Abstract
In targeted proteomics using liquid chromatography-tandem triple quadrupole mass spectrometry (LC/MS/MS) in the selected reaction monitoring (SRM) mode, selecting the best observable or visible peptides is a key step in the development of SRM assay methods of target proteins. A direct comparison of signal intensities among all candidate peptides by brute-force LC/MS/MS analysis is a concrete approach for peptide selection. However, the analysis requires an SRM method with hundreds of transitions. This study reports on the development of a method for predicting and identifying hopeless peptides to reduce the number of candidate peptides needed for brute-force experiments. Hopeless peptides are proteotypic peptides that are unlikely to be selected for targets in SRM analysis owing to their poor ionization characteristics. Targeted proteomics data from Escherichia coli demonstrated that the relative ionization efficiency between two peptides could be predicted from sequences of two peptides, when a multivariate regression model is used. Validation of the method showed that >20% of the candidate peptides could be successfully eliminated as hopeless peptides with a false positive rate of less than 2%.
Keywords: targeted proteomics, selected reaction monitoring, multivariate regression analysis, hopeless peptide, in silico prediction
INTRODUCTION
Targeted proteomics is a method that is used to determine the abundance of target proteins in biological samples.1–3) A crude protein fraction from a biological sample is digested to produce a mixture of proteotypic peptides (PTPs). The amounts of pre-selected peptides derived from the target proteins are determined by the selected reaction monitoring (SRM) mode of liquid chromatography-tandem triple quadrupole mass spectrometry (LC/MS/MS).4) In usual SRM assay methods, 2–4 PTPs are selected for the analysis of each target protein, the amounts of which are determined by 3–4 SRM transitions per peptide.5,6) Selecting the best-observable or visible peptides is a key step in the development of the SRM assay method for the selective and sensitive analysis of target proteins. This is because numerous peptides with various lengths, sequences, and ionization efficiencies are produced when a protein is digested with trypsin. For example, in the analysis of a phosphoglucokinase in yeast (Pgk1p from Saccharomyces cerevisiae) using the SRM method, 4 suitable peptides were selected from more than 30 candidate PTPs (6–25 amino acid residues) produced by trypsin digestion (Supplementary Table S1).7,8)
After establishing comprehensive SRM assay methods, such as the SRMAtlas of human and yeast proteins,9–13) these methods could be reused owing to their basic compatibility among triple quadrupole mass spectrometers.14) However, SRM assay methods for the targeted proteome analysis of non-model organisms, such as various industrially important bacteria for biomaterial productions, are under continuous development.15,16) For the efficient development of SRM assay methods, heuristic rules have been proposed for selecting suitable peptides.17,18) In silico tools such as PeptideSieve, CONSeQuence, and PeptideRank have also been reported to predict the best-observable, visible, or flyer peptides from a sequence of the target protein.19–21) These algorithms were developed based on training data containing lists of observable peptides in shotgun proteomics datasets. However, a literature-reported SRM assay method showed that these rules do not always explain the selected peptides. For example, the selection rules recommended using peptides within 8–20 residues and to avoid peptides that contained His residues. However, 4% and 10% of the peptides violated these rules in the yeast SRM assay method for the central metabolism-related enzymes.7,8) Moreover, the selected peptides in the SRM assay method do not coincide with the results of in silico predictions. A peptide, VLENTEIGDSIFDK, which is employed in the SRM assay method for Pgk1p, is ranked at 25th and 15th by CONSeQuence and PeptideRank, respectively (Supplementary Table S1).
These results suggest that predicting the best-observable peptides still includes a measure of uncertainty, and a brute-force experiment using LC/MS/MS is the most reliable approach for selecting suitable peptides from large numbers of candidate peptides in the development of an SRM assay method.4,6,22) For example, an SRM method with more than 200 channels is required for an experimental survey of all y series product ions produced from divalent precursor ions [M+2H]2+ derived from candidate peptides of S. cerevisiae Pgk1p.
In this study, a method for predicting and identifying hopeless PTPs was investigated. Hopeless peptides refer to peptides that are unlikely to be selected as targets of SRM analysis owing to their poor signal intensity in the SRM chromatogram. The prediction of hopeless PTPs will reduce the number of candidate peptides to be investigated in a brute-force experiment. For this purpose, an SRM assay dataset was obtained from 203 lines of E. coli that overexpress central metabolism related enzymes. Using the total peak area data for 3,856 peptides derived from 203 different proteins, a multivariate regression model was constructed that permits the relative total peak areas between two peptides to be predicted. The prediction method developed in this study was able to reduce the number of candidate peptides by >20% with a false positive rate of less than 2%.
