Abstract
Long non-coding RNAs (lncRNAs) have been identified as novel biomarkers for the diagnosis, staging and prognosis for gastric cancer. However, various studies have reported a series of significances based on different diagnostic values. Therefore, the current study performed a systematic review and meta-analysis to evaluate the diagnostic accuracy of lncRNAs for gastric cancer, and to discuss lncRNA types and sources of heterogeneity. The Cochrane Central Register of Controlled Trials, MEDLINE, PubMed, EMBASE, the Chinese Biomedical Literature Database, the China Academic Journals Full-text Database and the Chinese Scientific Journals Database were systematically searched for potential studies. Studies were included if they were associated with lncRNAs, gastric cancer and reported diagnostic outcomes. Analysis of diagnostic values was used to summarize the overall test performance of lncRNAs. Ten studies were included in this meta-analysis. The ranges of the diagnostic value of lncRNAs for gastric cancer were as follows: Sensitivity was 0.45–0.83, and pooled sensitivity was 0.63; specificity was 0.60–0.93, and pooled specificity was 0.75; positive likelihood ratio was 1.80–6.92, and pooled positive likelihood ratio was 2.51; negative likelihood ratio was 0.23–0.67, and pooled negative likelihood ratio was 0.50; diagnostic odds ratio was 3.33–13.75, and pooled diagnostic odds ratio was 5.47. An overall area under the curve value of the summary receiver operating characteristic curve was 0.7550. LncRNAs did not have a high accuracy for identifying gastric cancer at present, but may be a useful screening tool for diagnosing gastric cancer due to their correlation with gastric cancer biological features. LncRNAs are potential biomarkers for gastric cancer if the screening strategy is altered, or they are combined with other biomarkers to diagnose gastric cancer.
Keywords: long non-coding RNAs, gastric cancer, meta-analysis, sensitivity, specificity
Introduction
Gastric cancer is one of the most common causes of cancer mortality worldwide, and results in ~738,000 deaths globally each year (1). As early-stage gastric cancer has non-specific symptoms, the mortality rate remains high due to late presentation. For advanced disease, the overall 5-year survival following surgical resection is 30–40%, compared with 70–90% in early-stage disease (2). Therefore, early diagnosis and treatment is an important way to raise the 5-year survival rate and improve prognosis. Currently, diagnosis of gastric cancer relies on endoscopic and pathological examinations and the measurement of serum biomarkers, including carcinoembryonic antigen, carbohydrate antibody 19–9, carbohydrate antibody 72–4 (CA72-4) and carbohydrate antibody 50. However, these tumor markers have limited utility due to the lack of sufficiently high diagnostic sensitivity and specificity (3). Despite endoscopic and pathological examinations being considered as the ‘gold standard’ for diagnosing gastric cancer, they may be associated with advanced tumor stages and significant expense, manpower issues and risk of patient injury, and are not available for early cancer screening (4). In order to optimize therapeutic strategies, predict the outcome of treatment and extend the survival period of patients, novel diagnostic markers for early stage must urgently be investigated.
Long non-coding RNAs (lncRNAs) represent a novel class of non-protein coding molecules that have recently been implicated in the pathogenesis of numerous types of cancer, and previous studies have revealed interactions between lncRNAs and other molecules that are responsible for important gene regulatory functions in various contexts including gastric cancer (5,6). Recently, lncRNAs were considered as new markers of gastric cancer diagnosis in several clinical reports (7,8), but with varying diagnostic accuracy and using differing lncRNA types. Based on the present research situation, the present study undertook a meta-analysis and systematic review to assess the overall accuracy of these lncRNAs for the diagnosis of gastric cancer.
Materials and methods
Search strategy and study selection
A comprehensive literature search to identify studies that evaluated the diagnostic accuracy of lncRNAs for gastric cancer was conducted. Databases including MEDLINE (1946 to August 2015), the Cochrane Database of Systematic Reviews (2005 to August 2015), the Chinese Biomedical Literature Database (1978 to August 2015), the China Academic Journals Full-text Database (1979 to August 2015), and the Chinese Scientific Journals Database (1989 to August 2015) were searched without language restrictions.
