Abstract
The centromere, a chromosomal locus that acts as a microtubule attachment site, is epigenetically specified by the enrichment of CENP‐A nucleosomes. Centromere maintenance during the cell cycle requires HJURP‐mediated CENP‐A deposition, a process regulated by the Mis18 complex (Mis18α/Mis18β/Mis18BP1). Spatial and temporal regulation of Mis18 complex assembly is crucial for its centromere association and function. Here, we provide the molecular basis for the assembly and regulation of the Mis18 complex. We show that the N‐terminal region of Mis18BP1 spanning amino acid residues 20–130 directly interacts with Mis18α/β to form the Mis18 complex. Within Mis18α/β, the Mis18α MeDiY domain can directly interact with Mis18BP1. Mis18α/β forms a hetero‐hexamer with 4 Mis18α and 2 Mis18β. However, only two copies of Mis18BP1 interact with Mis18α/β to form a hetero‐octameric assembly, highlighting the role of Mis18 oligomerization in limiting the number of Mis18BP1 within the Mis18 complex. Furthermore, we demonstrate the involvement of consensus Cdk1 phosphorylation sites on Mis18 complex assembly and thus provide a rationale for cell cycle‐regulated timing of Mis18 assembly and CENP‐A deposition.
Keywords: Cdk1, CENP‐A deposition, centromere, HJURP, Mis18 complex
Subject Categories: Cell Cycle, Structural Biology
Introduction
Equal and identical distribution of chromosomes to each daughter cell during cell division is essential for maintaining genome integrity. A central player regulating this process is the kinetochore, a large proteinaceous structure assembled at a specialized region of the chromosome called the centromere 1, 2, 3. Kinetochores physically couple chromosomes with spindle microtubules to facilitate chromosome segregation. Consequently, correct kinetochore assembly and function depends on centromeres being maintained at the right place on chromosomes 3.
In most eukaryotes, the centromeric chromatin is epigenetically defined by the enrichment of nucleosomes containing the histone H3 variant CENP‐A 4, 5, 6, 7. To maintain centromere identity, new CENP‐A must be deposited in each cell cycle. The timing of CENP‐A deposition varies among species (G1—humans and G2—Schizosaccharomyces pombe); however, the molecular mechanisms by which it is achieved share considerable similarity 3, 5, 8, 9, 10, 11, 12, 13. This process is initiated by the centromere targeting of the Mis18 complex (composed of Mis18α, Mis18β, and Mis18BP1) along with canonical histone chaperones, RbAp46/48 14, 15, 16. Centromere association of the Mis18 complex directly or indirectly makes the underlying chromatin permissive for CENP‐A deposition. Mis18 proteins have also been shown to affect histone acetylation and DNA methylation at centromeres 14, 17. Mis18 centromere association subsequently allows HJURP, a CENP‐A‐specific chaperone, to associate with centromeres resulting in CENP‐A loading 18, 19. Finally, chromatin remodelers like MgcRacGAP, RSF, Ect2, and Cdc42 have been suggested to fully stabilize the centromeric chromatin by a poorly understood maturation process 20, 21. Timely CENP‐A deposition critical for centromere maintenance and function is determined by the kinase activities of Cdk1 and Plk1, which influence the centromere association of the Mis18 complex through negative regulation and positive regulation, respectively 22, 23.
The Mis18 proteins contain two structurally distinct domains (Yippee/MeDiY and a C‐terminal α‐helix), both of which can self‐oligomerize. Previously, we and others have shown that oligomerization of Mis18 proteins is required for their centromere association and function both in S. pombe and humans 24, 25. The human Mis18 paralogs, Mis18α and Mis18β, directly interact with Mis18BP1 and CENP‐C, respectively, facilitating Mis18 complex formation and centromere association 14, 26. However, the molecular basis for the cell cycle‐dependent regulation of Mis18 complex formation, and thus, CENP‐A deposition has yet to be defined.
Here, we show that Mis18BP1, through its highly conserved N‐terminal region comprising amino acids 20–130 (Mis18BP120–130), binds Mis18α/β hetero‐oligomer via the Mis18α/β MeDiY hetero‐dimer to form the Mis18 complex. Characterization of the oligomeric structures of the Mis18 subcomplexes revealed that the Mis18α/β complex is a hetero‐hexamer with four copies of Mis18α and two copies of Mis18β, while the Mis18 holo‐complex is a hetero‐octamer with two additional copies of Mis18BP1. We identify two conserved consensus Cdk1 phosphorylation sites (T40 and S110) within Mis18BP120–130 and show that phospho‐mimicking mutations of these residues disrupt Mis18 complex formation. This explains the molecular basis for Cdk1‐mediated timing of Mis18 assembly and CENP‐A deposition.
Results and Discussion
The N‐terminal region of Mis18BP1 comprising amino acids 20–130 directly interacts with Mis18α/β to form the Mis18 complex
Mis18BP1 contains two evolutionary conserved domains, the SANTA (residues 385–472) and SANT (residues 878–927) domains (Figs 1A and EV1A). It has previously been shown that a region overlapping the SANTA domain, Mis18BP1475–878, interacts with CENP‐C while Mis18BP11–376 can interact with Mis18α 15, 22, 26. The amino acid sequence analysis revealed the presence of a highly conserved 110 amino acid stretch within the first 130 amino acids of Mis18BP1 (residues 20–130; Mis18BP120–130; Fig 1A). We hypothesized that Mis18BP120–130 might be the minimal region that interacts with Mis18α/β. To test this, we expressed TetR‐eYPF‐Mis18α in HeLa 3‐8 cells containing a synthetic alphoid DNA array with tetracycline operator sequences (alphoidtetO array) integrated in a chromosome arm 27 and analyzed the ability of Mis18α to recruit full‐length Mis18BP1 (mCherry‐Mis18BP1fl) as well as two different N‐terminal fragments: mCherry‐Mis18BP120–130 and mCherry‐Mis18BP1336–483 (covering the SANTA domain) to the tethering site. Mis18α tethered to the ectopic alphoidtetO array recruited Mis18BP1fl and Mis18BP120–130 more robustly compared to the Mis18BP1336–483 (Fig 1B). As expected, Mis18α tethered to the synthetic array recruited Mis18β along with Mis18BP120–130 (Fig EV1B). These data suggest that the Mis18BP120–130 is sufficient to interact with Mis18α/β to form the Mis18 complex in vivo.
Figure 1. Mis18BP120–130 is sufficient to interact with Mis18α/β.
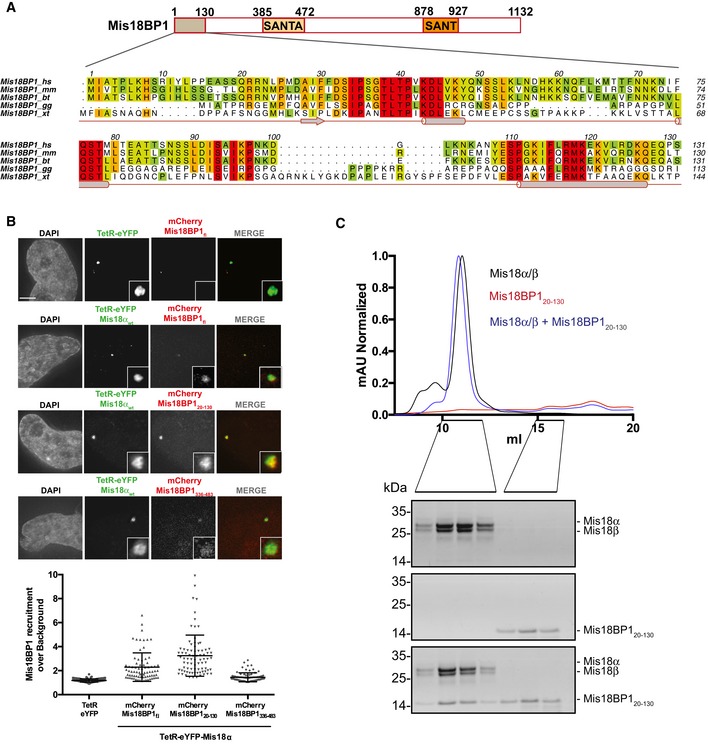
- Schematic representation of Mis18BP1 domain architecture, amino acid conservation (conservation score is mapped from red to cyan, where red corresponds to highly conserved and cyan to poorly conserved), and secondary structure (Conserved Domain Database [CDD] and PsiPred, http://bioinf.cs.ucl.ac.uk/psipred). Alignments include Homo sapiens (hs), Mus musculus (mm), Bos taurus (bt), Gallus gallus (gg), and Xenopus tropicalis (xt). Multiple sequence alignments were performed with MUSCLE 31 and edited with Aline 32.