MATERIALS AND METHODS
Sample preparation
Escherichia coli K-12 strains overexpressing the central metabolism-related enzymes were obtained from the E. coli ASKA library. The ASKA library is a complete set of an E. coli K-12 ORF archive including E. coli strains that overexpress each ORF.23) Each E. coli strain was cultured in 15 mL of Luria–Bertani (LB) medium containing 30 mg/mL chloramphenicol, with shaking at 150 rpm at 37°C. When the OD600 level reached 0.3, isopropyl β-D-1-thiogalactopyranoside (IPTG, final conc. 1 mM) was added to the culture. Crude proteins were extracted from E. coli cells in the exponential growth phase (OD600=1.0) by a previously described method using a cell lysis buffer containing 50 mM Hepes (pH 7.5), 5% glycerol, 15 mM dithiothreitol, 100 mM KCl, 5 mM ethylenediamine-tetraacetic acid, and complete protease inhibitors cocktail (Roche, Basel, Switzerland, 1 droplet/50 mL).7,8) Protein concentrations were determined by the Bradford method.24) Trypsin digestion was performed by the method described by Uchida et al.17) The peptide solutions were desalted using GL-Tip GC micropipette tips (GL Science, Tokyo, Japan).
Acquisition of test dataset
For each target protein, the multiple SRM series of single charged y series product ions, produced from a [M+2H]2+ precursor ion of all tryptic peptides that were comprised of 7–30 residues, were constructed using scripts written with Perl5.6. Miss cleavages at [RK|P] sites were considered in this study. In addition to y-ions whose m/z values were larger than that of the precursor ion, additional y-ions with first, second, third largest m/z values were employed in the SRM transitions. Peptide samples (3 μL) were analyzed by the SRM method using a nano-liquid chromatography-ultrafast mass spectrometry (nanoLC-UFMS) system (LC-20ADnano and LCMS-8040, Shimadzu, Kyoto, Japan), a nanospray interface (N8040, AMR, Tokyo, Japan), and a spray tip (Fortis tip 150–20, AMR). The analytical conditions were as follows: HPLC column, L-column ODS (pore size: 5 μm, 0.1×150 mm, CERI, Tokyo, Japan); trap column, L-column ODS (pore size: 5 μm, 0.3×5 mm, CERI); solvent system, acetonitrile (0.1% formic acid) : water (0.1% formic acid); gradient program, 10 : 90, v/v at 0 min, 10 : 90 at 10 min, 40 : 60 at 45 min, 95 : 5 at 55 min, and 90 : 10 at 65 min; and flow rate, 400 nL/min. MS detection parameters were as follows: interface temperature, 350°C; DL temperature, 120°C; heat block temperature, 200°C; drying gas flow, off; CID gas pressure, 290; interface voltage, +1.6 keV; and detection mode, MRM positive.14) The data were recorded with the aid of LabSolutions LCMS version 5.6 (Shimadzu). Chromatographic data was processed using Skyline version 3.1.25)
Data analyses
Multivariate regression analyses were performed by lm and step functions on R3.1.3. The AAindex (amino acid index) dataset was obtained from the KEGG database (http://www.genome.jp/aaindex/).26) Other data processing was performed by scripts written with Perl5.6.
RESULTS AND DISCUSSION
Hopeless proteotypic peptides
Three types of proteotypic peptides (PTPs)—suitable, promising, and hopeless—are introduced in this study. For the case of the Pgk protein in Escherichia coli (UniProt ID: P0A799), an in silico analysis using the amino acid sequence (504 aa) indicated that 19 PTPs within 7–30 residues were produced by trypsin digestion (Table 1). To compare signal intensities among the PTPs, a tryptic peptide sample was prepared from an E. coli strain over-expressing Pgk and analyzed by LC/MS/MS using a brute-force approach (Fig. 1). The signal intensity of the peptide was determined as the total peak area of multiple SRM series of single charged y-series product ions produced from a precursor ion [M+2H]2+ (See Materials and Methods). An SRM analysis showed that a signal derived from SLYEADLVDEAK was one of the most intense signals among the 19 candidate peptides (Table 1). The signal intensities of the candidate peptides were not correlated with the ranks predicted by CONSeQuence and PeptideRank20,21) (Table 1). These results suggest that the brute-force experiment using LC/MS/MS is promising in terms of developing a new SRM assay method. As mentioned above, 2–4 ‘suitable’ peptides for the SRM assay method were selected considering their signal intensity, retention time, and overlap with interfering peaks. Here, the peptides whose total peak areas were more than 20% of that of the most intense peptide, were considered to be ‘promising’ candidates for use in SRM assay methods. For example, the literature-reported SRM assay method selected two suitable peptides, VATEFSETAPATLK and LTVLDSLSK, from the list of promising peptides.16)
Table 1. Tryptic peptides derived from Escherichia coli Pgk protein.