The following search strategy was adopted: (‘long non-coding RNAs’ OR ‘lncRNAs’) and (‘gastric cancer’ OR ‘GC’ OR ‘stomach’). Duplicate articles identified in both Medline and EMBASE were manually deleted using Reference Manager (Thomson Reuters EndNote X5, New York, NY, USA). Results were arbitrated by two investigators (Zi-Yi Zhao and Shui-Qin Li), on the basis of the title and abstract, and the full paper of each potentially eligible study was then obtained. To obtain further relevant studies, the reference lists of identified trials were also examined.
Eligible criteria
All studies that reported data on patients with a confirmed diagnosis of gastric cancer made using lncRNAs were considered for inclusion. Only studies that reported sufficient data to allow construction of 2×2 tables were included. Irrelevant studies were excluded, such as animal studies.
Quality assessment
Two independent reviewers (Shui-Qin Li and Li Li) used the Quality Assessment of studies of Diagnostic Accuracy included in the Systematic reviews (QUADAS) instrument (9) to assess the quality of selected articles (Table I). Any disagreements were resolved by discussion and consensus, if necessary after contacting the authors for clarification. Study quality was assessed using the QUADAS-list, with each item scored as ‘yes’, ‘no’ or ‘unclear’. The calculation of summary scores of quality was not performed as the interpretation was problematic and potentially misleading (10).
Table I.
Items of quality assessment selected from QUADAS checklist.
| Item | Yes | No | Unclear |
|---|---|---|---|
| 1. Was the spectrum of patients representative of those who will receive the test in practice? | (−) | (−) | (−) |
| 2. Is the reference standard likely to correctly classify the target condition? | (−) | (−) | (−) |
| 3. Is the time period between reference standard and index test short enough to be reasonably sure that the target condition did not change between the two tests? | (−) | (−) | (−) |
| 4. Did the whole sample or a random selection of the sample, receive verification using a reference standard of diagnosis? | (−) | (−) | (−) |
| 5. Did patients receive the same reference standard regardless of the index test result? | (−) | (−) | (−) |
| 6. Was the reference standard independent of the index test (i.e. the index test did not form part of the reference standard)? | (−) | (−) | (−) |
| 7. Were the reference standard results interpreted without knowledge of the results of the index test? | (−) | (−) | (−) |
| 8. Were the index test results interpreted without knowledge of the results of the reference standard? | (−) | (−) | (−) |
| 9. Were the same clinical data available when test results were interpreted as would be available when the test is used in practice? | (−) | (−) | (−) |
| 10. Were uninterruptible/intermediate test results reported? | (−) | (−) | (−) |
| 11. Were withdrawals from the study explained? | (−) | (−) | (−) |
Data extraction and analysis
One reviewer (Qiong-Ying Hu) extracted and another reviewer (Zi-Yi Zhao) checked the data. The data extraction form was accompanied by a background document that stated how each item on the form must be interpreted. All data collection was performed according to a protocol with the following information being extracted from each study: First author, year of publication, population characteristics, study design, inclusion and exclusion criteria, number of subjects and the method of gastric cancer determination.
The primary endpoints of lncRNAs as biomarkers were sensitivity and specificity (the number of true positive, false negative, true negative and false positive results) for the comparison of patients diagnosed with gastric cancer vs. controls. Sensitivity and specificity were also extracted and calculated from each study and the data were presented as diagnostic odds ratio (DOR) and summary receiver operating characteristic (SROC) curves to determine the performance of lncRNAs in identifying gastric cancer using the area under the curve (AUC) value. Meta-Disc1.4 software was used to analyze the plots and curve (11).
