Abstract
Oryza sativa, the common cultivated rice, is one of the most important crops for human consumption, but production is increasingly threatened by abiotic stresses. Although many efforts have resulted in breeding rice cultivars that are relatively tolerant to their local environments, climate changes and population increase are expected to soon call for new, fast generation of stress tolerant rice germplasm, and current within-species rice diversity might not be enough to overcome such needs. The Oryza genus contains other 23 wild species, with only Oryza glaberrima being also domesticated. Rice domestication was performed with a narrow genetic diversity, and the other Oryza species are a virtually untapped genetic resource for rice stress tolerance improvement. Here we review the origin of domesticated Oryza sativa from wild progenitors, the ecological and genomic diversity of the Oryza genus, and the stress tolerance variation observed for wild Oryza species, including the genetic basis underlying the tolerance mechanisms found. The summary provided here is important to indicate how we should move forward to unlock the full potential of these germplasms for rice improvement.
Keywords: Oryza, rice, wild species, abiotic stress, domestication
Introduction
Continuous population and consumption growth are placing enormous demands on natural resources and agriculture. Today, approximately a billion people are chronically malnourished and it is uncertain whether our current agricultural system will be able to feed the expected world population, projected to reach nine billion by 2050. The efficient use of resources and reduced food waste can save much food, and increased crop production is fundamental to meet the world's future food needs (Godfray et al., 2010; Foley et al., 2011; Palmgren et al., 2014). However, there is an urgent need to reduce agriculture's environment footprint, and farming land should not be expanded at the expense of natural ecosystems. We are faced with the challenge of increasing food production without degrading land, water and biodiversity in an environment becoming increasingly exposed to a myriad of abiotic stresses (Foley et al., 2011; Mueller et al., 2012; Sang and Ge, 2013; Palmgren et al., 2014).
Rice is one of the world's most important staple crops, feeding more than 2.7 billion people worldwide (Muthayya et al., 2014), and also a model for genomic research in monocots. It is cultivated on 150 million hectares of land, and its annual yield is close to 610 million tons (http://irri.org/). Due to global adverse climate changes, rice growth and productivity in recent years has been seriously affected by abiotic stresses such as cold, drought, heat, flood, and salt (Zhang et al., 2015). Plants have evolved complex but not well understood responses. A complicated signaling network is effectively and timely initiated, which ultimately reprograms the expression of a large set of stress-responsive genes (Hong et al., 2016), leading to a series of morphological, physiological, and biochemical changes (Scafaro et al., 2011; Lei et al., 2013). The acclimation processes that result from stress perception aims at protecting plants from damages and increases the chance of survival (Hu and Xiong, 2014). However, it is important to highlight that acclimation to stressful conditions does not occur in all plant species, and depending on stress intensity and duration, growth and yield can be severely affected. In nature, there is a wide variability to stress response and tolerance, making worthwhile the search for “tolerance genes” in wild relatives of cultivated species.
Abiotic stress is a major concern for rice production, and an increasing threat to food security considering climate change, population increase and area of arable land available. Drought, flooding and extreme temperatures should become more frequent according to predictions (Wang, 2005; Bita and Gerats, 2013; Hirabayashi et al., 2013), which are likely to increase pressure on grain harvest. Food security is an issue to the human population as a whole, but especially in rural areas of Asia and Africa, which represent about 35% of the total rice harvest area. In Asia, where 700 million people live in extreme poverty, 30% of them are in regions that are prone to abiotic stresses such as flooding, drought and excess soil salinity (Ismail et al., 2013). These stresses often occur in combination, and stress responsive pathways often show extensive cross-talk (Mittler 2006).
Flooding is a widespread environmental stress. The rapid decline in the oxygen (O2) diffusion rate during flooding is accompanied by a reduction in cellular O2 levels and an energy crisis, which are particularly severe when photosynthesis is limited or absent (Bailey-Serres and Voesenek, 2008). Although rice is considered a flood tolerant crop, only limited cultivars display tolerance to prolonged submergence (Niroula et al., 2012), with most dying within 14 days of complete submergence.
Salinity is one of the most devastating abiotic stresses in rice, and the salt-affected soils currently account for about 20% of the total paddy rice planting area (Zhou et al., 2016). Soil salinity has adverse effects on plant germination, strength, and yield (Munns and Tester, 2008). On exposure to salt stress, the ionic balance (especially Na+/K+ ratio) and distribution is the ultimate manifestation of several physiologic processes in response to salt stress (Chen Y et al., 2013), creating an imbalance in the supplies of water and other nutrient solutes (Sengupta and Majumder, 2010). Most plants can adapt to low or moderate salinities, but their growth is severely limited above 200 mM NaCl. Therefore, plant survival and growth depends on adaptations to re-establish ionic homeostasis (Hasegawa et al., 2000).
Cold stress is one of the major environmental factors limiting the growth, productivity, and geographical distribution of crops, mostly in temperate and high altitude areas, due to the tropical origin of the rice species (Cruz et al., 2013; Zhang et al., 2014). Low temperature can affect growth and development of rice plants during any developmental stage, from germination to grain filling. During germination, the most common symptoms of cold temperature damage are low percentage and delayed germination (Cruz et al., 2013; Dametto et al., 2015), resulting in yield decreases up to 25% of the final yield and in increased weed competition (Fujino et al., 2004). During the vegetative stage, it can severely affect seedling establishment, leading to yellowing of the leaves, growth retardation, and decreased tillering (Cruz et al., 2013). Low temperatures that occur at critical reproductive stages can adversely affect grain quality (incomplete grain maturation) or cause yield reductions (Jena et al., 2012; Cruz et al., 2013; Zhang et al., 2014).
Availability of irrigation water is a limiting factor in attaining the full potential yield in many crops (Singh et al., 2015). Drought is one of the most widespread and damaging environmental stress factors in plants (Krannich et al., 2015), especially in rice, which is sensitive to drought stress because it is acclimated to either rain-fed or fully irrigated fields. The effect of drought may vary with the different genotypes, development stages, and degree and duration of drought stress (Wang et al., 2011). Rice plants are highly sensitive to drought stress during vegetative stage (resulting in reduced height, tiller number, and leaf area), at the panicle initiation and booting stages (Jiang et al., 2004; Wang et al., 2011). In China, the average annual drought affected area is up to 27 million hectares and rice production has decreased by 70-80 billion Kg since the 1990s (Luo, 2010).
A decline in rice production caused by heat stress is one of the biggest concerns resulting from future climate change. Maximum and minimum daily temperatures, and the number of hot days and warm nights in a year, are estimated to increase over most land areas (IPCC, 2014). In addition, climate variability is predicted to increase, leading to frequent episodes of heat stress, often coinciding with key developmental stages in crops, such as flowering (Hirabayashi et al., 2015). It is predicted that rice yields would be reduced by up to 10% with an average daily temperature increase of 1 °C (Peng et al., 2004). During early growth stages of rice, the occurrence of heat stress inhibits seedling establishment, leading to non-uniform growth, and reduced yield. The physiological and genetic basis of the heat response during the seedling stage is poorly understood (Lei et al., 2013). High temperatures at anthesis cause spikelet sterility due to the failure of anther dehiscence and the reduction in the number of germinating pollen grains on the stigma, leading to reductions in grain yield (Jagadish et al., 2010).
Therefore, the development of cultivated rice with abiotic stress tolerance is needed to stabilize the production level of rice. The method currently practiced for improving abiotic stress tolerance in rice cultivars is to explore germplasm for desirable traits (Scafaro et al., 2010). Recent attempts (until now limited to O. sativa species) have been successful, with backcrossing of Oryza sativa ssp. japonica and indica leading to substantial improvements in abiotic stress tolerance (Cheng et al., 2007). As there are about 24 known species within the Oryza genus, a large source of genetic material remains virtually untapped. The search for variability in the ancestors of cultivated rice, which undergoes a long history of artificial selection, has great potential value for rice breeding. Stress tolerance traits are likely to be found in wild rice species (Tian et al., 2015), which are recognized as an important genetic resource for cultivated rice improvement (Sakai and Itoh, 2010). In this review, we focus on the rice domestication history, the diversification of the Oryza genus, the mechanisms and underlying genetic basis for abiotic stress tolerance in species within the Oryza diversity, which illustrates the feasibility of using rice wild relatives as a source for breeding stress tolerant O. sativa.
Diversity in the Oryza genus
Rice belongs to the genus Oryza, of the tribe Oryzeae, subfamily Ehrhartoideae and family Poaceae (Grass Phylogeny Working Group, 2001). The tribe Oryzeae includes nine or ten genera and up to 70 species, and is very likely monophyletic; in this tribe, Oryza L. and Leersia Sw. are the two largest genera (reviewed by Kellogg, 2009). The analysis of both nuclear and chloroplast genes revealed that Oryza and Leersia are sister genera. The divergence of Oryza–Leersia from the other genera occurred approximately 20.5 MYA and the divergence of Oryza from Leersia 14.2 MYA (Ge et al., 2002; Guo and Ge, 2005).
The Oryza genus consists of 24 species spread worldwide (Table 1) (Kellogg, 2009; Jacquemin et al., 2013). The phylogeny of the Oryza genus spans approximately 15 million years (MY) of evolutionary history, a process that created diverse ecological adaptations (Vaughan et al., 2003; Ammiraju et al., 2010). The Oryza species have 11 different genome types (AA, BB, CC, BBCC, CCDD, EE, FF, GG, KKLL, HHJJ, and HHKK), and a 3.6-genome size variation (Lu et al., 2009; Jacquemin et al., 2013; Atwell et al., 2014).