| Peptides1) | Total peak area2) | Rank by Peptide-Rank3) | Rank by CONSeQuence4) | Predicted hopeless peptide5) |
|---|---|---|---|---|
| SLYEADLVDEAK | 176134453 | 11 | 7 | |
| VLPAVAMLEER | 144832556 | 6 | 8 | |
| VATEFSETAPATLK | 143831394 | 3 | 3 | |
| ASLPTIELALK | 134061614 | 2 | 6 | |
| FADVACAGPLLAAELDALGK | 127059698 | 9 | 1 | |
| ADLNVPVK | 101454545 | 15 | 17 | |
| LTVLDSLSK | 91342029 | 12 | 13 | |
| TILWNGPVGVFEFPNFR | 49526961 | 5 | 5 | |
| ADEQILDIGDASAQELAEILK | 46055365 | 16 | 4 | |
| ISYISTGGGAFLEFVEGK | 42893285 | 10 | 10 | |
| LLTTCNIPVPSDVR | 42865623 | 4 | 2 | |
| MTDLDLAGK | 38311752 | 7 | 18 | |
| DYLDGVDVAEGELVVLENVR | 31981496 | 17 | 12 | |
| YAALCDVFVMDAFGTAHR | 4227097 | 14 | 11 | hopeless |
| EPARPMVAIVGGSK | 1364859 | 1 | 16 | |
| IADQLIVGGGIANTFIAAQGHDVGK | 703672 | 8 | 9 | hopeless |
| DDETLSK | 132247 | 19 | 19 | |
| AQASTHGIGK | 106236 | 13 | 14 | hopeless |
| VMVTSHLGRPTEGEYNEEFSLLPVVNYLK | 1000 | 18 | 15 | hopeless |
1) Peptides less than 7 and more than 30 residues were removed from the candidates. 2) A total peak area of multiple SRM series of all y series product ions produced from a precursor ion [M+2H]2+. 3) Bartonella henselae was selected as the model organism. 4) Predicted from the result of score mode. 5) Threshold for an S score of more than 4.
Fig. 1. Selected reaction monitoring (SRM) chromatogram of 19 tryptic peptides with residues in the range of 7–30 derived from phosphoglucokinase in Escherichia coli by nano-liquid chromatography-ultrafast mass spectrometry. A crude peptide sample was prepared from an E. coli strain over-expressing Pgk. Signal intensities of peptides were determined from total peak areas of multiple SRM series of single charged y-series product ions produced from a precursor ion [M+2H]2+.
In this study, PTPs whose total peak areas were less than 20% of that of the most intense peptide were defined as ‘hopeless’ candidates for SRM assay methods. More strictly, the total peak area of hopeless peptide was required to be less than that of four or more other peptides, since 3–4 peptides are typically employed for the SRM assay methods.17) This indicates that there would be no hopeless candidates in the case of a small protein. For example, the YAALCDVFVMDAFGTAHR peptide from Pgk is a hopeless peptide, since its signal intensity was only 2.4% of the most intense peptide (Table 1). The findings also indicate that a sequence-based prediction of hopeless peptides would reduce the number of candidate peptides investigated by the brute-force experiment. It was also suggested that false positives should be avoided in the prediction, since suitable peptides would be overlooked by an error to consider a promising peptide as being classified as hopeless.
