Chlamydiae are known as major bacterial pathogens of humans, causing the ancient disease trachoma, but they are also frequently found in the environment where they infect ubiquitous protists such as amoebae. All known chlamydiae require a eukaryotic host cell to thrive. Using the environmental chlamydia Protochlamydia amoebophila within its natural host, Acanthamoeba castellanii, we investigated gene expression dynamics in vivo and throughout the complete chlamydial developmental cycle for the first time. This allowed us to infer how a major virulence mechanism, the type III secretion system, is regulated and employed, and we show that the physiology of chlamydiae undergoes a complete shift regarding carbon metabolism and energy generation. This study provides comprehensive insights into the infection strategy of chlamydiae and reveals a unique adaptation to life within a eukaryotic host cell.
KEYWORDS: Protochlamydia, RNA-seq, chlamydia, developmental cycle, gene expression, host-microbe interaction, metabolism, symbiont, type III secretion system
ABSTRACT
Chlamydiae are obligate intracellular bacteria comprising well-known human pathogens and ubiquitous symbionts of protists, which are characterized by a unique developmental cycle. Here we comprehensively analyzed gene expression dynamics of Protochlamydia amoebophila during infection of its Acanthamoeba host by RNA sequencing. This revealed a highly dynamic transcriptional landscape, where major transcriptional shifts are conserved among chlamydial symbionts and pathogens. Our data served to propose a time-resolved model for type III protein secretion during the developmental cycle, and we provide evidence for a biphasic metabolism of P. amoebophila during infection, which involves energy parasitism and amino acids as the carbon source during initial stages and a postreplicative switch to endogenous glucose-based ATP production. This fits well with major transcriptional changes in the amoeba host, where upregulation of complex sugar breakdown precedes the P. amoebophila metabolic switch. The biphasic chlamydial metabolism represents a unique adaptation to exploit eukaryotic host cells, which likely contributed to the evolutionary success of this group of microbes.
IMPORTANCE Chlamydiae are known as major bacterial pathogens of humans, causing the ancient disease trachoma, but they are also frequently found in the environment where they infect ubiquitous protists such as amoebae. All known chlamydiae require a eukaryotic host cell to thrive. Using the environmental chlamydia Protochlamydia amoebophila within its natural host, Acanthamoeba castellanii, we investigated gene expression dynamics in vivo and throughout the complete chlamydial developmental cycle for the first time. This allowed us to infer how a major virulence mechanism, the type III secretion system, is regulated and employed, and we show that the physiology of chlamydiae undergoes a complete shift regarding carbon metabolism and energy generation. This study provides comprehensive insights into the infection strategy of chlamydiae and reveals a unique adaptation to life within a eukaryotic host cell.
INTRODUCTION
Chlamydiae represent an ancient group of obligate intracellular bacteria (1). Hundreds of millions of years of evolution in association with eukaryotic host cells gave rise to successful pathogens of humans and diverse, globally distributed environmental chlamydiae associated with protists and animals (2–4). The human pathogen Chlamydia trachomatis (family Chlamydiaceae) is a frequent cause of sexually transmitted diseases and sight-threatening infections, affecting more than 180 million people each year (5, 6). While the chlamydiae are currently represented by a limited number of families, molecular evidence suggests the existence of a tremendous diversity in diverse habitats (>250 unexplored families), including potential new emerging pathogens (3, 7–11). Despite this diversity, all chlamydiae undergo a biphasic developmental cycle (2, 12). A detailed knowledge of this defining feature is the key to understanding the biology, evolution, and host interactions of this unique group of microbes.
The chlamydial developmental cycle consists of two morphologically and physiologically distinct stages termed elementary bodies (EBs) and reticulate bodies (RBs) (2, 12). Infection of eukaryotic host cells occurs at the EB stage. After host cell entry, EBs differentiate quickly into replicating RBs, which reside within a cytoplasmic vacuole called an inclusion, and undergo several rounds of cell division. RBs asynchronously convert back to EBs, which are subsequently released into the extracellular environment by host cell lysis or nonlytic extrusion (12, 13). The developmental cycle has mainly been studied for chlamydial pathogens, such as C. trachomatis and C. pneumoniae. Although there are slight variations between organisms, the cycle is well conserved among all known chlamydiae and likely has evolved before their diversification (1, 14–18).
DNA microarray and quantitative PCR studies have been instrumental in elucidating the molecular basis of the developmental cycle of C. trachomatis and C. pneumoniae, which found that infection and differentiation processes are accompanied by marked temporal changes of gene expression (19–24). The recent sequencing of entire transcriptomes (RNA-seq) using next-generation sequencing platforms improved the detection of early chlamydial genes (25), facilitated the identification of transcription start sites, and led to the discovery of novel transcripts (26, 27). However, this powerful technology has not yet been applied to study the chlamydial developmental cycle over the entire course of infection. In addition, although this method has the potential to interrogate microbe and host transcriptomes simultaneously (28), technical challenges have hampered its application for the analysis of chlamydiae, with only a single study available so far (25).
Protochlamydia amoebophila (hereafter referred to as Protochlamydia; a member of the family Parachlamydiaceae) is a well-studied symbiont (29–31). Of key importance, Protochlamydia in its natural Acanthamoeba host represents an in vivo infection model to study the dynamics of host-microbe interactions, and the model does not rely on the use of immortalized cell lines. Studying these environmental counterparts of the Chlamydiaceae has led to a number of discoveries with important implications for our understanding of the basic biology of all chlamydiae, including the discovery of peptidoglycan and metabolic activity of EBs (18, 32–37). Yet, a detailed analysis of the Protochlamydia developmental cycle and the underlying gene expression dynamics has thus far been strikingly lacking.
In this study, we describe the course of the infection and investigate the transcriptome of Protochlamydia and its amoeba host at key time points by RNA-seq. Highlighting conserved chlamydial and unique protochlamydial developmental events we aimed at understanding general chlamydial biology, including the role of protein secretion and the metabolic strategy accompanying the transitions between developmental stages. Our analyses revealed the conservation of a pronounced temporal gene expression profile during the developmental cycle among amoeba-associated chlamydiae and chlamydial pathogens and support a model in which a major shift in the metabolism of Protochlamydia—from energy parasitism to endogenous energy production—occurs during differentiation.
RESULTS AND DISCUSSION
Three main temporal classes of gene expression.
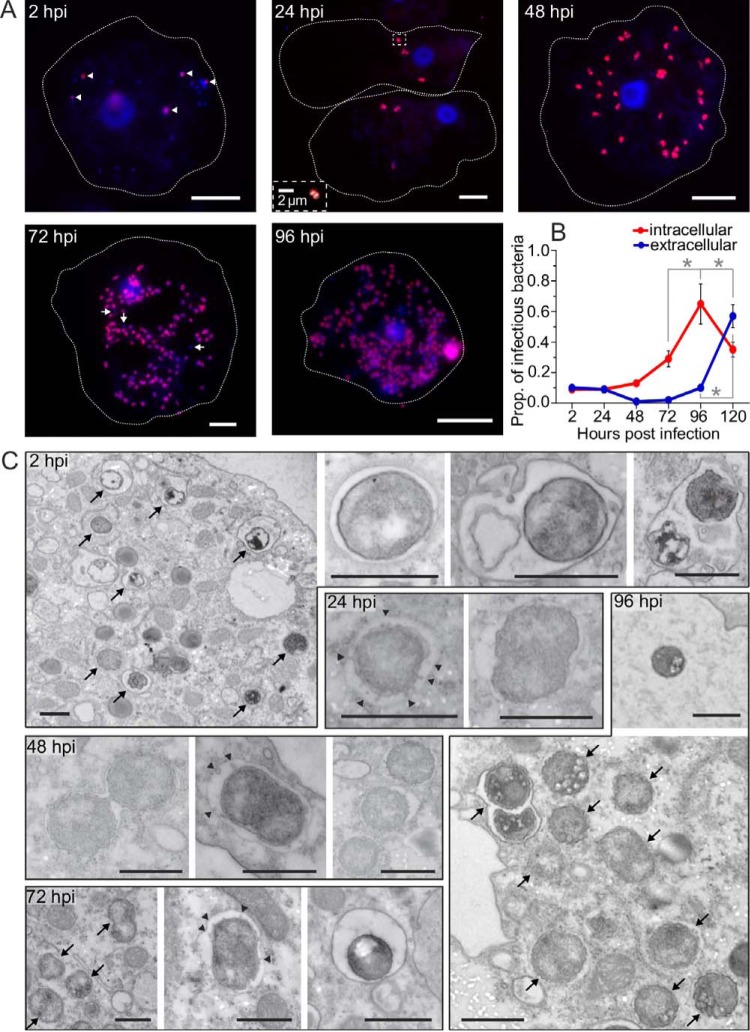
We investigated the developmental cycle of the amoeba endosymbiont Protochlamydia by monitoring the course of infection using fluorescence in situ hybridization, 4′,6′-diamidino-2-phenylindole (DAPI) staining, and transmission electron microscopy, and by quantifying the production of infectious EBs (Fig. 1). In summary, Protochlamydia in its Acanthamoeba host exhibits the characteristic chlamydial developmental cycle (see Text S1 in the supplemental material for details). Completion of the cycle takes 96 h, slightly longer than what has been reported for chlamydiae infecting humans and animals (20, 21). It is, however, shorter than the 6 to 15 days observed for other chlamydiae (14, 17, 38).
FIG 1 .
Developmental cycle of Protochlamydia amoebophila. (A) Fluorescence in situ hybridization in combination with DAPI staining was used to differentiate between RBs (pink) and EBs (blue). At 2 h postinfection EBs clearly dominate, but a few cells have already started to convert to RBs (white arrowheads). Exclusively RBs were detected at 48 hpi. The first EBs (white arrows) were seen at 72 hpi. The inset for the 24 hpi image is an enlargement of dividing RBs (to enhance clarity, the DAPI signal is shown in white). Dotted white lines indicate the outlines of amoeba host cells. If not indicated otherwise, all bars represent 10 µm. (B) The course of EB production and release was quantified by collecting intra- and extracellular bacteria, respectively, at indicated time points, and subsequent reinfection of fresh amoebae. The first intracellular EBs were present at 72 hpi, the first release of EBs was observed at 96 hpi. Values that are significantly different (P < 0.05) at the various time points by one-way analysis of variance (ANOVA) and Tukey’s posttest are indicated by an asterisk (n = 3). Prop., proportion. (C) Transmission electron micrographs visualizing developmental events at the ultrastructural level. Black arrows point to bacterial cells in overview images. Black arrowheads indicate vesicles that were observed in the inclusion lumen from 24 hpi on. Bars = 1 µm.
Supplemental methods, results, and discussion. Download TEXT S1, PDF file, 0.1 MB (155.6KB, pdf) .