Table 1. Oryza species, including their genome types, life span, geographical distributions, habitats and available accessions.
| Species | Genomea/Life historyb | Distributionc | Usual habitatd | Accessionse |
|---|---|---|---|---|
| Oryza alta | CCDD / Perennial | Belize, Brazil, Colombia, Guyana and Paraguay | Savanna and sometimes in woodland; wet sites; open | 27 |
| Oryza australiensis | EE / Perennial | Northern Australia | Undulating plains of Eucalyptus and Leptochloa or box woodland; wet sites; open | 52 |
| Oryza barthii | AA / Annual | Benin, Botswana, Burkina Faso, Cameroon, Central Africa Republic, Chad, Côte d’Ivoire, Ethiopia, The Gambia, Ghana, Guinea, Mali, Mauritania, Namibia, Niger, Nigeria, Senegal, Sierra Leone, Sudan, Tanzania, and Zambia | Mopane or savanna woodland, savanna or fadama; deep water; open | 405 |
| Oryza brachyantha | FF / Annual | Burkina Faso, Cameroon, Central African Republic, Chad, Democratic Republic of Congo, Guinea, Mali, Niger, Senegal, Sierra Leone, Sudan, Tanzania, and Zambia | Flat ironstone rocks, granite/lateritic outcrops; water up to 0.5 m deep, but more often in shallow water; open | 36 |
| Oryza coarctata | KKLL / Perennial | India, Sri Lanka, Bangladesh and Myanmar | Salt marsh ecosystems; inundated (twice a day) with saline river or seawater; open | 6f |
| Oryza eichingeri | CC / Perennial | Central Africa Republic, Côte d’Ivoire Democratic Republic of Congo, Kenya, Rwanda, Sri Lanka, Tanzania, and Uganda | Undisturbed forest, gallery or evergreen forest, or forest margins; damp or flooded sites; shade or semi-shade | 37 |
| Oryza glaberrima | AA / Annual | West Africa | Upland to deep water; open | 9073 |
| Oryza glumaepatula | AA / Perennial | Bolivia, Brazil, Colombia, Costa Rica, Cuba, Dominican Republic, French Guiana, Guyana, Honduras, Mexico, Panama, Surinam, and Venezuela | Swamps and marshes; usually deep water; open | 76 |
| Oryza grandiglumis | CCDD / Perennial | Argentina, Bolivia, Brazil, Colombia, Ecuador, French Guiana, Paraguay, and Peru | Savanna or woodland; water at river's edges or wet sites; open or shade | 16 |
| Oryza granulata | GG / Perennial | India, Cambodian, Vietnam, Thailand, southern China, Malaysia, Philippine, Nepal and Sri Lanka | Forest floor; damp sites; shade | 29 |
| Oryza latifolia | CCDD / Perennial | Argentina, Belize, Bolivia, Brazil, Colombia, Costa Rica, Cuba, Dominican Republic, Ecuador, El Salvador, French Guiana, Guatemala, Guyana, Haiti, Honduras, Mexico, Nicaragua, Panama, Paraguay, Peru, Puerto Rico, Surinam, Trinidad and Venezuela | Low forest, rainforest, secondary growth forest, open woodland, undulating savanna, pasture, cultivated fields, open swamp, hill slopes, high ridges, coastal belts; wet or damp sites; open or semi-shade | 86 |
| Oryza longiglumis | HHJJ / Perennial | Indonesia and Papua New Guinea | Forest areas; seasonally wet; shade or semi-shade | 6 |
| Oryza longistaminata | AA / Perennial | Angola, Benin, Botswana, Burkina Faso, Burundi, Cameroon, Central Africa Republic, Chad, Congo, Côte d’Ivoire, Democratic Republic of Congo, Ethiopia, Gabon, The Gambia, Ghana, Kenya, Madagascar, Malawi, Mali, Martinique, Mozambique, Namibia, Niger, Nigeria, Rwanda, Senegal, Seychelles, Sierra Leone, Somalia, South Africa, Sudan, Tanzania, Uganda, Zambia, and Zimbabwe | Savanna or openings in rain or gallery forests; deep water; open | 309 |
| Oryza malampuzhaensis | BBCC / Perennial | Western Ghats of South India | Forest pools; seasonally dry; shade | 12 |
| Oryza meridionalis | AA / Annual | Australia, Indonesia, and Papua New Guinea | Edges of seasonally freshwater lagoons, temporary pools, and swamps; seasonally wet; open | 65 |
| Oryza meyeriana | GG / Perennial | Indonesia, Malaysia, Philippines, and Thailand | Forest areas; damp sites; shade | 25 |
| Oryza minuta | BBCC / Perennial | Philippines and Papua New Guinea | Lowland areas beside streams and riverbanks; seasonally wet; shade or semi-shade | 86 |
| Oryza nivara | AA / Annual | Bangladesh, Cambodia, China, India, Laos, Malaysia, Myanmar, Nepal, Sri Lanka, Thailand, and Vietnam | Swampy areas; seasonally wet and shallow water; open | 2064 |
| Oryza officinalis | CC / Perennial | Australia, Bangladesh; Brunei, Cambodia, China, India, Indonesia, Laos, Malaysia, Myanmar, Nepal, Papua New Guinea, Philippines, Thailand, and Vietnam | Edge of or in forests, open vegetation, in abandoned cultivated rice fields, in Southeast Asia near the coast; wet sites; open or semi-shade | 317 |
| Oryza punctata | BB / Annual | Angola, Benin, Cameroon, Central Africa Republic, Chad, Congo, Côte d’Ivoire, Democratic Republic of Congo, Ethiopia, Ghana, Kenya, Madagascar, Malawi, Mozambique, Nigeria, Sudan, Swaziland, Tanzania, Uganda, Zambia, and Zimbabwe | Seasonally swamp areas, around water holes and pools, on riverbanks in areas that flood to 1 m depth; open | 98g |
| Oryza punctata | BBCC / Perennial | Angola, Benin, Cameroon, Central Africa Republic, Chad, Congo, Côte d’Ivoire, Democratic Republic of Congo, Ethiopia, Ghana, Kenya, Madagascar, Malawi, Mozambique, Nigeria, Sudan, Swaziland, Tanzania, Uganda, Zambia, and Zimbabwe | Forest areas; wet sites; shade | 98g |
| Oryza rhizomatis | CC / Perennial | Sri Lanka | Tropical forest and open, tall scrub with grassy openings; swampy or periodically flooded areas; open or semi-shade | 21 |
| Oryza ridleyi | HHJJ / Perennial | Cambodia, Indonesia, Laos, Malaysia, Myanmar, Papua New Guinea, and Thailand | Marshes or near streamsides in forest; seasonally wet; shade | 21 |
| Oryza rufipogon | AA / Perennial | Australia, Bangladesh, China, India, Indonesia, Laos, Malaysia, Myanmar, Nepal, Papua New Guinea, Philippines, Sri Lanka, Thailand, and Vietnam | Swamps and marshes; seasonally deepwater and wet year round; open | 1617 |
| Oryza sativa | AA / Annual | Southeast Asia | Upland to deep water; open | 212982 |
| Oryza schlechteri | HHKK / Perennial | Indonesia and Papua New Guinea | Forest areas; wet sites; shade or semi-shade | 3 |
According to a Jacquemin et al., 2013, a,b,c,d Lu and Jackson 2004, a,b,c,d Sengupta and Majumder 2010, a,c,d Vaughan et al., 2003
Number of accession entries per species in the Genesys Database (http://www.genesys-pgr.org/).
Includes accessions registered as Oryza/Porteresia coarctata.
Includes accessions from both annual and perennial O. punctata.
A phylogenomic study sampled and sequenced 142 single-copy genes to clarify the relationships among all diploid genome types of the rice genus (Zou et al., 2008). The analysis identified two episodes of rapid speciation that occurred approximately five and ten million years ago (MYA) and gave rise to almost the entire diversity of the genus. The first event occurred approximately ten MYA (Guo and Ge, 2005) and led to a rapid diversification of the G genome, F genome and a lineage that subsequently diversified into the rest of the rice genomes. Additionally, the H, J and K genomes that are now only present in tetraploid species also diverged around this time (Ge et al., 1999; Guo and Ge, 2005). The second event led to the diversification of the A, B, and C genomes approximately five MYA, confirming that the A and B genome species are sisters and the C genome clade is sister to that (Zou et al., 2008, Kellogg, 2009).
Divergence of the A genome took place over the past two million years, and the current most divergent species within this group are the perennial O. meridionalis (rhizomatous) and O. longistaminata (Zhu and Ge, 2005; Vaughan et al., 2008). O. sativa has two presumed wild ancestors (O. rufipogon and O. nivara), while the annual O. barthii is the progenitor of African domesticated rice O. glaberrima (please see next section). The two cultivated species and their respective progenitors shared an unknown ancestral of about 0.86 MYA (Sarla and Swamy, 2005; Zhu et al., 2014). Perennial O. glumaepatula is the only A genome group member with a current distribution in Latin America and presents a strong relation between genetic and geographic distances (Jacquemin et al., 2013; Vaughan et al., 2003). The A genome species O. meridionalis, O. longistaminata, O. glaberrima and O. barthii are considered candidates for tolerance to heat and drought stresses based on their distribution in temperature and moisture extremes (Atwell et al., 2014).
The CC genome group is formed by O. officinalis, O. rhizomatis and O. eichingeri. All three species are perennial, occurring in shade or semi-shade forest environments, among other habitats. O. officinalis (generally rhizomatous) has a high level of genetic diversity between populations, and O. rhizomatis (rhizomatous) is endemic to Sri Lanka (Vaughan et al., 2003; Lu and Jackson, 2004). The only member of the diploid BB genome group is O. punctata, which also exists in the allotetraploid form BBCC. Both BB and BBCC populations are distributed in Africa but occupying distinct niches. The diploid form is found in open habitats, while the allotetraploid in shade or semi-shade environments (Vaughan et al., 2003). Both forms of O. punctata are potential genetic reservoirs for tolerance to drought stress if considering their plasticity over moisture extremes (Atwell et al., 2014). O. eichingeri is believed to be the CC genome donor to the allotetraploid O. punctata, while the diploid O. punctata is the BB genome donor to the other allotetraploid BBCC species O. minuta and O. malampuzhaensis (rhizomatous). Perennial O. minuta and O. malampuzhaensis seem to have arisen from different polyploidy events if considering their distinct morphology, distribution and genetic diversity (Jacquemin et al., 2013; Vaughan et al., 2003). O. minuta diploid progenitors are O. punctata (BB), as already mentioned, and O. officinalis (CC), and its recent polyploidization is believed to have occurred within the last 400,000 years (Lu et al., 2009). O. malampuzhaensis, endemic to the Nallamalais of Eastern Ghats (India), is under severe threat considering its narrow distribution over a small geographical area, which leads to vulnerability to habitat destruction and fragmentation (Elangovan et al., 2012). The CCDD genome species O. latifolia, O. alta and O. grandiglumis are very closely related, with current distribution in Latin America. In fact, some studies suggest that CCDD group members are one complex species with different ecotypes (Vaughan et al., 2003). O. alta polyploidization was estimated to have happened less than 1.6 MYA. Additionally, O. grandiglumis and O. latifolia are considered candidates for tolerance to flooding stress (Lu et al., 2009; Atwell et al., 2014).
The GG genome species O. granulata and O. meyeriana are found in forest shade environments. O. granulata occupies the most basal position in the Oryza genus phylogeny, presents a high level of genetic diversity between populations, and is a potential genetic resource for tolerance to cold stress (Aggarwal et al., 1997; Vaughan et al., 2003; Atwell et al., 2014). The F genome species O. brachyantha has a compact genome (the smallest in the Oryza genus) and is distributed in West and East Africa (Chen J et al., 2013). Limited information is available for O. coarctata (KKLL), O. longiglumis (HHJJ), O. ridleyi (HHJJ) and O. schlechteri (HHKK) (Vaughan et al., 2003; Jacquemin et al., 2013). Regarding abiotic stresses, O. coarctata shows considerable adaptation to salinity, and O. ridleyi and O. schlechteri are considered good genetic reservoirs for tolerance to flooding (Lu et al., 2009; Sengupta and Majumder, 2010; Atwell et al., 2014). A summary of all Oryza species cited in this review, including their genotypes, life span, geographical distributions and habitats, is presented in Table 1.
