probands and parents with p.M448T mutation total RNA was isolated from peripheral blood using of MagNA Pure Compact RNA Isolation Kit (Roche, Basel, Switzerland) using MagNA Pure Compact Instrument. cDNA synthesis was performed with the Maxima First Strand cDNA Synthesis Kits for RT-qPCR (ThermoFisherScientific, Waltham, USA).
Bioinformatics analysis of mutations’ effects
The probable deleterious effects of the found missense variants were predicted using SIFT (http://sift.jcvi.org)19, MetaSVM1, MutationTaster220 and PolyPhen-221. TMPRSS3 model was generated with M4T server22, using the structure of the homologous transmembrane serine protease hepsin as a model (PDB code 1Z8G). The protein graphics was produced with the PyMOL Molecular Graphics System, Version 1.3, Schrödinger, LLC. Domain conservation consensus and TMPRSS3 sequence alignments were retrieved from Simple Modular Architecture Research Tool (SMART) database23.
Novel TMPRSS3 splice site variants were analyzed using the Alamut Visual Software 2.8 (Interactive Biosoftware, Rouen, France).
Results
In silico modelling of ISVS search for TMPRSS mutations in SNHL patients
By performing 10,000 ISVS simulations with parameters adjusted for TMPRSS gene and SNHL we found that, on average, by 4.75 steps of an ISVS experiment the success rate (i.e. the fraction of experiments where all affected individuals were detected) was 0.98. Average classification accuracy of pathogenic vs. non-pathogenic variants was 0.97 (at confidence 0.5) or 0.83 (at confidence 0.95). When SVM classification was used the latter value raised to 0.85.
ISVS Simulator software
In order to facilitate the more broad use of ISVS we developed software (ISVS Simulator) that enables user to test the performance of the method under scenarios different than the considered by us TMPRSS mutation screen in SNHL subjects. In particular, the application allows user to modify various parameters, such as disease cohort size, disease prevalence, fraction of disease cases explained by mutation in the given gene, expected numbers of distinct pathogenic and non-pathogenic mutations in the gene, cumulative frequency of non-pathogenic variants, parameters of beta distribution for variant allele frequencies and the number of simulations to be repeated in the single experiment.
The experiment results are presented in three tabs, located on the right side of the web page. First one, the “Fraction bi-allelic plot” presents the number of bi-allelic (in-trans) events associated with the given mutation (y-axis) in reference to the total number occurrences of this mutation (x-axis, Fig. 1). Every point of this plot represents a set of mutations that were observed in exactly x individuals where in y cases ISVS identified another mutation on the second allele. The tooltip associated with each point plot displays information about: (i) the x, y values (intrans: all); (ii) the total number of pathogenic and non-pathogenic mutations which yielded given (x, y) in ISVS simulations (# of pat: # of non-pat); (iii) cumulative value of the latter (cum # of pat: cum # of non-pat). This was calculated according to principle that if given (x, y) were observed only for pathogenic mutations and the same was seen for (x, y-1), (x, y-2), etc., than for the (x, y) point the cumulative number of pathogenic is conservative to sum the “# of pat” values with the value for (x, y-1), (x, y-2), etc. point(s); (iv) SVM likelihood ratio, i.e. probability that mutation is pathogenic/probability that mutation is non-pathogenic. The intensities of red and blue colors correspond to the number of observed pathogenic and non-pathogenic variants, respectively. A part of the plot can be enlarged by marking it with the mouse.
Figure 1.
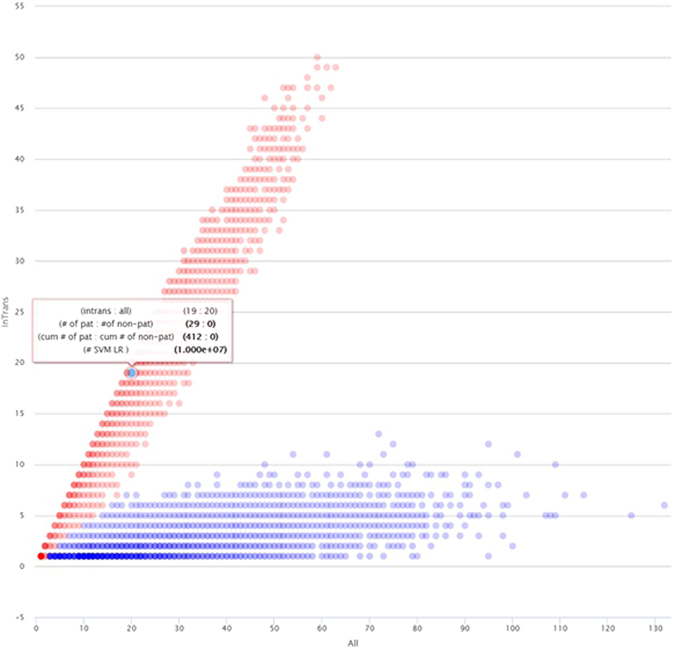
Screen-shot from ISVS simulator showing an example of “Fraction bi-allelic plot” obtained using default settings. Note information about given (x,y) point provided in the tooltip. This particular point indicates that if in a real ISVS experiment given variant was found in 20 patients of whom 19 also had a rare variant in the other allele (intrans:all = 19:20) there is considerable evidence (#SVM LR = 107) that the variant is pathogenic.