Construction of the multivariate regression model
A multivariate regression analysis was conducted to predict hopeless peptides from amino acid sequences. In this study, the total peak area determined by the SRM series of multiple y-ions produced from [M+2H]2+ was considered. The reason for this is that 82% and 100% of the literature report SRM methods for yeast and E. coli consist of transitions of y-ions produced from [M+2H]2+, respectively.7,8,16) The total peak area (the sum of the peak areas of all SRM transitions) was employed to represent the entire ionization efficiency of the peptides. A test dataset was obtained from the 203 lines of E. coli23) overexpressing the central metabolism-related enzymes (Supplementary Table S2). For each enzyme, an E. coli strain over-expressing the target protein was cultured in synthetic medium, from which a crude protein extract was obtained. Following the preparation of a tryptic peptide sample by reduction, alkylation and protease digestion, the total peak area of all tryptic peptides produced from the overexpressed protein were determined by the SRM series of multiple y-ions produced from [M+2H]2+ (Supplementary Table S2). Total peak area values were determined for all 3856 target peptides from 203 separate LC/MS/MS analyses for the 203 proteins. The values for the undetectable peptides were imputed with a value close to the noise level (1000) (Supplementary Table S2). Using the total peak area values as an objective variable, a multivariable regression model was constructed as follows:
| (1) |
where, Ix and Lx indicate the total peak area and the length of peptide x, respectively. NAA, x is the number of amino acid residues AA in the peptide x. Pp, x indicates values of parameter p of peptide x. aAA and bp are coefficients, as shown in Table 2. Parameters (P) were selected from the amino acid index (AAindex) considering Akaike information criterion (AIC) level.26) The coefficient of determination (R2) of the prediction model was 0.567. A comparison between the predicted and measured total peak area Ix (Fig. 2a) suggests that a direct prediction of the total peak area was difficult when the regression model was used. This can be attributed to a variation in the overexpressed protein levels among the samples of the training dataset.
Table 2. Coefficients of multivariate regression analysis.
| Terms | Total peak area | Relative total peak areas | ||
|---|---|---|---|---|
| Coefficient | p-Value | Coefficient | p-Value | |
| Intercept | 27.6 | <0.001 | −0.0211 | 0.0714 |
| Amino acid (AA) | ||||
| A | −1.87 | <0.001 | −1.48 | <0.001 |
| C | — | −5.92 | <0.001 | |
| D | — | 1.21 | <0.001 | |
| F | 9.67 | <0.001 | 7.80 | <0.001 |
| G | — | −2.49 | <0.001 | |
| I | 8.10 | <0.001 | 6.72 | <0.001 |
| K | −53.0 | <0.001 | −60.5 | <0.001 |
| L | 9.32 | <0.001 | 8.18 | <0.001 |
| M | −3.65 | <0.001 | −3.03 | <0.001 |
| N | −2.17 | <0.001 | −2.13 | <0.001 |
| P | 2.69 | 0.002 | 1.81 | <0.001 |
| R | −52.5 | <0.001 | −48.9 | <0.001 |
| S | −2.45 | 0.002 | −2.42 | <0.001 |
| T | −2.21 | 0.007 | −1.85 | <0.001 |
| V | 6.09 | <0.001 | 3.49 | <0.001 |
| W | 15.5 | <0.001 | 3.04 | <0.001 |
| Y | 4.28 | <0.001 | 5.10 | <0.001 |
| Parameters (P) | ||||
| Number of hydrophobic residue in N-terminal | −4.40 | <0.001 | −2.34 | <0.001 |
| Miss cleavage (KP and RP) | 4.97 | 0.049 | 15.7 | <0.001 |
| Peptide length | — | −1.21 | <0.001 | |
| Positive charge26) | −1.63 | <0.001 | −1.63 | <0.001 |
| Absolute entropy26) | −0.00913 | <0.001 | — | |
| Partition coefficient26) | −0.284 | <0.001 | — | |
| Retention coefficient in TFA26) | 0.0151 | <0.001 | — | |
| The number of bonds in the longest chain26) | — | −0.0942 | <0.001 | |
| Activation Gibbs energy of unfolding, pH 7.026) | — | 0.0476 | <0.001 | |
Fig. 2. Multivariate regression analyses for predicting the total peak area of peptides. (a) A comparison between total peak area predicted by Eq. (1) (Ix′) and measured data (Ix). (b) A comparison between relative total peak area predicted by Eq. (2) ((Ix/Iy)′) and measured data (Ix/Iy). Coefficients of determination (R2) of the prediction model were also represented.