Results
The literature search yielded 325 citations, of which 47 were excluded as they were duplicates. In total, 149 publications were excluded as they failed to meet the eligible criteria on the basis of title and abstract. Of the 129 potentially eligible studies, 119 publications were excluded because they were reviews (n=12), contained no diagnostic value studies (n=96) and the data were not able to be extracted (n=11). Finally, 10 focused on the target patient spectrum were included (Fig. 1). Table II presents the general characteristics of the included studies. Differing lncRNAs predicted various diagnostic values, and all the included studies reported cut-off values. From the raw data, the current study constructed diagnosis values of the patient population as a 2×2 table (Table III) using methods for diagnostic meta-analysis (12).
Figure 1.
Flow diagram of study selection process for the systematic review.
Table II.
Characteristics and quality assessment of the included studies with lncRNAs.
| First author, date | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Variable | Mei 2013 | Sun 2013 | Liu 2014 | Pang 2014 | Shao 2014 | Zhao 2014 | Chen 2015 | Sun 2015 | Zheng 2015 | Zhou 2015 |
| lncRNA types | SUMO1P3 | AC096655.1–002 | FER1L4 | LINC00152 | lncRNA-AA174084 | HULC | HIF1A-AS2 | lncRNA RP11-119F7.4 | lncRNA UCA1 | H19 |
| Patient spectrum | Gastric cancer | Gastric cancer | Gastric cancer | Gastric cancer | Gastric cancer | Gastric cancer | Gastric cancer | Gastric cancer | Gastric cancer | Gastric cancer |
| Sample | Tissues | Tissues | Tissues and plasma | Tissues | Tissues, plasma and gastric juice | Tissues | Tissues | Tissues | Tissues | Plasma |
| Detection method | RT-qPCRf | RT-qPCR | RT-qPCR | RT-qPCRf | RT-qPCR | RT-qPCR | RT-qPCR | RT-qPCR | RT-qPCR | RT-qPCR |
| Age group | ≥60 <60 | ≥65 <65 | ≥60 <60 | ≥60 <60 | ≥60 <60 | ≥60 <60 | >60 ≤60 | >60 ≤60 | >60 <60 | ≥60 <60 |
| Study design | CSS | CSS | CSS | CSS | CSS | CSS | CSS | CSS | CSS | CSS |
| Item 1 | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| Item 2 | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| Item 3 | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| Item 4 | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| Item 5 | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| Item 6 | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| Item 7 | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| Item 8 | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| Item 9 | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| Item 10 | No | No | No | No | No | No | No | No | No | No |
| Item 11 | Unclear | Unclear | Unclear | Unclear | Unclear | Unclear | Unclear | Unclear | Unclear | Unclear |
Items 1–10 selected to rate from QUADAS checklist in Table I. LncRNA, long non coding RNA; CSS, cross-sectional study; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; SUMO1P3, small ubiquitin-like modifier 1 pseudogene 3; FER1L4, lncRNA-Fer-1-like protein 4; HULC, highly upregulated in liver cancer; HIF1A-AS2, hypoxiainducible factor 1α antisense RNA-2; UCA1, urothelial carcinoma-associated 1.
Table III.
Diagnostic values of the patients who participated in the studies and were included in the meta-analysis.
| Author (year) | n | TP (a) | FP (b) | FN (c) | TN (d) | Sensitivity (%) | Specificity (%) | ROCAUC | Cutoff |
|---|---|---|---|---|---|---|---|---|---|
| Mei 2013 | 192 | 63 | 35 | 33 | 61 | 65.9 | 63.6 | 0.666 | 2.31 |
| Sun 2013 | 156 | 40 | 10 | 38 | 68 | 51.3 | 87.2 | 0.731 | 13.9555 |
| Liu 2014 | 122 | 41 | 12 | 20 | 49 | 67.2 | 80.3 | 0.778 | 15.43 |
| Pang 2014 | 142 | 44 | 23 | 27 | 48 | 62.5 | 68.1 | 0.645 | 4.385 |
| Shao 2014a | 268 | 76 | 36 | 58 | 98 | 57 | 73 | 0.676 | 11.62 |
| Shao 2014b | 84 | 18 | 3 | 21 | 42 | 46 | 93 | 0.848 | 0.88 |
| Zhao 2014 | 100 | 35 | 14 | 15 | 36 | 70.7 | 72.4 | 0.769 | 10.88 |
| Chen 2015 | 166 | 60 | 33 | 23 | 50 | 72.29 | 60.24 | 0.673 | 9.56 |
| Sun 2015 | 192 | 43 | 17 | 53 | 79 | 44.8 | 82.3 | 0.637 | 6.445 |
| Zheng 2015 | 224 | 75 | 22 | 37 | 90 | 67.2 | 80.3 | 0.721 | 13.74 |
| Zhou 2015 | 180 | 75 | 24 | 15 | 66 | 82.9 | 72.9 | 0.838 | Unclear |
Tissue
gastric juice.