- Representative fluorescence images (top) and quantification (bottom) for the analysis of mCherry‐Mis18BP1 recruitment to the alphoidtetO array by TetR‐eYFP‐Mis18α. HeLa 3‐8 cells co‐transfected with either TetR‐eYFP or TetR‐eYFP‐Mis18αwt and mCherry vectors containing the indicated versions of Mis18BP1. Middle bars show median whilst error bars show SEM. Mann–Whitney test vs. TetR‐eYFP; P ≤ 0.0001, n ≥ 77. Scale bar, 5 μm.
- SEC profiles and respective SDS–PAGE analysis of Mis18α/β, Mis18BP120–130, and Mis18α/β mixed with molar excess of Mis18BP120–130.
Figure EV1. Mis18BP1 amino acid conservation and Mis18 complex formation with the well conserved Mis18BP1 N‐terminal region (amino acids 20–130) in vivo .
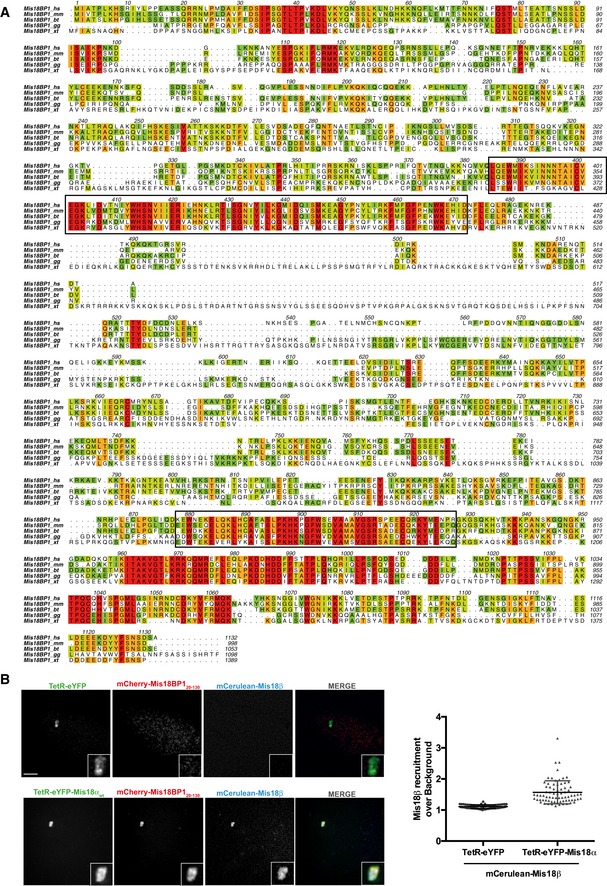
- Alignments include Homo sapiens (hs), Mus musculus (mm), Bos taurus (bt), Gallus gallus (gg), and Xenopus tropicalis (xt). Black boxes indicate SANTA and SANT domains, respectively. Multiple sequence alignment (conservation score is mapped from red to cyan, where red corresponds to highly conserved and cyan to poorly conserved) was performed with MUSCLE (MUltiple Sequence Comparison by Log‐Expectation, EMBL‐EBI) 31 and edited with Aline (sequence alignment editor) 32.
- Representative images (left) and quantification (right) for the recruitment of mCherry‐Mis18BP120–130 and mCerulean‐Mis18β to the alphoidtetO array in HeLa 3‐8 cells expressing TetR‐eYFP‐Mis18αwt. Middle bars show median whilst error bars show SEM. Mann–Whitney test vs. TetR‐eYFP; P ≤ 0.0001, n ≥ 63. Scale bar, 5 μm.
To confirm that Mis18BP120–130 can directly interact with Mis18α/β in vitro, individually purified recombinant proteins (Mis18α/β and Mis18BP120–130) were analyzed using size‐exclusion chromatography (SEC) before and after complex formation. While Mis18α/β and Mis18BP120–130 eluted at 11.0 and 15.7 ml, respectively, the complex containing Mis18α/β and Mis18BP120–130 eluted at 10.9 ml (Fig 1C). The elution volume of the Mis18α/Mis18β/Mis18BP120–130 complex is not very different from Mis18α/β, suggesting that the binding of Mis18BP120–130 does not significantly alter the hydrodynamic radius of the Mis18α/β. These data, together with the in vivo tethering assays, confirm that Mis18BP120–130 is sufficient to make a stable and direct interaction with Mis18α/β to form the Mis18 complex.
The Mis18α MeDiY domain can directly interact with Mis18BP120–130 in vitro
Mis18 proteins possess a globular MeDiY domain (Mis18αMeDiY: 77–187 and Mis18βMeDiY: 56–183) and a C‐terminal α‐helical domain (Fig 2A). Our previous structure‐function analysis of S. pombe Mis18 revealed that its centromere association and function requires Mis18 MeDiY dimerization 24. Stellfox et al 26 showed that mutations within the conserved C‐X‐X‐C motif of the Mis18αMeDiY domain perturbed its ability to interact with Mis18BP1, suggesting a direct role of Mis18α for Mis18 complex formation. However, as the C‐X‐X‐C motif mutation is expected to affect Zn binding required to stabilize the structure, it was not clear whether the inability of this mutant to interact with Mis18BP1 is a primary or secondary consequence.
Figure 2. Mis18αMeDiY domain directly interacts with Mis18BP120–130 .
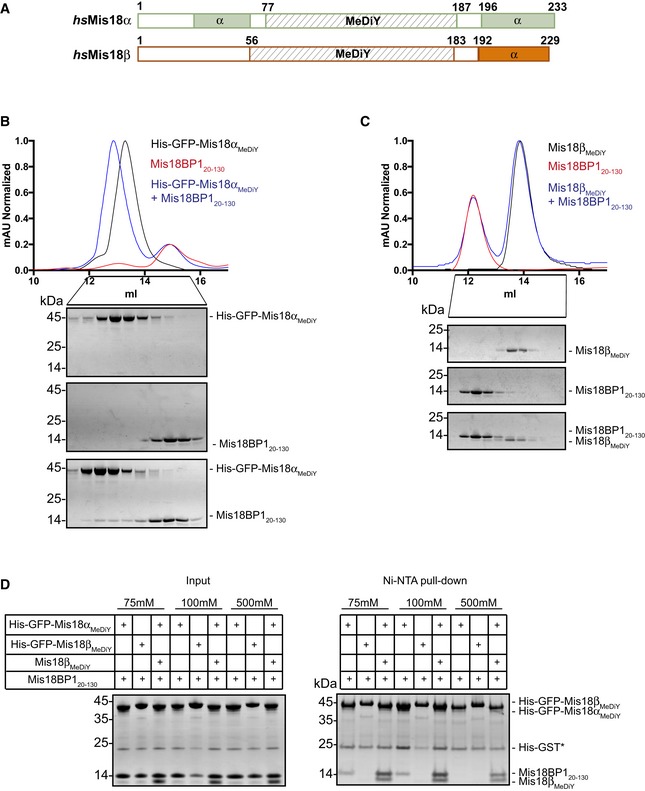
-
ASchematic representation of hsMis18α, hsMis18β domain architecture (using CDD and PsiPred).
-
B, CSEC profiles and respective SDS–PAGE analysis of (B) His‐GFP‐Mis18αMeDiY, Mis18BP120–130, and His‐GFP‐Mis18αMeDiY mixed with molar excess of Mis18BP120–130 and (C) Mis18βMeDiY, Mis18BP120–130, and Mis18βMeDiY with molar excess of Mis18BP120–130.
-
DSDS–PAGE analysis of the Ni‐NTA pull‐down assay where recombinant His‐GFP‐Mis18αMeDiY, His‐GFP‐Mis18βMeDiY, and Mis18βMeDiY were mixed with Mis18BP120–130 in different combinations in either 75, 100, or 500 mM NaCl. Left panel: inputs and right panel: Ni‐NTA pull‐down. * Contamination carried over from Mis18BP1 purification.
