The nuclear factor κB (NF-κB) pathway
Increased basal and chronic low-level activation of inflammation in patients with autoimmune disease has prompted many research lines trying to unravel the genetic control of the inflammatory cascades, being located at the forefront of such physiological disturbances. Accordingly, several abnormalities have been identified in cytokine-signaling pathways and immune receptors conducting the inflammatory response that commonly characterizes autoimmune reaction. Among the molecules involved in this kind of pathologies we identify Jak/Stat (1), Jun/Fos, p38, ERK1/3, and many others serine/threonine kinases involved in the MAP kinase transduction signal (2), and the NF-κB (3).
Multiple sclerosis (MS) exhibits many of the hallmarks of an inflammatory disease, including enhanced canonical and/or noncanonical NF-κB signaling, breakdown of the blood-brain barrier, and spontaneous recruitment of lymphocytes and macrophages to the lesion sites, which are multiple and abundant in the brain stem and spinal cord (4). The MS destructive process is an autoimmunity mixture of immune dysregulation, the regulatory failure and/or loss of tolerance more likely initiated by an inflammatory process that develops to a neurodegenerative component, which may progress independently of (or concomitantly to) the inflammatory cascade conducted through NF-κB. An alternative hypothesis proposes that MS is a neurodegenerative process exacerbated by sustained inflammatory activity (5). Being considered upstream to the primary activation of these disturbances, NF-κB is recognized as a proinflammatory cytokine response element located at the forefront of such impairment, according to its role into inflammatory pathways, controlling the activation, proliferation, and cytokine release following immune response, including (but not limited to) the focal inflammatory demyelinating lesions that, in MS, are characterized by perivascular infiltrates containing monocytes, a significant number of macrophages, and both T and B cells (6).
NF-κB is a DNA-binding transcriptional factor complex composed of a variety of homo and heterodimers of the subunits p50, p52, c-Rel, RelA (p65), and RelB, which interacts with promoter areas in proinflammatory genes (7). Many insults and inflammatory cascades can lead to the assembly and activation of NF-κB signaling, which, in turn, upregulates expression of genes for innate immunity and damage repair (8). The non-canonical NF-κB pathway is related to molecules that lead the activation of p52/RelB NF-κB heterodimer. The canonical p50/p65 NF-κB signaling involves IκBα phosphorylation by I kappa B kinases (IKK), triggering the subsequent ubiquitination and proteasomal degradation that allows subsequent nuclear translocation of NF-κB complexes (9) (Figure 1).
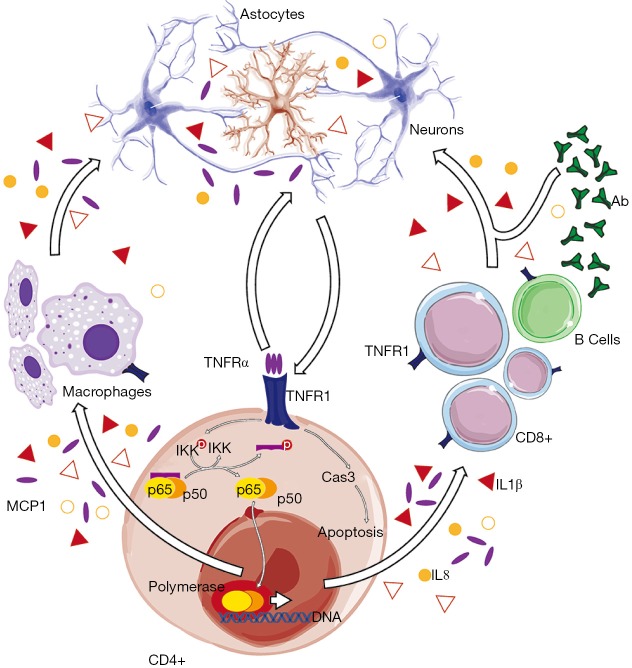
Figure 1.
