Abstract
DNA methylation of promoters is well known for its repressive effect on transcription initiation of protein‐coding genes, non‐coding RNAs, or transposon repeats. However, gene bodies represent the most conserved targets of DNA methylation among eukaryotes, and yet the function of intragenic methylation remains unclear. Recent research (Neri et al, 2017) suggests that intragenic methylation may serve to confine transcription initiation to canonical promoters in embryonic stem cells, thus preventing the production of aberrant transcripts.
Subject Categories: Chromatin, Epigenetics, Genomics & Functional Genomics; Stem Cells; Transcription
Cytosine methylation of promoters provides an epigenetic mark for transmitting patterns of gene transcription across cell division and for protecting the genome against transposons. However, comparative epigenomics has revealed that gene bodies are the most common location for this mark among eukaryotes, including animals, plants, and fungi (Feng et al, 2010; Zemach et al, 2010). In species with low genomic methylation—such as the honeybee or the sea squirt—gene bodies are the exclusive targets of DNA methylation. In mammalian genomes, where methylation occurs globally and covers 70–80% of all CpG motifs, intragenic sequences are still more methylated (80–90%) than intergenic sequences (Lister et al, 2009). How and why gene bodies are heavily methylated has been the subject of intense research over the last few years.
The mechanism that preferentially targets DNA methylation to gene bodies is now understood. First, the observation that intragenic methylation levels are often correlated with transcriptional strength indicates that DNA methylation is coupled to transcriptional elongation. Indeed, the de novo DNA methyltransferase (DNMT) enzymes of the DNMT3 family contain a PWWP domain, which recognizes the trimethylation of histone H3 lysine 36 residues (H3K36me3). H3K36me3, in turn, is a mark that is catalyzed by the SETD2 enzyme concomitantly with RNA polymerase II (RNA Pol II) transcriptional elongation (Wagner & Carpenter, 2012). Abrogation of this cross‐talk—either by deleting the PWWP domain of DNMT3 or by preventing H3K36me3 deposition through SETD2 depletion—specifically impairs gene body DNA methylation (Baubec et al, 2015; Morselli et al, 2015). Moreover, all eukaryotic species that have gene body methylation have retained DNMT3 enzymes or have evolved other types of DNMTs fused with PWWP domains, as observed in chlorophyte algae.
However, transcription‐coupled deposition of H3K36me3 is more widely conserved than DNA methylation: It is also found in species with a DNA methylation‐free genome, such as yeast or Drosophila. By promoting repressive chromatin within actively transcribed genes, H3K36me3 is universally required for preventing spurious transcription initiation that can stem from cryptic promoters or transposon remnants. In yeast, H3K36me3‐dependent repression is achieved through histone deacetylation (Carrozza et al, 2005), but this does not seem to be the case in mammalian cells. Recruitment of DNA methylation has therefore been postulated to reinforce the suppression of spurious transcription initiation. However, there is also evidence for DNA methylation to play a role in splicing control: Exons are markedly more methylated than introns and DNA methylation patterns abruptly change at exon–intron boundaries (Lister et al, 2009).
Whatever the function of gene body DNA methylation is, it comes at a cost. Methylated cytosines are prone to spontaneous deamination: If this occurs in the germline, this can change the coding sequences over evolutionary time. Accordingly, the average CpG content of genes is inversely correlated with their internal DNA methylation density: Human genes are globally methylated and relatively CpG‐poor, while genes from the silk moth are mostly unmethylated and CpG‐rich (Nanty et al, 2011). So gene body methylation has been conserved over a billion years of evolution despite its mutagenicity, indicating important functions. But the nature of these functions has been difficult to clarify, mostly because of the genome‐wide effects of DNA methylation deficiencies.
In mouse embryonic stem (ES) cells, DNMT3B is the main driver of de novo DNA methylation of actively transcribed genes via its PWWP domain, while DNMT3A plays very little role in this process (Baubec et al, 2015). Taking advantage of this specificity, Neri et al (2017) went on tracking the role of gene body DNA methylation using mouse Dnmt3B −/− ES cells. This mutant did not involve a targeted deletion of the PWWP H3K36me3‐reading domain though, meaning that DNA methylation was affected in all genomic compartments, not only gene bodies. Moreover, DNA methylation loss was subtle (15–20% decrease), as the Dnmt3B mutation was generated in serum‐grown ES cells—which express Dnmt3A and Dnmt1 at high levels—and further maintained in these conditions. The impact of DNMT3B on the general acquisition of DNA methylation could therefore not be assessed in this system, only its minor participation in the maintenance of methylation patterns. In reflection of these limited effects on the methylome, the transcriptome of Dnmt3B −/− ES cells was not strikingly different from wild‐type cells, as assessed by RNA‐seq.