Copyright © 2017 König et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
For RNA sequencing, we selected four time points that mark crucial developmental events during infection: 2 h postinfection (hpi) representing the start of EB-to-RB transition, 48 hpi as the peak of RB activity, 96 hpi covering secondary differentiation, and released EBs (Fig. 1 and Text S1). Despite different sequencing read numbers that could be mapped to the Protochlamydia genome at each time point (Table S1), the sequencing depths of all samples and the similarities between biological replicates were sufficient (Fig. S1 and Text S1) to identify 737 protein-coding genes that were differentially expressed (DE) at least once during the developmental cycle (corresponding to 51% of all genes with expression data) (Fig. 2). All other expressed genes were considered constitutively expressed genes (Fig. S2A). Hierarchical clustering of DE genes by expression profiles revealed three main temporal classes of gene expression (Fig. 2 and Data Set S1), whose expression profiles are broadly reflected in the EB proteome determined previously (39) (Fig. 2). Importantly, these three main temporal groups (termed early, mid, and late genes) correspond well with those found in microarray studies of Chlamydiaceae (23). Therefore, the pronounced transcriptional switches accompanying EB-to-RB transition and intracellular establishment, RB activity, and RB-to-EB transition are conserved among amoeba-associated chlamydiae and the Chlamydiaceae and represent a hallmark of all chlamydiae. However, given the proportionally larger number of differentially regulated genes upon entry and early developmental events we found for Protochlamydia (Fig. S3 and Text S1), amoeba-associated chlamydiae require a higher degree of adjustment when transitioning to the intracellular environment.
FIG 2 .
Temporal classes of gene expression during the Protochlamydia developmental cycle. A total of 797 genes were detected as differentially expressed; tRNA genes (20 genes), rRNA genes (2 genes), and genes detected only in a single replicate (38 genes) were excluded from further analysis. Clustering identified three main temporal classes of gene sets (colored bars to the left of the heatmap) that could be further divided into five large subclasses (colored bars to the right of the heatmap). The largest group of genes was most highly expressed early (n = 304), whereas the expression of the smallest group of genes peaked at midcycle when only RBs were present (n = 161). The third main gene cluster generally showed highest expression at the end of the cycle and the extracellular stage (n = 273; see Data Set S1 in the supplemental material). Gene products detected in the EB proteome in a previous study (n = 231) (39) are indicated in the bar plot next to the heatmap. To illustrate the course of gene expression for each subcluster, the expression values (log2 RPKM plotted on the y axis) were averaged per time point (x axis) and visualized as line plots (error bars indicate standard deviations). Selected gene names are shown for each of the temporal clusters. RPKM, reads per kilobase per million; hpi, h postinfection; extracell., extracellular; PG, peptidoglycan synthesis; T3SS, type III secretion system.
Similarity of Protochlamydia gene expression values between replicates and time points. The Euclidean distances between all normalized expression values (reads per kilobase per million [RPKM]) of each replicate and time point are shown, with darker colors meaning high similarity. The order of the samples is based on hierarchical cluster analysis of the distances using the complete linkage method—the clustering being illustrated by the dendrograms. The samples cluster by time point and therefore correctly reflect the experimental design. For subsequent analyses, genes were considered to be expressed at a particular time point when we obtained expression values in two or three replicates. The contribution of genes for which expression was detected in only two replicates is indicated in the right panel. Relatively low sequencing coverage at 2 h postinfection (hpi) explains the higher numbers of these genes at this early time point, and consequently also the larger distance between the 2 hpi replicates. Excl., excluding. Download FIG S1, PDF file, 0.2 MB (258.8KB, pdf) .
Copyright © 2017 König et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
(A) Gene expression heatmaps of constitutively expressed genes and (B) genes involved in peptidoglycan synthesis and cell division. (A) All genes that were detected to be expressed (in at least two replicates) but were found not to be significantly differentially expressed (n = 712) were termed constitutively expressed. Average gene-wise expression values (log2 reads per kilobase per million [RPKM]) were hierarchically clustered based on Euclidean distances using the complete linkage method. Five clusters representing five different expression levels were defined on the basis of the dendrogram’s branching pattern. Accordingly, we found 9 very highly expressed genes, 60 highly expressed genes, 131 genes with medium expression strength, 328 genes with medium-low expression strength, and 184 genes with low expression. Each cluster was tested for the presence of overrepresented COG classes and clusters, KEGG pathways, and GO terms for which only categories with a false discovery rate (FDR) below 0.05 are listed. In addition, a few selected genes are shown. Corresponding proteins that were detected in P. amoebophila EBs in a previous study (B. S. Sixt, C. Heinz, P. Pichler, E. Heinz, J. Montanaro, et al., Proteomics 11:1868–1892, 2011, https://doi.org/10.1002/pmic.201000510) are indicated by black lines (n = 234). (B) Average expression values (log2 RPKM) for genes involved in peptidoglycan synthesis and cell division (N. Jacquier, P. H. Viollier, and G. Greub, FEMS Microbiol Rev 39:262–275, 2015, https://doi.org/10.1093/femsre/fuv001) were hierarchically clustered (average linkage) based on Pearson correlation distances. Genes found to be differentially expressed between any two consecutive time points are marked with an asterisk. hpi, h postinfection; extracell., extracellular. Download FIG S2, PDF file, 0.7 MB (756.5KB, pdf) .
Copyright © 2017 König et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Gene expression dynamics during the Protochlamydia developmental cycle. The intersecting gene sets of statistically significantly up- or downregulated genes between successive time point pairs are shown. Each color represents a different set of differentially expressed (DE) genes, with the height of the bar indicating the number of genes. For example, at 2 h postinfection (hpi), there are a total of 250 significantly upregulated genes of which 153 genes are downregulated at the next time point (orange bars). Note that there are overlaps between gene sets of two consecutive time points. For instance, from the orange gene set (upregulated at 2 hpi and downregulated at 48 hpi), 22 genes were subsequently also downregulated at 96 hpi (pink boxes). White areas represent DE genes that were detected as significantly regulated only once during the cycle. Download FIG S3, PDF file, 0.2 MB (238KB, pdf) .
Copyright © 2017 König et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Sequencing and read mapping statistics. Download TABLE S1, PDF file, 0.1 MB (109.4KB, pdf) .
Copyright © 2017 König et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Gene expression values and annotation of Protochlamydia and A. castellanii. Download DATA SET S1, XLSX file, 1.8 MB (1.9MB, xlsx) .
Copyright © 2017 König et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Temporal partitioning of biological processes during development.
The three main temporal gene expression classes can be further subdivided into five distinct gene expression patterns termed here early I, early II, mid, late I, and late II (Fig. 2). The analysis of these gene sets provides detailed insights into the course of biological processes during chlamydial development.
Early I genes (n = 74) are highly expressed at 2 hpi and then strongly downregulated at midcycle. They remain at a lower expression level until late in the development, but transcripts are notably abundant in extracellular EBs (Fig. 2). Some of the genes referred to as “tardy” genes in a previous C. pneumoniae microarray study (20) follow a similar expression pattern, yet this transcriptional profile is strikingly pronounced in Protochlamydia. mRNA has been detected in EBs (20, 22, 26, 27, 40), which would be consistent with transcriptional activity, but EBs are generally considered transcriptionally silent (23). mRNA detected in EBs could thus represent “carryover” mRNA (19), for which it has been unclear whether it is actually used as the templates for translation (23). Here we propose that these transcripts are preserved in EBs to be readily available and functional during initial infection events. Consistent with this view, we find many genes encoding structural type III secretion proteins in this temporal class, as well as a number of predicted type III effectors (Fig. 2 and 3). Among those is a Protochlamydia-specific orthologue of the major enterobacterial virulence factor IpgD, which in Shigella facilitates host cell entry (41) and might be functionally similar to the Chlamydiaceae Tarp (42, 43). Taken together, the conspicuous expression pattern of the early I gene set points at a function in invasion of the amoeba host.
FIG 3 .
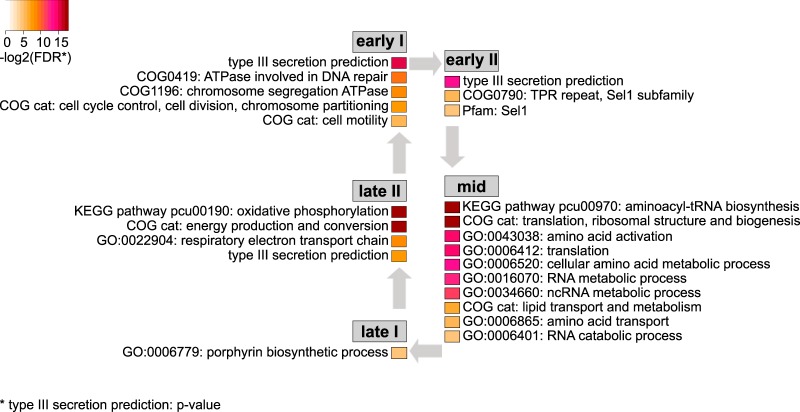
Enrichment of functional categories by temporal class. The overrepresentation of functional categories among genes assigned to temporal classes provides evidence for stage-specific activities during the Protochlamydia developmental cycle. Only functional categories that were significantly enriched with a false-discovery rate (FDR) of ≤0.05 are shown here; the color code indicates the degree of significance (dark red indicates highly significant). The significant enrichment of putatively type III secreted gene products was tested using Fisher’s exact test, and only P values of ≤0.05 are shown. cat, category.
A second set of early genes termed early II (n = 226) is upregulated at 2 hpi, subsequently downregulated, and—in contrast to early I genes—remains at a lower expression level for the rest of the cycle, including the EB stage (Fig. 2). These genes thus likely function mainly in EB-to-RB conversion and are important for establishing the intracellular niche. Consistently, this temporal class contains genes that in C. trachomatis follow a similar expression pattern (e.g., the chlamydia-specific inclusion membrane proteins and the transcriptional repressor of known late genes Euo) (19, 22, 23). This temporal class, however, was dominated in Protochlamydia by a large number of putative eukaryotic-like effectors with many exhibiting Sel1-like repeat domains or belonging to the recently identified large gene families PEX1 and PEX2 encoding proteins potentially involved in modification of host ubiquitination processes (Fig. 3 and Fig. S4) (33). Together, this indicates that while conserved chlamydial processes are at work during early niche establishment and RB-to-EB differentiation, Protochlamydia also employs a set of species-specific effector proteins during this stage.
The midcycle gene set comprises genes sharply upregulated at 48 hpi, when mainly RBs are present (Fig. 1). Transcripts for many of these genes were also detected at later stages at similar expression levels (Fig. 2) when the developmental cycle becomes more asynchronous. At this peak of RB activity, cellular processes like translation and metabolic activity (lipid and amino acid transport and metabolism) are most prominent (Fig. 3). Consistent with high metabolic activity, a number of genes responsible for cell division and peptidoglycan synthesis were upregulated, or by trend increased at midcycle (Fig. S2B). Analyzing the distribution of conserved and species-specific genes across the observed temporal gene expression classes revealed that the midcycle gene set was dominated by genes present in all chlamydiae, thereby confirming the role of chlamydial core genes in RB metabolism and proliferation (Fig. S5 and Text S1).
We found two subsets of late genes that are both characterized by an increased expression at the end of the cycle at 96 hpi and by generally high transcript levels at the EB stage (Fig. 2). Expression of late I genes (n = 105) already begins to increase at midcycle; the upregulation of late II genes (n = 168) is slightly delayed. These genes are thus involved in RB-to-EB transition, EB function, and EB maintenance.
Late I genes are enriched in genes involved in biosynthesis of porphyrins such as the cofactor heme required for the production of cytochromes (Fig. 3). This temporal class also includes genes involved in glucose and glycogen metabolism, such as glucokinase (glcK) and glycogen synthase (glgA), suggesting that glucose-based energy production is important at this stage. This trend is supported by the functions of late II genes, which are enriched in genes responsible for energy generation, including the electron transport chain for ATP synthesis (Fig. 3). Consistent with the presence of type III secretion systems on chlamydial EBs, we find many genes encoding structural proteins and known (CopB) and predicted effectors among the late I and late II gene sets (Fig. 3) and (Fig. S4).