To determine whether the Mis18αMeDiY domain is the minimal interaction surface of Mis18α/β required for Mis18BP120–130 binding, we purified recombinant Mis18αMeDiY and tested its ability to form a complex with Mis18BP120–130. The SEC elution volumes of Mis18αMeDiY on its own and in the presence of Mis18BP120–130 were 11.9 and 11.5 ml, respectively, demonstrating a direct interaction between Mis18αMeDiY and Mis18BP120–130 (Fig EV2A). However, due to their almost identical molecular weights (MW), Mis18αMeDiY (12.4 kDa) and Mis18BP120–130 (12.7 kDa) migrate similarly in the SDS–PAGE (Fig EV2A, bottom panel). To unambiguously validate the Mis18αMeDiY‐Mis18BP120–130 interaction, we purified His‐GFP‐Mis18αMeDiY and analyzed its ability to interact with Mis18BP120–130 in SEC (using Superdex 200 increase 10/300). While His‐GFP‐Mis18αMeDiY on its own eluted at 13.8 ml, the His‐GFP‐Mis18αMeDiY/Mis18BP120–130 complex eluted at 13.3 ml (Fig 2B), confirming a direct interaction between Mis18αMeDiY and Mis18BP120–130.
Figure EV2. Mis18αMeDiY can directly interact with Mis18BP120–130 .
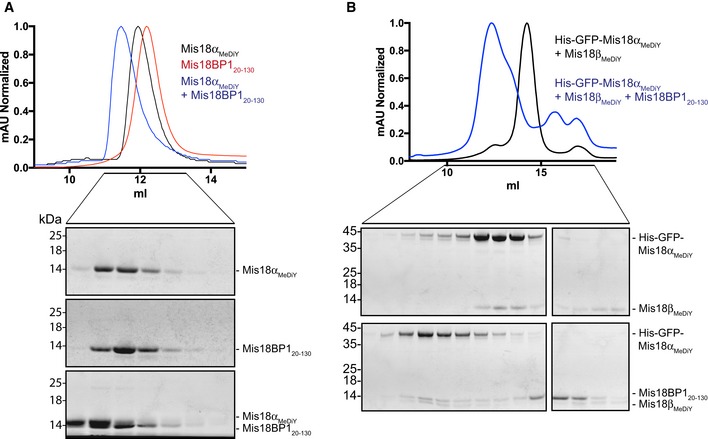
-
A, BSEC profiles and respective SDS–PAGE analyses of (A) Mis18αMeDiY, Mis18BP120–130, and Mis18αMeDiY mixed with two times molar excess of Mis18BP120–130. Mis18αMeDiY elutes at 11.9 ml while Mis18αMeDiY/Mis18BP120–130 elutes at 11.5 ml. Proteins were separated using a Superdex 75 10/300 column and (B) His‐GFP‐Mis18αMeDiY/Mis18βMeDiY and His‐GFP‐Mis18αMeDiY/Mis18βMeDiY mixed with two times molar excess of Mis18BP120–130 elutes at 14.3 ml and 12.4 ml, respectively. Proteins were analyzed using a Superdex 200 increase 10/300 column.
As Mis18αMeDiY and Mis18βMeDiY share 44% sequence similarity, we tested whether the Mis18βMeDiY can also directly interact with Mis18BP1. Size‐exclusion chromatography analysis of a sample containing Mis18βMeDiY and Mis18BP120–130 (using Superdex 75 10/300) showed that they elute at distinct elution volumes, 13.9 and 12.2 ml, respectively, demonstrating a lack of interaction (Fig 2C). This, together with a published report 26, shows that Mis18α, rather than Mis18β, mediates Mis18BP1 interaction to form the Mis18 complex.
Mis18BP120–130 binds Mis18αMeDiY/βMeDiY hetero‐dimer more robustly than Mis18αMeDiY
We had previously shown that Mis18αMeDiY prefers to hetero‐dimerize with Mis18βMeDiY over homo‐dimerizing with itself 28. Hence, we wanted to determine whether Mis18αMeDiY could still bind Mis18BP120–130 while part of the Mis18αMeDiY/βMeDiY hetero‐dimer. Ni‐NTA pull‐down assays were performed using purified His‐GFP‐Mis18αMeDiY, His‐GFP‐Mis18βMeDiY, and His‐GFP‐Mis18αMeDiY/Mis18βMeDiY with Mis18BP120–130 at varying salt concentrations (75–500 mM NaCl). Consistent with the SEC analysis shown in Figs 2B and C, and EV2A, when tested in isolation, only His‐GFP‐Mis18αMeDiY could bind Mis18BP120–130. His‐GFP‐Mis18βMeDiY failed to interact with Mis18BP120–130 even under a low ionic strength condition (Fig 2D). Interestingly, His‐GFP‐Mis18αMeDiY/βMeDiY hetero‐dimer interacted with Mis18BP120–130 (Figs 2D and EV2B). Moreover, in a high ionic strength buffer (containing 500 mM NaCl), only His‐GFP‐Mis18αMeDiY/Mis18βMeDiY bound Mis18BP120–130, demonstrating that Mis18BP1 preferentially binds Mis18αMeDiY/βMeDiY hetero‐dimer. These data indicate that either Mis18αMeDiY makes additional contacts with Mis18BP1 in the presence of Mis18βMeDiY or Mis18βMeDiY strengthens Mis18αMeDiY‐Mis18BP120–130 binding by directly interacting with Mis18BP120–130 in a Mis18αMeDiY‐dependent manner.
Mis18α/β forms a hetero‐hexamer comprising four copies of Mis18α and two copies of Mis18β
We and others have shown that homo‐ and hetero‐oligomerization of Mis18 proteins are crucial for Mis18 function in S. pombe 24 and in humans 25, respectively. However, the precise subunit composition of human Mis18α/β hetero‐oligomer and its consequence on Mis18 complex assembly and function remain elusive.
First, we determined the absolute molecular mass of untagged Mis18α/β complex using SEC combined with multi‐angle light scattering (SEC‐MALS). The measured MW of the untagged Mis18α/β complex was 151.2 ± 2.9 kDa (calculated MWs of Mis18α and Mis18β are 25.9 kDa and 24.7 kDa, respectively) and correlated well with the calculated MW of a hetero‐hexamer (151.8 kDa, using the average MW of Mis18 proteins 25.3 kDa; Figs 3A and EV3A). This contrasts with a previous report by Nardi et al 25, which used glycerol‐based gradient experiments to show that Mis18α/β is a hetero‐tetramer.
Figure 3. Mis18α/β is a hetero‐hexamer (4 Mis18α : 2 Mis18β) which binds two copies of Mis18BP1 to form a hetero‐octameric Mis18 complex.
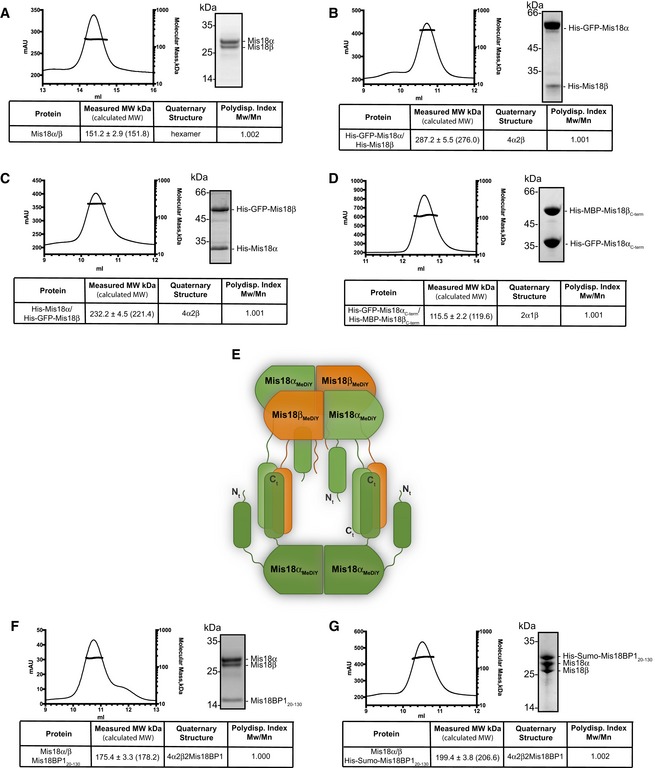
-
A–DSEC‐MALS analysis of (A) Mis18α/β complex, (B) His‐GFP‐Mis18α/His‐Mis18β, (C) His‐Mis18α/His‐GFP‐Mis18β, (D) His‐GFP‐Mis18αC‐term/His‐MBP‐Mis18βC‐term. Absorption at 280 nm (mAU, left y‐axis) and molecular mass (kDa, right y‐axis) are plotted against elution volume (ml, x‐axis). Measured molecular weight (MW) and the calculated subunit stoichiometry based on the predicted MW of different subunit compositions (Fig EV3) are shown in the tables below.