AUC, area under receiver operating characteristic curve; ROC, receiver operating characteristic; TP, true positive; FP, false positive; FN, false negative; TN, true negative.
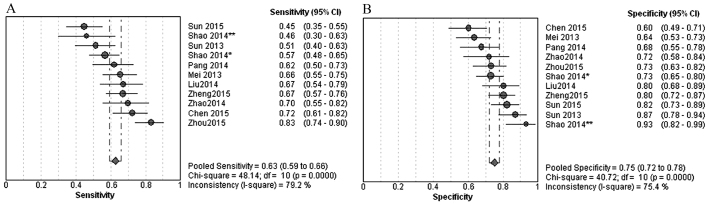
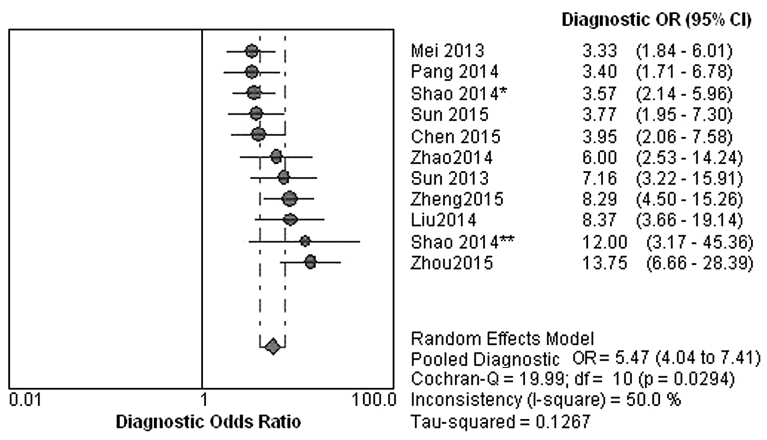
The ranges of sensitivity and specificity of diagnosis model with lncRNAs as identifying of gastric cancer were 0.45–0.83 (pooled 0.63; Fig. 2A) and 0.60–0.93 (pooled 0.75; Fig. 2B), respectively. The ranges of positive likelihood ratio (Fig. 3A) and negative likelihood ratio (Fig. 3B) were 1.80–6.92 (pooled 2.51) and 0.23–0.67 (pooled 0.50), respectively, with an overall AUC value of the SROC of 0.7550 (Fig. 4). The range of DOR was 3.33–13.75 (pooled 5.47; Fig. 5).
Figure 2.
(A) Sensitivity and (B) specificity of diagnostic models using long non-coding RNAs to identify gastric cancer. CI, confidence interval; df, degrees of freedom; LR, likelihood ratio.
Figure 3.
(A) Positive LR and (B) negative LR of diagnostic models using long non-coding RNAs to identify gastric cancer. LR, likelihood ratio; CI, confidence interval.
Figure 4.
SROC curve of diagnostic model with long non-coding RNAs as identifying gastric cancer. SROC, summary receiver operator characteristic curve; AUC, area under the curve; SE, standard error.
Figure 5.
Diagnostic OR of diagnostic model with long non-coding RNAs as identifying gastric cancer. OR, odds ratio; CI, confidence interval.
The studies reviewed were free of commercial funding. They provided a clear definition of what was considered to be a ‘positive’ result and the technology of the index test was unchanged from the time the study was performed. All studies were pre-specified objectively. Data on instrument variation and data on observer variation were not available. All papers failed to describe whether test operators had appropriate training. All studies clearly pointed out their treatments were performed after the index test and reference standard were tested.