A temporal model for chlamydial type III secretion.
The type III secretion system is an evolutionarily well conserved major virulence factor that was present in the last common ancestor of all known chlamydiae (1, 44, 45). Here we observed that genes involved in type III secretion, including structural components, chaperones, and effectors, belong to all five temporal gene sets and show strikingly different expression profiles (Fig. S4). While the basal apparatus and most components of the C-ring complex required for structural integrity and secretion activity (e.g., sctD, sctC) are expressed mid to late during RB/EB transition, notably lacking at this stage is a pronounced expression of the gene encoding the needle protein SctF. In fact, the transcript levels of the needle protein, the translocon CopB, most known chaperones, most putative effectors, as well as additional inner membrane components and the ATPase increase only later during maturation of EBs but then continue to be highly expressed early during infection (Fig. S4).
Expression patterns of genes encoding components of the type III secretion system and its (putative) effectors. All genes that met the following criteria were included: (i) genes that have been previously reported to encode components involved in Protochlamydia type III secretion (A. Collingro, P. Tischler, T. Weinmaier, T. Penz, E. Heinz, et al., Mol Biol Evol 28:3253–3270, 2011, https://doi.org/10.1093/molbev/msr161); (ii) genes that are orthologues of putative C. trachomatis effector proteins or structural components and chaperones of the chlamydial type III secretion system (H. J. Betts-Hampikian and K. A. Fields, Front Microbiol 1:114, 2010, https://doi.org/10.3389/fmicb.2010.00114); (iii) genes that were annotated to contain eukaryotic-like domains such as ankyrin repeats (ANK), Sel1-like repeats (Sel1), F-boxes (F-box), leucine-rich repeats (LRR, PEX1), tetratricopeptide repeats (TPR, PEX2), BTB/POZ domains (PEX1), and GDP-GTP exchange domains (RasGEF, RhoGEF); (iv) genes that were predicted to encode inclusion membrane proteins (put. inc) (E. Heinz, D. D. Rockey, J. Montanaro, K. Aistleitner, M. Wagner, et al., J Bacteriol 192:5093–5102, 2010, https://doi.org/10.1128/JB.00605-10); and (v) genes that encode putative effectors based on expression pattern and genomic location (data not shown). Differentially expressed genes are marked with an asterisk. Temporal classes are indicated by colored bars. The sct unified nomenclature for the naming of type III secretion system genes was used (C. J. Hueck, Microbiol Mol Biol Rev 62:379-433, 1998). RPKM, reads per kilobase per million; hpi, h postinfection; extracell., extracellular; PEX1, Protochlamydia expanded gene family 1; PEX2, Protochlamydia expanded gene family 2 (D. Domman, A. Collingro, I. Lagkouvardos, L. Gehre, T. Weinmaier, et al., Mol Biol Evol 31:2890–2904, 2014, https://doi.org/10.1093/molbev/msu227); put, putative; Ctr, C. trachomatis. Download FIG S4, PDF file, 1.3 MB (1.3MB, pdf) .
Copyright © 2017 König et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Expression of species-specific and chlamydial core genes. (A) Heatmap showing the distribution of temporarily regulated Protochlamydia genes at different levels of taxonomic conservation. The color indicates the percentage of differentially expressed (DE) genes per temporal class and taxonomic group. The absolute numbers of DE genes per temporal class and taxonomic group are indicated; the total numbers of DE genes per taxonomic group are shown in brackets. Note that the early gene set is dominated by species-specific genes (32%), whereas chlamydial core genes are most abundant at midcycle (44%). (B) Nearly all chlamydial core genes are expressed in Protochlamydia, with constitutively expressed genes contributing 62%. Download FIG S5, PDF file, 0.2 MB (223.2KB, pdf) .
Copyright © 2017 König et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Our gene expression-centric view of type III secretion thus suggests a stepwise assembly of the Protochlamydia type III secretion apparatus, which is consistent with knowledge about type III secretion system assembly in other bacteria (46) and models for Chlamydiaceae proposed earlier (45). Our time-resolved model for the role of type III secretion during the developmental cycle includes three major steps (Fig. 4). The first step is the formation of the main parts of novel type III secretion machineries mainly during RB-to-EB differentiation; because of the lack of a pronounced expression of the needle protein and other factors prior to this stage, we propose that type III secretion machineries are mostly incomplete at midcycle and therefore, secretion is less prominent when RBs likely devote most of their resources to proliferation (Fig. 3). At the tail end of RB-to-EB differentiation, SctF is upregulated, and most secretion machineries are then fully equipped. Thus, the second step is marked by functional type III secretion systems that are present on extracellular EBs and can be employed for host cell invasion. The third step is characterized by the peak of type III secretion activity occurring in early RBs, where we observed all structural components, including the needle protein to be continuously highly expressed. Importantly, the major role of type III secretion during early development is also well supported by the largest number of (putative) effectors upregulated at this stage (Fig. 4 and Fig. S4).
FIG 4 .
Course of Protochlamydia type III secretion system activity during the developmental cycle. This model is based on the observation that structural components of the type III secretion system and its (putative) effectors are expressed at different time points during the developmental cycle (Fig. S4). This suggests a scenario in which novel, fully assembled, and thus functional secretion systems occur only late in the developmental cycle, and type III secretion reaches its full capacity and highest activity during early stages of the infection. The indicated polarity of the active type III secretion system has been shown for C. trachomatis (54), but it is unclear whether this is also true for P. amoebophila, as the symbionts reside within single-cell inclusions. The color code for type III secretion components and effectors (nomenclature according to Hueck [105]) refers to the respective temporal gene expression classes (Fig. 2 and Fig. S4). Circles inside the cells represent chaperones. Differentially expressed components/effectors are labeled with asterisks; PEX1 and PEX2 refer to members of the expanded effector gene families in Protochlamydia (33); pc0309 is an ortholog of the putative chaperone encoded by CT274 (106). inc, inclusion membrane; hcm, host cell membrane; im, inner membrane; om, outer membrane.
It is well-known that type III secretion genes of bacterial pathogens are temporally regulated (47). Moreover, our model is consistent with gene expression data for the Chlamydiaceae, which indicate that the large majority of type III secretion component genes are temporally regulated and increasingly expressed from midcycle toward the end (19–21, 48, 49). The model is also supported by proteomic analyses of Protochlamydia and C. trachomatis EBs and RBs in which most type III secretion system components were identified on prereplicative RBs (50) and EBs (39, 51). The model is also generally in agreement with recent (cryo-)electron tomography analyses of Protochlamydia and C. trachomatis, demonstrating the presence of type III secretion systems on host-free EBs (52–54) and late RBs (55). Finally, our interpretation that type III secretion systems are more abundant on early RBs compared to EBs matches electron microscopy data for Chlamydia psittaci (56). The model is, however, not consistent with the apparent absence of the type III secretion machinery on C. trachomatis EBs noted immediately after host entry in a cryo-electron tomography-based study (54), which would imply that type III secretion is not required at this stage. The latter scenario is difficult to reconcile with the general notion that effector secretion is essential during early development (45, 57). We thus propose that type III secretion is active to facilitate invasion by EBs, but based on the pronounced early gene expression of type III secretion components and effectors—observed here for the first time—is substantially enhanced during establishment of the replicative niche by early RBs (Fig. 4).
Biphasic metabolism of Protochlamydia.
A number of previous observations provide evidence for developmental stage-specific differences of the chlamydial metabolism (reviewed in reference 58). Our comprehensive transcriptomic data set allows us to dissect the activity of individual pathways and metabolic modules during the Protochlamydia developmental cycle. As a result, we are able to propose a detailed model for the modulation of chlamydial physiology during infection and extracellular survival.
Stage-specific energy sources.
Chlamydiae are well-known energy parasites facilitated by nucleotide transport proteins, which import host-derived ATP in exchange for ADP (59, 60). Yet, they also harbor the genetic potential to synthesize ATP via oxidative phosphorylation. Our gene expression analysis, however, revealed that in Protochlamydia, genes involved in carbon and energy metabolism show a low transcription level at midcycle and are induced only late during development (Fig. 5). At the replicative stage, the import of host-derived ATP is thus likely the most important energy source. The ATP/ADP translocase (ntt1) is constitutively expressed at an exceptionally high level (among the top 1% of all expressed genes), and although it was not detected as significantly differentially expressed, its transcription profile clearly peaks at midcycle (Fig. 5). The same trend has been observed for C. trachomatis and C. pneumoniae (19, 20), and nucleotide transport proteins were most abundant in C. trachomatis RBs compared to EBs (51). In addition, ATP but not glucose-6-phosphate stimulated de novo protein synthesis in C. trachomatis RBs in a chemically defined axenic medium (61).
FIG 5 .
Expression maps of selected metabolic pathways of Protochlamydia. A pronounced expression of the ATP/ADP translocase (ntt1) at midcycle and an early-to-mid activity of genes involved in amino acid breakdown to pyruvate is observed, whereas pathways involved in central carbon metabolism and energy generation were generally only upregulated at later stages (with the ATPases indicated by purple boxes being notable exceptions). This suggests that a major metabolic shift occurs during the developmental cycle and provides evidence for a stage-specific metabolism. All genes marked with an asterisk were detected to be significantly differentially expressed. RPKM, reads per kilobase per million; hpi, h postinfection; extracell., extracellular.
Because late genes were enriched in respiratory chain genes (Fig. 3), we compiled detailed gene expression heatmaps for all genes known to be part of the chlamydial carbon and energy metabolism pathways (Fig. 5). Strikingly, we also found genes involved in glycolysis, the pentose phosphate pathway, and the tricarboxylic acid (TCA) cycle to be generally upregulated only late during development. In agreement, we observed a pronounced upregulation of genes responsible for glutamate and aspartate breakdown (gdhB, glutamate dehydrogenase; aspC, aspartate aminotransferase; Fig. 5), whose end products could fuel the TCA cycle.
A notable exception are the V-type and the F-type ATPases whose expression is induced at midcycle, a trend which was also seen for the respective C. trachomatis proteins (51). In the absence of other main components of the respiratory chain at this stage, the ATPases likely engage in hydrolysis of ATP (rather than ATP synthesis) and thus act as proton/sodium pumps maintaining the membrane potential—a function well-known for both types of ATPases and proposed for the ATPase in C. psittaci RBs (62–64). In Protochlamydia, the ATPases continue to be expressed at later stages, when all components of the oxidative phosphorylation pathway are in place. The proton/sodium gradient can then be generated by the other complexes of the respiratory chain, and the ATPases would function in ATP synthesis.
This model fits well with findings for C. trachomatis, where proteins involved in the endogenous energy metabolism were detected only in EBs, not in RBs (65), or were found to be more abundant in EBs (51). Moreover, glucose 6-phosphate but not ATP stimulated de novo protein synthesis in host-free C. trachomatis EBs, and ATP generation was observed at this stage (61). Furthermore, we have previously shown that C. trachomatis and Protochlamydia EBs require glucose 6-phosphate or glucose, respectively, for maintenance of infectivity during extracellular survival (35). In conclusion, the gene expression data presented here provide compelling evidence that a stage-specific energy metabolism indeed occurs in vivo and that it is well conserved among known chlamydiae.
Amino acids and pyruvate as main carbon source during replication.