The origin of Asian cultivated rice
There are two distinct groups of Asian cultivated rice, namely O. sativa ssp. japonica and indica, which can be differentiated by morphological and physiological traits, additionally to an incomplete sterility barrier (Sang and Ge, 2013). The origin and evolution of the japonica and indica subspecies is under considerable debate over the past several years. Two models of interactive domestication scenarios were proposed. The ‘snowballing model’ suggests a single domestication event that created an early cultivar with a set of domestication traits. This early cultivar, when hybridized with different wild rice populations, japonica-like and indica-like, would have enabled the fixation of critical domestication alleles in each one of them separately. Alternatively, in the ‘combination model’, both subspecies were domesticated independently from diverse wild rice ecotypes with subsequent hybridization, leading to the introgression and fixation of domestication alleles (Sang and Ge, 2007).
Considering archeological and genetic studies available to date, the ‘combination model’ is suggested as the closest scenario from what had happened during Asian rice domestication (Gross and Zhao, 2014). Briefly, the japonica subspecies was originated from O. rufipogon in the Yangtze River Valley in China, where rice cultivation possibly started around 8,000 years ago. Meanwhile, indica or proto-indica independent origin of cultivation probably took place in the Ganges plains in India, but proto-indica domestication was only complete after domesticated japonica was introduced and hybridized with indica, around 4,000 years ago (Fuller et al., 2009, 2011; Gross and Zhao, 2014). Both O. rufipogon and its annual derived O. nivara are native to India today. In most of the physiological and morphological traits, O. nivara is rather similar to the cultivated rice than O. rufipogon. Thus, O. nivara could have served as the indica progenitor, and domestication would have required fewer genetic modifications or mating system transitions (Li et al., 2006; Sang and Ge, 2013). If this hypothesis is correct, selection would have been less intense for indica because O. nivara already presented several cultivated rice traits. Thus, key domestication alleles had more chance to arise during japonica domestication from O. rufipogon, and semi-domesticated indica became the primary recipient of the domestication alleles when japonica was brought to India (Vaughan et al., 2008; Sang and Ge, 2013).
During the process of crop domestication many of the inherited traits involved in biotic and abiotic stress resistance may have been weakened or lost, since it is estimated that only 10-20% of wild species diversity is present in cultivated rice (Zhu et al., 2007; Palmgren et al., 2014). One of the most important resources for improvement of cultivated rice is the genetic reservoir hidden in wild rice species distributed across several biomes worldwide. It is also important to underline that isolated populations within species may also contain critical genes (Jacquemin et al., 2013; Atwell et al., 2014). Rice gene banks around the world exhibit an extensive seed collection, covering the genetic diversity present in farmers’ cultivars, landraces and Oryza species. The two largest gene banks are the International Rice Research Institute in the Philippines (4,370 wild species and hybrids accessions at IRRI, http://irri.org), and Oryzabase in Japan (1,703 entries, http://www.shigen.nig.ac.jp) (Jacquemin et al., 2013). Moreover, the Genesys database (http://www.genesys-pgr.or) allows searching accessions for many species in several seed banks, being a valuable resource for germplasm distribution (see total number of accessions available for each Oryza species in Table 1). A genus-wide comparative genome platform is essential to understand the genetic differences associated with abiotic factors. The sequencing of 16 Oryza genomes is either complete or underway with “gold standard” reference sequences available for the cultivated species O. sativa ssp. japonica and O. glaberrima, and for the wild species O. barthii and O. brachyantha (Jacquemin et al., 2013). The introgression of desirable traits via conventional breeding into cultivated rice should be more feasible from close relatives (although a viable hybrid O. coarctata X O. sativa has been reported – see below). Among the AA genome species, introgression lines can be obtained by backcrossing F1 hybrids that are partially sterile, and various levels of postzygotic barriers are known (Doi et al., 2008). The gene groups that cause these incompatibilities were genetically identified, but further studies are needed to break the reproductive isolation (Ji et al., 2005; Qiu et al., 2005). When O. sativa is crossed to non-AA genome species, viable but highly sterile F1 hybrids can be obtained using embryo culture. However, the aberrant chromosome pairing in meiosis makes it difficult to introgress genetic information from these species into cultivated rice (Doi et al., 2008). Transgenic approaches will be eventually needed for genes from taxonomically distant species (Vaughan et al., 2003; Palmgren et al., 2014). Therefore, it is of great significance to understand the genetic diversity within the wild relatives of Oryza in the context of their natural environment of origin, in order to identify the genetic basis of phenotypic variation between and within-species. This knowledge is an important resource to improve rice production under abiotic stresses (Sang and Ge, 2013; Gross and Zhao, 2014; Palmgren et al., 2014).
Abiotic stress tolerance in wild relatives: submergence
Rice is commonly grown in flooded soil containing a thin water layer. Although rice roots are well adapted to hypoxic conditions, most rice cultivars die rapidly if their shoots are submerged (Mackill et al., 2012). Several rainfed rice areas are at risk of flooding, a stress that decreases yield substantially (Ismail et al., 2013). Submergence inhibits aerobic metabolism and photosynthesis, leading to carbohydrate depletion and, depending on the stress intensity, plant death (Fukao and Bailey-Serres, 2004). However, rice plants are able to tolerate submergence using different strategies. In the low oxygen escape syndrome (or “escape strategy”) shoot elongation is stimulated by progressive flooding, keeping leaves in contact with the air and escaping the increasing levels of water (Bailey-Serres and Voesenek, 2008). In the “quiescent strategy”, hypoxia reduces elongation, represses carbohydrate degradation and stimulates anaerobic metabolism. When water recedes, plants resume growth. While the escape strategy only results in submergence tolerance if flooding occurs progressively, rice genotypes that use the quiescent strategy are able to survive up to 14 days of complete submergence (Bailey-Serres and Voesenek, 2008). Both strategies were elucidated at the molecular level: the quiescent strategy is linked to the SUB1 locus, while the escape strategy is dependent on the SNORKEL (SK) locus (for reviews see Bailey-Serres and Voesenek, 2008; Mickelbart et al., 2015).
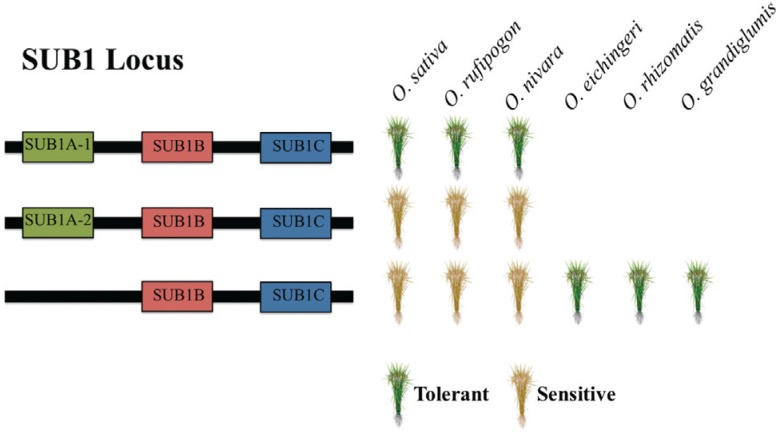
SUB1 is the major QTL associated with submergence tolerance in rice cultivars that use the quiescent strategy (Fukao et al., 2009). The SUB1 locus was mapped to chromosome 9, and is composed of a cluster of ethylene response factors (ERF) genes located in tandem, named SUB1A, SUB1B and SUB1C (Figure 1). Different rice genotypes have two genes in the cluster, SUB1B and SUB1C, whereas a third gene, SUB1A, is present only in a subset of them. Interestingly, tolerance to submergence is linked to a specific allele of the SUB1A gene, named SUB1A-1 (Xu et al., 2006). Accessions that lack SUB1A gene, or carry the SUB1A-2 allele, are sensitive to submergence (Figure 1). Introgression of functional copies of SUB1A-1 in sensitive genotypes is sufficient to generate tolerant plants (Xu et al., 2006).
Figure 1. Submergence tolerance and genetic architecture of SUB1 locus in cultivated and wild Oryza species. Genotypes from AA genome species Oryza sativa, Oryza rufipogon and Oryza nivara are tolerant or sensitive to submergence depending on the presence of SUB1A-1 allele of the SUB1A gene. Genotypes that either have SUB1A-2 allele or lack a SUB1A, carrying only SUB1B and SUB1C genes, are sensitive. Genotypes from CC genome species Oryza eichingeri, Oryza rhizomatis and the CCDD tetraploid species Oryza grandiglumis were shown to be tolerant to submergence while carrying a SUB1A gene-lacking SUB1 locus, indicating the locus does not contribute to the stress tolerance in these species.
The tolerant SUB1A-1 allele is derived from the aus subgroup of indica rice (Xu et al., 2006). Wild species from the Oryza genus commonly grow in constantly or seasonally wet habitats (Vaughan et al., 2003), and thus submergence tolerance could be found in other species. Niroula et al. (2012) tested 109 accessions of rice and wild relatives, including 12 species, for submergence tolerance, and found O. rufipogon and O. nivara (AA genome; Table 2) tolerant accessions that carry the SUB1A-1 allele, showing that SUB1 locus architecture determines submergence tolerance in these species, as in O. sativa (Figure 1; Niroula et al., 2012). Strikingly, accessions of O. rhizomatis and O. eichingeri (CC genome; Table 2) were also found to be submergence tolerant, but SUB1A sequences were absent from genomes of tested accessions, indicating that a novel, SUB1A-independent mechanism is responsible for submergence tolerance, at least in these two CC genome species (Figure 1; Niroula et al., 2012).
Table 2. Tolerance to abiotic stresses found in accessions of Oryza species and references.
| Species | Tolerance compared to O. sativa | Reference |
|---|---|---|
| Oryza coarctata | Salt | Sengupta and Majumder, 2009 |
| Oryza eichingeri | Submergence | Niroula et al., 2012 |
| Oryza glaberrima | Salt, Drought | Ndjiondjop et al., 2010, Platten et al., 2013 |
| Oryza glumaepatula | Submergence | Hattori et al., 2009 |
| Oryza grandiglumis | Submergence | Okishio et al., 2014, 2015 |
| Oryza meridionalis | Heat | Scafaro et al., 2010 |
| Oryza nivara | Drought | Singh et al., 2015 |
| Oryza officinalis | Drought, Heat (Early Morning Flowering) | Ishimaru et al., 2010, Feng et al., 2012 |
| Oryza rhizomatis | Submergence | Niroula et al., 2012 |
| Oryza rufipogon | Salinity, Cold, Drought, Submergence | Hattori et al., 2009, Tian et al., 2011, Xiao et al., 2014 |
Oryza grandiglumis is a tetraploid species with CCDD genome (Table 2) that grows in Amazonian floodplains, where water levels can reach up to 10 meters, and thus it was expected to show some degree of submergence tolerance (Okishio et al., 2014, 2015). Depending on the flooding conditions, O. grandiglumis showed distinct responses: when progressively submerged, the internodes elongated, resembling the escape strategy of O. sativa; however, when plants were completely submerged, growth was reduced, as in the quiescent strategy (Okishio et al., 2014). Interestingly, SUB1A is absent in O. grandiglumis, indicating that a SUB1A-independent mechanism for a quiescent strategy is present, as observed for O. rhizomatis and O. eichingeri (Figure 1). Genes similar to SNORKEL1 and SNORKEL2, responsible for the escape strategy in deepwater-adapted O. sativa, as well as O. rufipogon and O. glumeapatula (AA genome; Table 2), are absent in O. grandiglumis (Hattori et al., 2009, Okishio et al., 2015). These results indicate that O. grandiglumis is tolerant to both gradual and full submergence by unknown mechanisms (Okishio et al., 2014), indicating that CC genome Oryza species might provide new molecular mechanisms to improve cultivated rice.