The hideous loop. CD4+ is a subset of T cells that recruit many other immune cells to assist at the primary site of infection, tissue remodeling and/or damage, such as CD8+ cytotoxic T cells, B cells, astrocytes, and macrophages in CNS during inflammatory events. Then, the inflammatory cascade is amplified by proinflammatory cytokines released by these cells, further increasing cell recruitment, promoting activation of resident microglial cells, and compromising blood brain barrier. TNFR1 is expressed in most cell types and can be activated by binding of soluble TNFα. TNFR1 is located upstream to the canonical p50/p65 NF-κB pathway, which, in turn, involves IκBα phosphorylation by IKK, triggering the subsequent ubiquitination and proteasomal degradation that allows subsequent nuclear translocation of NF-κB complexes. Nuclear p50/p65 NF-κB promotes expression of genes codifying for powerful chemokines, such as the MCP1 and IL8, and many cytokines, including IL1β, and TNFα, maintaining and even worsen the inflammatory cascade in CNS. CNS, central nervous system; NF-κB, nuclear factor κB; IKK, I kappa B kinases; MCP1, monocyte chemoattractant protein-1; IL8, interleukin-8; IL1β, interleukin-1β; TNFα, tumor necrosis factor α; TNFR1, TNF receptor 1.
A number of studies have demonstrated that blood mononuclear cells from relapsing-remitting MS patients exhibit increased activation of the canonical p50/p65 NF-κB pathway. A predominant consequence of the increased NF-κB inflammatory signaling is either neuroprotective or deleterious effects, depending on the strength of the signal and the type of NF-κB dimers that are activated (10). Canonical p50/p65 NF-κB signaling is critical for immune responses downstream of T and B cell and toll-like receptors, cell adhesion molecules, enzymes, chemokines such as the monocyte chemoattractant protein-1 (MCP1) and interleukin-8 (IL8), and many cytokines, including interleukin-1β (IL1β) and tumor necrosis factor α (TNFα) (Figure 1). Clinical consequences of chronic inflammatory response activation include sustained cell and tissue remodeling, damage in the primary site, and the involvement of distant organs and tissues (9). Any change in NF-κB signaling pathway at the basal state can alter the immune response and enhance susceptibility of autoimmune diseases.
Genetic variations affecting NF-κB in MS
Many epidemiological and genome-wide association studies (GWAS) have been conducted trying to unravel autoimmune disease-relevant genomic regions affecting MS, but interpretation of the data generated is still far from conclusive. Epidemiological data have demonstrated that genetic factors are responsible for the substantially increased frequency of MS seen in the relatives of affected patients diagnosed with autoimmunity (11). Concordance rates of 26% in monozygotic twins compared with 2.3% in dizygotic twins emphasize the relevance of the genetic contribution in MS (12), in spite of the skepticism that meta-analyses have raised later (13). The huge effort in this sense is aimed at defining the underlying genetic factors and causal genes that may enable the development of strategies to prevent and even cure MS. So far, only a number of genetic changes located in genes coding for cytokine pathway and signal transduction have been linked to this disease (14), with no conclusive detail in which prevails over the others. Thereby, the genetic influences are believed to be exerted by several genes, which, in combination with the clinical heterogeneity of MS, is a major obstacle for genetic dissection of the disease.
Because NF-κB signaling is triggered by different environmental stimuli and located at the crosstalk of multiple signaling pathways involved in inflammation and autoimmune disease (15), sustained NF-κB activation may represent a critical intersection between genetic and environmental factors resulting in MS. However, the impact of autoimmunity-associated genetic variants on the biological function of this transcription factor is unveiled yet, and requires further investigations. Noteworthy, several variants affecting autoimmune disturbances and immune activation fall within signaling molecules involved in the NF-κB inflammatory cascade (16), suggesting that basal alterations in the whole pathway, rather than punctual changes in individual genes, may be at the forefront of increased susceptibility for autoimmune diseases (17). These genetic variations include (but are not limited to) some variants of the NF-κB1 and the tumor necrosis factor receptor 1 (TNFR1), which is located upstream to the canonical p50/p65 NF-κB pathway (Figure 1).
One of the strongest genome-wide association signals related to inflammation lies on chromosome 4, near the NF-κB gene (18), the span region that encodes both NF-κB and mannosidase beta. This region contains more than 90 genetic variants that are in linkage disequilibrium, making the identification of the likely causal variants challenging in the context of disease. Moreover, the region lacks protein-altering variants, so gene-regulatory studies aimed at predicting diverse and conflicting target genes, tissues, and different cell types are required.