The “ISVS summary” tab shows a summary statistics including: an average number of samples for which the targeted gene had to be sequenced, an average number of identified samples with bi-allelic pathogenic variants, an average number of pathogenic mutation carriers, and ISVS success rate, i.e. the fraction of simulations in which all individuals with pathogenic bi-allelic mutations were detected. In addition, the classification accuracies are reported for 0.5 and 0.95 confidence levels. Detailed classification results (including numbers of TP (true positive), TN (true negative) and classification accuracies) for confidence levels varying between 0.5 and 1 are also presented in the “Classification results” tab.
In silico analysis of the performance of ISVS
To study the performance and robustness of ISVS we repeated the above-mentioned experiment using different set of input parameters. To ensure the stability of results, for a given set of parameters ISVS simulation were repeated 10,000 times. We collected the following metrics: (i) successRate – the fraction of ISVS simulations in which all individuals affected by bi-allelic pathogenic mutation were detected (ii) patFractDetected – the average fraction of patients with bi-allelic pathogenic mutation who were properly identified; (iii) mutFractDetected – the average fraction of pathogenic variants that were properly identified; (iv) avgSteps – the average number of steps in ISVS experiment; (v) sequencedSampleNr – the average number of sequenced samples; (vi) screenedVarsNr – the average number of screened variants.
The aim of our analysis was to evaluate how the above-mentioned metrics depend on the change of the selected input parameters, including: disease cohort size (default = 2000), frequency of disease individuals within the population (default = 0.001), the fraction of disease individuals affected by bi-allelic variants in the GENE (default = 0.02), the number of distinct pathogenic mutations (default = 10), the number of distinct non-pathogenic (default = 20) and the cumulative frequency of non-pathogenic mutations (default = 0.05). Complete results from this analysis are presented in Supplementary Figures S1–3 (Supp. Fig. S1–3).
We observed that first three performance metrics (i.e. successRate, patFractDetected and mutFractDetected) all achieve very high (>0.9) values for most of the configurations. Smaller successRate (<0.9) were observed only when the disease cohort size was smaller than 1000 or the fraction of individuals with bi-allelic variants in disease cohort was smaller than 1%. This shows the limitations of ISVS method in terms of the minimal number of patients who need to be ascertained in order to effectively identify pathogenic variants and those subjects in whom these variants cause the disease.
Another finding is that the average number of ISVS steps (avgSteps) is limited never exceeding five in our simulations, regardless of the input parameters. We observed that the number of steps further decreases when the fraction of affected individuals by bi-allelic variants (or cumulative frequency of pathogenic mutations) grows. This is consistent with our expectations, since higher cumulative frequency of pathogenic variants enlarges the number of variant occurrences that “link” subsequent sequencing and screening steps in ISVS experiment.
We found that ISVS performance decreases with the increase of number and cumulative frequency of non-pathogenic variants. In particular, the increase of number and cumulative frequency of non-pathogenic variants causes sharp increase in the amount of variants which need to be screened and samples which need to be sequenced. Even more importantly, high numbers and high cumulative frequency of nonpathogenic variants decrease the accuracy of SVM classification. Sensitivity of classification is predominantly affected by high cumulative frequency of non-pathogenic variants whereas specificity appears especially sensitive to presence of high number of low frequency non-pathogenic mutations. (Supp. Fig. S3). These results indicate that ISVS is less useful for long genes which are likely to have relatively high numbers and high cumulative frequency of rare non-pathogenic variants.