-
EProposed model for Mis18α/β complex assembly. The Mis18α/β hetero‐hexamer contains 4 Mis18α and 2 Mis18β. The C‐terminal α‐helical domains form a hetero‐trimer (2 Mis18α and 1 Mis18β). While 2 Mis18αMeDiY domains engage with 2 Mis18βMeDiY domains to facilitate Mis18α/β hetero‐hexamerization, the remaining 2 Mis18αMeDiY self‐oligomerize. In this model, within each C‐terminal α‐helical assembly, Mis18α helices are shown in anti‐parallel arrangement. In the absence of any structural data, an alternate model with C‐terminal helices arranged in parallel orientation is equally probable.
-
F, GSEC‐MALS analysis of (F) Mis18α/Mis18β/Mis18BP120–130 and (G) Mis18α/Mis18β/His‐Sumo‐Mis18BP120–130.
Figure EV3. Predicted molecular weights for Mis18 complexes with different subunit stoichiometry.
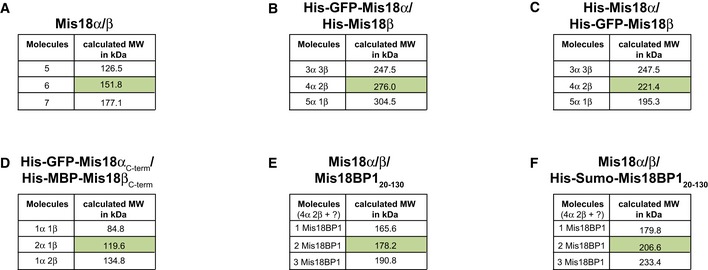
-
A–FPredicted MWs of (A) Mis18α/β, (B) His‐GFP‐Mis18α/His‐Mis18β, (C) His‐Mis18α/His‐GFP‐Mis18β, (D) His‐GFP‐Mis18αC‐term/His‐MBP‐Mis18βC‐term, (E) Mis18α/β/Mis18BP120–130, (F) Mis18α/β/His‐Sumo‐Mis18BP120–130 complexes with different subunit compositions. These values together with the measured MWs from SEC‐MALS analysis shown in Fig 3 were used to deduce the correct subunit composition.
As Mis18α and Mis18β are very similar in size (~25 kDa), it is almost impossible to accurately determine the subunit stoichiometry within the Mis18 hetero‐hexamer. Hence, we introduced a noticeable size variation by purifying His‐GFP‐Mis18α in a complex with His‐Mis18β. Interestingly, the measured MW of His‐GFP‐Mis18α/His‐Mis18β complex was 287.2 ± 5.5 kDa, matching the calculated MW of a hetero‐hexamer containing four copies of His‐GFP‐Mis18α and two copies of His‐Mis18β (276 kDa; Figs 3B and EV3B). The measured MW of Mis18α/β complex formed using His‐Mis18α and His‐GFP‐Mis18β revealed same stoichiometry as above (measured MW = 232.2 ± 4.5 kDa; calculated MW of 4 His‐Mis18α : 2 His‐GFP‐Mis18β = 221.4 kDa; Figs 3C and EV3C) and demonstrates that Mis18α/β is a hetero‐hexamer with a 4:2 stoichiometry.
Mis18α/β hetero‐hexamer is assembled from hetero‐trimers of C‐terminal α‐helical domains and hetero‐dimers of MeDiY domains
We had previously shown that the MeDiY domains of Mis18α and Mis18β form a homo‐dimer and a monomer, respectively, but can form a hetero‐dimer 28. In addition, Nardi et al 25 have shown that the C‐terminal α‐helical domains also have the ability to oligomerize. These observations together with our data that the Mis18α/β complex is a hetero‐hexamer (Fig 3A–C) prompted us to define the stoichiometry of the Mis18α/β C‐terminal helical assembly. Using individually purified His‐GFP‐Mis18αC‐term (Mis18α 188‐end) and His‐MBP‐Mis18βC‐term (Mis18β 184‐end), we reconstituted the C‐terminal helical assembly and analyzed their composition using SEC‐MALS (Figs 2A and 3D). The measured MW of His‐GFP‐Mis18αC‐term/His‐MBP‐Mis18βC‐term complex was 115.5 ± 2.2 kDa, which matches a calculated MW of a hetero‐trimeric assembly with 2 His‐GFP‐Mis18αC‐term and 1 His‐MBP‐Mis18βC‐term (119.6 kDa; Figs 3D and EV3D). This suggests that the formation of the full‐length hetero‐hexameric Mis18α/β assembly requires further oligomerization of Mis18α/β hetero‐trimers mediated by Mis18αMeDiY/βMeDiY hetero‐dimers (Fig 3E).
The Mis18 complex is a hetero‐octamer with two copies of Mis18BP1
The measured MW of the Mis18 holo‐complex (Mis18α/Mis18β/Mis18BP120–130) by SEC‐MALS (175.4 ± 3.3 kDa) is in agreement with the calculated MW of a hetero‐octamer containing a Mis18α/β hetero‐hexamer plus two copies of Mis18BP120–130 (178.2 kDa; Figs 3F and EV3E). Since the MW of Mis18BP120–130 is only 12.7 kDa, we reconstituted the Mis18 holo‐complex with His‐SUMO‐Mis18BP120–130. The measured MW of this complex (199.4 ± 3.8 kDa) matches the calculated MW (206.6 kDa; Figs 3G and EV3F) of a Mis18α/β hexamer bound to two copies of His‐SUMO‐Mis18BP120–130, confirming that the Mis18 holo‐complex is a hetero‐octamer with just two copies of Mis18BP1.
Although both Mis18αMeDiY and Mis18αMeDiY/βMeDiY hetero‐dimer can interact with Mis18BP1 (Figs 2B and D, and EV2), only two copies of Mis18BP1 bind to a Mis18α/β hexamer containing 2 Mis18αMeDiY and 2 Mis18αMeDiY/βMeDiY hetero‐dimers (Fig 3F and G). We propose that Mis18αMeDiY/βMeDiY hetero‐dimer is preferred over Mis18αMeDiY due its relatively stronger binding to Mis18BP1 (Figs 2D and 4A), thus limiting the number of Mis18BP1 binding sites to 2 in the holo‐complex. It is also possible that the Mis18BP1 binding sites of 2 Mis18αMeDiY domains that are not part of the Mis18αMeDiY/βMeDiY hetero‐dimer are occluded in the context of the full‐length Mis18α/β hetero‐hexamer (Fig 4A).
Figure 4. Hetero‐hexamerization of Mis18α/β mediated via the MeDiY domain is required for Mis18BP1 binding and CENP‐A loading in vivo .