Nevertheless, the authors could find evidence of cryptic initiation of transcription for 1,445 actively transcribed genes in Dnmt3B −/− ES cells. This was quantified by an increased representation of RNA‐seq reads mapping to downstream exons compared to the first exon of these genes. Further RNA Pol II chromatin immunoprecipitation (ChIP‐seq) and RNA immunoprecipitation with a CAP‐specific antibody (CAPIP‐seq) revealed that DNMT3B acts by restricting the entry of the stalled form of RNA Pol II into gene bodies. Single‐nucleotide mapping of the 5′ ends by DECAP‐seq showed that these spurious transcripts did not emanate from annotated, alternative promoters; rather, they initiated in “appealing” chromatin environment of constitutive nucleosome depletion, which implies occupancy of these cryptic start sites (TSSs) by transcription factors, even in wild‐type cells. Importantly, rescue experiments provided the crucial demonstration that it is the DNA methylation activity of DNMT3B—not solely its histone binding—that is required for protecting cryptic TSSs from the RNA Pol II initiation. Based on these results, the authors concluded that the recruitment of DNA methylation by H3K36me3 masks favorable but illegitimate contexts of transcription initiation within gene bodies, thereby forcing the usage of canonical, upstream promoters and ensuring transcription initiation fidelity (Fig 1). This role may have profound implications for development and cell physiology: The spurious intragenic transcripts produced in Dnmt3B −/− ES cells can be polyadenylated and associated with ribosomes, suggesting that they might lead to the production of aberrant proteins.
Figure 1. DNA methylation is preferentially targeted to gene bodies, and intragenic methylation restricts transcription initiation to canonical TSSs in mouse embryonic stem cells in a DNMT3B‐ and H3K36me3‐dependent manner.
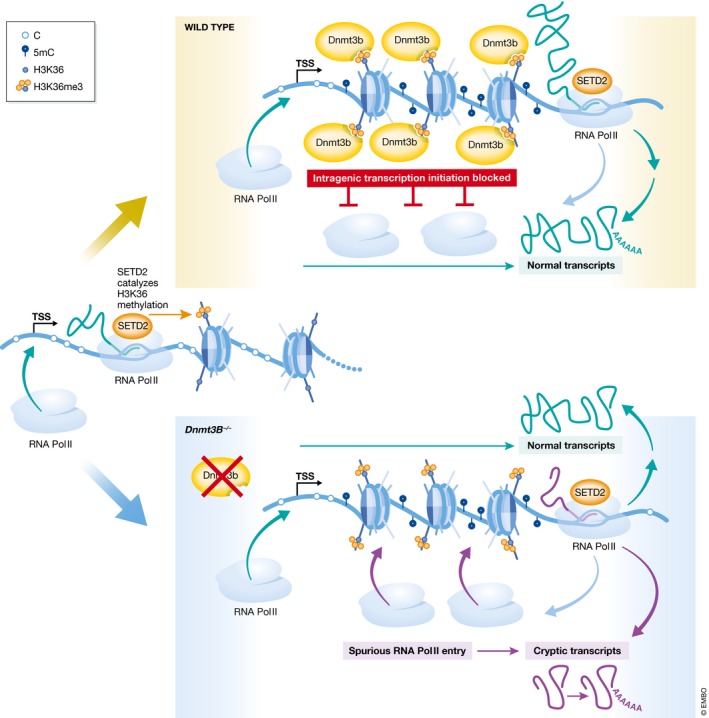
In DNMT3B KO embryonic stem cells, hypomethylated intragenic regions are entered by stalled RNA polymerase II, which leads to cryptic transcription initiation and generation of spurious transcripts, which can be polyadenylated and associated with ribosomes. Histone methyltransferase SETD2 is recruited by RNA Pol II during transcription elongation to catalyze H3K36me3 marks.