Although the immunometabolic regulatory capacity of pathogen-sensing systems downstream NF-κB activated under non-pathological circumstances is not understood, an important aspect of immune dysfunction in MS patients is the inability to maintain T cells homeostasis (19). In this respect, genetic alterations in NF-κB pathway have been associated with many inflammatory diseases and malignancies (20). Concurrently, increased basal activation of NF-κB in naïve CD4 T cells from patients with MS has prompted several research lines trying to unveil the genetic control of NF-κB signaling at the forefront of autoimmune inflammatory diseases and T-cell activity deregulation.
To determine whether sustained NF-κB basal activation in patients with MS susceptibility is due to genetic variations found in the NF-κB signaling cascade, Housley and coworkers have assessed the resilience of naïve CD4 T cells from different donors to activation ex vivo with TNFα and phorbol 12-myristate 13-acetate. Firstly, these authors validated that naïve CD4 T cells from patients with MS exhibit increased IκBα degradation and higher phospho-p65 and NF-κB activation than age-matched healthy controls, and that this increased activation is mitigated by treatments affecting NF-κB (18). Then, they investigated whether stimulated circulating immune cells from subjects with different genotypes at rs228614 (AG; average heterozygosis ± standard deviation =0.5±0.008) resulted in greater degradation of IκBα and phosphorylation of p65 NF-κB in naïve CD4 T cells with the risk genotype (GG). The authors used healthy control subjects carrying the risk or the protective variants, and the concordant impact of the rs228614 risk variant was identified after signaling induction. In addition, subjects with the GG risk genotype expressed p50 NF-κB at 20-fold higher level than the AA genotype, together with a loss of negative regulation of this pathway (18). They concluded that naïve CD4 T cells from subjects carrying MS risk variants affecting NF-κB1 exhibit increased p65 activation consistent with changes in NF-κB signaling that may alter the inflammatory response and MS susceptibility. Thus, the results provided demonstrate a central role for NF-κB in driving the pathophysiology of MS. However, the challenge of identifying the biological significance of variants that are non-coding for protein-altering modifications still uncovered. In this regard, the region spanned by the MS risk haplotype near NF-κB1 contains a number of regulatory elements that have been previously pointed out by these authors and others (21,22). Interpretation of these non-coding disease variants, which, in fact, comprise the vast majority of GWAS hits, remains a challenge, due to the limited understanding of the haplotype structure and mechanisms in physiological contexts. The implementation of results from high-density genotyping and epigenomic data has identified causal variants based on genetic evidence, integrated chromatin and transcription factor binding maps to distinguish the potential functions. In the context of MS, the results highlight the involvement of specific transcription factors, target loci, genes and pathways with important roles in autoimmunity. Still, systematic integration of fine-mapped genetic and epigenetic data implies a nuanced complexity to disease variant function. Understanding the regulatory mechanisms could have broad implications for MS diagnosis and treatment, given genetic links to regulators and transcriptional aberrations being potential candidates for therapeutic intervention.
Regulation and dysregulation of TNFR1
The cellular complexity of the central nervous system (CNS), in which glial cell types, including microglia and astrocytes, are interspersed among neurons of many subtypes, obliges to evaluate all these components separately, or even identify common pathways involved in CNS cell fate and behavior. For instance, TNF receptors type 1 (TNFR1) has been demonstrated to be required for astrocytic response to endotoxemia (23). Notably, neuronal expression of TNFR1 is negligible, suggesting that neurotoxic, neuroinflammatory, and neurodegenerative activities of this receptor likely occur through indirect signaling mechanisms and/or cells of the immune system located into the CNS. CD4+ is a subset of T cells that recruit many other immune cells to assist at the primary site of infection, tissue remodeling, and/or damage, such as CD8+ cytotoxic T cells, astrocytes, and macrophages in CNS during inflammatory events. Increased activity of CD4+ T cells secreting inflammatory cytokines and loss of regulatory T cell function in MS (24) justifies researches and experimental models focused on these cells and its potential contribution to autoimmune disease (Figure 1).