Another consequence of the observed expression of genes involved in breakdown of glucose predominantly at the postreplicative stage is that glucose cannot represent the major carbon source at the RB stage. In fact, chlamydiae acquire the majority of cell building blocks such as amino acids, nucleotides, and certain lipids from the host cell (66). Transport proteins required for the uptake of those compounds exist, and most of the 24 known amino acid transporters of Protochlamydia are expressed early and midcycle, providing all the substrates required for protein synthesis in RBs (Fig. 5). The importance of host-derived amino acids for chlamydial protein synthesis is well supported by a recent isotopologue profiling study of C. trachomatis (67). While no external carbon source might thus be required for protein, DNA, and RNA synthesis, chlamydiae clearly need carbon to synthesize branched-chain fatty acids for the generation of phospholipids (68). Consistent with this notion, lipid metabolism was induced at the replicative stage in Protochlamydia (Fig. 3). This includes the pyruvate dehydrogenase genes (pdhABC), which were upregulated early and at midcycle and catalyze the conversion of pyruvate to acetyl coenzyme A (acetyl-CoA), the precursor of fatty acid biosynthesis (Fig. 5). The pyruvate required for this step could be imported directly from the host as suggested for C. psittaci (69), although no pyruvate transporter has yet been identified in Protochlamydia or Chlamydiaceae. Pyruvate cannot be generated from host-derived nucleotides or host lipids, as Protochlamydia lacks the genetic repertoire for breakdown of those compounds. However, genes encoding components involved in degradation of amino acids are present and were indeed most highly expressed early (sdaA, l-serine dehydratase) and at midcycle (tdh, l-threonine 3-dehydrogenase; ald, alanine dehydrogenase; gcvT, T protein of the glycine cleavage complex; Fig. 5). These enzymes catabolize alanine, threonine, glycine, and serine to pyruvate, which may then serve as the substrate for branched-chain fatty acid synthesis. The increased number of gene copies encoding a specific transporter for alanine (dagA) and their early and midcycle expression further support a model in which amino acids provide the carbon required for fatty acid biosynthesis in Protochlamydia RBs (Fig. 6).
FIG 6 .
Biphasic metabolism of Protochlamydia during development in Acanthamoeba castellanii. This model is based on observed transcriptional patterns, enriched functional categories at different developmental stages (Fig. 2, 3, and 5 and Fig. S2A), and independent experimental evidence reported previously (see text for references). Activity of metabolic pathways as inferred from gene expression levels followed similar trends early and at midcycle, and at the two later stages, respectively. This suggests that ATP import and an amino acid-based anabolism prevails during the EB-to-RB transition and RB replication. Later stages are characterized by a glucose-based metabolism and a pronounced increase in the activity of the tricarboxylic acid (TCA) cycle and oxidative (ox.) phosphorylation pathway. Nucleotide transporters (Ntt’s) are shown in blue, and amino acid (AA) and oligopeptide transporters are shown in green. “Multi” indicates that multiple amino acid/peptide transporters with different substrate specificities are expressed. Question marks refer to hypothetical transporters not yet identified. Asterisks indicate an increased expression at the RB stage compared to the early time point. “RNA” denotes transcription, whereas “DNA” indicates DNA replication. Glc-6-P, glucose 6-phosphate; PPP, pentose phosphate pathway; glyconeog., gluconeogenesis.
Metabolism of EBs.
Many of the late genes involved in carbon and energy metabolism are still highly expressed at the EB stage in Protochlamydia. Remarkably, this is also the case for genes involved in transcription and protein synthesis. This includes the RNA polymerase, amino acid transporters, and many translation-related genes (Fig. 5 and Fig. S2A). Our transcriptome data thus support a number of early observations and recent findings suggesting that chlamydial EBs are not metabolically inert but maintain a limited metabolism during host-free survival (35, 36, 58, 61).
Biphasic versus bipartite metabolism.
In summary, transcriptional dynamics during the Protochlamydia developmental cycle provide compelling evidence for a biphasic metabolism with stage-specific carbon and energy sources (Fig. 6). This is well supported by a number of earlier findings, suggesting that a biphasic metabolism is generally conserved among known chlamydiae (51, 58, 61).
In addition to these stage-specific differences in physiology, a recent study demonstrated cosubstrate usage by C. trachomatis, with host amino acids required for bacterial protein biosynthesis and glucose 6-phosphate as the carbon source for lipopolysaccharide biosynthesis (67). Termed bipartite metabolism, this efficient use of host-derived metabolites (i.e., the use of two different carbon sources) has also been observed in Listeria monocytogenes (70) and Legionella pneumophila (71) and might thus represent a more general adaptation of intracellular bacteria (67).
Yet, for L. pneumophila, there is also evidence for a growth phase-dependent metabolism. Intracellular growth of L. pneumophila within A. castellanii requires amino acids as energy source and—similarly to our model for Protochlamydia—as the carbon source and directly for protein biosynthesis (72, 73). Although a temporal separation of amino acid and carbohydrate usage has not yet been shown in vivo for L. pneumophila, a pronounced transcriptional switch between replicative and transmissive phase within A. castellanii has been reported (74). In addition, in in vitro experiments, L. pneumophila used serine as a major carbon source for the synthesis of other amino acids and for energy generation during all growth phases, while glucose served as the carbon source primarily in the postreplicative phase (71, 75). A growth or developmental stage-specific metabolism might thus not be restricted to the chlamydiae but is perhaps more widespread among intracellular bacteria.
Transcriptional response of the Acanthamoeba host.
Close to 75% of the known A. castellanii genes (76) were detected to be expressed during infection with Protochlamydia (n = 12,044; Data Set S1). Although the different infection rates in our experiments (5 to 60%) entail capturing only a transcript mix of infected and uninfected amoebae, we still observed pronounced temporal patterns and characteristic expression profiles, with 3,582 Acanthamoeba genes found to be differentially expressed. Major transcriptional shifts occurred at 2 hpi when 1,722 genes were up- and subsequently downregulated and at 48 hpi when 1,747 genes were induced (Fig. S6).
Gene expression dynamics of the Acanthamoeba host during infection with Protochlamydia. Ten different temporal subclasses of genes can be distinguished (indicated by black lines) within three main classes (colored bars). Major gene expression shifts occurred early during infection (yellow) and at the peak of Protochlamydia RB activity (red). Only significantly differentially expressed genes are shown. The number of genes per temporal class is indicated. RPKM, reads per kilobase per million; hpi, h postinfection. Download FIG S6, PDF file, 1.2 MB (1.2MB, pdf) .
Copyright © 2017 König et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Enrichment analysis of putative gene functions at the early stage of infection indicated a strong increase in cell signaling (serine threonine kinases), transport activity (ABC transporter of unknown specificity), translation (tRNA-related functions), and assembly of the mitochondrial respiratory chain complex III (Table S2). These effects on the Acanthamoeba host are consistent with the observed pronounced expression of type III secretion effectors by Protochlamydia at this stage, interfering with diverse host signaling pathways and possibly inducing a stress response.
Overrepresented functions among A. castellanii genes. Download TABLE S2, PDF file, 0.1 MB (143.1KB, pdf) .
Copyright © 2017 König et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
When Protochlamydia is proliferating at the RB stage, the amoeba host transcriptome is characterized by increased expression of genes involved in breakdown of complex sugars (Table S2). This might account for the increased ATP demand of replicating Protochlamydia RBs. Notably downregulated at this stage are genes functioning in cell signaling (Ras proteins, serine threonine kinases), ubiquitination, translation (ribosome biogenesis), transcription (transcription factors), and replication (DNA replication initiation) (Table S2). Together, this suggests a general downregulation of central cellular processes due to replicating Protochlamydia, which is at the peak of its metabolic activity at this stage.
At later stages of the infection at which Protochlamydia EBs are formed, the amoeba transcriptome is still significantly altered and strikingly different compared to the onset of the infection. In particular, genes involved in cell signaling (histidine kinases, serine threonine kinases) are enriched at 96 hpi, whereas key cellular pathways are still less active, illustrating the fundamental impact of Protochlamydia on gene expression of its amoeba host (Fig. S6 and Table S2).
The response of human cells upon infection with Chlamydiaceae continues to be extensively studied (25, 77, 78), whereas studies on the impact of environmental chlamydiae on their host cells are strikingly lacking. Our transcriptomic data provide the first insights into this unexplored aspect of chlamydia-host interplay.
Conclusions.
Owing to their obligate intracellular lifestyle and the lack of routine genetic methods (79), chlamydiae are—compared to many other bacterial pathogens—inherently difficult to study. This particularly applies to environmental chlamydiae, which were discovered only about 2 decades ago. The present study represents the most comprehensive analysis of the transcriptional landscape of chlamydiae during infection and development thus far. We obtained detailed insights into gene expression dynamics of Protochlamydia and present models for metabolism and the role and assembly of the type III secretion system at different developmental stages. Our findings revealed striking parallels to what is known about pathogenic chlamydiae and provide a substantially novel perspective on the interplay between chlamydiae and protist hosts. In addition, the vast majority of chlamydial organisms lie outside the well-known human and animal pathogens, and thus the Protochlamydia model system serves to represent this vast diversity. Environmental chlamydiae constitute invaluable model systems to understand fundamental chlamydial biology, common themes and differences among known chlamydiae, and to shed light on the evolution of the intracellular lifestyle and pathogenesis in a ubiquitous bacterial phylum.
MATERIALS AND METHODS
Cell culture.
Acanthamoeba castellanii Neff (ATCC 50373) with or without the symbiont Protochlamydia amoebophila UWE25 (ATCC PRA-7) were maintained at 20°C in PYG medium [20 g/liter proteose peptone, 100 mM glucose, 2 g/liter yeast extract, 1 g/liter sodium citrate dihydrate, 4 mM MgSO4 ⋅ 7H2O, 1.32 mM Na2HPO4 ⋅ 2H2O, 2.5 mM KH2PO4, 0.05 mM Fe(NH4)2(SO4)2 ⋅ 6H2O; pH 6.5]. Cultures were regularly screened by fluorescence in situ hybridization and 4′,6′-diamidino-2-phenylindole (DAPI) staining (0.1 µg/ml) to exclude contamination.
Infection experiments.
Protochlamydia EBs were freshly purified from amoeba cultures grown in 500-cm2 culture flasks (Nalge Nunc International, Rochester, NY, USA), in which EBs had been allowed to accumulate in the medium for 1 week. Purification of EBs was conducted by filtering culture supernatants through 5-µm and 1.2-µm syringe filters (Sartorius, Göttingen, Germany) to remove residual host cells. Bacteria were collected by centrifugation (15,550 × g, 15 min, 20°C), resuspended in precooled SPG buffer (75 g/liter sucrose, 0.52 g/liter KH2PO4, 1.53 g/liter NaHPO4 ⋅ 7H2O, 1.53 g/liter Na2HPO4 ⋅ 2H2O, 0.75 g/liter glutamic acid; pH 7.2), homogenized using a 21-gauge injection needle (B. Braun, Melsungen, Germany), and stored overnight at 4°C in SPG buffer. For quantification of purified EBs, cell suspensions were filtered onto a polycarbonate membrane with a pore size of 0.2 µm (EMD Millipore, Billerica, MA, USA); cells were stained with DAPI and counted using an epifluorescence microscope (Axioplan 2 imaging; Carl Zeiss, Oberkochen, Germany).