Discussion
LncRNAs have important roles in differentiation, development and tumorigenesis of gastric cancer, and a number of publications have implicated lncRNAs in a wide range of the biological processes involved in gastric cancer pathogenesis. Yang et al (13) revealed that the expression levels of C21 or F96 were associated with gross appearance, lymphatic metastasis and distal metastasis. Lü et al (14) reported lncRNA BC032469 was able to upregulate human telomerase reverse transcriptase (hTERT) expression by sponging microRNA (miR)-1207-5p and promoting proliferation in gastric cancer. Li et al (15) thought TM4SF5-CTD-2354A18.1 miR-4697-3P may play a key role in the pathogenesis of gastric cancer. Du et al (16) identified that the lncRNA WT1-AS is significantly downregulated in gastric cancer and may be associated with tumor progression. Additional studies found that lncRNAs were associated with early diagnosis, metastasis and prognosis (17,18). Mei et al (19) revealed that lncRNA SUMO1P3 was significantly upregulated in gastric cancer tissues compared with paired-adjacent non-tumorous tissues, and the area under the ROC curve of SUMO1P3 was up to 0.666. Sun et al (20) indicated that lncRNA AC096655.1–002 may be a potential biomarker in the diagnosis of gastric carcinoma, with an area under the ROC curve value of 0.731, and a sensitivity and specificity of 0.513 and 0.872, respectively.
These previous investigations have revealed that lncRNAs are involved in cell proliferation, apoptosis, migration and invasion, and correlate with the malignant phenotype of gastric cancer. LncRNAs are present in the tissue, blood and gastric juice, and show potential value as biomarkers for the diagnosing of GC or for determining disease prognosis. However, a range of diagnostic values and differences in literature quality mean that there are challenges to address prior to lncRNAs being able to be used in clinical applications. In the current meta-analysis, the diagnostic value of lncRNAs as potential biomarkers for gastric cancer and the literature quality and bias of all included studies were assessed.
LncRNAs were found in various samples (e.g. tissue, blood, body fluid), and different sample types correspond to disparate diagnostic values. Seven included studies focused on tissue research, 2 studies compared with other sample types and only 1 study involved plasma samples. The lncRNA AA174084 was identified to be differentially expressed in tissue, plasma and gastric juice. Shao et al (21) compared 3 types of samples, and observed the highest specificity (93%) for lncRNA AA174084 in gastric juice. However, significant differences were not identified for all sample types; Shao et al (21) also found that plasma lncRNA AA174084 levels did not differ between gastric cancer patients and healthy individuals. However, plasma AA174084 levels in patients with gastric cancer decreased markedly on day 15 after surgery compared with preoperative levels and were associated with invasion and lymphatic metastasis, which may be informative of other disease characteristics (21). Similarly, the same sample types but different lncRNA types had various diagnostic values. Three previous studies (21–23) detected plasma lncRNA levels, but two of these studies (21,22) found no difference in plasma lncRNA levels between healthy individuals and patients with gastric cancer. In terms of the patient spectrum, sample, detection method, age group and study design, 10 included studies corresponded to the QUADAS checklist. Furthermore, every included study had reported diagnostic cut-off values except Zhou et al (23) Cut-off values are very important for diagnostic tests as they decide diagnostic values and differing cut-off values correspond to altered sensitivity and specificity.