Infiltration of lymphocytes and monocytes into the CNS is one of the most prominent features of MS. Then, the inflammatory cascade is much amplified by proinflammatory cytokines released by these cells, compromising blood brain barrier, increasing cell recruitment, and promoting activation of resident microglial cells. Being the role of aberrant immune responses evident, the fundamental impact of a spectrum of cytokines in determination of MS outcomes has been deeply analyzed (25). Among them, the strongest link between cytokines and disease activity has been reported for TNFα, a soluble protein derived from cells of monocyte and macrophage lineage with strong proinflammatory properties and important roles in immunity. TNFα is best known for orchestrating communication between immune cells, modulating their functions and enhancing inflammation, with the subsequent induction of oligodendrocyte apoptosis and demyelization during disturbances affecting the CNS (26). TNFα leading immune defenses are aimed at protecting a localized area from invasion or injury but they may be also involved in controlling whether target cells die or live (i.e., apoptosis or cell survival and proliferation). Thus, defective immune cells in autoimmune diseases may provoke destruction of cells and tissues being coordinated by anomalous TNFα activity and/or response (27).
Increased TNFα has been reported in MS (28). TNFα stimulation of NF-κB canonical pathway is involved in gene activity, influencing biological processes in inflammation, stress, and lymphoid organogenesis and B cell development (Figure 1). Also, activation of TNFR members leads to activation of NF-κB through the non-canonical pathway (29). Although this might support the use of TNFα inhibitors in MS, current evidences suggest that it may be not possible, or very tricky, determine the specific stimuli downstream TNFα signaling cascades leading to cell reaction. Notably, TNFα blockade by monoclonal antibodies or soluble receptors is effective in some autoimmune diseases, but may worsen and even induce MS (30). Selective targeting of TNFR1-mediated signaling, while sparing TNF receptors type 2 (TNFR2) activation, seems lessen adverse effects of anti-TNF therapies in the CNS, leading to the effective blockage of systematic NF-κB-induced inflammation upon increased TNFα (31).
TNFR1 and 2 are membrane receptors that specifically bind TNFα but differ in the expression profile in cells from different lineage, ligand affinity, cytoplasmic tail structure, and downstream signaling pathway activation. TNFR2 is expressed primarily by cells of the immune system (including microglia) and endothelial cells, shows fewer biological effects compared to those mediated by TNFR1, and its affinity for soluble TNFα is lower than those reported for the proinflammatory counterpart (32). TNFR1 is expressed in most cell types and can be activated by binding of soluble TNFα. When TNFα interacts with TNFR1 their trimerization is initiated, leading to recruitment of TNFR1-associated death domain proteins (TRADD) that, in turn, recruit TNF receptor-associated factor 2 (TRAF2), receptor-interacting protein 1 (RIP1), and several other phosphorylases and ubiquitin ligases needed for NF-κB activity (29). Following TNFα treatment the intracellular adaptors recruit and activate IKK complexes that, at the end, phosphorylate IκB proteins, leading to the subsequent polyubiquitination and degradation. This allows NF-κB molecules to translocate to the nucleus and regulate the transcription of many inflammation-associated target genes. However, the involvement of other pathways and the complexity of divergent TNFR1-mediated signaling allow many potential outcomes and contribute to the difficulties inherent in broad TNF signaling inhibition, particularly in areas including the CNS, where TNF signaling has been demonstrated to be at the forefront of both cell fate and immune activation. In this context, the genetic influences affecting TNFR1 are believed to provide important contribution to the clinical heterogeneity of MS. Their study in the context of disease may allow the genetic and molecular dissection of this disease.
Genetic variations affecting TNFR1 in MS
Genetic studies have uncovered many parts of the genome playing a role in the risk of autoimmune disease, or affecting neurodegenerative disorders in general population (33). The limited availability of experimental data endorsing the causal relationship genotype-immune activity has been a major hurdle in translating the genetic findings into functional biology. Combining experimental and population based data with distinct systems biology approaches should provide new insights on disease pathogenesis.
There is substantial overlap between loci that fall either within or proximal to NF-κB signaling genes, including the variants of NF-κB1 disclosed before, and TNFR1. Physiological studies in MS individuals indicate that most of these loci primarily influence inflammation, highlighting a key role for the immune cells in the mechanistic underpinnings of these association signals. In this context, the intersection of GWAS (16) with physiological and functional data (34) indicates that variants regulating TNFR1-mediated inflammation and gene transcription significantly influence MS risk. These findings have motivated efforts to catalogue the epigenomic and transcriptional landscape of immune cells activated through TNF stimulus, applying these findings to deliver biological insights into autoimmune disease pathogenesis.