Symbiont-free amoebae were harvested 3 days before infection and 6.4 × 107 cells per 500-cm2 culture flask per time point and replicate were seeded and incubated at 20°C until infection. To optimize infection efficiency, particularly at early time points, we used a multiplicity of infection (MOI) of 150 for 2 h postinfection (hpi), an MOI of 100 for 48 hpi, and an MOI of 15 for 96 hpi. Precultivated amoebae were harvested, transferred to 50-ml Greiner tubes, and purified Protochlamydia EBs were added, followed by repeated centrifugation (centrifuged at 130 × g twice for 5 min each time and then once for 10 min at 20°C) with vortexing between the centrifugation steps. Infected amoebae were then transferred back to the culture flasks and incubated in PYG medium at 20°C for 2 h before the infection was synchronized by gently washing the attached amoebae three times with Page’s amoebic saline (PAS) (0.12 g/liter NaCl, 0.004 g/liter MgSO4 ⋅ 7H2O, 0.004 g/liter CaCl2 ⋅ 2H2O, 0.142 g/liter Na2HPO4, 0.136 g/liter KH2PO4). PYG medium was added to the cultures, the culture was sampled at the 2 hpi time point, and the remaining culture flasks were incubated at 20°C for 48 and 96 h. Extracellular Protochlamydia EBs were purified as described above. All infection experiments were performed in biological triplicates. One culture of symbiont-free amoebae was harvested when the culture was sampled at 2 hpi.
Fluorescence in situ hybridization and transmission electron microscopy.
Fluorescence in situ hybridization using a combination of two Cy3-labeled probes (Chls-0523, E25-454; Thermo Fisher Scientific, Waltham, MA, USA) was performed as described elsewhere (35, 80, 81), and cells were stained with DAPI for 5 min. Images were recorded with a charge-coupled-device (CCD) camera (AxioCam HRc; Carl Zeiss) connected to an epifluorescence microscope and were processed using the AxioVision 4.6.3 software package (Carl Zeiss).
For transmission electron microscopy, the culture medium was replaced with fixative solution (2.5% glutaraldehyde in 3 mM cacodylate containing 0.1 M sucrose; pH 6.5). Amoebae were fixed for 1 h at room temperature, then collected, washed three times (0.1 M cacodylate containing 0.1 M sucrose [pH 7.2]), and mixed with one drop of 1% Biozym plaque agarose (Biozym, Hessisch Oldendorf, Germany) in washing buffer equilibrated at 35°C. Secondary fixation was conducted in 1% buffered osmium tetroxide for 1 h on ice, followed by dehydration in ethanol and infiltration with low-viscosity resin (Agar Scientific, Essex, United Kingdom). Ultrathin sections (70 nm) were cut using a Leica EM UC7 ultramicrotome (Leica, Wetzlar, Germany) and stained with 0.5% uranyl acetate and 3% lead citrate, and imaging was done with a Zeiss EM 902 transmission electron microscope (Carl Zeiss).
Infectious progeny production assay.
To monitor the production of infectious Protochlamydia EBs during the developmental cycle, amoebae were infected with Protochlamydia using an MOI of 10. The infection was synchronized by centrifugation (130 × g, 15 min, 20°C) and subsequent medium exchange. Cells were harvested at different time points postinfection, and extracellular bacteria were separated from amoebae by low-speed centrifugation (300 × g, 10 min, 4°C). The supernatant containing the extracellular bacteria was centrifuged (20,800 × g, 30 min, 4°C), the pellet was resuspended in precooled SPG medium, and stored at −80°C until further use. The pellet containing the infected amoebae was resuspended in precooled SPG medium and subjected to two freeze/thaw (−20°C/room temperature) steps, followed by vortexing with glass beads (diameter of 0.75 to 1 mm; Carl Roth, Karlsruhe, Germany) for 3 min to break up the amoebae. Amoeba cell debris and glass beads were removed by centrifugation (300 × g, 10 min, 4°C), and the supernatants containing intracellular bacteria were stored at −80°C. The total numbers of bacteria in intra- and extracellular suspensions were determined by counting DAPI signals as described above. The percentage of infectious Protochlamydia EBs was determined by infecting fresh amoebae and counting bacteria (inclusions) per amoeba at 12 hpi. The infection was carried out as described above. Bacteria were detected by indirect immunofluorescence and DAPI staining after methanol fixation as described previously (82). Mean relative infectivity of each fraction (intra/extracellular) per time point was expressed as the number of bacteria or amoeba/total number of bacteria (± standard error of the mean, three biological replicates).
RNA extraction and sequencing.
Preliminary tests showed that RNA extraction and sequencing of intact Protochlamydia-infected amoebae yielded an insufficient number of bacterial transcripts. Thus, to increase the coverage of the Protochlamydia transcriptome, a protocol for enrichment of bacteria prior to RNA extraction was developed. To minimize possible changes of the transcriptomes during enrichment, each sample was processed in less than 7 min, as the half-life of total mRNA from Escherichia coli was demonstrated to be in this range (83). Infected amoebae were harvested and collected (7,600 × g, 2 min, 20°C), and the pellets were resuspended in a sucrose buffer (35 mM Tris-HCl, 250 mM sucrose, 25 mM KCl, 10 mM MgCl2) supplemented with 50 µg/ml rifampin in order to inhibit active transcription during the enrichment procedure (84, 85). Amoebae were then disrupted by vortexing in the presence of glass beads (diameter of 0.75 to 1 mm; Carl Roth) for 1 min. The suspensions were subsequently filtered through a 5-µm filter, the flowthrough fractions containing the bacteria were collected by centrifugation (10,600 × g, 2 min, room temperature), and the pellets were immediately resuspended in TRIzol reagent (Thermo Fisher Scientific). Extracellular Protochlamydia EBs were pelleted (20,800 × g, 2 min, room temperature), and the pellets were resuspended in sucrose buffer and subsequently treated like the enriched bacteria.
Cells were mechanically disrupted by beat beating for 30 s at 4.5 m/s using lysing matrix A tubes and a FastPrep-24 instrument (MP Biomedicals, Santa Ana, CA, USA). Subsequent RNA extraction was performed according to the TRIzol guidelines. Residual DNA was digested using the Turbo DNA-free kit (Thermo Fisher Scientific) according to the manufacturer’s instructions. DNase-treated RNA was precipitated with ethanol and sodium acetate and dissolved in nuclease-free water (Thermo Fisher Scientific), and DNA contamination was controlled for via PCR targeting a short region of the bacterial 16S rRNA gene (SigF2/R2 primers; 11) using 35 PCR cycles. rRNA was removed using the Ribo-Zero magnetic kit for Gram-positive bacteria as recommended by the manufacturer (Illumina, San Diego, CA, USA). To enrich for mRNA from symbiont-free amoebae, the RNA was additionally treated with Dynabeads mRNA purification kit (Thermo Fisher Scientific). After another round of precipitation with ethanol, rRNA depletion and RNA quality were examined using the Experion automated electrophoresis system (Bio-Rad Laboratories, Hercules, CA, USA). RNA fragmentation was performed at 70°C for 5 min using the RNA fragmentation reagents from Thermo Fisher Scientific and was followed by another ethanol precipitation. For strand-specific cDNA library preparation, the NEBNext Ultra directional RNA library prep kit for Illumina in combination with the NEBNext multiplex oligonucleotides (New England Biolabs, Ipswich, MA, USA) was used starting at first-strand cDNA synthesis. Purification and size selection steps were done as recommended using Agencourt AMPure beads (Beckman Coulter, Brea, CA, USA). All libraries were sequenced using an Illumina HiSeq2000 system at the Vienna Biocenter Core Facilities (VBCF) Next-Generation Sequencing (NGS) Unit (http://www.vbcf.ac.at) with 50-bp read length.
Sequence read processing.
Sequencing reads were trimmed and cleaned before mapping (see Text S1 in the supplemental material). To map bacterial reads to the Protochlamydia genome (18), the Burrows-Wheeler Aligner (BWA) (86) was used; amoeba reads were mapped to the A. castellanii Neff genome (76), the rRNA genes (87–89), and the mitochondrial genome (90) using TopHat (91), both with default settings. Only unambiguously mapped reads were kept using SAMtools (92). Strand-specific reads per predicted gene were counted via HTSeq (93). Reads that could not be assigned to any gene but mapped to the genome were considered transcripts of intergenic regions (IGRs) and antisense transcripts.
Gene expression analyses.
Differentially expressed genes were determined between two consecutive time points (2 hpi to 48 hpi, 48 hpi to 96 hpi, 96 hpi to extracellular, extracellular to 2 hpi for Protochlamydia; uninfected to 2 hpi, 2 hpi to 48 hpi, 48 hpi to 96 hpi for A. castellanii) using the R software environment and the Bioconductor package edgeR (94–96). Genes were considered differentially expressed if their expression changed twofold with a false-discovery rate (FDR) smaller or equal 0.05, except for detecting gene expression changes between uninfected amoebae and infected amoebae 2 hpi, when a fivefold change threshold was used because only one sample of uninfected amoebae was sequenced.
Gene expression data were further analyzed using custom R scripts, integrated R tools, and R packages (94). To determine temporal expression patterns, sets of genes with similar expression profiles were identified by hierarchical clustering of gene expression values (log2 reads per kilobase per million [RPKM]) based on Pearson correlation distances. Obtained clusters were validated using the R package clValid (97). Mean centered expression values were used for visualization as heatmaps using the R package gplots (98).
To extend and improve the available Protochlamydia genome annotation by Horn et al. (18) for each gene, we collected Pfam domains (99), Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway and gene ontology (GO) term level 5 assignments using DAVID (100), COG (clusters of orthologous groups of proteins) cluster and class assignments using MaGe (101), and type III secretion effector predictions using Effective (102). Blast2GO (103) was used to assign GO terms to predicted proteins of A. castellanii. The Bioconductor software package GOseq (104) and the Blast2GO enrichment analysis tool were used to test for statistical enrichment of functional categories (FDR < 0.05) among differentially expressed genes per time point or temporal class. To test whether predicted type III secreted proteins were significantly enriched in any given gene set, two-tailed Fisher’s exact tests were conducted, and P values below 0.05 were considered statistically significant.
Availability of data.
Sequences were deposited at the Gene Expression Omnibus (GEO) database and are accessible through accession number GSE93891.
ACKNOWLEDGMENTS
This work was funded by the European Research Council (ERC StG EVOCHLAMY, project 281633) and the Austrian Science Fund (project I1628-B22) in the context of the InfectERA project Eugenpath.
We are grateful to David Berry for help with R scripts, to Daniela Gruber, Nikolaus Leisch, Mario Schimak, and Jacqueline Montanaro for assistance with electron microscopy, to Gitta Szabo for assistance in the lab, and to Astrid Collingro for helpful discussions. We thank the team at the VBCF for sequencing and Thomas Rattei for providing access to the Life Science Compute Cluster at the University of Vienna.