Thus, the relative total peak areas among the two peptides derived from an identical protein were used as an objective variable to develop the following modified model:
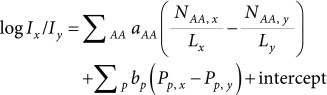 |
(2) |
Here, x and y indicate two peptides derived from an identical protein. The value of R2 of the prediction model was 0.658, indicating that, when the relative total peak area among two peptides was employed, the prediction was improved (Fig. 2b). Factors correlated to ionization efficiency could be estimated from the results of multivariate regression (Table 2). In addition to the so called hydrophobic residues (I, L, and V), aromatic amino acids (F, W, and Y) are also preferable for ionization efficiency. However, the positive charge derived from R, and K has a strong negative effect, probably due to the formation of multiple charged ions. The negative coefficient for M suggests that peptide abundance could be affected by the partial oxidation of methionine side chain. Furthermore, several factors related to peptide length, steric conformation, and hydrophobicity also contribute to ionization efficiency, as suggested in previous studies.19,27)
Prediction of hopeless peptide
Since the regression model, Eq. (2), predicts a relative total peak area between two peptides, a heuristic procedure was employed for selecting hopeless peptides as follows:
All sequences of proteotypic peptides within 7–30 residues were generated from a sequence of the target protein by in silico trypsin digestion.
The values of predicted relative total peak area (log(Ix/Iy)′) were calculated between peptide x and all other peptides y, using Eq. (2). The values were compared with a threshold value (thres) to determine the score (S) of peptide x, as the number of cases with log(Ix/Iy)′<thres. A peptide with a larger S would be hopeless because the total peak area of this peptide was significantly smaller than that of many other PTPs.
A peptide was considered to be hopeless if its S score was larger than 0.2×N. Here, N is the total number of proteotypic peptides produced from a target protein. In the case of a small protein (0.2×N<4), a peptide with S>4 was considered to be hopeless, since 3–4 peptides are employed for the SRM assay methods.17)
Since there is one variable (thres) in the procedure, a relationship between thres, total number of false positives, and the total number of predicted hopeless peptides was investigated. In the case of predicting the hopeless peptides for 203 E. coli proteins used in the training dataset, 33.1% (1275/3856) of the PTPs were predicted to be hopeless when the threshold level was thres=−2.0. A comparison with the predicted hopeless peptides and the measured promising peptides indicated that the false positive rate was 3.0% (50/1645). When a more rigorous threshold such as thres=−2.5 was employed, 27.1% (=1045/3856) of the total PTPs were still predicted to be hopeless, and the number of false positive rate was reduced 1.1% (18 cases in total) (Supplementary Table S3).
The identical 203 E. coli proteins were also analyzed by the CONSeQuence web tool to predict hopeless peptides. The results showed that 20.4% (=787/3856) of the PTPs were predicted to be hopeless (with CONS levels=0). However, a relatively large false positive rate (18.1%=297/1645) was found (Supplementary Table S3). The results suggest that the method developed in this study is capable of efficiently removing hopeless peptides from candidate PTPs with a low false positive rate.
Validation by other datasets
The prediction method was also validated using by the literature reported SRM assay methods for 393 proteins of E. coli.16) The prediction of hopeless peptides for the 393 proteins of E. coli by the developed method showed that 23.3% (1952/8371) of tryptic peptide can be classified as hopeless with a threshold level at thres=−2.5 (Table 3). A comparison of the predicted hopeless peptides with the literature reported SRM assay method revealed that the false positive rate was 0.4% (3/670 suitable peptides), although the training and validation datasets were obtained using different mass spectrometers (Shimadzu LCMS8040 and Sciex5500 QTrap, respectively).
Table 3. Performance of the prediction method.