A number of polymorphisms have been detected in TNFR1 gene in association with MS (16). It is recognized that genetic modifications affecting TNFR1 gene expression or enzyme activity modulate TNFα signaling, which depends on the association of adaptor proteins that bind to the cytoplasmic domains of TNFR receptors. The cytoplasmic tail of TNFR1 contains a death domain that couples this receptor to distinct signaling pathways. One pathway leads to activation of NF-κB and the subsequent transcription of proinflammatory and anti-apoptotic genes, whereas a different pathway leads to apoptosis via activation of caspases (35). Therefore, TNFR1 will activate proinflammatory or apoptotic signals depending on the cell type and the state of the cell. Although more in-depth research is needed to characterize the effects of polymorphism affecting TNFR1 gene, many studies have provided evidence that the TNFR1 susceptibility locus is involved in the risk of MS. The SNP rs4149584, for example, is positioned in TNFR1 exon 4 and conducts an arginine-to-glutamine substitution (R92Q) with low frequency but greater impact in basal inflammation, increased susceptibility to autoimmune disease, and association with MS (36). In line with this, the impressive work of Housley and coworkers confirmed, first, the association of common variants within TNFR1 gene (e.g., rs1800693 Average Het ± std err: 0.462±0.133) with increased response to the inflammatory stimuli, altered intracellular accumulation of TNFR1 (34), and risk of developing MS (37) in the context of changes affecting NF-κB signaling. This variant falls 10 base pairs upstream of TNFR1 exon 6 in a splice acceptor site that introduces a stop codon and loss of exon 6 in ~10% of TNFR1 mRNAs homozygous C risk allele carriers (38). This transcriptional change results in an exaggerated cytokine production response from monocytes after stimulation with TNFα, and exacerbates inflammatory responses. As well as with the genetic variant affecting (or associated with) NF-κB, the study by Housley and coworkers demonstrates that the likely causal impact of rs1800693 variations in inflammation and response to TNFα involves increased activation of the NF-κB pathway (18). Current authors’ findings suggest that altered TNFα induced NF-κB signaling was due to rs1800693 SNP exerting regulatory effects through changes in TNFR1. This agrees with the specific impact of TNFR1 blockage in naïve CD4 cells, with little if any contribution from TNFR2 (31), and endorses the link between the increased risk of MS and polymorphisms affecting TNFR1 expression or activity, impaired NF-κB response to TNFα, changes in the expression and secretion profile of inflammatory cytokines, and altered intracellular accumulation of TNFR1 within the cytoplasm of CD14+ cells and monocytes.
Final disclosure
As for many other autoimmune diseases, the pathogenesis of MS is very complex, involving changes in cell composition and hundreds of genes and proteins that evolve along time and disease progression. There are many insights that point out the genetic basis of these disturbances which, coupled with external incomes, may trigger disease activation. In order to integrate GWAS knowledge and generate a comprehensive model of MS pathogenesis, pathway and networks analysis represents a promising strategy in the field of autoimmunity. So far, numerous studies demonstrate that genes functioning in the same pathway can individually or collectively influence susceptibility to neurodegenerative diseases and traits such as inflammation in MS. The careful interpretation and biological evaluation of the relatively modest effect on disease risk reported for each gene in genome association studies may provide new targets for biomarker and drug development. Mechanistic hypotheses guiding confirmatory testing in additional study datasets and cell lines are much needed in order to make possible the causal implementation of this knowledge.
Acknowledgements
We would like to thank Dr. Pere Tubert Juhé for his help with Figure 1.
Provenance: This is a Guest Editorial commissioned by Section Editor Ning Ding (Department of Respiratory Medicine, Hospital of Nanjing Medical University, Nanjing, China).