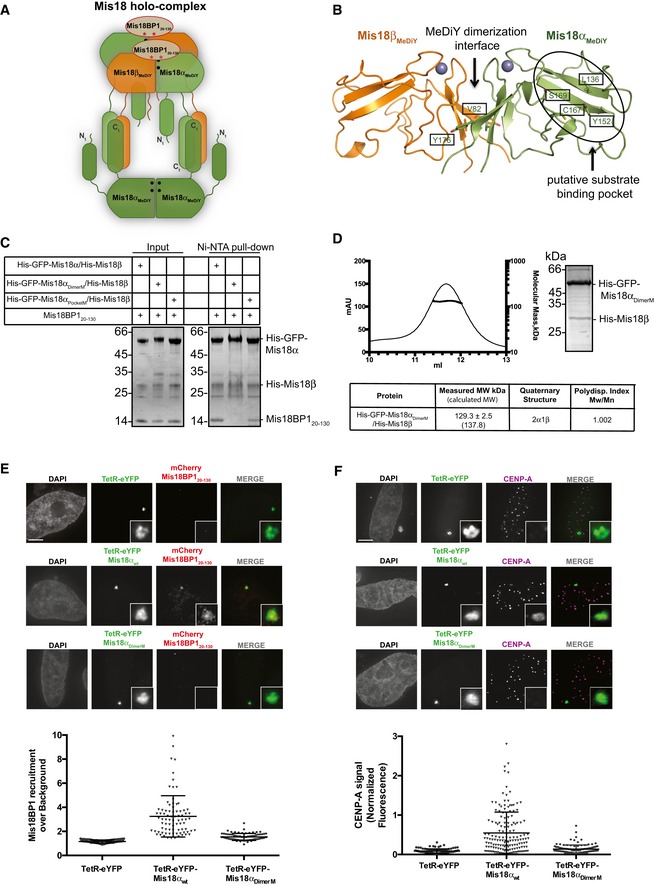
-
AProposed model for Mis18 complex assembly. Our data showing that the Mis18 holo‐complex contains just two copies of Mis18BP1 (Fig 3F and G) and that Mis18αMeDiY/βMeDiY hetero‐dimer binds Mis18BP120‐130 more robustly as compared to Mis18αMeDiY (Fig 2D) suggest that Mis18BP1 prefers Mis18αMeDiY/βMeDiY hetero‐dimer over Mis18αMeDiY. Filled circles (•) and asterisks (*) represent the sites of Mis18α residues involved in MeDiY dimerization (V82 and Y176; Fig 4B) and consensus Cdk1 phosphorylation on Mis18BP120–130 (T40 and S110; Fig 5A), respectively.
-
BCartoon representation of the homology‐modeled human Mis18αMeDiY/βMeDiY hetero‐dimer (with Phyre2 web server, www.sbg.bio.ic.ac.uk/phyre2/, using S. pombe Mis18 MeDiY domain—PDB: 5HJ0). Key amino acid residues forming the dimeric interface and putative substrate‐binding pocket are shown in stick representation. Residues mutated in this study are highlighted by boxes.
-
CSDS–PAGE analysis of the Ni‐NTA pull‐down assay where recombinant His‐GFP‐Mis18αwt/His‐Mis18β, His‐GFP‐Mis18αPocketM/His‐Mis18β, and His‐GFP‐Mis18αDimerM/His‐Mis18β were mixed with Mis18BP120–130. Left panel: inputs and right panel: Ni‐NTA pull‐down.
-
DSEC‐MALS profile of His‐GFP‐Mis18αDimerM/His‐Mis18β. Absorption at 280 nm (mAU, left y‐axis) and molecular mass (kDa, right y‐axis) plotted against elution volume (ml, x‐axis).
-
E, FRepresentative fluorescence images (top) and quantification (bottom) for the ability of TetR‐eYFP, TetR‐eYFP‐Mis18αwt, and TetR‐eYFP‐Mis18αDimerM (E) to recruit mCherry‐Mis18BP120–130 to the alphoidtetO array (Mann–Whitney test vs. TetR‐eYFP; P ≤ 0.0001, n ≥ 78) and (F) to deposit CENP‐A at the tethering site (Mann–Whitney test vs. TetR‐eYFP; P ≤ 0.0001, n ≥ 106). Middle bars show median whilst error bars show SEM. Scale bars, 5 μm. Data for TetR‐eYFP‐Mis18αwt/mCherry‐Mis18BP120–130‐transfected cells (Fig 1B) have been used as a control.
MeDiY dimerization interface of Mis18α is required for Mis18 oligomerization and Mis18BP1 binding
Our previous structural analysis of the S. pombe MeDiY domain identified a putative substrate‐binding pocket and a dimerization interface, both required for Mis18 function 24. To evaluate whether equivalent surfaces in human Mis18α are required for Mis18BP1 binding, we introduced mutations V82E/Y176D (Mis18αDimerM) at the MeDiY dimeric interface of Mis18α/β and L136A/Y152A/C167A/S169K (Mis18αPocketM) in the putative substrate‐binding pocket using a S. pombe‐based homology‐modeled structure (Fig 4B). We then analyzed Mis18BP1 binding to His‐GFP‐Mis18α/His‐Mis18β (Mis18αwt/β), His‐GFP‐Mis18αDimerM/His‐Mis18β (Mis18αDimerM/β), and His‐GFP‐Mis18αPocketM/His‐Mis18β (Mis18αPocketM/β) complexes using a Ni‐NTA pull‐down assay. While Mis18αwt/β and Mis18αPocketM/β interacted with Mis18BP120–130, Mis18αDimerM/β failed to do so efficiently (Fig 4C). This suggests that the Mis18αMeDiY interface proposed to mediate dimerization with Mis18βMeDiY is also required for Mis18BP1 binding (in agreement with Fig 2D). Size‐exclusion chromatography analysis of Mis18αDimerM/β, unlike Mis18αwt/β, showed the presence of several distinct populations including an aggregated and a smaller MW species (Fig EV4). The measured MW of the smaller MW species (the sample eluting at 11.7 ml) using SEC‐MALS was 129.3 ± 2.5 kDa (Figs 4D and EV4). This correlates with the calculated MW of a complex containing 2 His‐GFP‐Mis18αDimerM and 1 His‐Mis18β (137.8 kDa). This strengthens the notion that the Mis18α/β hetero‐trimer is the core oligomeric unit formed through the interactions of the C‐terminal helices which then assemble into a hetero‐hexamer via the MeDiY‐dimerization interface (Figs 3D and 4A). We conclude that the Mis18αMeDiY dimerization interface is required both for Mis18BP1 binding and for the higher order oligomerization of the Mis18α/β hetero‐trimer (Fig 4A).
Figure EV4. Characterization of Mis18αDimerM/Mis18β.
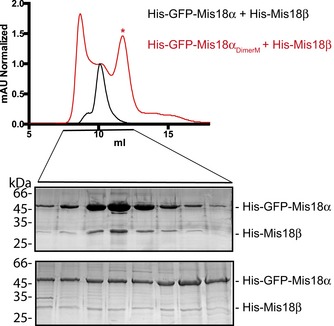
SEC profiles and SDS–PAGE analyses of His‐GFP‐Mis18α/His‐Mis18β and His‐GFP‐Mis18αDimerM/His‐Mis18β complexes. SEC analyses were carried out using a Superdex 200 increase 10/300 column. Sample used for SEC‐MALS is indicated with an asterisk (*).
We next evaluated the contribution of Mis18αMeDiY dimerization interface on Mis18BP1 binding in vivo by tethering TetR‐eYFP‐Mis18αDimerM to the alphoidtetO array and probing the recruitment of mCherry‐Mis18BP120–130. In agreement with the in vitro data, the TetR‐eYFP‐Mis18αDimerM failed to recruit mCherry‐Mis18BP120–130 (Fig 4E). Furthermore, this mutant, unlike the TetR‐eYFP‐Mis18αwt, was unable to deposit CENP‐A at the tethering site (Fig 4F). This confirms that the Mis18αMeDiY dimerization interface is required for Mis18BP1 binding and CENP‐A deposition.
Cdk1 consensus sites of Mis18BP1 are directly involved in Mis18α/β binding
Mis18 complex assembly and its centromere localization have been suggested to be regulated by Cdk1 in a cell cycle‐dependent manner 22, 23. Interestingly, the amino acid sequence analysis revealed two consensus Cdk1 phosphorylation sites, T40 and S110, within Mis18BP120–130 (Fig 5A), which we show here is able to interact with Mis18α/β (Fig 1). Notably, Mis18BP1 S110 has previously been shown to be phosphorylated in vivo 23. We hypothesized that phosphorylation of Mis18BP1 T40 or/and S110 negatively regulates its association with Mis18α/β and hence Mis18 complex assembly.
Figure 5. Consensus Cdk1 phosphorylation sites of Mis18BP1, T40, and S110, are directly involved in Mis18α/β binding.