REFERENCES
- 1.Subtil A, Collingro A, Horn M. 2014. Tracing the primordial Chlamydiae: extinct parasites of plants? Trends Plant Sci 19:36–43. doi: 10.1016/j.tplants.2013.10.005. [DOI] [PubMed] [Google Scholar]
- 2.Horn M. 2008. Chlamydiae as symbionts in eukaryotes. Annu Rev Microbiol 62:113–131. doi: 10.1146/annurev.micro.62.081307.162818. [DOI] [PubMed] [Google Scholar]
- 3.Lagkouvardos I, Weinmaier T, Lauro FM, Cavicchioli R, Rattei T, Horn M. 2014. Integrating metagenomic and amplicon databases to resolve the phylogenetic and ecological diversity of the Chlamydiae. ISME J 8:115–125. doi: 10.1038/ismej.2013.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kuo C-C, Stephens RS. 2015. Chlamydiaceae. In Whitman WB, Rainey F, Kämpfer P, Trujillo M, Chun J, DeVos P, Hedlund B, Dedysh S (ed), Bergey’s manual of systematics of archaea and bacteria. John Wiley & Sons, Ltd, Chichester, United Kingdom. [Google Scholar]
- 5.World Health Organization 2012. Global incidence and prevalence of selected curable sexually transmitted infections − 2008. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 6.Burton MJ, Mabey DCW. 2009. The global burden of trachoma: a review. PLoS Negl Trop Dis 3:e460. doi: 10.1371/journal.pntd.0000460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Greub G. 2009. Parachlamydia acanthamoebae, an emerging agent of pneumonia. Clin Microbiol Infect 15:18–28. doi: 10.1111/j.1469-0691.2008.02633.x. [DOI] [PubMed] [Google Scholar]
- 8.Casson N, Michel R, Müller K-D, Aubert JD, Greub G. 2008. Protochlamydia naegleriophila as etiologic agent of pneumonia. Emerg Infect Dis 14:168–172. doi: 10.3201/eid1401.070980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baud D, Goy G, Osterheld M-C, Croxatto A, Borel N, Vial Y, Pospischil A, Greub G. 2014. Role of Waddlia chondrophila placental infection in miscarriage. Emerg Infect Dis 20:460–464. doi: 10.3201/eid2003.131019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Friedman MG, Dvoskin B, Kahane S. 2003. Infections with the chlamydia-like microorganism Simkania negevensis, a possible emerging pathogen. Microbes Infect 5:1013–1021. doi: 10.1016/S1286-4579(03)00188-6. [DOI] [PubMed] [Google Scholar]
- 11.Haider S, Collingro A, Walochnik J, Wagner M, Horn M. 2008. Chlamydia-like bacteria in respiratory samples of community-acquired pneumonia patients. FEMS Microbiol Lett 281:198–202. doi: 10.1111/j.1574-6968.2008.01099.x. [DOI] [PubMed] [Google Scholar]
- 12.AbdelRahman YM, Belland RJ. 2005. The chlamydial developmental cycle. FEMS Microbiol Rev 29:949–959. doi: 10.1016/j.femsre.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 13.Hatch TP. 1999. Developmental biology, p 26–37. In Stephens RS. (ed), Chlamydia: intracellular biology, pathogenesis, and immunity. ASM Press, ; Washington, DC. [Google Scholar]
- 14.Kahane S, Kimmel N, Friedman MG. 2002. The growth cycle of Simkania negevensis. Microbiology 148:735–742. doi: 10.1099/00221287-148-3-735. [DOI] [PubMed] [Google Scholar]
- 15.Michel R, Steinert M, Zöller L, Hauröder B, Henning K. 2004. Free-living amoebae may serve as hosts for the Chlamydia-like bacterium Waddlia chondrophila isolated from an aborted bovine foetus. Acta Protozool 43:37–42. [Google Scholar]
- 16.Greub G, Raoult D. 2002. Crescent bodies of Parachlamydia acanthamoeba and its life cycle within Acanthamoeba polyphaga: an electron micrograph study. Appl Environ Microbiol 68:3076–3084. doi: 10.1128/AEM.68.6.3076-3084.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sixt BS, Kostanjšek R, Mustedanagic A, Toenshoff ER, Horn M. 2013. Developmental cycle and host interaction of Rhabdochlamydia porcellionis, an intracellular parasite of terrestrial isopods. Environ Microbiol 15:2980–2993. doi: 10.1111/1462-2920.12252. [DOI] [PubMed] [Google Scholar]
- 18.Horn M, Collingro A, Schmitz-Esser S, Beier CL, Purkhold U, Fartmann B, Brandt P, Nyakatura GJ, Droege M, Frishman D, Rattei T, Mewes H-W, Wagner M. 2004. Illuminating the evolutionary history of chlamydiae. Science 304:728–730. [DOI] [PubMed] [Google Scholar]
- 19.Belland RJ, Zhong G, Crane DD, Hogan D, Sturdevant D, Sharma J, Beatty WL, Caldwell HD. 2003. Genomic transcriptional profiling of the developmental cycle of Chlamydia trachomatis. Proc Natl Acad Sci U S A 100:8478–8483. doi: 10.1073/pnas.1331135100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mäurer AP, Mehlitz A, Mollenkopf HJ, Meyer TF. 2007. Gene expression profiles of Chlamydophila pneumoniae during the developmental cycle and iron depletion-mediated persistence. PLoS Pathog 3:e83. doi: 10.1371/journal.ppat.0030083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nicholson TL, Olinger L, Chong K, Schoolnik G, Stephens RS. 2003. Global stage-specific gene regulation during the developmental cycle of Chlamydia trachomatis. J Bacteriol 185:3179–3189. doi: 10.1128/JB.185.10.3179-3189.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shaw EI, Dooley CA, Fischer ER, Scidmore MA, Fields KA, Hackstadt T. 2000. Three temporal classes of gene expression during the Chlamydia trachomatis developmental cycle. Mol Microbiol 37:913–925. doi: 10.1046/j.1365-2958.2000.02057.x. [DOI] [PubMed] [Google Scholar]
- 23.Tan M. 2012. Temporal gene regulation during the chlamydial developmental cycle. In Tan M, Bavoil P (ed), Intracellular pathogens I: Chlamydiales, 10th ed. ASM Press, ; Washington, DC. [Google Scholar]
- 24.Mathews SA, Volp KM, Timms P. 1999. Development of a quantitative gene expression assay for Chlamydia trachomatis identified temporal expression of σ factors. FEBS Lett 458:354–358. doi: 10.1016/S0014-5793(99)01182-5. [DOI] [PubMed] [Google Scholar]
- 25.Humphrys MS, Creasy T, Sun Y, Shetty AC, Chibucos MC, Drabek EF, Fraser CM, Farooq U, Sengamalay N, Ott S, Shou H, Bavoil PM, Mahurkar A, Myers GSA. 2013. Simultaneous transcriptional profiling of bacteria and their host cells. PLoS One 8:e80597. doi: 10.1371/journal.pone.0080597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Albrecht M, Sharma CM, Dittrich MT, Müller T, Reinhardt R, Vogel J, Rudel T. 2011. The transcriptional landscape of Chlamydia pneumoniae. Genome Biol 12:R98. doi: 10.1186/gb-2011-12-10-r98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Albrecht M, Sharma CM, Reinhardt R, Vogel J, Rudel T. 2010. Deep sequencing-based discovery of the Chlamydia trachomatis transcriptome. Nucleic Acids Res 38:868–877. doi: 10.1093/nar/gkp1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Westermann AJ, Gorski SA, Vogel J. 2012. Dual RNA-seq of pathogen and host. Nat Rev Microbiol 10:618–630. doi: 10.1038/nrmicro2852. [DOI] [PubMed] [Google Scholar]
- 29.Collingro A, Toenshoff ER, Taylor MW, Fritsche TR, Wagner M, Horn M. 2005. ‘Candidatus Protochlamydia amoebophila’, an endosymbiont of Acanthamoeba spp. Int J Syst Evol Microbiol 55:1863–1866. doi: 10.1099/ijs.0.63572-0. [DOI] [PubMed] [Google Scholar]
- 30.Lagkouvardos I, Shen J, Horn M. 2014. Improved axenization method reveals complexity of symbiotic associations between bacteria and acanthamoebae. Environ Microbiol Rep 6:383–388. doi: 10.1111/1758-2229.12162. [DOI] [PubMed] [Google Scholar]
- 31.Fritsche TRT, Horn M, Wagner M, Herwig RP, Schleifer K-H, Gautom RK. 2000. Phylogenetic diversity among geographically dispersed Chlamydiales endosymbionts recovered from clinical and environmental isolates of Acanthamoeba spp. Appl Environ Microbiol 66:2613–2619. doi: 10.1128/AEM.66.6.2613-2619.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Collingro A, Tischler P, Weinmaier T, Penz T, Heinz E, Brunham RC, Read TD, Bavoil PM, Sachse K, Kahane S, Friedman MG, Rattei T, Myers GSA, Horn M. 2011. Unity in variety—the pan-genome of the Chlamydiae. Mol Biol Evol 28:3253–3270. doi: 10.1093/molbev/msr161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Domman D, Collingro A, Lagkouvardos I, Gehre L, Weinmaier T, Rattei T, Subtil A, Horn M. 2014. Massive expansion of ubiquitination-related gene families within the chlamydiae. Mol Biol Evol 31:2890–2904. doi: 10.1093/molbev/msu227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pilhofer M, Aistleitner K, Biboy J, Gray J, Kuru E, Hall E, Brun YV, VanNieuwenhze MS, Vollmer W, Horn M, Jensen GJ. 2013. Discovery of chlamydial peptidoglycan reveals bacteria with murein sacculi but without FtsZ. Nat Commun 4:2856. doi: 10.1038/ncomms3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sixt BS, Siegl A, Müller C, Watzka M, Wultsch A, Tziotis D, Montanaro J, Richter A, Schmitt-Kopplin P, Horn M. 2013. Metabolic features of Protochlamydia amoebophila elementary bodies—a link between activity and infectivity in Chlamydiae. PLoS Pathog 9:e1003553. doi: 10.1371/journal.ppat.1003553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Haider S, Wagner M, Schmid MC, Sixt BS, Christian JG, Häcker G, Pichler P, Mechtler K, Müller A, Baranyi C, Toenshoff ER, Montanaro J, Horn M. 2010. Raman microspectroscopy reveals long-term extracellular activity of chlamydiae. Mol Microbiol 77:687–700. doi: 10.1111/j.1365-2958.2010.07241.x. [DOI] [PubMed] [Google Scholar]
- 37.Domman D, Horn M. 2015. Following the footsteps of chlamydial gene regulation. Mol Biol Evol 32:3035–3046. doi: 10.1093/molbev/msv193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Greub G, Raoult D. 2002. Parachlamydiaceae: potential emerging pathogens. Emerg Infect Dis 8:625–630. doi: 10.3201/eid0806.010210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sixt BS, Heinz C, Pichler P, Heinz E, Montanaro J, Op den Camp HJM, Ammerer G, Mechtler K, Wagner M, Horn M. 2011. Proteomic analysis reveals a virtually complete set of proteins for translation and energy generation in elementary bodies of the amoeba symbiont Protochlamydia amoebophila. Proteomics 11:1868–1892. doi: 10.1002/pmic.201000510. [DOI] [PubMed] [Google Scholar]
- 40.Douglas AL, Hatch TP. 2000. Expression of the transcripts of the sigma factors and putative sigma factor regulators of Chlamydia trachomatis L2. Gene 247:209–214. doi: 10.1016/S0378-1119(00)00094-9. [DOI] [PubMed] [Google Scholar]
- 41.Niebuhr K, Giuriato S, Pedron T, Philpott DJ, Gaits F, Sable J, Sheetz MP, Parsot C, Sansonetti PJ, Payrastre B. 2002. Conversion of PtdIns(4,5)P2 into PtdIns(5)P by the S. flexneri effector IpgD reorganizes host cell morphology. EMBO J 21:5069–5078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lane BJ, Mutchler C, Al Khodor S, Grieshaber SS, Carabeo RA. 2008. Chlamydial entry involves TARP binding of guanine nucleotide exchange factors. PLoS Pathog 4:e1000014. doi: 10.1371/journal.ppat.1000014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Clifton DR, Fields KA, Grieshaber SS, Dooley CA, Fischer ER, Mead DJ, Carabeo RA, Hackstadt T. 2004. A chlamydial type III translocated protein is tyrosine-phosphorylated at the site of entry and associated with recruitment of actin. Proc Natl Acad Sci U S A 101:10166–10171. doi: 10.1073/pnas.0402829101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bertelli C, Collyn F, Croxatto A, Rückert C, Polkinghorne A, Kebbi-Beghdadi C, Goesmann A, Vaughan L, Greub G. 2010. The Waddlia genome: a window into chlamydial biology. PLoS One 5:e10890. doi: 10.1371/journal.pone.0010890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Betts-Hampikian HJ, Fields KA. 2010. The chlamydial type III secretion mechanism: revealing cracks in a tough nut. Front Microbiol 1:114. doi: 10.3389/fmicb.2010.00114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Galán JE, Lara-Tejero M, Marlovits TC, Wagner S. 2014. Bacterial type III secretion systems: specialized nanomachines for protein delivery into target cells. Annu Rev Microbiol 68:415–438. doi: 10.1146/annurev-micro-092412-155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wilson JW. 2006. Bacterial protein secretion mechanisms, p 274–320. In Nickerson CA, Schurr M (ed), Molecular paradigms of infectious disease—a bacterial perspective, 1st ed. Springer, New York, NY, US. [Google Scholar]