We also studied the performance of ISVS method when the disease cohort is a mixture of two ethnically different populations. We assumed that the same set of pathogenic and non-pathogenic variants is present in both groups but the allele frequencies of those variants differ between populations. In such a setting, we analyzed the performance of ISVS for various proportions of two populations sizes, i.e. from 1:0, 0.9:0.1, …, up to 1:1. As expected we observed a small decrease in the average fraction of detected pathogenic mutations or the average fraction of properly identified affected individuals, when one population is much larger than the other. However, when the sizes of both populations become more balanced, the performance of ISVS returns to the level observed for single ethnically homogenous population.
TMPRSS3 mutations in Polish SNHL population
The practical performance of ISVS procedure as applied to TMPRSS3 mutation screening among subjects with SNHL is shown in Fig. 2 and the summary of potentially pathogenic variants found is shown in Table 1. We found 4 previously described as well as 9 novel variants. TMPRSS3 variants which were not reported to date are: p.C73Y, p.S124L, p.C194Mfs*17, c.782 + 2 T > A, c.953-5 A > G, p.L325Q, p.D334Mfs*24, p.R436G, p.M448T whereas the known variants include p.H70Tfs*19, p.R109W, p.A138E, p.A426T. Sanger chromatograms showing novel variants are presented in Supp. Fig. S4.
Figure 2.
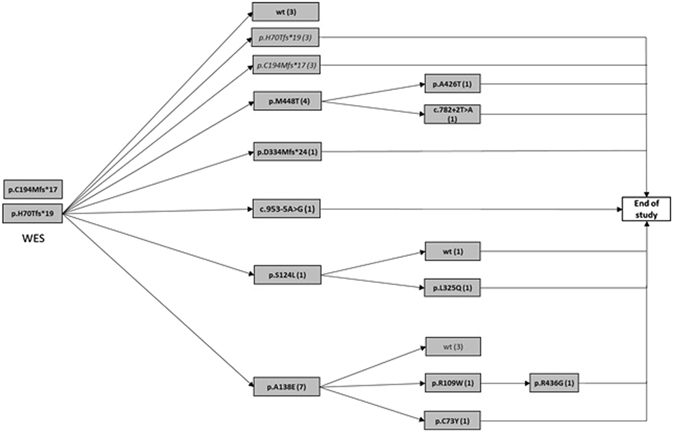
Summary of findings during cascade TMPRSS mutation screening by ISVS in a cohort of HI subjects. WES – whole exome sequencing; arrows – cycles of Real Time PCR screening; filled boxes - variants found by Sanger sequencing, (number of samples with each variant is given in parentheses); italics – variants observed at a previous stage.
Table 1.
Prevalence of rare variants in the TMPRSS3 gene detected among Polish HI patients and controls.
| Variant | het | hom | PATIENTS | CONTROLS | P value | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| allele mut | allele sum | freq. % | het | hom | allele mut | allele sum | freq. % | ||||
| p.H70Tfs*19 | 20 | 3 | 26 | 4376 | 0.59 | 0 | 0 | 0 | 1058 | 0 | 0.012 |
| p.A138E | 17 | 2 | 21 | 4604 | 0.46 | 2 | 0 | 2 | 1918 | 0.1 | 0.029 |
| p.M448T^ | 10 | 0 | 10 | 4596 | 0.22 | 0 | 0 | 0 | 1222 | 0 | 0.13 |
| p.S124L^ | 5 | 0 | 5 | 4524 | 0.11 | 0 | 0 | 0 | 1046 | 0 | 0.59 |
| p.C194Mfs*17^ | 4 | 0 | 4 | 4594 | 0.09 | 2 | 0 | 2 | 1932 | 0.1 | 1.0 |
| c.953-5 A > G^ | 4 | 0 | 4 | 4560 | 0.09 | 0 | 0 | 0 | 1058 | 0 | 1.0 |
| p.R109W | 2 | 0 | 2 | 4506 | 0.04 | 0 | 0 | 0 | 998 | 0 | 1.0 |
| p.D334Mfs*24^ | 2 | 0 | 2 | 4618 | 0.04 | 0 | 0 | 0 | 1080 | 0 | 1.0 |
| p.C73Y^ | 1 | 0 | 1 | 4562 | 0.02 | 0 | 0 | 0 | 1046 | 0 | 1.0 |
| c.782 + 2 T > A^ | 1 | 0 | 1 | 4206 | 0.02 | 0 | 0 | 0 | 1042 | 0 | 1.0 |
| p.L325Q^ | 1 | 0 | 1 | 4566 | 0.02 | 0 | 0 | 0 | 1018 | 0 | 1.0 |
| p.A426T | 1 | 0 | 1 | 4104 | 0.02 | 0 | 0 | 0 | 1112 | 0 | 1.0 |
| p.R436G^ | 1 | 0 | 1 | 4604 | 0.02 | 0 | 0 | 0 | 986 | 0 | 1.0 |
^Mutation not reported to date; het- heterozygous, hom- homozygous, allele mut- number of mutated alleles, allele sum - number of tested alleles.