Salinity
Soil salinization is a worldwide problem for agriculture. It affects 6% of total Earth's land, as a result of natural accumulation over long periods of time (Rengasamy, 2002). However, agricultural activity contributes to secondary salinization: 2% of all dry land is becoming salinized, and more than 20% of irrigated soils are affected, mostly because of irrigation water containing small amounts of sodium chloride (Tester and Davenport, 2003).
Plants vary in their sensitivity to salt stress, and rice is the most sensitive among cereals. Salinity reduces growth rate, including cellular and leaf expansion, number of tillers and photosynthesis, and can lead to premature senescence of older leaves (Munns and Tester, 2008, Sirault et al., 2009). The deleterious effects of salt in plants can be a result of osmotic stress (caused by salt in the soil), or of ionic stress (toxic effect of Na+ accumulation in plant tissues; Munns and Tester, 2008). The tolerance to ionic stress is dealt with by Na+ exclusion from xylem vessels to avoid shoot accumulation, or by tissue tolerance, when Na+ levels reach toxic levels and plant leaf cells compartmentalize salt to reduce damage (Roy et al., 2014).
The genetic basis of tolerance to ionic stress is much better understood than to osmotic stress (Roy et al., 2014). A range of transporters involved in reducing Na+ accumulation in shoots and in subcellular compartmentalization was described, such as the high affinity potassium transporter (HKT), salt overly sensitive (SOS) and Na+/H+ exchanger (NHX) gene families (Mickelbart et al., 2015). HKT members are crucial determinants of tissue concentration of Na+. OsHKT1;5 is the causative gene of Saltol, the major quantitative trait locus (QTL) for salt accumulation in O. sativa genotypes (Ren et al., 2005). OsHKT1;5 is a plasma membrane transporter that regulates partitioning of Na+ between roots and shoots by efflux of Na+ from the xylem to adjacent parenchyma cells (Hauser and Horie, 2010). Four amino acid changes in OsHKT1;5 resulted in increased Na+ efflux activity in salt tolerant indica cultivar Nona Bokra compared to the salt sensitive cultivar Koshihikari (Ren et al., 2005). Although other QTLs were described (Negrão et al., 2011), Saltol is the only one that has been cloned so far.
In a large screening effort which included several O. sativa cultivars, landraces and O. glaberrima (AA genome, Table 2) genotypes, it was shown that salinity sensitivity is correlated with Na+ concentration in leaf blades. OsHKT1;5 genotype was shown to be a major determinant for tolerance: the more active the efflux transporter, which directs the Na+ exclusion from the transpiration stream, the less Na+ is translocated to leaves (Platten et al., 2013). Interestingly, these authors found tolerant O. glaberrima accessions with low Na+ concentration in leaves, but carrying OsHKT1;5 alleles that are associated with salt sensitivity and Na+ accumulation in leaves of O. sativa genotypes. These results indicate that O. glaberrima genotypes could exclude Na+ from shoots using a mechanism independent of OsHKT1;5 (Platten et al., 2013).
Other species from the genus Oryza were explored for salt tolerance genes. O. rufipogon was shown to be salt tolerant when compared to rice sensitive cultivars (Zhou et al., 2016). Introgression lines derived from O. rufipogon X O. sativa cross revealed 15 QTLs for salinity tolerance, 13 of them derived from the O. rufipogon parent (Tian et al., 2011). Over-expression of bHLH transcription factors OrbHLH001 and OrbHLH2 from O. rufipogon resulted in Arabidopsis and O. sativa salt tolerant lines (Zhou et al., 2009; Li et al., 2010; Chen Y et al., 2013). These authors showed that OrbHLH001 is able to positively regulate the K+ transporter OsAKT1, suggesting that salt tolerance results from maintenance of K+ homeostasis under high Na+ conditions (Chen Y et al., 2013). Based on heterologous expression in Arabidopsis, OrbHLH2 was suggested to positively regulate genes from the CBF/DREB pathway (Zhou et al., 2009). Indeed, transcriptomic studies of the O. rufipogon response to high salinity showed that transcription factors are among the top up-regulated genes (Zhou et al., 2016).
Oryza coarctata, a promising source of salinity and submergence genes for rice
Oryza coarctata (also known as Porteresia coarctata, Lu and Ge, 2003) is an allotetraploid wild rice with extreme salt and submergence tolerance. It is unique among wild rice species, since it has a KKLL genome (Table 1; Lu et al., 2009). O. coarctata grows in coastal region of India and Bangladesh, where it experiences lunar tides and is submerged with saline seawater every 12 hours (Sengupta and Majumder, 2010, Garg et al., 2014). It has also been established as an important resource for prospecting genes to improve cultivated rice (Garg et al., 2014). A hybrid derived from a cross between O. sativa and O. coarctata has been reported (Jena, 1994), and salt tolerant rice cultivars with introgressed O. coarctata traits for salt tolerance are currently under development in the International Rice Research Institute (IRRI; www.irri.org). Still, little is known about the physiological and molecular details of this wild rice adaptation to high salinity conditions.
O. coarctata growth, relative water content and photosynthesis are unaffected by high concentrations (400 mM) of NaCl, conditions in which O. sativa salt tolerant cultivars do not develop properly (Sengupta and Majumder, 2009). Leaves of O. coarctata contain “salt hairs”, outgrowths of the epidermis that increase their number under high salinity and secrete excessive salt. When salt concentration in the growth media is high (300-400 mM), the salt hairs in the abaxial surface collapse and fall off from the leaf surface (Sengupta and Majumder, 2009). Still, total Na+ concentration in leaves of O. coarctata does not increase under salt stress, indicating that O. coarctata avoids Na+ toxicity in mesophyll cells by compartmentalization of salt in epidermal hairs, a mechanism similar to what is known for other halophyte grasses (Sengupta and Majumder, 2009, 2010). The secreted salt is a significant proportion of the Na+ reaching leaves, and important to maintain a low Na:K ratio (Sengupta and Majumder, 2010). A tonoplast-localized transporter from the NHX family of O. coarctata was recently cloned. PcNHX1 transcription is regulated during the day, presumably reflecting the circadian variation in tide experienced by the plant, and is also rapidly induced by NaCl treatment, compartmentalizing Na+ into vacuoles (Kizhakkedath et al., 2015). Thus, O. coarctata is adapted to high salinity environments by using multiple mechanisms to cope with salt stress, including decreased root-to-shoot translocation and increased compartmentalization in the vacuole and secretion in salt hairs.
Proteomic analyses identified up-regulated proteins by Na+ treatment in O. coarctata, including transcription factors of the CBF/DREB pathway of abiotic stress response (Shinozaki and Yamaguchi-Shinozaki, 2000); a cellulose synthase-like, which could help maintaining cellulose synthesis during salt stress (Endler et al., 2015); and an L-myo-inositol 1-phosphate synthase, important for inositol synthesis. Inositol metabolism is, in fact, one of the most studied aspects of O. coarctata salt tolerance. Its derivative, pinitol, is a known osmoprotectant in many plant species, and accumulates in O. coarctata under high salinity (Sengupta et al., 2008; Sengupta and Majumder, 2010). Strikingly, cloning and characterization of O. coarctata L-myo-inositol 1-phosphate synthase (INO1) showed that enzyme activity is maintained properly even in high salt concentrations, and that its expression in plants and bacteria confers high, albeit variable, salt tolerance to these organisms (Majee et al., 2004; Das-Chatterjee et al., 2006; Sengupta and Majumder, 2010). This highlights the potential of wild species to provide useful proteins for rice (and other crops) improvement. Moreover, inositol methyl transferase (IMT1), which methylates inositol into pinitol, is up-regulated in the same conditions as INO1 in O. coarctata, indicating that inositol synthesis and conversion to pinitol are key steps for salt tolerance in this species (Sengupta et al., 2008).
More recently, a study evaluated O. coarctata transcriptomic changes under salt and submergence stresses (alone or combined, compared to control conditions), and found several transcription factors up-regulated in leaves under stress conditions, such as NAC, WRKY and MYB gene family members, indicating extensive transcriptional regulation in stress responses (Garg et al., 2014). Gene Ontology analyses showed enrichment of ABA-responsive genes under salinity stress, and of carbohydrate metabolism and anaerobic respiration genes under submergence stress. In plants under submergence stress, genes related to ethylene and gibberellin responses were also identified, along with Alcohol Dehydrogenase, a marker for anoxia stress, indicating that a SUB1A-related response might be present in O. coarctata (Garg et al., 2014; see above for discussion on submergence stress mechanisms). However, demonstration of the presence of SUB1A-like ERF transcription factors and of their role in submergence response in O. coarctata is lacking. Moreover, suberin and cellulose synthesis-related transcripts were identified as up-regulated in both stresses, indicating that these processes might be key for stress tolerance, as observed in other species (Garg et al., 2014; Endler et al., 2015; Barberon et al., 2016).
Cold
The japonica cultivars of O. sativa are usually adapted to temperate climates, a process that was driven by domestication, while indica cultivars are generally tropical (Kovach et al., 2007; Ma et al., 2015). Thus, temperate japonica cultivars are more tolerant to low temperatures than indica, and introgression of cold tolerance traits from temperate into tropical genotypes is desirable (Cruz et al., 2013; Dametto et al., 2015). However, little is known about the molecular basis for low temperature tolerance in O. sativa, and even less about the variation of cold tolerance among its wild relatives.
The Oryza genus has a pan-tropical distribution, growing in regions with an average low temperature of 15 °C or above during the growth season, with only a few exceptions (Atwell et al., 2014). Hence, it is possible that other species of Oryza might be better adapted to lower temperatures, being able to provide alleles to improve cold tolerance. Atwell et al. (2014) used the distribution of each species in different climates to estimate the best candidates for stress tolerance, and O. granulata is suggested as a possible source for cold tolerance. O. eichingeri (CC genome, Table 2) also grows in low temperature environments, and could be considered a good candidate (Atwell et al., 2014). However, screenings of cold tolerance are still lacking for most Oryza species.
One genotype of O. rufipogon, named Dongxiang wild rice, is able to withstand overwintering in its natural habitat and temperature as low as 3 °C for three days in laboratory conditions (Xiao et al., 2014; Mao et al., 2015). Using an experimental population derived from Dongxiang wild rice X Nanjing 11 (a cold sensitive cultivar) crosses, a CBF3/DREB1G gene was found to co-localize to a previously identified cold-related QTL (Xiao et al., 2014). CBF/DREBs are known regulators of cold and other abiotic stress responses (Mao and Chen, 2012; Mickelbart et al., 2015). Interestingly, CBF3/DREB1G is up-regulated as early as three hours after cold treatment in both Dongxiang wild rice, but only after 12 hours in the sensitive one. Genes known to be downstream of CBF/DREB1 in the cold response are up-regulated accordingly in both tolerant genotypes (Xiao et al., 2014). Other QTLs unique to Dongxiang wild rice were described (Mao et al., 2015).