From the present meta-analysis, the ranges of sensitivity and specificity of diagnostic models with lncRNAs as an identifier of gastric cancer were 0.45–0.83 and 0.60~0.93, respectively. The pooled sensitivity and specificity were 0.63 and 0.75, respectively. This does not appear to be ideal for diagnostic biomarkers. However, certain lncRNAs were potentially indicators for associated gastric cancer biological features. Mei et al (19) revealed that the expression level of SUMO1P3 was significantly correlated with tumor size, differentiation, lymphatic metastasis and invasion. Sun et al (20) reported that the expression level of AC096655.1–002 was significantly correlated with lymph node metastasis, distant metastasis, tumor-node-metastasis stage and differentiation. Liu et al (22) indicated that low FER1L4 levels were associated with tumor size, histologic grade, general classification, depth of invasion, lymphatic metastasis, distant metastasis and tumor-node-metastasis (TNM) stage. Pang et al (24) revealed that the high expression level of LINC00152 was correlated with invasion. Zhao et al (25) demonstrated that HULC may have an important role in the growth and tumorigenesis of gastric cancer, which provides a novel biomarker for gastric cancer and perhaps a potential target for gastric cancer prevention, diagnosis and therapeutic treatment. Chen et al (26) suggested overexpression of HIF1A-AS2 overexpression was correlated with gastric cancer TNM stage, tumor invasion, lymph node metastasis and poor prognosis. Sun et al (27) revealed that RP11-119F7.4 expression was significantly associated with the macroscopic type and Lauren grade, and indicated that lncRNA RP11-119F7.4 may be involved in carcinogenesis and function as a biomarker for diagnosis and prognostic significance in patients with gastric cancer. Zheng et al (28) revealed that high UCA1 expression correlated with poor differentiation, tumor size, invasion depth and TNM stage in gastric cancer. A multivariate survival analysis indicated that UCA1 may be an independent prognostic marker (29). Zhou et al (23) reported that H19 expression enabled the differentiation of early stage gastric cancer from controls. In addition, plasma levels of H19 were significantly lower in postoperative samples than preoperative samples.
In conclusion, as a diagnostic biomarker, the diagnostic values of lncRNAs for gastric cancer are not yet sufficient. There remain a number of challenges to be faced before lncRNAs may be used in clinical diagnosis. LncRNAs are associated with every phase of gastric cancer development, including initiation, progression, migration, invasion and metastasis, but their underlying biological mechanisms require further elucidation. Altering screening strategies, or combining other biomarkers to build new diagnostic frameworks may obtain better diagnostic values for lncRNAs as clinical biomarkers for gastric cancer.
Acknowledgements
The present study was supported by National Natural Science Foundation of China (No. 81601835).
Glossary
Abbreviations
- lncRNA
Long non-coding RNA
- DOR
diagnostic odds ratio
- SROC
summary receiver operating characteristic
- AUC
area under the curve
References