In the control population the aforementioned variants were either absent or had low allelic frequency (0.1% for p.A138E and p.C194Mfs*17) (Table 1).
In Table 2 we show frequency of second TMPRSS3 alteration among patients with rare variants in this gene. According to ISVS Simulator, for two known mutations (p.H70Tfs*19, p.A138E) as well as four novel variants (p.M448T, p.S124L, p.C194Mfs*17, c.953-5 A > G) the odds for pathogenicity exceeded 1000:1. For another two variants p.D334Mfs*24 (novel) and p.R109W the odds were ~90:1. It should be noted that only two of the abovementioned variants with pathogenicity supported by ISVS Simulator were associated with SNHL when their prevalence was compared with population controls (p.H70Tfs*19, p.A138E, Table 1).
Table 2.
Statistical assessment of the disease-association of the detected TMPRSS3 mutations using ISVS Simulator.
| Mutation | BAM/all | #pat/#non-pat | SVM LR | Second mutations found in patients | |
|---|---|---|---|---|---|
| Observed | Cumulat. | ||||
| p.H70Tfs*19 | 20/23 | 92/0 | 929/0 | 107 | p.A138E (7), p.M448T (4), p.H70Tfs*19 (3), p.C194Mfs*17 (3), wt (3), p.S124L (1), c.953-5 A > G (1), p.D334Mfs*24 (1) |
| p.A138E | 16/19 | 349/0 | 1076/0 | 107 | p.H70Tfs*19 (7), wt (3), p.A138E (2), p.C73Y (1), p.R109W (1), p.S124L (1), p.C194Mfs*17 (1), c.953-5 A > G (1), p.D334Mfs*24 (1), p.M448T (1) |
| p.M448T^ | 10/10 | 361/0 | 2599/0 | 107 | p.H70Tfs*19 (4), c.953-5 A > G (2), p.S124L (1), p.A138E (1), c.782 + 2 T > A (1), p.A426T (1), |
| p.S124L^ | 4/5 | 1689/0 | 1689/0 | 1390 | p.H70Tfs*19 (1), p.A138E (1), p. L325Q (1), p.M448T (1), wt (1) |
| p.C194Mfs*17^ | 4/4 | 2204/1 | 2204/1 | 7126 | p.H70Tfs*19 (3), p.A138E (1) |
| c.953-5 A > G^ | 4/4 | 2204/1 | 2204/1 | 7126 | p.M448T (2), p.H70Tfs*19 (1), p.A138E (1) |
| p.R109W | 2/2 | 4709/57 = 82 | 4709/57 = 82 | 90 | p.A138E (1), p.R436G (1) |
| p.D334Mfs*24^ | 2/2 | 4709/57 = 82 | 4709/57 = 82 | 90 | p.H70Tfs*19 (1), p.A138E (1) |
| p.L325Q^ | 1/1 | 7429/655 = 11 | 7429/655 = 11 | 10 | p.S124L (1) |
| p.R436G^ | 1/1 | 7429/655 = 11 | 7429/655 = 11 | 10 | p.R109W (1) |
| p.A426T | 1/1 | 7429/655 = 11 | 7429/655 = 11 | 10 | p.M448T (1) |
| c.782 + 2 T > A^ | 1/1 | 7429/655 = 11 | 7429/655 = 11 | 10 | p.M448T (1) |
| p.C73Y^ | 1/1 | 7429/655 = 11 | 7429/655 = 11 | 10 | p.A138E (1) |
BAM/all– number with bi-allelic mutations/total number with given mutation. in brackets - number of patients with a given mutation. The settings for ISVS Simulator: number of patients = 2 200; disease prevalence 1/1000; fraction of disease cases explained by mutation in the gene: 0.02; Number of iterations: 10 000; Number of pathogenic mutations: 10; Number of non-pathogenic mutations: 20; Cumulative frequency of non-pathogenic variants: 0.05.