Recently, a SNP associated with temperate japonica cold tolerance was described. COLD1 (chilling-tolerance divergence) is a plasma membrane- and endoplasmic reticulum-localized regulator of G protein that activates Ca2+ influx during cold sensing. One SNP that results in an amino acid change was found to be responsible for the difference in cold tolerance. The SNP found in japonica is shared with accessions of O. rufipogon, but not with O. nivara or O. barthii (AA genome; Ma et al., 2015). Thus, the COLD1 sequence found in tolerant japonica cultivars represents an ancient allele from O. rufipogon that was selected during domestication (Ma et al., 2015). However, it is important to note that the SNP in COLD1 explains only part of the cold tolerance in rice, a trait for which many minor effect QTLs are expected to contribute (Cruz et al., 2013; Ma et al., 2015; Mao et al., 2015). Thus, other QTLs should be characterized and used in combination in order to develop highly tolerant lines, and newly identified genes from wild species might also be useful.
Drought
Drought tolerance is a complex trait, with many genes and processes involved (Singh et al., 2015). Rice in particular demands great amounts of water for proper development, owing to its shallow roots compared to other crops (Kondo et al., 2000). QTLs for drought tolerance were identified within O. sativa variability, and causative genes have been cloned (Vikram et al., 2011; Uga et al., 2013). DRO1 (DEEPER ROOTING 1), a previously unknown protein, is responsible for downward growth of rice roots, and introgression of DRO1 in otherwise shallow root rice genotypes increases root angle and drought tolerance (Uga et al., 2013). Still, wild rice species might be a source of stronger drought tolerance genes and mechanisms, since they are adapted to a much wider spectrum of environments (Vaughan et al., 2003; Atwell et al., 2014).
Species that are present in low moisture regions were suggested as more likely candidates for drought tolerance, namely: O. barthii, O. australiensis, O. glaberrima, O. longistaminata and O. punctata (Atwell et al., 2014). At least three of these (O. australiensis, glaberrima and longistaminata), plus O. meridionalis, present thick leaves and high mesophyll conductance to CO2 diffusion, indicating that they might be drought tolerant, since these traits can be associated with a higher water use efficiency (Scafaro et al., 2011; Giuliani et al., 2013). Indeed, field evaluation of O. glaberrima showed that some accessions could be used as donors in crossing with O. sativa for drought tolerance breeding (Ndjiondjop et al., 2010).
The O. rufipogon genotype Dongxiang was also used to breed drought tolerance in O. sativa. An introgressed line was shown to be more tolerant when compared to the O. sativa recurrent parent, with higher survival rate, along with higher proline and soluble sugar accumulation (Zhang et al., 2014). However, it is clear that the trait is genotype-specific, since different O. rufipogon accessions can have widely different sensitivity levels. Feng et al. (2012) tested tolerance of eight accessions of O. rufipogon and one of O. officinalis, and observed that accessions from tropical origin are more tolerant. Interestingly, the single O. officinalis (CC genome; Table 2) accession performed even better under drought conditions (Feng et al., 2012).
Another study analyzed leaf rolling score and relative water content in several O. sativa, O. rufipogon and O. nivara genotypes from India, and associated these traits with sequence diversity of OsDREB1F, a known drought stress-responsive transcription factor (Wang et al., 2008; Singh et al., 2015). At least five truncated versions of OsDREB1F were found to be associated with drought sensitivity. Interestingly, one protein variant, present in four O. nivara accessions, was associated with high relative water content and low leaf rolling score. The variant harbors an amino acid mutation in a putative activation domain, which, based on molecular modeling, is likely to affect the tertiary structure of the protein (Singh et al., 2015). The over-expression of OsDREB1F in O. sativa confers tolerance to drought, low temperature and salt stresses (Wang et al., 2008). Thus, the protein variant from O. nivara genotypes is likely to confer tolerance to multiple stresses, possible due to effects on tolerance to the osmotic adjustment component that is common in these conditions.
Heat
Rice is a pan-tropical species, and thus well adapted to high temperatures compared to other grasses. However, the increasing global average temperatures and the more frequent occurrence of heat waves (IPCC, 2007) could affect rice growth, both during vegetative and reproductive stages. Species from the Oryza genus were suggested as heat tolerant (Atwell et al., 2014), but few have been physiologically characterized.
O. meridionalis (AA genome, Table 1), is endemic to hot regions of northern Australia, and is described as heat tolerant (Scafaro et al., 2010). When compared to O. sativa, O. meridionalis leaf elongation rate is slower under 27 °C, but faster at 45 °C. The photosynthesis temperature optimum of O. meridionalis is 3 °C above that of O. sativa, which is accounted for by a higher RuBisCO activation state under heat stress (Scafaro et al., 2012). Proteomics analyses showed that Calvin Cycle and heat shock-related proteins increased their abundance in O. meridionalis leaves at high temperatures, including Rubisco activase (Scafaro et al., 2010). Thus, higher Rubisco activase accumulation is directly involved in heat tolerance of O. meridionalis, maintaining RuBisCO carboxylation at higher temperatures (Scafaro et al., 2010, 2012).
Temperatures higher than 32-36 °C at anthesis cause spikelet sterility and yield reduction, mainly due to reduced anther dehiscence and germinating pollen on the stigma (Ishimaru et al., 2010, and references therein). Flowering in cultivated rice occurs between mid-morning to noon, when heat has already built up since the beginning of the day (Nishiyama and Blanco, 1980). A useful trait to avoid flowering at high temperatures is early morning flowering (EMF). The wild rice species O. officinalis shows EMF, which can be used to increase cultivated rice fertilization and yield by escaping heat stress (Ishimaru et al., 2010). Indeed, introgression lines were produced from O. sativa X O. officinalis crosses, and these showed increased fertility due to the shift in anthesis timing (Ishimaru et al., 2010). QTL analyses revealed that an O. officinalis EMF candidate gene is located in chromosome 3 and reduces the flowering opening time by 1.5 to 2 hours in both temperate and tropical cultivars, thus demonstrating the usefulness of this trait to reduce effects of heat stress on spikelet sterility (Hirabayashi et al., 2015).
Concluding remarks
Early domestication of crops was a bottleneck for gene diversity. While selecting important traits for cultivation, the process inadvertently lost others that might be interesting for humans today. As an example, a wheat NAC transcription factor, NAM-B1, was shown to regulate iron, zinc and protein levels in grains. Interestingly, the NAM-B1 allele from wild emmer wheat is functional, accelerating senescence and increasing remobilization of nutrients to developing seeds, while the modern varieties carry a non-functional allele, and thus have decreased levels of iron, zinc and protein (Uauy et al., 2006). Considering that iron and zinc deficiencies in humans are a common dietary problem (Sperotto et al., 2012; Ricachenevsky et al., 2015), introgression of the functional NAM-B1 allele is likely to improve wheat nutritional quality.
The use of wild relatives as a reservoir of new alleles to confer stress tolerance to cultivated species has been successful before. In wheat, introgression of the Nax2 locus (containing the TmHKT1;5-A allele) from Triticum monoccocum conferred improved salt tolerance, with reduced Na+ accumulation in leaves and a 25% increase in grain yield in high salt soils, and resulted in a wide distribution of Nax2 bearing germplasm to producers (James et al., 2012; Munns et al., 2012; Mickelbart et al., 2015). A soybean wild relative, Glycine soja, has also been shown to be salt tolerant (Chen P et al., 2013). A Cation H+ Exchange (CHX) transporter was identified as the causative gene of the tolerance phenotype, and gain-of-function transgenic plants expressing the transporter were shown to be more salt tolerant (Qi et al., 2014).
Considering the wide variety of environments in which species from the Oryza genus are found, it is expected that they also vary in abiotic stress tolerance (Atwell et al., 2014). Thus, screening for stress tolerance in these species, and within multiple genotypes of each species, should yield new alternatives for rice improvement. Efforts such as the International Oryza Map Alignment Project and the genome sequencing of 16 species (Jacquemin et al., 2013) should fast track the unlocking of rice wild relatives potential for breeding and transgenic approaches. The combination of these data with large scale screening for tolerance in wild genotypes, together with the available and newly generated QTL maps and genome re-sequencing of tolerant individuals will be key to guarantee food security, considering the prospects of climate change and overpopulation.
Acknowledgments
The authors would like to thank CNPq (Conselho Nacional de Desenvolvimento Científico e Tecnológico) for funding. Raul Antonio Sperotto was also funded by Centro Universitário UNIVATES.