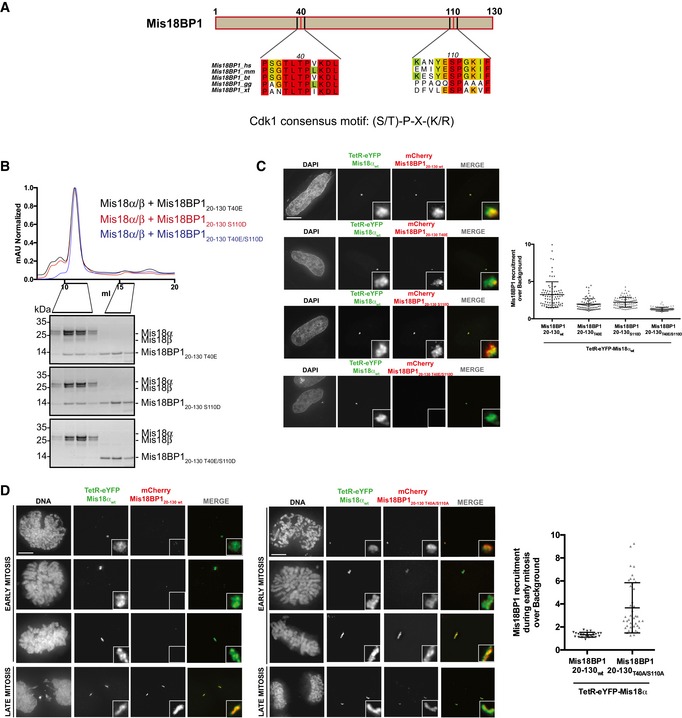
- Multiple sequence alignment of amino acid stretches containing the consensus Cdk1 phosphorylation sites within Mis18BP120–130. Alignments include Homo sapiens (hs), Mus musculus (mm), Bos taurus (bt), Gallus gallus (gg), and Xenopus tropicalis (xt). Multiple sequence alignment (conservation score is mapped from red to cyan, where red corresponds to highly conserved and cyan to poorly conserved) was performed with MUSCLE 31 and edited with Aline 32.
- SEC profiles and respective SDS–PAGE analysis of Mis18α/β mixed with molar excess of Cdk1 phospho‐mimic mutants, Mis18BP120–130 T40E, Mis18BP120–130 S110D, and Mis18BP120–130 T40E/S110D.
- Representative images (left) and quantification (right) for the recruitment of different mCherry‐Mis18BP120–130 Cdk1 phospho‐mimic mutants (T40E, S110D or T40E/S110D) to the alphoidtetO array in HeLa 3‐8 cells expressing TetR‐eYFP‐Mis18α. Middle bars show median whilst error bars show SEM. Mann–Whitney test vs. TetR‐eYFP; P ≤ 0.0001, n ≥ 78. Scale bar, 10 μm. Data for TetR‐eYFP‐Mis18αwt/mCherry‐Mis18BP120–130‐transfected cells (Fig 1B) have been used as a control.
- Representative images (left) for the cell cycle‐dependent and independent recruitment of mCherry‐Mis18BP120–130 wt and mCherry‐Mis18BP120–130 T40A/S110A, respectively, to the alphoidtetO array in HeLa 3‐8 cells expressing TetR‐eYFP‐Mis18α. Quantification (right) of Mis18BP120–130 wt and Mis18BP120–130 T40A/S110A recruitment to the array during early mitosis. Middle bars show median whilst error bars show SEM. Mann–Whitney test vs. Mis18BP120–130 wt; P ≤ 0.0001, n = 23 (mCherry‐Mis18BP120–130 wt) and n = 41 (mCherry‐Mis18BP120–130 T40A/S110A). Scale bar, 10 μm.
We evaluated the effect of Mis18BP1 phospho‐mimicking mutations T40E and S110D, either alone (Mis18BP120–130 T40E and Mis18BP120–130 S110D) or in combination (Mis18BP120–130 T40E/S110D), on Mis18 complex formation using SEC. Both Mis18BP120–130 T40E and Mis18BP120–130 S110D interacted with Mis18α/β. However, Mis18BP120–130 T40E/S110D failed to form a complex with Mis18α/β (Fig 5B), demonstrating a direct role of conserved Cdk1 phosphorylation sites for Mis18 complex formation in vitro.
Consistent with the in vitro data, Mis18BP120–130 T40E and Mis18BP120–130 S110D were recruited to the alphoidtetO array by TetR‐eYFP‐Mis18α, albeit less efficiently as compared to Mis18BP120–130 wt (Fig 5C). The recruitment of Mis18BP120–130 T40E/S110D mutant by TetR‐eYFP‐Mis18α was almost completely abolished, highlighting the involvement of the consensus Cdk1 phosphorylation sites of Mis18BP1, T40 and S110, in forming a direct Mis18α/β binding interface.
To further study the cell cycle regulation of Mis18BP1, we tested the recruitment of mCherry‐Mis18BP120–130 to the alphoidtetO array by TetR‐eYFP‐Mis18α during different stages of mitosis (Fig 5D). Interestingly, in agreement with a previously reported suggestion 22, Mis18α was unable to recruit Mis18BP120–130 wt during early stages of mitosis when Cdk1 levels are high. On the contrary, a non‐phosphorylatable mutant of Mis18BP1 (mCherry‐Mis18BP120–130 T40A/S110A) was recruited by TetR‐eYFP‐Mis18α to the array throughout the cell cycle. Consistent with this, recombinantly purified Mis18BP120–130 T40A/S110A interacted with Mis18α/β as analyzed by SEC (Fig EV5).
Figure EV5. Mis18BP120–130 T40A/S110A binds Mis18α/β in vitro .
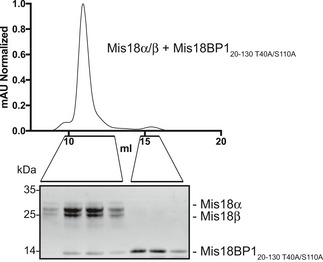
SEC profile and respective SDS–PAGE analysis of Mis18α/β mixed with two times molar excess of Cdk1 non‐phosphorylatable mutant, Mis18BP120–130 T40A/S110A. SEC run was carried out using a Superdex 200 increase 10/300 column.
The temporal regulation of Mis18 complex formation defines the timing of HJURP‐mediated CENP‐A deposition essential for centromere inheritance and function 18, 22, 23. Oligomerization of Mis18 proteins and Cdk1 activity are both key regulators of Mis18 complex assembly 22, 23, 24, 25. Our work presented here, in agreement with a parallel study by Pan et al 29, shows that the Mis18α/β complex is a hetero‐hexamer made of 2 Mis18α/β hetero‐trimers, each with 2 Mis18α and 1 Mis18β that are held together by the hetero‐trimeric C‐terminal α‐helical assembly. One Mis18αMeDiY from each Mis18α/β trimer hetero‐dimerizes with Mis18βMeDiY to form a Mis18α/β hetero‐hexamer. This arrangement results in 2 Mis18αMeDiY/βMeDiY hetero‐dimers that bind two copies of Mis18BP1 and 2 Mis18αMeDiY that possibly form a homo‐dimer (Fig 4A). However, our data show that Mis18αMeDiY can bind Mis18BP1, albeit less efficiently compared to Mis18αMeDiY/βMeDiY hetero‐dimer contradicting the suggestion by Pan et al 29 that neither Mis18αMeDiY nor Mis18βMeDiY can interact with Mis18BP1 on their own. The apparent contradiction could be due to the higher ionic strength buffer used by Pan et al 29 in their binding assays. Whether Mis18α/β hetero‐hexamer can bind more than two copies of Mis18BP1 mediated via the free Mis18αMeDiY under specific circumstances is yet to be determined. Finally, we identified two highly conserved Cdk1 consensus sites 70 amino acids apart within the Mis18 binding region of Mis18BP1 (T40 and S110). While mutating both these amino acid residues to phospho‐mimicking residues completely abolished the ability of Mis18BP1 to bind Mis18α/β in vitro and in vivo, individual phospho‐mimic mutants (T40E or S110D) failed do so efficiently. This suggests that Mis18BP1 binds Mis18α/β possibly via a bipartite binding mode. Overall, our findings together with the recent independent study by Pan et al 29 provide key insights into the molecular basis for cell cycle‐dependent Mis18 complex assembly and function.
Materials and Methods
Expression and purification of recombinant human proteins
Human Mis18α, Mis18αMeDiY, Mis18αC‐term, Mis18β, Mis18βMeDiY, and Mis18βC‐term were amplified from codon‐optimized sequences (GeneArt) while Mis18BP120–130 was amplified from a human cDNA library (MegaMan human transcriptome library, Agilent). Amplifications were then cloned into pET His6 msfGFP TEV, pET His6 TEV, pET His6 Sumo TEV, pET His6 MBP TEV, pGEX‐6P‐1 (GE Healthcare), pEC‐K‐3C‐His‐GST, and pEC‐K‐3C‐His LIC vectors. pET His6 msfGFP TEV (9GFP Addgene plasmid # 48287), pET His6 TEV (9B Addgene plasmid # 48284), pET His6 Sumo TEV (14S Addgene plasmid # 48291), and pET His6 MBP TEV (9C Addgene Plasmid #48286) were a gift from Scott Gradia.