Prediction of biological effect of novel TMPRSS3 variants
Frameshift or splice-site variants The c.579dupA and c.999delC mutations are predicted to cause frameshift and premature termination of protein (p.C194Mfs*17 and p.D334Mfs*24, respectively). The c.782 + 2 T > A and c.953-5 A > G mutations are likely to affect splicing of TMPRSS3 mRNA since both in minor and major class introns at the + 2 position (second position at the 5′ end of an intron) T is always present whereas at position -5 (the 5th base from the 3’ end of an intron) C, T or A but not G occur24. Both mutations are predicted as most probably affecting splicing variants (Table 3).
Table 3.
Splice site mutation predictions with the usage of Alamut bioinformatics algorithms. SSF- SpliceSiteFinder-like; HSF- Human Splicing Finder; – no data.
| Variant | Alamut bioinformatics algorithms | Interpretation | ||||
|---|---|---|---|---|---|---|
| SSF | MaxEntScan | NNSPLICE | GeneSplicer | HSF | ||
| c.782 + 2 T > A | 100% | 100% | 100% | 100% | 100% | Broken WT Donor Site, most probably affecting splicing |
| c.953-5A > G | — | 61% | — | 46% | <1% | Broken WT Acceptor Site, most probably affecting splicing |
Missense variants Three out of five missenses are predicted to be deleterious/disease-causing by all five prediction tools used (SIFT, PolyPhen-2, MutationTaster2, MetaSVM and MetaLR, Table 4); p.S124L was predicted as benign by all the tools, p.M448T was “diseases causing” only according to MutationTaster2. In addition to the use of the abovementioned prediction programs we also evaluated properties of each variant by protein modelling.
Table 4.
Bioinformatics analysis of novel missense variants in TMPRSS3 gene.
| TMPRSS3 mutation | SIFT | PolyPhen-2 | MutationTaster2 | MetaSVM | MetaLR |
|---|---|---|---|---|---|
| p.C73Y | damaging | probably damaging | disease causing | damaging | damaging |
| p.S124L | tolerated | benign | polymorphism | tolerated | tolerated |
| p.L325Q | damaging | probably damaging | disease causing | damaging | damaging |
| p.R436G | damaging | probably damaging | disease causing | damaging | damaging |
| p.M448T | tolerated | benign | disease causing | tolerated | tolerated |
p.C73Y (c.218 G > A) The LDLRA domain contains two loops interconnected by three disulfide bonds, formed by highly conserved Cys residues. The conservation of amino acids in TMPRSS3 LDLR domain is presented in Supp. Fig. S5A. According to the predicted topology model, C73 forms a disulfide bond with C85, establishing the domain scaffold (InterPro, ref: doi: 10.1093/nar/gku1243). Mutation of C73 to Y disables the putative disulfide bond formation and is likely to affect conformation of the domain and may cause protein destabilization (Supp. Fig. S5D).
p.S124L (c.371 C > T) S124 is located in the SRCR domain of TMPRSS3. A polar residue is conserved at this position what may suggest that the residue is involved in the stabilization of the adjacent loop comprising amino acids 162-170, and therefore may be involved in structural stabilization of the domain (Supp. Fig. S5B,D).
p.L325Q (c.974 T > A) and p.R436G (c.1306 C > G) Both mutations are placed in the proteolytic domain but are located in a substantial distance from the active site (Supp. Fig. S5C,D). L325 is a highly conserved, hydrophobic amino acid, buried inside the protein structure. Substitution of Leu into a charged Gln may negatively affect the stability of this domain. R436 is located at a protein surface and its side chain is exposed to solvent. A positively charged residue is conserved at this position, suggesting a putative role in the domain structural stability.
p.M448T (c.1343 T > C) The novel p.M448T (c.1343 T > C) mutation is located close to the splice site. Since in homologous proteins there are variants of Ile or Thr at this position we speculated that this variant may affect mRNA amount rather than the protein structure. In order to check this we performed direct sequencing of TMPRSS3 cDNA and DNA encompassing the p.M448T in a sample isolated from peripheral blood of heterozygous subjects. cDNA sequencing revealed lower dose of p.M448T allele vs. the wild type allele, in contrast of gDNA sequencing showed equal doses of both alleles (Supp. Fig. S6). This suggests that p.M448T may have a deleterious effect on splicing and/or mRNA stability.