Footnotes
Associate Editor: Marcio de Castro Silva Filho
References
- Aggarwal RK, Brar DS, Khush GS. Two new genomes in the Oryza complex identified on the basis of molecular divergence analysis analysis using total genomic DNA hydridization. Mol Gen Genet. 1997;254:1–12. doi: 10.1007/s004380050384. [DOI] [PubMed] [Google Scholar]
- Ammiraju JSS, Song X, Luo M, Sisneros N, Angelova A, Kudrna D, Kim HR, Yu Y, Goicoechea JL, Lorieux M, et al. The Oryza BAC resource: A genus-wide and genome scale tool for exploring rice genome evolution and leveraging useful genetic diversity. Breed Sci. 2010;60:536–543. [Google Scholar]
- Atwell BJ, Wang H, Scafaro AP. Could abiotic stress tolerance in wild relatives of rice be used to improve Oryza sativa? Plant Sci. 2014;215-216:48–58. doi: 10.1016/j.plantsci.2013.10.007. [DOI] [PubMed] [Google Scholar]
- Bailey-Serres J, Voesenek LACJ. Flooding stress: Acclimations and genetic diversity. Annu Rev Plant Biol. 2008;59:313–339. doi: 10.1146/annurev.arplant.59.032607.092752. [DOI] [PubMed] [Google Scholar]
- Barberon M, Vermeer JE, De Bellis D, Wang P, Naseer S, Andersen TG, Humbel BM, Nawrath C, Takano J, Salt DE, et al. Adaptation of root function by nutrient-induced plasticity of endodermal differentiation. Cell. 2016;164:447–459. doi: 10.1016/j.cell.2015.12.021. [DOI] [PubMed] [Google Scholar]
- Bita CE, Gerats T. Plant tolerance to high temperature in a changing environment: Scientific fundamentals and production of heat stress-tolerant crops. Front Plant Sci. 2013;4:273–273. doi: 10.3389/fpls.2013.00273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Huang Q, Gao D, Wang J, Lang Y, Liu T, Li B, Bai Z, Goicoechea JL, Liang C, et al. Whole-genome sequencing of Oryza brachyantha reveals mechanisms underlying Oryza genome evolution. Nat Commun. 2013;4:1595–1595. doi: 10.1038/ncomms2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Li F, Ma Y, Chong K, Xu Y. Overexpression of OrbHLH001, a putative helix-loop-helix transcription factor, causes increased expression of AKT1 and maintains ionic balance under salt stress in rice. J Plant Physiol. 2013;170:93–100. doi: 10.1016/j.jplph.2012.08.019. [DOI] [PubMed] [Google Scholar]
- Chen P, Yan K, Shao H, Zhao S. Physiological mechanisms for high salt tolerance in wild soybean (Glycine soja) from Yellow River Delta, China: Photosynthesis, osmotic regulation, ion flux and antioxidant capacity. PLoS One. 2013;8:e83227. doi: 10.1371/journal.pone.0083227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng SH, Zhuang JY, Fan YY, Du JH, Cao LY. Progress in research and development on hybrid rice: A superdomesticate in China. Ann Bot. 2007;100:959–966. doi: 10.1093/aob/mcm121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz RP, Sperotto RA, Cargnelutti D, Adamski JM, Terra TF, Fett JP. Avoiding damage and achieving cold tolerance in rice plants. Food Energy Sec. 2013;2:96–119. [Google Scholar]
- Dametto A, Sperotto RA, Adamski JM, Blasi EAR, Cargnelutti D, de Oliveira LFV, Ricachenevsky FK, Fregonezi JN, Mariath JEA, Cruz RP, et al. Cold tolerance in rice germinating seeds revealed by deep RNAseq analysis of contrasting indica genotypes. Plant Sci. 2015;238:1–12. doi: 10.1016/j.plantsci.2015.05.009. [DOI] [PubMed] [Google Scholar]
- Das-Chatterjee A, Goswami L, Maitra S, Dastidar KG, Ray S, Majumder AL. Introgression of a novel salt-tolerant L-myo-inositol 1-phosphate synthase from Porteresia coarctata (Roxb.) Tateoka (PcINO1) confers salt tolerance to evolutionary diverse organisms. FEBS Lett. 2006;580:3980–3988. doi: 10.1016/j.febslet.2006.06.033. [DOI] [PubMed] [Google Scholar]
- Doi K, Yasui H, Yoshimura A. Genetic variation in rice. Curr Opin Plant Biol. 2008;11:144–148. doi: 10.1016/j.pbi.2008.01.008. [DOI] [PubMed] [Google Scholar]
- Elangovan M, Kiran Babu P, Tonapi VA, Subba Rao LV, Sivaraj N. Cultivated grasses and their wild relatives in Andhra Pradesh and their conservation concerns. Indian J Plant Genet Resour. 2012;25:166–173. [Google Scholar]
- Endler A, Kesten C, Schneider R, Zhang Y, Ivakov A, Froehlich A, Funke N, Persson S. A mechanism for sustained cellulose synthesis during salt stress. Cell. 2015;162:1353–1364. doi: 10.1016/j.cell.2015.08.028. [DOI] [PubMed] [Google Scholar]
- Feng F, Xu X, Du X, Tong H, Luo L, Mei H. Assessment of drought resistance among wild rice accessions using a protocol based on single-tiller propagation and PVC-tube cultivation. Aust J Crop Sci. 2012;6:1204–1211. [Google Scholar]
- Foley JA, Ramnkutty N, Brauman KA, Cassidy ES, Gerber JS, Johnston M, Mueller ND, O’Connell C, Ray DK, West PC, et al. Solutions for a cultivated planet. Nature. 2011;478:337–342. doi: 10.1038/nature10452. [DOI] [PubMed] [Google Scholar]
- Fujino K, Sekiguchi H, Sato T, Kiuchi H, Nonoue Y, Takeuchi Y, Ando T, Lin SY, Yano M. Mapping of quantitative trait loci controlling low-temperature germinability in rice (Oryza sativa L.) Theor Appl Genet. 2004;108:794–799. doi: 10.1007/s00122-003-1509-4. [DOI] [PubMed] [Google Scholar]
- Fuller DQ, Qin L, Zheng YF, Zhao ZJ, Chen XG, Hosoya LA, Sun G-P. The domestication process and domestication rate in rice, spikelet bases from the Lower Yangtze. Science. 2009;323:1607–1610. doi: 10.1126/science.1166605. [DOI] [PubMed] [Google Scholar]
- Fuller DQ, van Etten J, Manning K, Castillo C, Kingwell-Banham E, Weisskopf A, Qin L, Sato YI, Hijmans RJ. The contribution of rice agriculture and livestock pastoralism to prehistoric methane levels. Holocene. 2011;21:743–759. [Google Scholar]
- Fukao T, Bailey-Serres J. Plant responses to hypoxia-is survival a balancing act? Trends Plant Sci. 2004;9:449–456. doi: 10.1016/j.tplants.2004.07.005. [DOI] [PubMed] [Google Scholar]
- Fukao T, Harris T, Bailey-Serres J. Evolutionary analysis of the Sub1 gene cluster that confers submergence tolerance to domesticated rice. Ann Bot. 2009;103:143–150. doi: 10.1093/aob/mcn172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg R, Verma M, Agrawal S, Shankar R, Majee M, Jain M. Deep transcriptome sequencing of wild halophyte rice, Porteresia coarctata, provides novel insights into the salinity and submergence tolerance factors. DNA Res. 2014;21:69–84. doi: 10.1093/dnares/dst042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge S, Sang T, Lu B-R, Hong D-Y. Phylogeny of rice genomes with emphasis on origins of allotetraploid species. Proc Natl Acad Sci U S A. 1999;96:14400–14405. doi: 10.1073/pnas.96.25.14400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge S, Li A, Lu B-R, Zhang S-Z, Hong D-Y. A phylogeny of the rice tribe Oryzeae (Poaceae) based on matK sequence data. Amer J Bot. 2002;89:1967–1972. doi: 10.3732/ajb.89.12.1967. [DOI] [PubMed] [Google Scholar]
- Giuliani R, Koteyeva N, Voznesenskaya E, Evans MA, Cousins AB, Edwards GE. Coordination of leaf photosynthesis, transpiration, and structural traits in rice and wild relatives (genus Oryza) Plant Physiol. 2013;162:1632–1651. doi: 10.1104/pp.113.217497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grass Phylogeny Working Group Phylogeny and subfamily classification of the Poaceae. Ann Missouri Bot Gard. 2001;88:373–457. [Google Scholar]
- Godfray HC, Beddington JR, Crute IR, Haddad L, Lawrence D, Muir JF, Pretty J, Robinson S, Thomas SM, Toulmin C. Food security: The challenge of feeding 9 billion people. Science. 2010;327:812–818. doi: 10.1126/science.1185383. [DOI] [PubMed] [Google Scholar]
- Gross B, Zhao Z. Archaeological and genetic insights into the origins of domesticated rice. Proc Natl Acad. Sci U S A. 2014;111:6190–6197. doi: 10.1073/pnas.1308942110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y-L, Ge S. Molecular phylogeny of Oryzeae (Poaceae) based on DNA sequences from chloroplast, mitochondrial, and nuclear genome. Amer J Bot. 2005;92:1548–1558. doi: 10.3732/ajb.92.9.1548. [DOI] [PubMed] [Google Scholar]
- Hattori Y, Nagai K, Furukawa S, Song XJ, Kawano R, Sakakibara H, Wu J, Matsumoto T, Yoshimura A, Kitano H, et al. The ethylene response factors SNORKEL1 and SNORKEL2 allow rice to adapt to deep water. Nature. 2009;460:1026–1030. doi: 10.1038/nature08258. [DOI] [PubMed] [Google Scholar]
- Hasegawa PM, Bressan RA, Zhu JK, Bohnert HJ. Plant cellular and molecular responses to high salinity. Annu Rev Plant Physiol Plant Mol Biol. 2000;51:463–499. doi: 10.1146/annurev.arplant.51.1.463. [DOI] [PubMed] [Google Scholar]
- Hauser F, Horie T. A conserved primary salt tolerance mechanism mediated by HKT transporters: A mechanism for sodium exclusion and maintenance of high K+/Na+ ratio in leaves during salinity stress. Plant Cell Environ. 2010;33:552–565. doi: 10.1111/j.1365-3040.2009.02056.x. [DOI] [PubMed] [Google Scholar]
- Hirabayashi H, Sasaki K, Kambe T, Gannaban RB, Miras MA, Mendioro MS, Simon EV, Lumanglas PD, Fujita D, Takemoto-Kuno Y, et al. qEMF3, a novel QTL for the early-morning flowering trait from wild rice, Oryza officinalis, to mitigate heat stress damage at flowering in rice, O. sativa. J Exp Bot. 2015;66:1227–1236. doi: 10.1093/jxb/eru474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirabayashi Y, Mahendran R, Koirala S, Konoshima L, Yamazaki D, Watanabe S, Kim H, Kanae S. Global flood risk under climate change. Nature Clim Change. 2013;3:816–821. [Google Scholar]
- Hong Y, Zhang H, Huang L, Li D, Song F. Overexpression of a stress-responsive NAC transcription factor gene ONAC022 improves drought and salt tolerance in rice. Front Plant Sci. 2016;7:4–4. doi: 10.3389/fpls.2016.00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H, Xiong L. Genetic engineering and breeding of drought resistant crops. Annu Rev Plant Biol. 2014;65:715–741. doi: 10.1146/annurev-arplant-050213-040000. [DOI] [PubMed] [Google Scholar]
- IPCC . In: Climate change 2007: The Physical Science Basis. Contribution of Working Group I to the Fourth Assessment Report of the Intergovernmental Panel on Climate Change. Solomon S, Qin D, Manning M, Chen Z, Marquis M, Averyt KB, Tignor M, Miller HL, editors. Cambridge University Press; Cambridge, MA: 2007. 996 p [Google Scholar]