- 48.Fields KA, Mead DJ, Dooley CA, Hackstadt T. 2003. Chlamydia trachomatis type III secretion: evidence for a functional apparatus during early-cycle development. Mol Microbiol 48:671–683. doi: 10.1046/j.1365-2958.2003.03462.x. [DOI] [PubMed] [Google Scholar]
- 49.Slepenkin A, Motin V, de la Maza LM, Peterson EM. 2003. Temporal expression of type III secretion genes of Chlamydia pneumoniae. Infect Immun 71:2555–2562. doi: 10.1128/IAI.71.5.2555-2562.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Skipp PJS, Hughes C, McKenna T, Edwards R, Langridge J, Thomson NR, Clarke IN. 2016. Quantitative proteomics of the infectious and replicative forms of Chlamydia trachomatis. PLoS One 11:e0149011. doi: 10.1371/journal.pone.0149011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Saka HA, Thompson JW, Chen Y-S, Kumar Y, Dubois LG, Moseley MA, Valdivia RH. 2011. Quantitative proteomics reveals metabolic and pathogenic properties of Chlamydia trachomatis developmental forms. Mol Microbiol 82:1185–1203. doi: 10.1111/j.1365-2958.2011.07877.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nans A, Kudryashev M, Saibil HR, Hayward RD. 2015. Structure of a bacterial type III secretion system in contact with a host membrane in situ. Nat Commun 6:10114. doi: 10.1038/ncomms10114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pilhofer M, Aistleitner K, Ladinsky MS, König L, Horn M, Jensen GJ. 2014. Architecture and host interface of environmental chlamydiae revealed by electron cryotomography. Environ Microbiol 16:417–429. doi: 10.1111/1462-2920.12299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nans A, Saibil HR, Hayward RD. 2014. Pathogen-host reorganisation during Chlamydia invasion revealed by cryo-electron tomography. Cell Microbiol 16:1457–1472. doi: 10.1111/cmi.12310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dumoux M, Nans A, Saibil HR, Hayward RD. 2015. Making connections: snapshots of chlamydial type III secretion systems in contact with host membranes. Curr Opin Microbiol 23:1–7. doi: 10.1016/j.mib.2014.09.019. [DOI] [PubMed] [Google Scholar]
- 56.Matsumoto A. 1982. Electron microscopic observations of surface projections on Chlamydia psittaci reticulate bodies. J Bacteriol 150:358–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fields KA, Hackstadt T. 2006. The Chlamydia type III secretion system: structure and implications for pathogenesis, p 219–233. In Bavoil P, Wyrick P (ed), Chlamydia: genomics and pathogenesis. Horizon Scientific Press, ; Poole, United Kingdom. [Google Scholar]
- 58.Omsland A, Sixt BS, Horn M, Hackstadt T. 2014. Chlamydial metabolism revisited: interspecies metabolic variability and developmental stage-specific physiologic activities. FEMS Microbiol Rev 38:779–801. doi: 10.1111/1574-6976.12059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Haferkamp I, Schmitz-Esser S, Wagner M, Neigel N, Horn M, Neuhaus HE. 2006. Tapping the nucleotide pool of the host: novel nucleotide carrier proteins of Protochlamydia amoebophila. Mol Microbiol 60:1534–1545. doi: 10.1111/j.1365-2958.2006.05193.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tjaden J, Winkler HH, Schwöppe C, Van Der Laan M, Möhlmann T, Neuhaus HE. 1999. Two nucleotide transport proteins in Chlamydia trachomatis, one for net nucleoside triphosphate uptake and the other for transport of energy. J Bacteriol 181:1196–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Omsland A, Sager J, Nair V, Sturdevant DE, Hackstadt T. 2012. Developmental stage-specific metabolic and transcriptional activity of Chlamydia trachomatis in an axenic medium. Proc Natl Acad Sci U S A 109:19781–19785. doi: 10.1073/pnas.1212831109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hatch TP, Al-Hossainy E, Silverman JA. 1982. Adenine nucleotide and lysine transport in Chlamydia psittaci. J Bacteriol 150:662–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Weber J, Senior AE. 1997. Catalytic mechanism of F1-ATPase. Biochim Biophys Acta 1319:19–58. doi: 10.1016/S0005-2728(96)00121-1. [DOI] [PubMed] [Google Scholar]
- 64.Radolf JD, Samuels DS. 2010. Borrelia: molecular biology, host interaction and pathogenesis. Horizons Science Press, ; Poole, United Kingdom. [Google Scholar]
- 65.Skipp P, Robinson J, O’Connor CD, Clarke IN. 2005. Shotgun proteomic analysis of Chlamydia trachomatis. Proteomics 5:1558–1573. doi: 10.1002/pmic.200401044. [DOI] [PubMed] [Google Scholar]
- 66.Saka HA, Valdivia RH. 2010. Acquisition of nutrients by Chlamydiae: unique challenges of living in an intracellular compartment. Curr Opin Microbiol 13:4–10. doi: 10.1016/j.mib.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mehlitz A, Eylert E, Huber C, Lindner B, Vollmuth N, Karunakaran K, Goebel W, Eisenreich W, Rudel T. 2017. Metabolic adaptation of Chlamydia trachomatis to mammalian host cells. Mol Microbiol 103:1004–1019. doi: 10.1111/mmi.13603. [DOI] [PubMed] [Google Scholar]
- 68.Yao J, Abdelrahman YM, Robertson RM, Cox JV, Belland RJ, White SW, Rock CO. 2014. Type II fatty acid synthesis is essential for the replication of Chlamydia trachomatis. J Biol Chem 289:22365–22376. doi: 10.1074/jbc.M114.584185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Weiss E. 1967. Transaminase activity and other enzymatic reactions involving pyruvate and glutamate in Chlamydia (psittacosis-trachoma group). J Bacteriol 93:177–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Grubmüller S, Schauer K, Goebel W, Fuchs TM, Eisenreich W. 2014. Analysis of carbon substrates used by Listeria monocytogenes during growth in J774A.1 macrophages suggests a bipartite intracellular metabolism. Front Cell Infect Microbiol 4:156. doi: 10.3389/fcimb.2014.00156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Häuslein I, Manske C, Goebel W, Eisenreich W, Hilbi H. 2016. Pathway analysis using 13C-glycerol and other carbon tracers reveals a bipartite metabolism of Legionella pneumophila. Mol Microbiol 100:229–246. doi: 10.1111/mmi.13313. [DOI] [PubMed] [Google Scholar]
- 72.Schunder E, Gillmaier N, Kutzner E, Eisenreich W, Herrmann V, Lautner M, Heuner K. 2014. Amino acid uptake and metabolism of Legionella pneumophila hosted by Acanthamoeba castellanii. J Biol Chem 289:21040–21054. doi: 10.1074/jbc.M114.570085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Eisenreich W, Heuner K. 2016. The life stage-specific pathometabolism of Legionella pneumophila. FEBS Lett 590:3868–3886. doi: 10.1002/1873-3468.12326. [DOI] [PubMed] [Google Scholar]
- 74.Brüggemann H, Hagman A, Jules M, Sismeiro O, Dillies M-A, Gouyette C, Kunst F, Steinert M, Heuner K, Coppée J-Y, Buchrieser C. 2006. Virulence strategies for infecting phagocytes deduced from the in vivo transcriptional program of Legionella pneumophila. Cell Microbiol 8:1228–1240. doi: 10.1111/j.1462-5822.2006.00703.x. [DOI] [PubMed] [Google Scholar]
- 75.Gillmaier N, Schunder E, Kutzner E, Tlapák H, Rydzewski K, Herrmann V, Stämmler M, Lasch P, Eisenreich W, Heuner K. 2016. Growth-related metabolism of the carbon storage poly-3-hydroxybutyrate in Legionella pneumophila. J Biol Chem 291:6471–6482. doi: 10.1074/jbc.M115.693481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Clarke M, Lohan AJ, Liu B, Lagkouvardos I, Roy S, Zafar N, Bertelli C, Schilde C, Kianianmomeni A, Bürglin TR, Frech C, Turcotte B, Kopec KO, Synnott JM, Choo C, Paponov I, Finkler A, Tan CSH, Hutchins AP, Weinmeier T, Rattei T, Chu JS, Gimenez G, Irimia M, Rigden DJ, Fitzpatrick DA, Lorenzo-Morales J, Bateman A, Chiu CH, Tang P, Hegemann P, Fromm H, Raoult D, Greub G, Miranda-Saavedra D, Chen N, Nash P, Ginger ML, Horn M, Schaap P. 2013. Genome of Acanthamoeba castellanii highlights extensive lateral gene transfer and early evolution of tyrosine kinase signaling. Genome Biol 14:R11. doi: 10.1186/gb-2013-14-2-r11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Roan NR, Starnbach MN. 2008. Immune-mediated control of Chlamydia infection. Cell Microbiol 10:9–19. doi: 10.1111/j.1462-5822.2007.01069.x. [DOI] [PubMed] [Google Scholar]
- 78.Elwell C, Mirrashidi K, Engel J. 2016. Chlamydia cell biology and pathogenesis. Nat Rev Microbiol 14:385–400. doi: 10.1038/nrmicro.2016.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bastidas RJ, Valdivia RH. 2016. Emancipating Chlamydia: advances in the genetic manipulation of a recalcitrant intracellular pathogen. Microbiol Mol Biol Rev 80:411–427. doi: 10.1128/MMBR.00071-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Daims H, Stoecker K, Wagner M. 2005. Fluorescence in situ hybridisation for the detection of prokaryotes, p 213–239. In Osbourne AM, Smith CJ (ed), Advanced methods in molecular microbial ecology. BIOS Scientific Publishers, Abingdon, United Kingdom. [Google Scholar]
- 81.Poppert S, Essig A, Marre R, Wagner M, Horn M. 2002. Detection and differentiation of chlamydiae by fluorescence in situ hybridization. Appl Environ Microbiol 68:4081–4089. doi: 10.1128/AEM.68.8.4081-4089.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Heinz E, Rockey DD, Montanaro J, Aistleitner K, Wagner M, Horn M. 2010. Inclusion membrane proteins of Protochlamydia amoebophila UWE25 reveal a conserved mechanism for host cell interaction among the chlamydiae. J Bacteriol 192:5093–5102. doi: 10.1128/JB.00605-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Selinger DW, Saxena RM, Cheung KJ, Church GM, Rosenow C. 2003. Global RNA half-life analysis in Escherichia coli reveals positional patterns of transcript degradation. Genome Res 13:216–223. doi: 10.1101/gr.912603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Dreses-Werringloer U, Padubrin I, Zeidler H, Köhler L. 2001. Effects of azithromycin and rifampin on Chlamydia trachomatis infection in vitro. Antimicrob Agents Chemother 45:3001–3008. doi: 10.1128/AAC.45.11.3001-3008.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sarov I, Becker Y. 1971. Deoxyribonucleic acid-dependent ribonucleic acid polymerase activity in purified trachoma elementary bodies: effect of sodium chloride on ribonucleic acid transcription. J Bacteriol 107:593–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.MacKay RM, Doolittle WF. 1981. Nucleotide sequences of Acanthamoeba castellanii 5S and 5.8S ribosomal ribonucleic acids: phylogenetic and comparative structural analyses. Nucleic Acids Res 9:3321–3334. doi: 10.1093/nar/9.14.3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gast RJ, Ledee DR, Fuerst PA, Byers TJ. 1996. Subgenus systematics of Acanthamoeba: four nuclear 18S rDNA sequence types. J Eukaryot Microbiol 43:498–504. doi: 10.1111/j.1550-7408.1996.tb04510.x. [DOI] [PubMed] [Google Scholar]