Discussion
Here we describe ISVS - a novel strategy for detection of bi-allelic pathogenic mutations in a large cohort of patients affected by autosomal recessive disease. ISVS takes advantage of the fact that patients with an autosomal recessive disease who carry a pathogenic variant in a single chromosome should have another pathogenic variant (in the other chromosome). ISVS consists of cyclic rounds of sequencing of the exons/splice sites of a gene combined with focused screening of patients’ cohort for all variants with likely pathogenicity found at the sequencing step. This strategy allows a cost efficient search for ultra-rare, potentially pathogenic variants of a gene implicated in any AR disease. ISVS is particularly effective if a given gene accounts only for minority of cases (due to locus heterogeneity and/or relatively many cases being caused by polygenic or non-genetic factors). Early onset non syndromic hearing loss (NSHL), blindness or mental retardation are examples of prevalent diseases where ISVS can be applied. Importantly, through simulations (using freely available ISVS Simulator software) it is possible to test statistical significance of the associations between the found variants and the disease without referring to a control group. We demonstrated that our simple (two-feature based) score has a good discriminative power and can be used as a new predictor of variant pathogenicity. Although including additional features, such as functional annotation scores, mutation frequencies across populations, could potentially increase classification performance, we decided not to use it in order to keep the classification model simple and allow easy use in combination with other independent pathogenicity predictors without the risk of any hidden circularity. As discussed by Grimm et al.25, the most valuable prediction scores are those that are unrelated and orthogonal to other existing methods.
Using ISVS Simulator we assessed the method’s performance contingent on a number of population/mutation parameters. The simulations revealed that within wide range of tested parameters ISVS is highly accurate, i.e. identifies majority (>90%) of pathogenic mutations present in the disease cohort. Moreover, the number of steps in ISVS is usually limited (i.e. up to five steps), regardless of selected input data. We also showed that ISVS works well when a disease cohort is a mixture of two ethnically diverse populations, although a large disproportion in the cohort sizes may have negative effect on the overall method’s performance.
We found that the ISVS performance decreases significantly with the increase of number and cumulative frequency of non-pathogenic variants. In particular, there is an increase in number of samples required to be sequenced, increase in number of variants selected for screening and decrease of the accuracy of SVM classification of pathogenic vs nonpathogenic variants. These results indicate that ISVS is less useful for very long genes which are likely to have relatively high numbers and high cumulative frequency of rare non-pathogenic variants. However, this limitation does not preclude generally high ISVS usefulness due to the fact that majority of genes are relatively short: the coding sequence of 95% of all human genes is shorter than 4400 bp with the median length (1302 bp) similar to the length of the TMPRSS3 gene (1365 bp) which we used to validate ISVS in practice11.
The general knowledge of ISVS performance together with possibility to perform in silico under different scenarios should help to decide if performing any particular ISVS experiment would be cost effective in comparison to alternative approaches, e.g. direct sequencing of entire cohort.
To further validate ISVS, we applied it for screening of a cohort of Polish NSHL patients with the aim to identify and characterize pathogenic TMPRSS3 mutations in this population. TMPRSS3 screening in NSHL is suitable for ISVS for the following reasons: (i) NSHL has diverse causes but in a significant proportion of cases it is due to genetic defects with AR inheritance. (ii) AR NSHL displays pronounced locus heterogeneity with 85 genes/loci listed in OMIM (http://omim.org/phenotypicSeries/PS220290), (iii) the TMPRSS3 gene mutations are a relatively rare cause of NSHL in Caucasians4, with so far no reports from Polish population. Furthermore, as mentioned previously the length of the TMPRSS3 gene is typical for majority of human genes.
By the ISVS approach we found four known (p.H70Tfs*19, p.R109W, p.A138E, p.A426T) and nine novel TMPRSS3 variants: p.C73Y, p.S124L, p.C194Mfs*17, c.782 + 2 T > A, c.953-5 A > G, p.L325Q, p.D334Mfs*24, p.R436G, p.M448T. For three of the known variants (p.H70Tfs*19, p.R109W, p.A138E) and five of the novel variants (p.S124L, p.C194Mfs*17, c.953-5 A > G, p.D334Mfs*24, p.M448T) the disease association was supported by ISVS Simulator with odds of 90:1 or more.
It should be emphasized that by directly comparing variant prevalence in patients with controls, despite sizable numbers of subjects in both cohorts, the evidence for disease association was obtained only for two variants (p.H70Tfs*19, p.A138E, previously known) and it was relatively weak (P < 0.05).