- Ishimaru T, Hirabayashi H, Ida M, Takai T, San-Oh YA, Yoshinaga S, Ando I, Ogawa T, Kondo M. A genetic resource for early-morning flowering trait of wild rice Oryza officinalis to mitigate high temperature-induced spikelet sterility at anthesis. Ann Bot. 2010;106:515–520. doi: 10.1093/aob/mcq124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismail AM, Singh US, Singh S, Dar MH, Mackill DJ. The contribution of Submergence-Tolerant (Sub1) rice varieties to food security in flood-prone rainfed lowland areas in Asia. Field Crops Res. 2013;152:83–93. [Google Scholar]
- Jacquemin J, Bhatia D, Singh K, Wing RA. The International Oryza Map Alignment Project: Development of a genus-wide comparative genomics platform to help solve the 9 billion-people question. Curr Opin Plant Biol. 2013;16:1–10. doi: 10.1016/j.pbi.2013.02.014. [DOI] [PubMed] [Google Scholar]
- Jagadish SVK, Muthurajan R, Oane R, Wheeler TR, Heuer S, Bennett J, Craufurd PQ. Physiological and proteomic approaches to address heat tolerance during anthesis in rice (Oryza sativa L.) J Exp Bot. 2010;61:143–156. doi: 10.1093/jxb/erp289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James RA, Blake C, Zwart AB, Hare RA, Rathjen AJ, Munns RE. Impact of ancestral wheat sodium exclusion genes Nax1 and Nax2 on grain yield of durum wheat on saline soils. Funct Plant Biol. 2012;39:609–618. doi: 10.1071/FP12121. [DOI] [PubMed] [Google Scholar]
- Jena KK. Production of intergeneric hybrid between Oryza sativa L. and Porteresia coarctata . Curr Sci. 1994;67:9–10. [Google Scholar]
- Jena KK, Kim SM, Suh JP, Yang CI, Kim YJ. Identification of cold-tolerant breeding lines by Quantitative Trait Loci associated with cold tolerance in rice. Crop Sci. 2012;51:517–523. [Google Scholar]
- Ji Q, Lu JF, Chao Q, Gu MH, Xu ML. Delimiting a rice wide-compatibility gene S-5(n) to a 50 kb region. Theor Appl Genet. 2005;111:1495–1503. doi: 10.1007/s00122-005-0078-0. [DOI] [PubMed] [Google Scholar]
- Jiang XL, Zheng JG, Yuan Y. Effects of water stresses on grain yield at different rice growth stage. Southwest China J Agric Sci. 2004;4:435–438. [Google Scholar]
- Kellogg EA. The evolutionary history of Ehrhartoideae, Oryzeae, and Oryza . Rice. 2009;2:1–14. [Google Scholar]
- Kizhakkedath P, Jegadeeson V, Venkataraman G, Parida A. A vacuolar antiporter is differentially regulated in leaves and roots of the halophytic wild rice Porteresia coarctata (Roxb.) Tateoka. Mol Biol Rep. 2015;42:1091–1105. doi: 10.1007/s11033-014-3848-4. [DOI] [PubMed] [Google Scholar]
- Kondo M, Murty MVR, Aragones DV. Characteristics of root growth and water uptake from soil in upland rice and maize under water stress. Soil Sci Plant Nutr. 2000;46:721–732. [Google Scholar]
- Kovach MJ, Sweeney MT, McCouch SR. New insights into the history of rice domestication. Trends Genet. 2007;23:578–587. doi: 10.1016/j.tig.2007.08.012. [DOI] [PubMed] [Google Scholar]
- Krannich CT, Maletzki L, Kurowsky C, Horn R. Network candidate genes in breeding for drought tolerant crops. Int J Mol Sci. 2015;16:16378–16400. doi: 10.3390/ijms160716378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei D, Tan L, Liu F, Chen L, Sun C. Identification of heat-sensitive QTL derived from common wild rice (Oryza rufipogon Griff.) Plant Sci. 2013;201-202:121–127. doi: 10.1016/j.plantsci.2012.12.001. [DOI] [PubMed] [Google Scholar]
- Li C, Zhou A, Sang T. Genetic analysis of rice domestication syndrome with the wild annual species, Oryza nivara . New Phytol. 2006;170:185–194. doi: 10.1111/j.1469-8137.2005.01647.x. [DOI] [PubMed] [Google Scholar]
- Li F, Guo S, Zhao Y, Chen D, Chong K, Xu Y. Overexpression of a homopeptide repeat-containing bHLH protein gene (OrbHLH001) from Dongxiang Wild Rice confers freezing and salt tolerance in transgenic Arabidopsis. Plant Cell Rep. 2010;29:977–986. doi: 10.1007/s00299-010-0883-z. [DOI] [PubMed] [Google Scholar]
- Lu BR, Ge S. Oryza coarctata: the name that best reflects the relationships of Porteresia coarctata (Poaceae: Oryzeae) Nord J Bot. 2003;23:555–558. [Google Scholar]
- Lu F, Ammiraju JS, Sanyal A, Zhang S, Song R, Chen J, Li G, Sui Y, Song X, Cheng Z, et al. Comparative sequence analysis of MONOCULM1 -orthologous regions in 14 Oryza genomes. Proc Natl Acad Sci U S A. 2009;106:2071–2076. doi: 10.1073/pnas.0812798106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo LJ. Breeding for water-saving and drought-resistance rice (WDR) in China. J Exp Bot. 2010;61:3509–3517. doi: 10.1093/jxb/erq185. [DOI] [PubMed] [Google Scholar]
- Ma Y, Dai X, Xu Y, Luo W, Zheng X, Zeng D, Pan Y, Lin X, Liu H, Zhang D, et al. COLD1 confers chilling tolerance in rice. Cell. 2015;160:1209–1221. doi: 10.1016/j.cell.2015.01.046. [DOI] [PubMed] [Google Scholar]
- Mao D, Yu L, Chen D, Li L, Zhu Y, Xiao Y, Zhang D, Chen C. Multiple cold resistance loci confer the high cold tolerance adaptation of Dongxiang wild rice (Oryza rufipogon) to its high-latitude habitat. Theor Appl Genet. 2015;128:1359–1371. doi: 10.1007/s00122-015-2511-3. [DOI] [PubMed] [Google Scholar]
- Mackill DJ, Ismail AM, Singh US, Labios RV, Paris TR. Development and rapid adoption of submergence-tolerant (Sub1) rice varieties. Adv Agron. 2012;115:303–356. [Google Scholar]
- Majee M, Maitra S, Dastidar KG, Pattnaik S, Chatterjee A, Hait NC, Das KP, Majumder AL. A novel salt-tolerant L-myo-inositol-1-phosphate synthase from Porteresia coarctata (Roxb.) Tateoka, a halophytic wild rice: Molecular cloning, bacterial overexpression, characterization, and functional introgression into tobacco-conferring salt tolerance phenotype. J Biol Chem. 2004;279:28539–28552. doi: 10.1074/jbc.M310138200. [DOI] [PubMed] [Google Scholar]
- Mao DH, Chen CY. Colinearity and similar expression pattern of rice DREB1s reveal their functional conservation in the cold- responsive pathway. PLoS One. 2012;7:e47275. doi: 10.1371/journal.pone.0047275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mickelbart MV, Hasegawa PM, Bailey-Serres J. Genetic mechanisms of abiotic stress tolerance that translate to crop yield stability. Nat Rev Genet. 2015;16:237–251. doi: 10.1038/nrg3901. [DOI] [PubMed] [Google Scholar]
- Mittler R. Abiotic stress, the field environment and stress combination. Trends Plant Sci. 2006;11:15–19. doi: 10.1016/j.tplants.2005.11.002. [DOI] [PubMed] [Google Scholar]
- Mueller ND, Gerber JS, Johnston M, Ray DK, Ramankutty N, Foley JA. Closing yield gaps through nutrient and water management. Nature. 2012;490:254–257. doi: 10.1038/nature11420. [DOI] [PubMed] [Google Scholar]
- Munns R, James RA, Xu B, Athman A, Conn SJ, Jordans C, Byrt CS, Hare RA, Tyerman D, Teste M, et al. Wheat grain yield on saline soils is improved by an ancestral Na+ transporter gene. Nat Biotech. 2012;30:360–364. doi: 10.1038/nbt.2120. [DOI] [PubMed] [Google Scholar]
- Munns R, Tester M. Mechanisms of salinity tolerance. Annu Rev Plant Biol. 2008;59:651–681. doi: 10.1146/annurev.arplant.59.032607.092911. [DOI] [PubMed] [Google Scholar]
- Muthayya S, Sugimoto JD, Montgomery S, Maberly GF. An overview of global rice production, supply, trade, and consumption. Ann New York Acad Sci. 2014;1324:7–14. doi: 10.1111/nyas.12540. [DOI] [PubMed] [Google Scholar]
- Ndjiondjop MN, Cisse F, Girma G, Sow M, Bocco R, Djedatin G, Blandine F. Morpho-agronomic and molecular characterisation of Oryza glaberrima germplasm from Mali. Afr J Biotechnol. 2010;9:7409–7417. [Google Scholar]
- Negrão S, Courtois B, Ahmadi N, Abreu I, Saibo N, Oliveira MM. Recent updates on salinity stress in rice: From physiological to molecular responses. Crit Rev Plant Sci. 2011;30:329–377. [Google Scholar]
- Niroula RK, Pucciariello C, Ho VT, Novi G, Fukao T, Perata P. SUB1A-dependent and -independent mechanisms are involved in the flooding tolerance of wild rice species. Plant J. 2012;72:282–293. doi: 10.1111/j.1365-313X.2012.05078.x. [DOI] [PubMed] [Google Scholar]
- Nishiyama I, Blanco L. Avoidance of high temperature sterility by flower opening in the early morning. Jpn Agric Res Q. 1980;14:116–117. [Google Scholar]
- Okishio T, Sasayama D, Hirano T, Akimoto M, Itoh K, Azuma T. Growth promotion and inhibition of the Amazonian wild rice species Oryza grandiglumis to survive flooding. Planta. 2014;240:459–469. doi: 10.1007/s00425-014-2100-8. [DOI] [PubMed] [Google Scholar]
- Okishio T, Sasayama D, Hirano T, Akimoto M, Itoh K, Azuma T. Ethylene is not involved in adaptive responses to flooding in the Amazonian wild rice species Oryza grandiglumis . J Plant Physiol. 2015;174:49–54. doi: 10.1016/j.jplph.2014.09.012. [DOI] [PubMed] [Google Scholar]
- Palmgren MG, Edenbrandt AK, Vedel SE, Andersen MM, Landes X, Østerberg JT, Falhof J, Olsen LI, Christensen SB, Sandøe P. Are we ready for back-to-nature crop breeding? Trends Plant Sci. 2014;20:155–164. doi: 10.1016/j.tplants.2014.11.003. [DOI] [PubMed] [Google Scholar]
- Peng S, Huang J, Sheehy JE, Laza RC, Visperas RM, Zhong X, Centeno GS, Khush GS, Cassman KG. Rice yields decline with higher night temperature from global warming. Proc Natl Acad Sci U S A. 2004;101:9971–9975. doi: 10.1073/pnas.0403720101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platten JD, Egdane JA, Ismail AM. Salinity tolerance, Na+ exclusion and allele mining of HKT1;5 in Oryza sativa and O. glaberrima: Many sources, many genes, one mechanism? BMC Plant Biol. 2013;13:32–32. doi: 10.1186/1471-2229-13-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi X, Li MW, Xie M, Liu X, Ni M, Shao G, Song C, Kay-Yuen Yim A, Tao Y, Wong FL, et al. Identification of a novel salt tolerance gene in wild soybean by whole-genome sequencing. Nat Commun. 2014;5:4340–4340. doi: 10.1038/ncomms5340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu SQ, Liu KD, Jiang JX, Song X, Xu CG, Li XH, Zhang QF. Delimitation of the rice wide compatibility gene S5(n) to a 40-kb DNA fragment. Theor Appl Genet. 2005;111:1080–1086. doi: 10.1007/s00122-005-0033-0. [DOI] [PubMed] [Google Scholar]