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- 1.Hirahara K, Schwartz D, Gadina M, et al. Targeting cytokine signaling in autoimmunity: back to the future and beyond. Curr Opin Immunol 2016;43:89-97. 10.1016/j.coi.2016.10.001 [DOI] [PubMed] [Google Scholar]
- 2.Patterson H, Nibbs R, McInnes I, et al. Protein kinase inhibitors in the treatment of inflammatory and autoimmune diseases. Clin Exp Immunol 2014;176:1-10. 10.1111/cei.12248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Delft MA, Huitema LF, Tas SW. The contribution of NF-κB signalling to immune regulation and tolerance. Eur J Clin Invest 2015;45:529-39. 10.1111/eci.12430 [DOI] [PubMed] [Google Scholar]
- 4.Franciotta D, Salvetti M, Lolli F, et al. B cells and multiple sclerosis. Lancet Neurol 2008;7:852-8. 10.1016/S1474-4422(08)70192-3 [DOI] [PubMed] [Google Scholar]
- 5.Hemmer B, Hartung HP. Toward the development of rational therapies in multiple sclerosis: what is on the horizon? Ann Neurol 2007;62:314-26. 10.1002/ana.21289 [DOI] [PubMed] [Google Scholar]
- 6.Ortiz GG, Pacheco-Moisés FP, Bitzer-Quintero OK, et al. Immunology and oxidative stress in multiple sclerosis: clinical and basic approach. Clin Dev Immunol 2013;2013:708659. 10.1155/2013/708659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sun SC. Non-canonical NF-κB signaling pathway. Cell Res 2011;21:71-85. 10.1038/cr.2010.177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Perkins ND. Integrating cell-signalling pathways with NF-kappaB and IKK function. Nat Rev Mol Cell Biol 2007;8:49-62. 10.1038/nrm2083 [DOI] [PubMed] [Google Scholar]
- 9.Hoffmann A, Levchenko A, Scott ML, et al. The IkappaB-NF-kappaB signaling module: temporal control and selective gene activation. Science 2002;298:1241-5. 10.1126/science.1071914 [DOI] [PubMed] [Google Scholar]
- 10.Srinivasan M, Lahiri DK. Significance of NF-κB as a pivotal therapeutic target in the neurodegenerative pathologies of Alzheimer's disease and multiple sclerosis. Expert Opin Ther Targets 2015;19:471-87. 10.1517/14728222.2014.989834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hemminki K, Li X, Sundquist J, et al. Risk for multiple sclerosis in relatives and spouses of patients diagnosed with autoimmune and related conditions. Neurogenetics 2009;10:5-11. 10.1007/s10048-008-0156-y [DOI] [PubMed] [Google Scholar]
- 12.Ebers GC, Bulman DE, Sadovnick AD, et al. A population-based study of multiple sclerosis in twins. N Engl J Med 1986;315:1638-42. 10.1056/NEJM198612253152603 [DOI] [PubMed] [Google Scholar]
- 13.Hawkes CH, Macgregor AJ. Twin studies and the heritability of MS: a conclusion. Mult Scler 2009;15:661-7. 10.1177/1352458509104592 [DOI] [PubMed] [Google Scholar]
- 14.International Multiple Sclerosis Genetics Consortium .; Wellcome Trust Case Control Consortium 2., Sawcer S, et al. Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. Nature 2011;476:214-9. 10.1038/nature10251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Park MH, Hong JT. Roles of NF-κB in Cancer and Inflammatory Diseases and Their Therapeutic Approaches. Cells 2016;5. pii: E15. 10.3390/cells5020015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.De Jager PL, Jia X, Wang J, et al. Meta-analysis of genome scans and replication identify CD6, IRF8 and TNFRSF1A as new multiple sclerosis susceptibility loci. Nat Genet 2009;41:776-82. 10.1038/ng.401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cotsapas C, Voight BF, Rossin E, et al. Pervasive sharing of genetic effects in autoimmune disease. PLoS Genet 2011;7:e1002254. 10.1371/journal.pgen.1002254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Housley WJ, Fernandez SD, Vera K, et al. Genetic variants associated with autoimmunity drive NF-κB signaling and responses to inflammatory stimuli. Sci Transl Med 2015;7:291ra93. 10.1126/scitranslmed.aaa9223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eggert M, Goertsches R, Seeck U, et al. Changes in the activation level of NF-kappa B in lymphocytes of MS patients during glucocorticoid pulse therapy. J Neurol Sci 2008;264:145-50. 10.1016/j.jns.2007.08.026 [DOI] [PubMed] [Google Scholar]