- 1.Hsieh YH, Chiu WC, Lin CF, Chan HL, Liang HY, Lee Y, McIntyre RS, Chen VC. Antidepressants and gastric cancer: A nationwide population-based nested case-control study. PLoS One. 2015;10:e0143668. doi: 10.1371/journal.pone.0143668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shin VY, Ng EK, Chan VW, Kwong A, Chu KM. A three-miRNA signature as promising non-invasive diagnostic marker for gastric cancer. Mol Cancer. 2015;14:202. doi: 10.1186/s12943-015-0473-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang L, Wang J, Li J, Zhang H, Guo S, Yan M, Zhu Z, Lan B, Ding Y, Xu M, et al. Identification of serum biomarkers for gastric cancer diagnosis using a human proteome microarray. Mol Cell Proteomics. 2016;15:614–623. doi: 10.1074/mcp.M115.051250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hu QY, Jiang H, Su J, Jia YQ. MicroRNAs as biomarkers for hepatocellular carcinoma: A diagnostic meta-analysis. Clin Lab. 2013;59:1113–1120. doi: 10.7754/Clin.Lab.2012.121102. [DOI] [PubMed] [Google Scholar]
- 5.Smolle MA, Bullock MD, Ling H, Pichler M, Haybaeck J. Long non-coding rnas in endometrial carcinoma. Int J Mol Sci. 2015;16:26463–26472. doi: 10.3390/ijms161125962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kiang KM, Zhang XQ, Leung GK. Long non-coding RNAs: The key players in glioma pathogenesis. Cancers (Basel) 2015;7:1406–1424. doi: 10.3390/cancers7030843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fan ZY, Liu W, Yan C, Zhu ZL, Xu W, Li JF, Su L, Li C, Zhu ZG, Liu B, Yan M. Identification of a five-lncRNA signature for the diagnosis and prognosis of gastric cancer. Tumour Biol. 2016;37:13265–13277. doi: 10.1007/s13277-016-5185-9. [DOI] [PubMed] [Google Scholar]
- 8.Jin C, Shi W, Wang F, Shen X, Qi J, Cong H, Yuan J, Shi L, Zhu B, Luo X, et al. Long non-coding RNA HULC as a novel serum biomarker for diagnosis and prognosis prediction of gastric cancer. Oncotarget. 2016;7:51763–51772. doi: 10.18632/oncotarget.10107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Whiting PF, Rutjes AW, Westwood ME, Mallett S, Deeks JJ, Reitsma JB, Leeflang MM, Sterne JA, Bossuyt PM, QUADAS-2 Group QUADAS-2: A revised tool for the quality assessment of diagnostic accuracy studies. Ann Intern Med. 2011;155:529–536. doi: 10.7326/0003-4819-155-8-201110180-00009. [DOI] [PubMed] [Google Scholar]
- 10.Whiting P, Harbord R, Kleijnen J. No role for quality scores in systematic reviews of diagnostic accuracy studies. BMC Med Res Methodol. 2005;5:19. doi: 10.1186/1471-2288-5-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lin R, Yan J, Liu W, Fan T, Tang L. The predictive value of cervical lymph node metastasis through sentinel lymph node biopsy in patients with oral cancer: A meta-analysis. J Cancer Res Ther. 2016;12(Suppl):C256–C259. doi: 10.4103/0973-1482.200751. [DOI] [PubMed] [Google Scholar]
- 12.Chu H, Guo H. A unification of models for meta-analysis of diagnostic accuracy studies. Biostatistics. 2009;10:201–203. doi: 10.1093/biostatistics/kxn040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang Z, Zhi Q, Wang D, Zhang L, Preston B, Brandon C, Kuang Y, Miao R, Shi Y, Guo X. Long non-coding RNA C21orF96 promotes the migration, invasion and lymph node metastasis in gastric cancer. Anticancer Agents Med Chem. 2016;16:1101–1108. doi: 10.2174/1871520616666151116122921. [DOI] [PubMed] [Google Scholar]
- 14.Lü MH, Tang B, Zeng S, Hu CJ, Xie R, Wu YY, Wang SM, He FT, Yang SM. Long non-coding RNA BC032469, a novel competing endogenous RNA, upregulates hTERT expression by sponging miR-1207-5p and promotes proliferation in gastric cancer. Oncogene. 2016;35:3524–3534. doi: 10.1038/onc.2015.413. [DOI] [PubMed] [Google Scholar]
- 15.Li DF, Yang MF, Shi SL, Du YL, Wang H, Zhou YL, Luo YL, Ren LL, Nie YQ. TM4SF5-CTD-2354A18.1-miR-4697-3P may play a key role in the pathogenesis of gastric cancer. Bratisl Lek Listy. 2015;116:608–615. doi: 10.4149/bll_2015_118. [DOI] [PubMed] [Google Scholar]
- 16.Du T, Zhang B, Zhang S, Jiang X, Zheng P, Li J, Yan M, Zhu Z, Liu B. Decreased expression of long non-coding RNA WT1-AS promotes cell proliferation and invasion in gastric cancer. Biochim Biophys Acta. 2016;1862:12–19. doi: 10.1016/j.bbadis.2015.10.001. [DOI] [PubMed] [Google Scholar]