- Ren ZH, Gao JP, Li LG, Cai XL, Huang W, Chao DY, Zhu MZ, Wang ZY, Luan S, Lin HX. A rice quantitative trait locus for salt tolerance encodes a sodium transporter. Nat Genet. 2005;37:1141–1146. doi: 10.1038/ng1643. [DOI] [PubMed] [Google Scholar]
- Rengasamy P. Transient salinity and subsoil constraints to dryland farming in Australian sodic soils: An overview. Aust J Exp Agric. 2002;42:351–361. [Google Scholar]
- Ricachenevsky FK, Menguer PK, Sperotto RA, Fett JP. Got to hide your Zn away: Molecular control of Zn accumulation and biotechnological applications. Plant Sci. 2015;236:1–17. doi: 10.1016/j.plantsci.2015.03.009. [DOI] [PubMed] [Google Scholar]
- Roy SJ, Negrão S, Tester M. Salt resistant crop plants. Curr Opin Biotechnol. 2014;26:115–124. doi: 10.1016/j.copbio.2013.12.004. [DOI] [PubMed] [Google Scholar]
- Sakai H, Itoh T. Massive gene losses in Asian cultivated rice unveiled by comparative genome analysis. BMC Genom. 2010;11:121–121. doi: 10.1186/1471-2164-11-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sang T, Ge S. The puzzle of rice domestication. J Integr Plant Biol. 2007;49:760–768. [Google Scholar]
- Sang T, Ge S. Understanding rice domestication and implications for cultivar improvement. Curr Opin Plant Biol. 2013;16:139–146. doi: 10.1016/j.pbi.2013.03.003. [DOI] [PubMed] [Google Scholar]
- Sarla N, Swamy BPM. Oryza glaberrima: A source for the improvement of Oryza sativa. Curr Sci. 2005;89:955–963. [Google Scholar]
- Scafaro AP, Haynes PA, Atwell BJ. Physiological and molecular changes in Oryza meridionalis Ng., a heat-tolerant species of wild rice. J Exp Bot. 2010;61:191–202. doi: 10.1093/jxb/erp294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scafaro AP, Von Caemmerer S, Evans JR. Temperature response of mesophyll conductance in cultivated and wild Oryza species with contrasting mesophyll cell wall thickness. Plant Cell Environ. 2011;34:1999–2008. doi: 10.1111/j.1365-3040.2011.02398.x. [DOI] [PubMed] [Google Scholar]
- Scafaro AP, Yamori W, Carmo-Silva AE, Salvucci ME, von Caemmerer S, Atwell BJ. Rubisco activity is associated with photosynthetic thermotolerance in a wild rice (Oryza meridionalis. Physiol Plant. 2012;146:99–109. doi: 10.1111/j.1399-3054.2012.01597.x. [DOI] [PubMed] [Google Scholar]
- Sengupta S, Majumder AL. Insight into the salt tolerance factors of a wild halophytic rice, Porteresia coarctata: A physiological and proteomic approach. Planta. 2009;229:911–929. doi: 10.1007/s00425-008-0878-y. [DOI] [PubMed] [Google Scholar]
- Sengupta S, Majumder AL. Porteresia coarctata (Roxb.) Tateoka, a wild rice: A potential model for studying salt-stress biology in rice. Plant Cell Environ. 2010;33:526–542. doi: 10.1111/j.1365-3040.2009.02054.x. [DOI] [PubMed] [Google Scholar]
- Sengupta S, Patra B, Ray S, Majumder AL. Inositol methyl transferase from a halophytic wild rice, Porteresia coarctata Roxb. (Tateoka): Regulation of pinitol synthesis under abiotic stress. Plant Cell Environ. 2008;31:1442–1459. doi: 10.1111/j.1365-3040.2008.01850.x. [DOI] [PubMed] [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K. Molecular responses to dehydration and low temperature: Differences and cross-talk between two stress signaling pathways. Curr Opin Plant Biol. 2000;3:217–223. [PubMed] [Google Scholar]
- Singh BP, Jayaswal PK, Singh B, Singh PK, Kumar V, Mishra S, Singh N, Panda K, Singh NK. Natural allelic diversity in OsDREB1F gene in the Indian wild rice germplasm led to ascertain its association with drought tolerance. Plant Cell Rep. 2015;34:993–1004. doi: 10.1007/s00299-015-1760-6. [DOI] [PubMed] [Google Scholar]
- Sirault XRR, James RA, Furbank RT. A new screening method for osmotic component of salinity tolerance in cereals using infrared thermography. Funct Plant Biol. 2009;36:970–977. doi: 10.1071/FP09182. [DOI] [PubMed] [Google Scholar]
- Sperotto RA, Ricachenevsky FK, Waldow Vde A, Fett JP. Iron biofortification in rice: It's a long way to the top. Plant Science. 2012;190:24–39. doi: 10.1016/j.plantsci.2012.03.004. [DOI] [PubMed] [Google Scholar]
- Tester M, Davenport R. Na+ tolerance and Na+ transport in higher plants. Ann Bot. 2003;91:503–527. doi: 10.1093/aob/mcg058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian XJ, Long Y, Wang J, Zhang JW, Wang YY, Li WM, Peng YF, Yuan QH, Pei XW. De novo transcriptome assembly of common wild rice (Oryza rufipogon Griff.) and discovery of drought-response genes in root tissue based on transcriptomic data. PLoS ONE. 2015;10:e0131455. doi: 10.1371/journal.pone.0131455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian L, Tan L, Liu F, Cai H, Sun C. Identification of quantitative trait loci associated with salt tolerance at seedling stage from Oryza rufipogon . J Genet Genomics. 2011;38:593–601. doi: 10.1016/j.jgg.2011.11.005. [DOI] [PubMed] [Google Scholar]
- Uauy C, Distelfeld A, Fahima T, Blechl A, Dubcovsky J. A NAC Gene regulating senescence improves grain protein, zinc, and iron content in wheat. Science. 2006;314:1298–1301. doi: 10.1126/science.1133649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uga Y, Sugimoto K, Ogawa S, Rane J, Ishitani M, Hara N, Kitomi Y, Inukai Y, Ono K, Kanno N, et al. Control of root system architecture by DEEPER ROOTING 1 increases rice yield under drought conditions. Nat Genet. 2013;45:1097–1102. doi: 10.1038/ng.2725. [DOI] [PubMed] [Google Scholar]
- Vaughan DA, Morishima H, Kadowaki K. Diversity in the Oryza genus. Curr Opin Plant Biol. 2003;6:139–146. doi: 10.1016/s1369-5266(03)00009-8. [DOI] [PubMed] [Google Scholar]
- Vaughan DA, Lu BR, Tomooka N. The evolving story of rice evolution. Plant Sci. 2008;174:394–408. [Google Scholar]
- Vikram P, Mallikarjuna Swamy BP, Dixit S, Ahmed HU, Sta Cruz MT, Singh AK, Kumar A. qDTY1.1 a major QTL for rice grain yield under reproductive-stage drought stress with a consistent effect in multiple elite genetic backgrounds. BMC Genet. 2011;12:89–89. doi: 10.1186/1471-2156-12-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao N, Huang WN, Li AH, Gao Y, Li HY, Pan CH, Ji H, Zhang XX, Dai X, Dai ZY, et al. Fine mapping of the qLOP2 and qPSR2-1 loci associated with chilling stress tolerance of wild rice seedlings. Theor Appl Genet. 2014;128:173–185. doi: 10.1007/s00122-014-2420-x. [DOI] [PubMed] [Google Scholar]
- Xu K, Xu X, Fukao T, Canlas P, Maghirang-Rodriguez R, Heuer S, Ismail AM, Bailey-Serres J, Ronald PC, Mackill DJ. Sub1A is an ethylene-response-factor- like gene that confers submergence tolerance to rice. Nature. 2006;442:705–708. doi: 10.1038/nature04920. [DOI] [PubMed] [Google Scholar]
- Wang D, Pan Y, Zhao X, Zhu L, Fu B, Li Z. Genome-wide temporal-spatial gene expression profiling of drought responsiveness in rice. BMC Genomics. 2011;12:149–149. doi: 10.1186/1471-2164-12-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G. Agricultural drought in a future climate: Results from 15 global climate models participating in the IPCC 4th assessment. Clim Dynam. 2005;25:739–753. [Google Scholar]
- Wang Q, Guan Y, Wu Y, Chen H, Chen F, Chu C. Overexpression of a rice OsDREB1F gene increases salt, drought, and low temperature tolerance in both Arabidopsis and rice. Plant Mol Biol. 2008;67:589–602. doi: 10.1007/s11103-008-9340-6. [DOI] [PubMed] [Google Scholar]
- Zhang F, Luo X, Zhou Y, Xie J. Genome-wide identification of conserved microRNA and their response to drought stress in Dongxiang wild rice (Oryza rufipogon Griff.) Biotechnol Lett. 2015;38:711–721. doi: 10.1007/s10529-015-2012-0. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Chen Q, Wang S, Hong Y, Wang Z. Rice and cold stress: Methods for its evaluation and summary of cold tolerance-related quantitative trait loci. Rice. 2014;7:24–24. doi: 10.1186/s12284-014-0024-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Li F, Wang JL, Ma Y, Chong K, Xu YY. Basic helix-loop-helix transcription factor from wild rice (OrbHLH2) improves tolerance to salt- and osmotic stress in Arabidopsis. J Plant Physiol. 2009;166:1296–1306. doi: 10.1016/j.jplph.2009.02.007. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Yang P, Cui F, Zhang F, Luo X, Xie J. Transcriptome analysis of salt stress responsiveness in the seedlings of Dongxiang wild rice (Oryza rufipogon Griff.) PLoS ONE. 2016;11:e0146242. doi: 10.1371/journal.pone.0146242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q, Ge S. Phylogenetic relationships among A-genome species of the genus Oryza revealed by intron sequences of four nuclear genes. New Phytol. 2005;167:249–265. doi: 10.1111/j.1469-8137.2005.01406.x. [DOI] [PubMed] [Google Scholar]
- Zhu Q, Zheng X, Luo J, Gaut BS, Ge S. Multilocus analysis of nucleotide variation of Oryza sativa and its wild relatives: Severe bottleneck during domestication of rice. Mol Biol Evol. 2007;24:875–888. doi: 10.1093/molbev/msm005. [DOI] [PubMed] [Google Scholar]
- Zhu T, Xu PZ, Liu JP, Peng S, Mo XC, Gao LZ. Phylogenetic relationships and genome divergence among the AA- genome species of the genus Oryza as revealed by 53 nuclear genes and 16 intergenic regions. Mol Phylogenet Evol. 2014;70:348–361. doi: 10.1016/j.ympev.2013.10.008. [DOI] [PubMed] [Google Scholar]
- Zou X-H, Zhang F-M, Zhang J-G, Zang L-L, Tang L, Wang J, Sang T, Ge S. Analysis of 142 genes resolves the rapid diversification of the rice genus. Genome Biol. 2008;9:R49–R49. doi: 10.1186/gb-2008-9-3-r49. [DOI] [PMC free article] [PubMed] [Google Scholar]
Internet Resources
- IPCC . Climate Change 2014: Synthesis Report. Contribution of Working Groups I, II and III to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change. IPCC; Geneva, Switzerland: 2014. 151 p. https://www.ipcc.ch/pdf/assessment-report/ar5/syr/SYR_AR5_FINAL_full.pdf. [Google Scholar]
- Lu BR, Jackson M. Wild Rice Taxonomy. IRRI; 2004. [accessed February 2016]. http://www.knowledgebank.irri.org/wildRiceTaxonomy/default.htm. [Google Scholar]