| thres | Total number of peptides | Number of predicted hopeless peptide | Number of promising or suitable peptides for SRM assay | Number of false positive hits | False positive rate (%) |
|---|---|---|---|---|---|
| E. coli (203 proteins, Shimadzu LCMS8040, this study) | |||||
| −2 | 3856 | 12751) | 1645 from [M+2H]2+ 2) | 50 | 3.0 |
| −2.5 | 3856 | 10451) | 1645 from [M+2H]2+ 2) | 18 | 1.1 |
| −3 | 3856 | 8691) | 1645 from [M+2H]2+ 2) | 6 | 0.4 |
| E. coli (394 proteins, Sciex 5500QTAP)16) | |||||
| −2.5 | 8371 | 19471) | 670 from [M+2H]2+ 3) | 3 | 0.4 |
| Synechocystis sp. PCC 6803 (106 proteins, ThermoScientific, TSQ Vantage)15) | |||||
| −2.5 | 1919 | 4701) | 252 from [M+2H]2+ & [M+3H]3+ 3) | 11 | 4.3 |
| 216 from [M+2H]2+ 3) | 1 | 0.5 | |||
| 36 from [M+3H]3+ 3) | 10 | 27.8 | |||
| S. cerevisiae (204 proteins, Sciex 4000QTRAP)7) | |||||
| −2.5 | 4716 | 12161) | 411 from [M+2H]2+ & [M+3H]3+ 3) | 32 | 7.8 |
| 331 from [M+2H]2+ 3) | 1 | 0.3 | |||
| 80 from [M+3H]3+ 3) | 31 | 38.8 | |||
1) Numbers of hopeless peptides determined by the method developed in this study. 2) Numbers of promising peptides. 3) Numbers of suitable peptides employed in the literature reported SRM assay methods.
In the case of the SRM assay methods for 106 proteins in a model cyanobacteria (Synechocystis sp. PCC 6803, constructed for Thermo Scientific TSQ Vantage),15) two types of precursor ions, including [M+2H]2+ and [M+3H]3+, were employed for the analysis of suitable peptides. The numbers of suitable peptides selected for the SRM assay from [M+2H]2+ and [M+3H]3+ were 216 and 36, respectively.
The results showed that 24.5% (470/1919) of the tryptic peptides was predicted to be hopeless with a threshold level at thres=−2.5 (Table 3). A comparison between the predicted hopeless peptides and suitable peptides reported in the literature indicated that the false positive rate was 0.5% (1/216 suitable peptides) when suitable peptides analyzed using [M+2H]2+ were considered. On the contrary, the false positive rate increased to 27.8% (10/36) for suitable peptides analyzed using [M+3H]3+. This is because the training dataset only includes data derived from [M+2H]2+. Similar trends were also observed for the literature-reported SRM assay methods for S. cerevisiae enzymes developed by Sciex 4000QTRAP (Table 3).7)
CONCLUSION
In this study, a method for predicting hopeless peptides was investigated using a test dataset including total peak area values for 3,856 peptides derived from 203 E. coli proteins. The method developed in this study successfully predicted hopeless peptides without suitable peptides being overlooked. This indicates that the number of SRM channels required for a brute-force experiment could be decreased by >20% with a false positive rate of less than 2%. The required number of SRM channels could be further reduced by development of more efficient prediction methods by introducing a more sophisticated regression model using larger amounts of training data, and considering additional multivalent ions such as [M+3H]3+ and the contribution of other product ions, such as b series ions.
Acknowledgments
We wish to thank Mr. Ichiro Hirano and Dr. Tairo Ogura (Shimadzu Co., Kyoto, Japan) for the technical supports to this work. This study was supported, in part, by JSPS KAKENHI Grant Number 16H06559 and based on results obtained from a project commissioned by the New Energy and Industrial Technology Development Organization (NEDO).
Mass Spectrom (Tokyo) 2017; 6(1): A0056
Supplementary Data
References
- 1) M. Bantscheff, S. Lemeer, M. M. Savitski, B. Kuster. Quantitative mass spectrometry in proteomics: Critical review update from 2007 to the present. Anal. Bioanal. Chem. 404: 939–965, 2012. [DOI] [PubMed] [Google Scholar]
- 2) S. Ohtsuki, C. Ikeda, Y. Uchida, Y. Sakamoto, F. Miller, F. Glacial, X. Decleves, J. M. Scherrmann, P. O. Couraud, Y. Kubo, M. Tachikawa, T. Terasaki. Quantitative targeted absolute proteomic analysis of transporters, receptors and junction proteins for validation of human cerebral microvascular endothelial cell line hCMEC/D3 as a human blood-brain barrier model. Mol. Pharm. 10: 289–296, 2013. [DOI] [PubMed] [Google Scholar]