- 17.Xia H, Chen Q, Chen Y, Ge X, Leng W, Tang Q, Ren M, Chen L, Yuan D, Zhang Y, et al. The lncRNA MALAT1 is a novel biomarker for gastric cancer metastasis. Oncotarget. 2016;7:56209–56218. doi: 10.18632/oncotarget.10941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang E, He X, Yin D, Han L, Qiu M, Xu T, Xia R, Xu L, Yin R, De W. Increased expression of long non-coding RNA TUG1 predicts a poor prognosis of gastric cancer and regulates cell proliferation by epigenetically silencing of p57. Cell Death Dis. 2016;7:e2109. doi: 10.1038/cddis.2015.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mei D, Song H, Wang K, Lou Y, Sun W, Liu Z, Ding X, Guo J. Up-regulation of SUMO1 pseudogene 3 (SUMO1P3) in gastric cancer and its clinical association. Med Oncol. 2013;30:709. doi: 10.1007/s12032-013-0709-2. [DOI] [PubMed] [Google Scholar]
- 20.Sun W, Wu Y, Yu X, Liu Y, Song H, Xia T, Xiao B, Guo J. Decreased expression of long non-coding RNA AC096655.1–002 in gastric cancer and its clinical significance. Tumour Biol. 2013;34:2697–2701. doi: 10.1007/s13277-013-0821-0. [DOI] [PubMed] [Google Scholar]
- 21.Shao Y, Ye M, Jiang X, Sun W, Ding X, Liu Z, Ye G, Zhang X, Xiao B, Guo J. Gastric juice long non-coding RNA used as a tumor marker for screening gastric cancer. Cancer. 2014;120:3320–3328. doi: 10.1002/cncr.28882. [DOI] [PubMed] [Google Scholar]
- 22.Liu Z, Shao Y, Tan L, Shi H, Chen S, Guo J. Clinical significance of the low expression of FER1L4 in gastric cancer patients. Tumour Biol. 2014;35:9613–9617. doi: 10.1007/s13277-014-2259-4. [DOI] [PubMed] [Google Scholar]
- 23.Zhou X, Yin C, Dang Y, Ye F, Zhang G. Identification of the long non-coding RNA H19 in plasma as a novel biomarker for diagnosis of gastric cancer. Sci Rep. 2015;5:11516. doi: 10.1038/srep11516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pang Q, Ge J, Shao Y, Sun W, Song H, Xia T, Xiao B, Guo J. Increased expression of long intergenic non-coding RNA LINC00152 in gastric cancer and its clinical significance. Tumour Biol. 2014;35:5441–5447. doi: 10.1007/s13277-014-1709-3. [DOI] [PubMed] [Google Scholar]
- 25.Zhao Y, Guo Q, Chen J, Hu J, Wang S, Sun Y. Role of long non-coding RNA HULC in cell proliferation, apoptosis and tumor metastasis of gastric cancer: A clinical and in vitro investigation. Oncol Rep. 2014;31:358–364. doi: 10.3892/or.2013.2850. [DOI] [PubMed] [Google Scholar]
- 26.Chen WM, Huang MD, Kong R, Xu TP, Zhang EB, Xia R, Sun M, De W, Shu YQ. Antisense long non-coding RNA HIF1A-AS2 Is upregulated in gastric cancer and associated with poor prognosis. Dig dis Sci. 2015;60:1655–1662. doi: 10.1007/s10620-015-3524-0. [DOI] [PubMed] [Google Scholar]
- 27.Sun J, Song Y, Chen X, Zhao J, Gao P, Huang X, Xu H, Wang Z. Novel long non-coding RNA RP11-119F7.4 as a potential biomarker for the development and progression of gastric cancer. Oncol Lett. 2015;10:115–120. doi: 10.3892/ol.2015.3186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zheng Q, Wu F, Dai WY, Zheng DC, Zheng C, Ye H, Zhou B, Chen JJ, Chen P. Aberrant expression of UCA1 in gastric cancer and its clinical significance. Clin Transl Oncol. 2015;17:640–646. doi: 10.1007/s12094-015-1290-2. [DOI] [PubMed] [Google Scholar]
- 29.Sun XD, Huan C, Qiu W, Sun DW, Shi XJ, Wang CL, Jiang C, Wang GY, Lv GY. Clinical significance of UCA1 to predict metastasis and poor prognosis of digestive system malignancies: A meta-analysis. Gastroenterol Res Pract. 2016;2016:3729830. doi: 10.1155/2016/3729830. [DOI] [PMC free article] [PubMed] [Google Scholar]