Our results concerning six of the novel mutations have been validated by prediction of mutations’ effects. The variants p.C194Mfs*17, c.782 + 2 T > A, c.953-5 A > G, p.D334Mfs*24 are expected to cause protein truncation due to splice site mutation or frameshift. Both in silico and cDNA analyses suggest that p.M448T affects splicing rather than acts as a missense mutation (interestingly, p.M448T was the 3rd most prevalent mutation in our study). The pathogenicity of p.C73Y may result from the destruction of the C73-C89 disulfide bond, which is likely to affect the structure of LDLRA domain of the TMPRSS3 protein. An overview of the location of novel TMPRSS3 variants relative to protein domains and the previously described TMPRSS3 mutations is shown in Supp. Fig. S7.
According to our results, the frequency of TMPRSS3 mutations in Polish SNHL population is at least 1.91% (43/2247), which is close to the prevalence of 1% of childhood NSHL in Caucasians and 2.5% in a Korean cohort4, 9, 26. In other studies, among Koreans the prevalence of 5.9 and 8.3% of TMPRSS3 mutations in total AR NSHL and postlingual AR NSHL patients, respectively, was reported10. Similar prevalence was found in Tunisian27 and Turkish population28.
In conclusion, we propose ISVS as a strategy and ISVS Simulator software as a tool for detection of genetic variants causing AR diseases. We validate the ISVS approach providing the TMPRSS3 mutation profile of Polish SNHL patients.
Electronic supplementary material
Acknowledgements
This research was financially supported by the grants 2011/03/D/NZ5/05592 and 2014/13/B/NZ2/01248 from National Science Centre, Poland.
Author Contributions
U.L., T.G, A.Pol., R.P. - The concept of work and thesis research T.G. - Development of IGVS simulator software. M.O., H.S. - Provide clinical material. U.L., T.G, A.Pol., A.Podg., A.F., BS.P., R.P. - Research work T.G, R.P. - Statistical analysis M.F. - Generation of TMPRSS3 structural model and structural analysis of mutated variants. U.L., A.Pol., M.O., R.P. - Participation in phenotyping and clinical data collection. U.L., T.G, A.Pol., M.F., R.P. - Participation in the writing of manuscript. U.L., T.G, R.P. - Preparing and sending the manuscript U.L., T.G, A.Pol., A.Podg., P.S, A.F., B.S.P., M.F., M.O., H.S., R.P. - Review of the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-02315-w
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Dong C, et al. Comparison and integration of deleteriousness prediction methods for nonsynonymous SNVs in whole exome sequencing studies. Hum. Mol. Genet. 2015;24:2125–2137. doi: 10.1093/hmg/ddu733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Richards S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. Off. J. Am. Coll. Med. Genet. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smith RJH, Bale JF, White KR. Sensorineural hearing loss in children. Lancet Lond. Engl. 2005;365:879–890. doi: 10.1016/S0140-6736(05)71047-3. [DOI] [PubMed] [Google Scholar]
- 4.Wattenhofer M, et al. Mutations in the TMPRSS3 gene are a rare cause of childhood nonsyndromic deafness in Caucasian patients. J. Mol. Med. Berl. Ger. 2002;80:124–131. doi: 10.1007/s00109-001-0310-6. [DOI] [PubMed] [Google Scholar]
- 5.Ahmed ZM, et al. Characterization of a new full length TMPRSS3 isoform and identification of mutant alleles responsible for nonsyndromic recessive deafness in Newfoundland and Pakistan. BMC Med. Genet. 2004;5:24. doi: 10.1186/1471-2350-5-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ben-Yosef T, et al. Novel mutations of TMPRSS3 in four DFNB8/B10 families segregating congenital autosomal recessive deafness. J. Med. Genet. 2001;38:396–400. doi: 10.1136/jmg.38.6.396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duman D, Sirmaci A, Cengiz FB, Ozdag H, Tekin M. Screening of 38 genes identifies mutations in 62% of families with nonsyndromic deafness in Turkey. Genet. Test. Mol. Biomark. 2011;15:29–33. doi: 10.1089/gtmb.2010.0120. [DOI] [PubMed] [Google Scholar]
- 8.Ganapathy A, et al. Non-syndromic hearing impairment in India: high allelic heterogeneity among mutations in TMPRSS3, TMC1, USHIC, CDH23 and TMIE. PloS One. 2014;9:e84773. doi: 10.1371/journal.pone.0084773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee J, et al. Genetic analysis of TMPRSS3 gene in the Korean population with autosomal recessive nonsyndromic hearing loss. Gene. 2013;532:276–280. doi: 10.1016/j.gene.2013.07.108. [DOI] [PubMed] [Google Scholar]