- 3) P. Picotti, R. Aebersold. Selected reaction monitoring-based proteomics: Workflows, potential, pitfalls and future directions. Nat. Methods 9: 555–566, 2012. [DOI] [PubMed] [Google Scholar]
- 4) K. Bluemlein, M. Ralser. Monitoring protein expression in whole-cell extracts by targeted label- and standard-free LC-MS/MS. Nat. Protoc. 6: 859–869, 2011. [DOI] [PubMed] [Google Scholar]
- 5) C. M. Colangelo, L. Chung, C. Bruce, K. H. Cheung. Review of software tools for design and analysis of large scale MRM proteomic datasets. Methods 61: 287–298, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6) Y. Mohammed, D. Domanski, A. M. Jackson, D. S. Smith, A. M. Deelder, M. Palmblad, C. H. Borchers. PeptidePicker: A scientific workflow with web interface for selecting appropriate peptides for targeted proteomics experiments. J. Proteomics 106: 151–161, 2014. [DOI] [PubMed] [Google Scholar]
- 7) R. Costenoble, P. Picotti, L. Reiter, R. Stallmach, M. Heinemann, U. Sauer, R. Aebersold. Comprehensive quantitative analysis of central carbon and amino-acid metabolism in Saccharomyces cerevisiae under multiple conditions by targeted proteomics. Mol. Syst. Biol. 7: 464, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8) P. Picotti, B. Bodenmiller, L. N. Mueller, B. Domon, R. Aebersold. Full dynamic range proteome analysis of S. cerevisiae by targeted proteomics. Cell 138: 795–806, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9) P. Picotti, M. Clement-Ziza, H. Lam, D. S. Campbell, A. Schmidt, E. W. Deutsch, H. Rost, Z. Sun, O. Rinner, L. Reiter, Q. Shen, J. J. Michaelson, A. Frei, S. Alberti, U. Kusebauch, B. Wollscheid, R. L. Moritz, A. Beyer, R. Aebersold. A complete mass-spectrometric map of the yeast proteome applied to quantitative trait analysis. Nature 494: 266–270, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10) P. Picotti, H. Lam, D. Campbell, E. W. Deutsch, H. Mirzaei, J. Ranish, B. Domon, R. Aebersold. A database of mass spectrometric assays for the yeast proteome. Nat. Methods 5: 913–914, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11) U. Kusebauch, D. S. Campbell, E. W. Deutsch, C. S. Chu, D. A. Spicer, M. Y. Brusniak, J. Slagel, Z. Sun, J. Stevens, B. Grimes, D. Shteynberg, M. R. Hoopmann, P. Blattmann, A. V. Ratushny, O. Rinner, P. Picotti, C. Carapito, C. Y. Huang, M. Kapousouz, H. Lam, T. Tran, E. Demir, J. D. Aitchison, C. Sander, L. Hood, R. Aebersold, R. L. Moritz. Human SRMAtlas: A resource of targeted assays to quantify the complete human proteome. Cell 166: 766–778, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12) U. Kusebauch, E. W. Deutsch, D. S. Campbell, Z. Sun, T. Farrah, R. L. Moritz. Using PeptideAtlas, SRMAtlas, and PASSEL: Comprehensive resources for discovery and targeted proteomics. Curr. Protoc. Bioinformatics 46: 13.25.1–13.25.28, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13) M. Matsumoto, F. Matsuzaki, K. Oshikawa, N. Goshima, M. Mori, Y. Kawamura, K. Ogawa, E. Fukuda, H. Nakatsumi, T. Natsume, K. Fukui, K. Horimoto, T. Nagashima, R. Funayama, K. Nakayama, K. I. Nakayama. A large-scale targeted proteomics assay resource based on an in vitro human proteome. Nat. Methods 14: 251–258, 2017. [DOI] [PubMed] [Google Scholar]
- 14) F. Matsuda, T. Ogura, A. Tomita, I. Hirano, H. Shimizu. Nano-scale liquid chromatography coupled to tandem mass spectrometry using the multiple reaction monitoring mode based quantitative platform for analyzing multiple enzymes associated with central metabolic pathways of Saccharomyces cerevisiae using ultra fast mass spectrometry. J. Biosci. Bioeng. 119: 117–120, 2015. [DOI] [PubMed] [Google Scholar]
- 15) L. Vuorijoki, J. Isojarvi, P. Kallio, P. Kouvonen, E. M. Aro, G. L. Corthals, P. R. Jones, D. Muth-Pawlak. Development of a quantitative SRM-based proteomics method to study iron metabolism of Synechocystis sp. PCC 6803. J. Proteome Res. 15: 266–279, 2016. [DOI] [PubMed] [Google Scholar]