We note that initiation from cryptic TSSs did not reduce the usage of the main, upstream TSSs, while former models suggested the existence of exclusive competition between alternative TSSs of the same gene (Maunakea et al, 2010). Moreover, the very limited over‐usage of downstream versus first exon indicates that very few Dnmt3B −/− cells (likely one out of several thousands of cells) experience intragenic transcription events and/or that they occur in transient bursts. This limited effect could be due to the mild defect of gene body methylation in serum‐grown Dnmt3B −/− ES cells. We therefore tested the occurrence of spurious intragenic initiation when DNA methylation is drastically abrogated, using available RNA‐seq datasets from mutant ES cells for the three DNA methyltransferases (Dnmt‐tKO; Karimi et al, 2011) and from ES cells grown in conditions that chemically counteract DNA methylation activity (two inhibitors and vitamin C; Walter et al, 2016). However, by applying the same quantification of the ratio of intermediate to first exon usage to highly expressed genes, we could not detect signs of increased intragenic transcription (https://doi.org/10.6084/m9.figshare.4780483). While these results will require further exploration in the future, this may imply that the role of gene body DNA methylation on suppressing spurious transcription initiation may be specific to the lack of DNMT3B, while other DNMTs are still present. In this regard, it would be relevant to analyze the production of intragenic transcripts in the context of the ICF (immunodeficiency, centromeric instability, and facial anomalies) syndrome, a human pathology that is linked to DNMT3B mutations.
See also: F Neri et al (March 2017)
References
- Baubec T, Colombo DF, Wirbelauer C, Schmidt J, Burger L, Krebs AR, Akalin A, Schubeler D (2015) Genomic profiling of DNA methyltransferases reveals a role for DNMT3B in genic methylation. Nature 520: 243–247 [DOI] [PubMed] [Google Scholar]
- Carrozza MJ, Li B, Florens L, Suganuma T, Swanson SK, Lee KK, Shia WJ, Anderson S, Yates J, Washburn MP, Workman JL (2005) Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell 123: 581–592 [DOI] [PubMed] [Google Scholar]
- Feng S, Cokus SJ, Zhang X, Chen PY, Bostick M, Goll MG, Hetzel J, Jain J, Strauss SH, Halpern ME, Ukomadu C, Sadler KC, Pradhan S, Pellegrini M, Jacobsen SE (2010) Conservation and divergence of methylation patterning in plants and animals. Proc Natl Acad Sci USA 107: 8689–8694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi MM, Goyal P, Maksakova IA, Bilenky M, Leung D, Tang JX, Shinkai Y, Mager DL, Jones S, Hirst M, Lorincz MC (2011) DNA methylation and SETDB1/H3K9me3 regulate predominantly distinct sets of genes, retroelements, and chimeric transcripts in mESCs. Cell Stem Cell 8: 676–687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti‐Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, Edsall L, Antosiewicz‐Bourget J, Stewart R, Ruotti V, Millar AH, Thomson JA, Ren B, Ecker JR (2009) Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 462: 315–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maunakea AK, Nagarajan RP, Bilenky M, Ballinger TJ, D'Souza C, Fouse SD, Johnson BE, Hong C, Nielsen C, Zhao Y, Turecki G, Delaney A, Varhol R, Thiessen N, Shchors K, Heine VM, Rowitch DH, Xing X, Fiore C, Schillebeeckx M et al (2010) Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature 466: 253–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morselli M, Pastor WA, Montanini B, Nee K, Ferrari R, Fu K, Bonora G, Rubbi L, Clark AT, Ottonello S, Jacobsen SE, Pellegrini M (2015) In vivo targeting of de novo DNA methylation by histone modifications in yeast and mouse. eLife 4: e06205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanty L, Carbajosa G, Heap GA, Ratnieks F, van Heel DA, Down TA, Rakyan VK (2011) Comparative methylomics reveals gene‐body H3K36me3 in Drosophila predicts DNA methylation and CpG landscapes in other invertebrates. Genome Res 21: 1841–1850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neri F, Rapelli S, Krepelova A, Incarnato D, Parlato C, Basile G, Maldotti M, Anselmi F, Oliviero S (2017) Intragenic DNA methylation prevents spurious transcription initiation. Nature 543: 72–77 [DOI] [PubMed] [Google Scholar]
- Wagner EJ, Carpenter PB (2012) Understanding the language of Lys36 methylation at histone H3. Nat Rev Mol Cell Biol 13: 115–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter M, Teissandier A, Perez‐Palacios R, Bourc'his D (2016) An epigenetic switch ensures transposon repression upon dynamic loss of DNA methylation in embryonic stem cells. eLife 5: pii: e11418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemach A, McDaniel IE, Silva P, Zilberman D (2010) Genome‐wide evolutionary analysis of eukaryotic DNA methylation. Science 328: 916–919 [DOI] [PubMed] [Google Scholar]