Mis18α MeDiY dimer‐disrupting mutant (Mis18αDimerM; V82E/Y176D) and putative substrate‐binding pocket mutant (Mis18αPocketM; L136A/Y152A/C167A/S169K) and Mis18BP1 mutants, Mis18BP120–130 T40E, Mis18BP120–130 S110D, Mis18BP120–130 T40E/S110D, and Mis18BP120–130 T40A/S110A were generated following the Quikchange site‐directed mutagenesis protocol (Stratagene).
Proteins were expressed, either individually or together with their binding partners, using E. coli BL21 gold grown in LB media. Cultures were induced with 0.35 mM IPTG at 18°C overnight. Cell lysis was carried out by sonicating cells re‐suspended in a lysis buffer containing 20 mM Tris (pH 8.0), 250 mM NaCl (or 500 mM for Mis18BP120–130), 35 mM imidazole, and 2 mM βME. Lysis buffer was supplemented with 10 μg/ml DNase, 1 mM PMSF, and cOmplete (EDTA‐free, Roche). Proteins were purified from the clarified lysates by affinity chromatography using a 5 ml HisTrap HP column (GE Healthcare). The protein‐bound resin was washed with lysis buffer, followed by a buffer containing 20 mM Tris (pH 8.0), 1 M NaCl, 50 mM KCl, 10 mM MgCl2, 2 mM ATP, 35 mM imidazole, 2 mM βME, and with a final lysis buffer wash. Proteins were eluted using a lysis buffer supplemented with 500 mM imidazole and dialyzed overnight into 20 mM Tris (pH 8.0), 75–100 mM NaCl, and 2 mM DTT. All proteins were subjected to anion exchange chromatography using the HiTrap Q column (GE Healthcare). Appropriate fractions were pooled, concentrated, and injected into a Superdex 200 increase 10/300 or Superdex 75 10/300 column (GE Healthcare) equilibrated with 20 mM Tris (pH 8.0), 100–250 mM NaCl, and 2 mM DTT. Fractions were analyzed on SDS–PAGE and stained with Coomassie blue.
Ni‐NTA pull‐down assay was performed in 20 mM Tris (pH 8.0), 75–500 mM NaCl, 10% glycerol, 0.5% NP‐40, 35 mM imidazole, and 2 mM βME. Proteins were mixed with two times molar excess of Mis18BP120–130 and made up to 200 μl with buffer before incubated for 30 min at 4°C with 60–120 μl of slurry that had been equilibrated in buffer. Beads were then washed four times with 1 ml of buffer, and bound protein was eluted by boiling in SDS–PAGE loading dye before being analyzed by SDS–PAGE.
SEC‐MALS
Size‐exclusion chromatography (ÄKTAMicro™, GE Healthcare) coupled to UV, static light scattering, and refractive index detection (Viscotek SEC‐MALS 20 and Viscotek RI Detector VE3580; Malvern Instruments) was used to determine the absolute molecular mass of proteins and protein complexes in solution. Injections of 100 μl of about 1 mg/ml material were run on a Superdex 200 10/300 GL (GE Healthcare) size‐exclusion column pre‐equilibrated in 50 mM HEPES (pH 8.0), 150 mM NaCl, and 1 mM TCEP at 22°C with a flow rate of 0.5 ml/min. Light scattering, refractive index (RI), and A280 nm were analyzed by a homo‐polymer model (OmniSEC software, v5.02; Malvern Instruments) using the following parameters for: ∂A280 nm/∂c = 0.97 AU ml mg−1 (Mis18α/β complex), 0.86 AU ml mg−1 (His‐GFP‐Mis18α/His‐Mis18β and His‐Mis18α/His‐GFP‐Mis18β), 0.82 AU ml mg−1 (Mis18α/Mis18β/Mis18BP120–130), 0.71 AU ml mg−1 (Mis18α/Mis18β/His‐Sumo‐Mis18BP120–130), 1.10 AU ml mg−1 (His‐GFP‐Mis18αC‐term/His‐MBPMis18βC‐term), 0.86 AU ml mg−1 (His‐GFP‐Mis18α2M/His‐Mis18β), ∂n/∂c = 0.185 ml g−1, and buffer RI value of 1.335. The mean standard error in the mass accuracy determined for a range of protein–protein complexes spanning the mass range of 6–600 kDa is ±1.9%.
Construction of TetR and mCherry/mCerulean fusion constructs
Human Mis18β, Mis18BP1fl, Mis18BP120–130, and Mis18BP1336–483 were amplified from a human cDNA library (MegaMan human transcriptome library, Agilent) and cloned into the pCDNA3‐mCherry or ‐mCerulean LIC cloning vectors (Addgene plasmids # 30125 and 30128, respectively, a gift from Scott Gradia). Mis18α was cloned into the TetR‐eYFP‐IRES‐Puro vector. Mutant constructs, TetR‐eYFP‐Mis18αDimerM, mCherry‐Mis18BP120–130 T40E, mCherry‐Mis18BP120–130 S110D, mCherry‐Mis18BP120–130 T40E/S110D, and mCherry‐Mis18BP120–130 T40A/S110A, were generated using the Quikchange mutagenesis protocol (Stratagene).
Cell culture and transfection
The HeLa 3‐8 overexpressing CENP‐A‐SNAP integration cell line, containing a synthetic α‐satellite (alphoid) DNA array integration with tetO sites (alphoidtetO array) integrated in a chromosome arm, was maintained in DMEM (Gibco) supplemented with 5% fetal bovine serum (Biowest) and penicillin/streptomycin (Gibco). Cells were grown at 37°C and 5% CO2. Transfections were performed in parallel with XtremeGene‐9 (Roche) following manufacturer's instructions. Briefly, 24 h after plating, cells grown in 12‐well plates were incubated with transfection complexes containing: 0.25 μg of each vector, 100 μl of Opti‐MEM (Invitrogen), and 3 μl of XtremeGene‐9 reagent for 36 h.
Immunofluorescence and quantification
Thirty‐six hours after transfection, cells growing on coverslips were fixed with 2.6% paraformaldehyde in PBS 1× buffer for 10 min at 37°C. Cells were then treated with permeabilization buffer (PBS containing 0.2% Triton X‐100, Sigma) for 5 min at room temperature and blocked with permeabilization buffer containing 3% BSA for 1 h at 37°C. Mouse anti‐CENP‐A (AN1; 1/500 dilution) and Cy‐5‐conjugated donkey anti‐mouse secondary antibody (Jackson Immunoresearch) were used for centromere staining.
Micrographs were acquired in the Centre Optical Instrumentation Laboratory on a DeltaVision Elite system (Applied Precision) using an inverted Olympus IX‐71 stand, with an Olympus UPlanSApo 100× oil immersion objective (numerical aperture (NA) 1.4) and a Lumencor light source. Camera (Photometrics Cool Snap HQ), shutter and stage were controlled through SoftWorx (Applied Precision). The 0.2‐μm‐spaced z‐stacks were deconvolved using SoftWorx and analyzed using ImageJ software (NIH, Bethesda). For CENP‐A signal quantification, a custom‐made macro in ImageJ modified from Bodor et al 30 was used. Briefly, the CENP‐A signal (Cy‐5) found at the alphoidTetO array (identified by the eYFP signal) was determined for every z‐section within a 7‐square pixel box. The mean signal intensity in the array was obtained and the background subtracted using the minimum intensity values within the section. The average intensity of the CENP‐A signal from endogenous centromeres was used to normalize. At least three biological independent experiments were performed for each assay.