- 16) T. S. Batth, P. Singh, V. R. Ramakrishnan, M. M. Sousa, L. J. Chan, H. M. Tran, E. G. Luning, E. H. Pan, K. M. Vuu, J. D. Keasling, P. D. Adams, C. J. Petzold. A targeted proteomics toolkit for high-throughput absolute quantification of Escherichia coli proteins. Metab. Eng. 26: 48–56, 2014. [DOI] [PubMed] [Google Scholar]
- 17) Y. Uchida, M. Tachikawa, W. Obuchi, Y. Hoshi, Y. Tomioka, S. Ohtsuki, T. Terasaki. A study protocol for quantitative targeted absolute proteomics (QTAP) by LC-MS/MS: Application for inter-strain differences in protein expression levels of transporters, receptors, claudin-5, and marker proteins at the blood-brain barrier in ddY, FVB, and C57BL/6J mice. Fluids Barriers CNS 10: 21, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18) J. Kamiie, S. Ohtsuki, R. Iwase, K. Ohmine, Y. Katsukura, K. Yanai, Y. Sekine, Y. Uchida, S. Ito, T. Terasaki. Quantitative atlas of membrane transporter proteins: Development and application of a highly sensitive simultaneous LC/MS/MS method combined with novel in-silico peptide selection criteria. Pharm. Res. 25: 1469–1483, 2008. [DOI] [PubMed] [Google Scholar]
- 19) P. Mallick, M. Schirle, S. S. Chen, M. R. Flory, H. Lee, D. Martin, J. Ranish, B. Raught, R. Schmitt, T. Werner, B. Kuster, R. Aebersold. Computational prediction of proteotypic peptides for quantitative proteomics. Nat. Biotechnol. 25: 125–131, 2007. [DOI] [PubMed] [Google Scholar]
- 20) C. E. Eyers, C. Lawless, D. C. Wedge, K. W. Lau, S. J. Gaskell, S. J. Hubbard. CONSeQuence: Prediction of reference peptides for absolute quantitative proteomics using consensus machine learning approaches. Mol. Cell. Proteomics 10: M110.003384, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21) E. Qeli, U. Omasits, S. Goetze, D. J. Stekhoven, J. E. Frey, K. Basler, B. Wollscheid, E. Brunner, C. H. Ahrens. Improved prediction of peptide detectability for targeted proteomics using a rank-based algorithm and organism-specific data. J. Proteomics 108: 269–283, 2014. [DOI] [PubMed] [Google Scholar]
- 22) M. S. Bereman, B. MacLean, D. M. Tomazela, D. C. Liebler, M. J. MacCoss. The development of selected reaction monitoring methods for targeted proteomics via empirical refinement. Proteomics 12: 1134–1141, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23) M. Kitagawa, T. Ara, M. Arifuzzaman, T. Ioka-Nakamichi, E. Inamoto, H. Toyonaga, H. Mori. Complete set of ORF clones of Escherichia coli ASKA library (a complete set of E. coli K-12 ORF archive): Unique resources for biological research. DNA Res. 12: 291–299, 2005. [DOI] [PubMed] [Google Scholar]
- 24) M. M. Bradford. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72: 248–254, 1976. [DOI] [PubMed] [Google Scholar]
- 25) B. MacLean, D. M. Tomazela, N. Shulman, M. Chambers, G. L. Finney, B. Frewen, R. Kern, D. L. Tabb, D. C. Liebler, M. J. MacCoss. Skyline: An open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 26: 966–968, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26) S. Kawashima, M. Kanehisa. AAindex: Amino acid index database. Nucleic Acids Res. 28: 374, 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27) W. S. Sanders, S. M. Bridges, F. M. McCarthy, B. Nanduri, S. C. Burgess. Prediction of peptides observable by mass spectrometry applied at the experimental set level. BMC Bioinformatics 8(Suppl. 7): S23, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



![Fig. 1. Selected reaction monitoring (SRM) chromatogram of 19 tryptic peptides with residues in the range of 7–30 derived from phosphoglucokinase in Escherichia coli by nano-liquid chromatography-ultrafast mass spectrometry. A crude peptide sample was prepared from an E. coli strain over-expressing Pgk. Signal intensities of peptides were determined from total peak areas of multiple SRM series of single charged y-series product ions produced from a precursor ion [M+2H]2+.](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/c4d7/5451515/4fa343e9c28a/massspectrometry-6-1-A0056-figure01.jpg)