- 89.Lai S, Henney HR. 1993. Nucleotide sequence of a portion of the 26S rDNA of Acanthamoeba. Nucleic Acids Res 21:4401. doi: 10.1093/nar/21.18.4401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Burger G, Plante I, Lonergan KM, Gray MW. 1995. The mitochondrial DNA of the amoeboid protozoon, Acanthamoeba castellanii: complete sequence, gene content and genome organization. J Mol Biol 245:522–537. doi: 10.1006/jmbi.1994.0043. [DOI] [PubMed] [Google Scholar]
- 91.Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. 2012. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup . 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Anders S, Pyl PT, Huber W. 2015. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.R Development Core Team 2011. R: a language and environment for statistical computing. R Foundation for Statistical Computing, ; Vienna, Austria. [Google Scholar]
- 95.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G, Smith C, Smyth G, Tierney L, Yang JYH, Zhang J. 2004. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Brock G, Pihur V, Datta S, Datta S. 2008. clValid: an R package for cluster validation. J Stat Softw 25:1–22. [Google Scholar]
- 98.Warnes G, Bolker B, Bonebakker L, Gentleman R, Huber W, Liaw A, Lumley T, Maechler M, Magnusson A, Moeller S. 2009. gplots: various R programming tools for plotting data. R package version 2. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- 99.Bateman A, Birney E, Cerruti L, Durbin R, Etwiller L, Eddy SR, Griffiths-Jones S, Howe KL, Marshall M, Sonnhammer EL. 2002. The Pfam protein families database. Nucleic Acids Res 30:276–280. doi: 10.1093/nar/30.1.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Huang DW, Sherman BT, Lempicki RA. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 101.Vallenet D, Labarre L, Rouy Z, Barbe V, Bocs S, Cruveiller S, Lajus A, Pascal G, Scarpelli C, Médigue C. 2006. MaGe: a microbial genome annotation system supported by synteny results. Nucleic Acids Res 34:53–65. doi: 10.1093/nar/gkj406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Jehl M-A, Arnold R, Rattei T. 2011. Effective—a database of predicted secreted bacterial proteins. Nucleic Acids Res 39:D591–D595. doi: 10.1093/nar/gkq1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M. 2005. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21:3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- 104.Young MD, Wakefield MJ, Smyth GK, Oshlack A. 2010. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol 11:R14. doi: 10.1186/gb-2010-11-2-r14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hueck CJ. 1998. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol Mol Biol Rev 62:379–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Spaeth KE, Chen Y-S, Valdivia RH. 2009. The chlamydia type III secretion system C-ring engages a chaperone-effector protein complex. PLoS Pathog 5:e1000579. doi: 10.1371/journal.ppat.1000579. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental methods, results, and discussion. Download TEXT S1, PDF file, 0.1 MB (155.6KB, pdf) .
Copyright © 2017 König et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Similarity of Protochlamydia gene expression values between replicates and time points. The Euclidean distances between all normalized expression values (reads per kilobase per million [RPKM]) of each replicate and time point are shown, with darker colors meaning high similarity. The order of the samples is based on hierarchical cluster analysis of the distances using the complete linkage method—the clustering being illustrated by the dendrograms. The samples cluster by time point and therefore correctly reflect the experimental design. For subsequent analyses, genes were considered to be expressed at a particular time point when we obtained expression values in two or three replicates. The contribution of genes for which expression was detected in only two replicates is indicated in the right panel. Relatively low sequencing coverage at 2 h postinfection (hpi) explains the higher numbers of these genes at this early time point, and consequently also the larger distance between the 2 hpi replicates. Excl., excluding. Download FIG S1, PDF file, 0.2 MB (258.8KB, pdf) .
Copyright © 2017 König et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
(A) Gene expression heatmaps of constitutively expressed genes and (B) genes involved in peptidoglycan synthesis and cell division. (A) All genes that were detected to be expressed (in at least two replicates) but were found not to be significantly differentially expressed (n = 712) were termed constitutively expressed. Average gene-wise expression values (log2 reads per kilobase per million [RPKM]) were hierarchically clustered based on Euclidean distances using the complete linkage method. Five clusters representing five different expression levels were defined on the basis of the dendrogram’s branching pattern. Accordingly, we found 9 very highly expressed genes, 60 highly expressed genes, 131 genes with medium expression strength, 328 genes with medium-low expression strength, and 184 genes with low expression. Each cluster was tested for the presence of overrepresented COG classes and clusters, KEGG pathways, and GO terms for which only categories with a false discovery rate (FDR) below 0.05 are listed. In addition, a few selected genes are shown. Corresponding proteins that were detected in P. amoebophila EBs in a previous study (B. S. Sixt, C. Heinz, P. Pichler, E. Heinz, J. Montanaro, et al., Proteomics 11:1868–1892, 2011, https://doi.org/10.1002/pmic.201000510) are indicated by black lines (n = 234). (B) Average expression values (log2 RPKM) for genes involved in peptidoglycan synthesis and cell division (N. Jacquier, P. H. Viollier, and G. Greub, FEMS Microbiol Rev 39:262–275, 2015, https://doi.org/10.1093/femsre/fuv001) were hierarchically clustered (average linkage) based on Pearson correlation distances. Genes found to be differentially expressed between any two consecutive time points are marked with an asterisk. hpi, h postinfection; extracell., extracellular. Download FIG S2, PDF file, 0.7 MB (756.5KB, pdf) .
Copyright © 2017 König et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Gene expression dynamics during the Protochlamydia developmental cycle. The intersecting gene sets of statistically significantly up- or downregulated genes between successive time point pairs are shown. Each color represents a different set of differentially expressed (DE) genes, with the height of the bar indicating the number of genes. For example, at 2 h postinfection (hpi), there are a total of 250 significantly upregulated genes of which 153 genes are downregulated at the next time point (orange bars). Note that there are overlaps between gene sets of two consecutive time points. For instance, from the orange gene set (upregulated at 2 hpi and downregulated at 48 hpi), 22 genes were subsequently also downregulated at 96 hpi (pink boxes). White areas represent DE genes that were detected as significantly regulated only once during the cycle. Download FIG S3, PDF file, 0.2 MB (238KB, pdf) .
Copyright © 2017 König et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Sequencing and read mapping statistics. Download TABLE S1, PDF file, 0.1 MB (109.4KB, pdf) .
Copyright © 2017 König et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Gene expression values and annotation of Protochlamydia and A. castellanii. Download DATA SET S1, XLSX file, 1.8 MB (1.9MB, xlsx) .
Copyright © 2017 König et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Expression patterns of genes encoding components of the type III secretion system and its (putative) effectors. All genes that met the following criteria were included: (i) genes that have been previously reported to encode components involved in Protochlamydia type III secretion (A. Collingro, P. Tischler, T. Weinmaier, T. Penz, E. Heinz, et al., Mol Biol Evol 28:3253–3270, 2011, https://doi.org/10.1093/molbev/msr161); (ii) genes that are orthologues of putative C. trachomatis effector proteins or structural components and chaperones of the chlamydial type III secretion system (H. J. Betts-Hampikian and K. A. Fields, Front Microbiol 1:114, 2010, https://doi.org/10.3389/fmicb.2010.00114); (iii) genes that were annotated to contain eukaryotic-like domains such as ankyrin repeats (ANK), Sel1-like repeats (Sel1), F-boxes (F-box), leucine-rich repeats (LRR, PEX1), tetratricopeptide repeats (TPR, PEX2), BTB/POZ domains (PEX1), and GDP-GTP exchange domains (RasGEF, RhoGEF); (iv) genes that were predicted to encode inclusion membrane proteins (put. inc) (E. Heinz, D. D. Rockey, J. Montanaro, K. Aistleitner, M. Wagner, et al., J Bacteriol 192:5093–5102, 2010, https://doi.org/10.1128/JB.00605-10); and (v) genes that encode putative effectors based on expression pattern and genomic location (data not shown). Differentially expressed genes are marked with an asterisk. Temporal classes are indicated by colored bars. The sct unified nomenclature for the naming of type III secretion system genes was used (C. J. Hueck, Microbiol Mol Biol Rev 62:379-433, 1998). RPKM, reads per kilobase per million; hpi, h postinfection; extracell., extracellular; PEX1, Protochlamydia expanded gene family 1; PEX2, Protochlamydia expanded gene family 2 (D. Domman, A. Collingro, I. Lagkouvardos, L. Gehre, T. Weinmaier, et al., Mol Biol Evol 31:2890–2904, 2014, https://doi.org/10.1093/molbev/msu227); put, putative; Ctr, C. trachomatis. Download FIG S4, PDF file, 1.3 MB (1.3MB, pdf) .
Copyright © 2017 König et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Expression of species-specific and chlamydial core genes. (A) Heatmap showing the distribution of temporarily regulated Protochlamydia genes at different levels of taxonomic conservation. The color indicates the percentage of differentially expressed (DE) genes per temporal class and taxonomic group. The absolute numbers of DE genes per temporal class and taxonomic group are indicated; the total numbers of DE genes per taxonomic group are shown in brackets. Note that the early gene set is dominated by species-specific genes (32%), whereas chlamydial core genes are most abundant at midcycle (44%). (B) Nearly all chlamydial core genes are expressed in Protochlamydia, with constitutively expressed genes contributing 62%. Download FIG S5, PDF file, 0.2 MB (223.2KB, pdf) .
Copyright © 2017 König et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Gene expression dynamics of the Acanthamoeba host during infection with Protochlamydia. Ten different temporal subclasses of genes can be distinguished (indicated by black lines) within three main classes (colored bars). Major gene expression shifts occurred early during infection (yellow) and at the peak of Protochlamydia RB activity (red). Only significantly differentially expressed genes are shown. The number of genes per temporal class is indicated. RPKM, reads per kilobase per million; hpi, h postinfection. Download FIG S6, PDF file, 1.2 MB (1.2MB, pdf) .
Copyright © 2017 König et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Overrepresented functions among A. castellanii genes. Download TABLE S2, PDF file, 0.1 MB (143.1KB, pdf) .
Copyright © 2017 König et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.