- 10.Chung J, et al. A novel mutation of TMPRSS3 related to milder auditory phenotype in Korean postlingual deafness: a possible future implication for a personalized auditory rehabilitation. J. Mol. Med. Berl. Ger. 2014;92:651–663. doi: 10.1007/s00109-014-1128-3. [DOI] [PubMed] [Google Scholar]
- 11.Lek M, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Keerthi SS, Shevade SK, Bhattacharyya C, Murthy KK. A fast iterative nearest point algorithm for support vector machine classifier design. IEEE Trans. Neural Netw. 2000;11:124–136. doi: 10.1109/72.822516. [DOI] [PubMed] [Google Scholar]
- 13.Kelley PM, et al. Novel mutations in the connexin 26 gene (GJB2) that cause autosomal recessive (DFNB1) hearing loss. Am. J. Hum. Genet. 1998;62:792–799. doi: 10.1086/301807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Estivill X, et al. Connexin-26 mutations in sporadic and inherited sensorineural deafness. Lancet Lond. Engl. 1998;351:394–398. doi: 10.1016/S0140-6736(97)11124-2. [DOI] [PubMed] [Google Scholar]
- 15.Yang H, Liu R, Wang C-C. Searching the co-occurrence of pathogenic mutations for Leber’s hereditary optic neuropathy and hearing loss in more than 26,000 whole mitochondrial genomes. Mitochondrial DNA Part DNA Mapp. Seq. Anal. 2016;27:3399–3402. doi: 10.3109/19401736.2015.1018239. [DOI] [PubMed] [Google Scholar]
- 16.Ploski R, et al. Does p.Q247X in TRIM63 cause human hypertrophic cardiomyopathy? Circ. Res. 2014;114:e2–5. doi: 10.1161/CIRCRESAHA.114.302662. [DOI] [PubMed] [Google Scholar]
- 17.1000 Genomes Project Consortium. et al. An integrated map of genetic variation from 1,092 human genomes. Nature491, 56–65 (2012). [DOI] [PMC free article] [PubMed]
- 18.Exome Variant Server, NHLBI GO Exome Sequencing Project (ESP), Seattle, WA (URL: http://evs.gs.washington.edu/EVS/) [(10, 2016) accessed].
- 19.Ng PC, Henikoff S. Predicting deleterious amino acid substitutions. Genome Res. 2001;11:863–874. doi: 10.1101/gr.176601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schwarz JM, Cooper DN, Schuelke M, Seelow D. MutationTaster2: mutation prediction for the deep-sequencing age. Nat. Methods. 2014;11:361–362. doi: 10.1038/nmeth.2890. [DOI] [PubMed] [Google Scholar]
- 21.Adzhubei IA, et al. A method and server for predicting damaging missense mutations. Nat. Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fernandez-Fuentes N, Rai BK, Madrid-Aliste CJ, Fajardo JE, Fiser A. Comparative protein structure modeling by combining multiple templates and optimizing sequence-to-structure alignments. Bioinforma. Oxf. Engl. 2007;23:2558–2565. doi: 10.1093/bioinformatics/btm377. [DOI] [PubMed] [Google Scholar]
- 23.Schultz J, Milpetz F, Bork P, Ponting CP. SMART, a simple modular architecture research tool: identification of signaling domains. Proc. Natl. Acad. Sci. USA. 1998;95:5857–5864. doi: 10.1073/pnas.95.11.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Patel AA, Steitz JA. Splicing double: insights from the second spliceosome. Nat. Rev. Mol. Cell Biol. 2003;4:960–970. doi: 10.1038/nrm1259. [DOI] [PubMed] [Google Scholar]
- 25.Grimm DG, et al. The evaluation of tools used to predict the impact of missense variants is hindered by two types of circularity. Hum. Mutat. 2015;36:513–523. doi: 10.1002/humu.22768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hutchin T, et al. Assessment of the genetic causes of recessive childhood non-syndromic deafness in the UK - implications for genetic testing. Clin. Genet. 2005;68:506–512. doi: 10.1111/j.1399-0004.2005.00539.x. [DOI] [PubMed] [Google Scholar]
- 27.Masmoudi S, et al. Novel missense mutations of TMPRSS3 in two consanguineous Tunisian families with non-syndromic autosomal recessive deafness. Hum. Mutat. 2001;18:101–108. doi: 10.1002/humu.1159. [DOI] [PubMed] [Google Scholar]
- 28.Wattenhofer M, et al. A novel TMPRSS3 missense mutation in a DFNB8/10 family prevents proteolytic activation of the protein. Hum. Genet. 2005;117:528–535. doi: 10.1007/s00439-005-1332-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