- 20.Sun XF, Zhang H. NFKB and NFKBI polymorphisms in relation to susceptibility of tumour and other diseases. Histol Histopathol 2007;22:1387-98. [DOI] [PubMed] [Google Scholar]
- 21.Farh KK, Marson A, Zhu J, et al. Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature 2015;518:337-43. 10.1038/nature13835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heinz S, Romanoski CE, Benner C, et al. Effect of natural genetic variation on enhancer selection and function. Nature 2013;503:487-92. 10.1038/nature12615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Srinivasan K, Friedman BA, Larson JL, et al. Untangling the brain's neuroinflammatory and neurodegenerative transcriptional responses. Nat Commun 2016;7:11295. 10.1038/ncomms11295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Han MH, Steinman L. Systems biology for identification of molecular networks in multiple sclerosis. Mult Scler 2009;15:529-30. 10.1177/1352458509103318 [DOI] [PubMed] [Google Scholar]
- 25.Ozenci V, Kouwenhoven M, Link H. Cytokines in multiple sclerosis: methodological aspects and pathogenic implications. Mult Scler 2002;8:396-404. 10.1191/1352458502ms837rr [DOI] [PubMed] [Google Scholar]
- 26.Arnett HA, Mason J, Marino M, et al. TNF alpha promotes proliferation of oligodendrocyte progenitors and remyelination. Nat Neurosci 2001;4:1116-22. 10.1038/nn738 [DOI] [PubMed] [Google Scholar]
- 27.Faustman D, Davis M. TNF receptor 2 pathway: drug target for autoimmune diseases. Nat Rev Drug Discov 2010;9:482-93. 10.1038/nrd3030 [DOI] [PubMed] [Google Scholar]
- 28.Ozenci V, Kouwenhoven M, Huang YM, et al. Multiple sclerosis is associated with an imbalance between tumour necrosis factor-alpha (TNF-alpha)- and IL-10-secreting blood cells that is corrected by interferon-beta (IFN-beta) treatment. Clin Exp Immunol 2000;120:147-53. 10.1046/j.1365-2249.2000.01175.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alvarez SE, Harikumar KB, Hait NC, et al. Sphingosine-1-phosphate is a missing cofactor for the E3 ubiquitin ligase TRAF2. Nature 2010;465:1084-8. 10.1038/nature09128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Atzeni F, Talotta R, Salaffi F, et al. Immunogenicity and autoimmunity during anti-TNF therapy. Autoimmun Rev 2013;12:703-8. 10.1016/j.autrev.2012.10.021 [DOI] [PubMed] [Google Scholar]
- 31.McCoy MK, Tansey MG. TNF signaling inhibition in the CNS: implications for normal brain function and neurodegenerative disease. J Neuroinflammation 2008;5:45. 10.1186/1742-2094-5-45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grell M. Tumor necrosis factor (TNF) receptors in cellular signaling of soluble and membrane-expressed TNF. J Inflamm 1995-1996;47:8-17. [PubMed] [Google Scholar]
- 33.Ramanan VK, Saykin AJ. Pathways to neurodegeneration: mechanistic insights from GWAS in Alzheimer's disease, Parkinson’s disease, and related disorders. Am J Neurodegener Dis 2013;2:145-75. [PMC free article] [PubMed] [Google Scholar]
- 34.Gregory AP, Dendrou CA, Attfield KE, et al. TNF receptor 1 genetic risk mirrors outcome of anti-TNF therapy in multiple sclerosis. Nature 2012;488:508-11. 10.1038/nature11307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tracey D, Klareskog L, Sasso EH, et al. Tumor necrosis factor antagonist mechanisms of action: a comprehensive review. Pharmacol Ther 2008;117:244-79. 10.1016/j.pharmthera.2007.10.001 [DOI] [PubMed] [Google Scholar]
- 36.Caminero A, Comabella M, Montalban X. Role of tumour necrosis factor (TNF)-α and TNFRSF1A R92Q mutation in the pathogenesis of TNF receptor-associated periodic syndrome and multiple sclerosis. Clin Exp Immunol 2011;166:338-45. 10.1111/j.1365-2249.2011.04484.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.International Multiple Sclerosis Genetics Consortium (IMSGC) ., Beecham AH, Patsopoulos NA, et al. Analysis of immune-related loci identifies 48 new susceptibility variants for multiple sclerosis. Nat Genet 2013;45:1353-60. 10.1038/ng.2770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ottoboni L, Frohlich IY, Lee M, et al. Clinical relevance and functional consequences of the TNFRSF1A multiple sclerosis locus. Neurology 2013;81:1891-9. 10.1212/01.wnl.0000436612.66328.8a [DOI] [PMC free article] [PubMed] [Google Scholar]