Author contributions
AAJ conceived the project. FS, BM‐P, MAA, OM, AAJ, and WCE designed experiments. FS, BM‐P, MAA, MAW, and OM performed experiments and analyzed data. FS, BM‐P, MAA, OM, WCE, and AAJ wrote the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Expanded View Figures PDF
Review Process File
Acknowledgements
We thank the staff of the Edinburgh Protein Production Facility and the Centre Optical Instrumentation Laboratory for their help. The Wellcome Trust generously supported this work through a Wellcome Trust Career Development Grant (095822) and a Senior Research Fellowship (202811) to AAJ, a Principal Research Fellowship to WCE (073915), a Centre Core Grant (092076 and 203149) and an instrument grant (091020) to the Wellcome Trust Centre for Cell Biology, a Multi‐User Equipment grant 101527/Z/13/Z to the EPPF and a Wellcome‐UoE ISSF award toward the procurement of SEC‐MALS equipment for the EPPF. The European Commission supported this work through a Career Integration Grant to AAJ (334291). FS is supported by the Darwin Trust of Edinburgh.
EMBO Reports (2017) 18: 894–905
References
- 1. Fukagawa T, Earnshaw WC (2014) The centromere: chromatin foundation for the kinetochore machinery. Dev Cell 30: 496–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Catania S, Allshire RC (2014) Anarchic centromeres: deciphering order from apparent chaos. Curr Opin Cell Biol 26: 41–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. McKinley KL, Cheeseman IM (2016) The molecular basis for centromere identity and function. Nat Rev Mol Cell Biol 17: 16–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Stellfox ME, Bailey AO, Foltz DR (2013) Putting CENP‐A in its place. Cell Mol Life Sci 70: 387–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chen CC, Mellone BG (2016) Chromatin assembly: journey to the CENter of the chromosome. J Cell Biol 214: 13–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Black BE, Jansen LE, Foltz DR, Cleveland DW (2010) Centromere identity, function, and epigenetic propagation across cell divisions. Cold Spring Harb Symp Quant Biol 75: 403–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jansen LE, Black BE, Foltz DR, Cleveland DW (2007) Propagation of centromeric chromatin requires exit from mitosis. J Cell Biol 176: 795–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Foltz DR, Jansen LE, Bailey AO, Yates JR III, Bassett EA, Wood S, Black BE, Cleveland DW (2009) Centromere‐specific assembly of CENP‐a nucleosomes is mediated by HJURP. Cell 137: 472–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lando D, Endesfelder U, Berger H, Subramanian L, Dunne PD, McColl J, Klenerman D, Carr AM, Sauer M, Allshire RC et al (2012) Quantitative single‐molecule microscopy reveals that CENP‐A(Cnp1) deposition occurs during G2 in fission yeast. Open Biol 2: 120078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hayashi T, Ebe M, Nagao K, Kokubu A, Sajiki K, Yanagida M (2014) Schizosaccharomyces pombe centromere protein Mis19 links Mis16 and Mis18 to recruit CENP‐A through interacting with NMD factors and the SWI/SNF complex. Genes Cells 19: 541–554 [DOI] [PubMed] [Google Scholar]
- 11. Subramanian L, Toda NR, Rappsilber J, Allshire RC (2014) Eic1 links Mis18 with the CCAN/Mis6/Ctf19 complex to promote CENP‐A assembly. Open Biol 4: 140043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hayashi T, Fujita Y, Iwasaki O, Adachi Y, Takahashi K, Yanagida M (2004) Mis16 and Mis18 are required for CENP‐A loading and histone deacetylation at centromeres. Cell 118: 715–729 [DOI] [PubMed] [Google Scholar]
- 13. Hirai H, Arai K, Kariyazono R, Yamamoto M, Sato M (2014) The kinetochore protein Kis1/Eic1/Mis19 ensures the integrity of mitotic spindles through maintenance of kinetochore factors Mis6/CENP‐I and CENP‐A. PLoS One 9: e111905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fujita Y, Hayashi T, Kiyomitsu T, Toyoda Y, Kokubu A, Obuse C, Yanagida M (2007) Priming of centromere for CENP‐A recruitment by human hMis18alpha, hMis18beta, and M18BP1. Dev Cell 12: 17–30 [DOI] [PubMed] [Google Scholar]
- 15. Dambacher S, Deng W, Hahn M, Sadic D, Frohlich J, Nuber A, Hoischen C, Diekmann S, Leonhardt H, Schotta G (2012) CENP‐C facilitates the recruitment of M18BP1 to centromeric chromatin. Nucleus 3: 101–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Maddox PS, Hyndman F, Monen J, Oegema K, Desai A (2007) Functional genomics identifies a Myb domain‐containing protein family required for assembly of CENP‐A chromatin. J Cell Biol 176: 757–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kim IS, Lee M, Park KC, Jeon Y, Park JH, Hwang EJ, Jeon TI, Ko S, Lee H, Baek SH et al (2012) Roles of Mis18alpha in epigenetic regulation of centromeric chromatin and CENP‐A loading. Mol Cell 46: 260–273 [DOI] [PubMed] [Google Scholar]
- 18. Dunleavy EM, Roche D, Tagami H, Lacoste N, Ray‐Gallet D, Nakamura Y, Daigo Y, Nakatani Y, Almouzni‐Pettinotti G (2009) HJURP is a cell‐cycle‐dependent maintenance and deposition factor of CENP‐A at centromeres. Cell 137: 485–497 [DOI] [PubMed] [Google Scholar]
- 19. Pidoux AL, Choi ES, Abbott JK, Liu X, Kagansky A, Castillo AG, Hamilton GL, Richardson W, Rappsilber J, He X et al (2009) Fission yeast Scm3: a CENP‐A receptor required for integrity of subkinetochore chromatin. Mol Cell 33: 299–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lagana A, Dorn JF, De Rop V, Ladouceur AM, Maddox AS, Maddox PS (2010) A small GTPase molecular switch regulates epigenetic centromere maintenance by stabilizing newly incorporated CENP‐A. Nat Cell Biol 12: 1186–1193 [DOI] [PubMed] [Google Scholar]
- 21. Perpelescu M, Nozaki N, Obuse C, Yang H, Yoda K (2009) Active establishment of centromeric CENP‐A chromatin by RSF complex. J Cell Biol 185: 397–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. McKinley KL, Cheeseman IM (2014) Polo‐like kinase 1 licenses CENP‐A deposition at centromeres. Cell 158: 397–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Silva MC, Bodor DL, Stellfox ME, Martins NM, Hochegger H, Foltz DR, Jansen LE (2012) Cdk activity couples epigenetic centromere inheritance to cell cycle progression. Dev Cell 22: 52–63 [DOI] [PubMed] [Google Scholar]
- 24. Subramanian L, Medina‐Pritchard B, Barton R, Spiller F, Kulasegaran‐Shylini R, Radaviciute G, Allshire RC, Arockia Jeyaprakash A (2016) Centromere localization and function of Mis18 requires Yippee‐like domain‐mediated oligomerization. EMBO Rep 17: 496–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nardi IK, Zasadzinska E, Stellfox ME, Knippler CM, Foltz DR (2016) Licensing of centromeric chromatin assembly through the Mis18alpha‐Mis18beta heterotetramer. Mol Cell 61: 774–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Stellfox ME, Nardi IK, Knippler CM, Foltz DR (2016) Differential binding partners of the Mis18alpha/beta YIPPEE domains regulate Mis18 complex recruitment to centromeres. Cell Rep 15: 2127–2135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ohzeki J, Bergmann JH, Kouprina N, Noskov VN, Nakano M, Kimura H, Earnshaw WC, Larionov V, Masumoto H (2012) Breaking the HAC barrier: histone H3K9 acetyl/methyl balance regulates CENP‐A assembly. EMBO J 31: 2391–2402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Subramanian L, Medina‐Pritchard B, Barton R, Spiller F, Kulasegaran‐Shylini R, Radaviciute G, Allshire RC, Jeyaprakash AA (2016) Centromere localisation and function of Mis18 requires ‘Yippee‐like’ domain‐mediated oligomerisation. EMBO Rep 17: 496–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pan D, Klare K, Petrovic A, Take A, Walstein K, Singh P, Rondelet A, Bird AW, Musacchio A (2017) CDK‐regulated dimerization of M18BP1 on a Mis18 hexamer is necessary for CENP‐A loading. Elife 6: e23352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bodor DL, Mata JF, Sergeev M, David AF, Salimian KJ, Panchenko T, Cleveland DW, Black BE, Shah JV, Jansen LE (2014) The quantitative architecture of centromeric chromatin. Elife 3: e02137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bond CS, Schuttelkopf AW (2009) ALINE: a WYSIWYG protein‐sequence alignment editor for publication‐quality alignments. Acta Crystallogr D Biol Crystallogr 65: 510–512 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expanded View Figures PDF
Review Process File


