Abstract
Background
Normally, one miRNA interacts with the mRNA of one gene. However, there are miRNAs that can bind to many mRNAs, and one mRNA can be the target of many miRNAs. This significantly complicates the study of the properties of miRNAs and their diagnostic and medical applications.
Results
The search of 2,750 human microRNAs (miRNAs) binding sites in 12,175 mRNAs of human genes using the MirTarget program has been completed. For the binding sites of the miR-619-5p the hybridization free energy of the bonds was equal to 100% of the maximum potential free energy. The mRNAs of 201 human genes have complete complementary binding sites of miR-619-5p in the 3’UTR (214 sites), CDS (3 sites), and 5’UTR (4 sites). The mRNAs of CATAD1, ICA1L, GK5, POLH, and PRR11 genes have six miR-619-5p binding sites, and the mRNAs of OPA3 and CYP20A1 genes have eight and ten binding sites, respectively. All of these miR-619-5p binding sites are located in the 3’UTRs. The miR-619-5p binding site in the 5’UTR of mRNA of human USP29 gene is found in the mRNAs of orthologous genes of primates. Binding sites of miR-619-5p in the coding regions of mRNAs of C8H8orf44, C8orf44, and ISY1 genes encode the WLMPVIP oligopeptide, which is present in the orthologous proteins. Binding sites of miR-619-5p in the mRNAs of transcription factor genes ZNF429 and ZNF429 encode the AHACNP oligopeptide in another reading frame. Binding sites of miR-619-5p in the 3’UTRs of all human target genes are also present in the 3’UTRs of orthologous genes of mammals. The completely complementary binding sites for miR-619-5p are conservative in the orthologous mammalian genes.
Conclusions
The majority of miR-619-5p binding sites are located in the 3’UTRs but some genes have miRNA binding sites in the 5’UTRs of mRNAs. Several genes have binding sites for miRNAs in the CDSs that are read in different open reading frames. Identical nucleotide sequences of binding sites encode different amino acids in different proteins. The binding sites of miR-619-5p in 3’UTRs, 5’UTRs and CDSs are conservative in the orthologous mammalian genes.
Electronic supplementary material
The online version of this article (doi:10.1186/s12864-017-3811-6) contains supplementary material, which is available to authorized users.
Keywords: miR-619-5p, miRNA, mRNA, Gene, Human, Orthologous genes
Background
miRNAs participate in the regulation of the expression of protein-coding genes at the post-transcriptional stage [1]. miRNAs, as a part of the RNA-induced silencing complex, bind to mRNAs and interfere with translation or promote mRNA destruction [2]. In the last two decades, properties of miRNAs and their influences on the expression of the genes involved in all key cellular processes have been established. The actions of miRNAs on the cell cycle [3], apoptosis [4], differentiation [5], and growth and development of plants [6] and animals [7] have been shown. Connections between miRNA expression and the development of various diseases have been established. miRNA concentrations change in cancer [8] and cardiovascular diseases [9]. Metabolic perturbations change miRNA concentrations in cells [10]. The aforementioned roles do not encompass all of the biological processes in which miRNAs participate, which further proves the importance of their biological functions. Despite the significant success in the study of miRNA properties, there are obstacles in identifying the target genes of miRNAs. Normally, one miRNA interacts with the mRNA of one gene. However, there are miRNAs that can bind to many mRNAs, and one mRNA can be the target of many miRNAs, which significantly complicates the study of the properties of miRNAs and their diagnostic and medical applications. There are more than 2,500 miRNAs in the human genome, and they are believed to act on 60% or more genes. Therefore, it is difficult to draw specific conclusions about the participation of miRNAs in specific biological processes, and until then the connections between the majority of miRNAs and their target genes will remain unknown. Recently, a set of unique miRNAs (umiRNA) were identified that have hundreds of target genes and bind to mRNAs with high affinity [11–14]. The binding sites of these umiRNAs are located in the 3’UTRs, CDSs, and 5’UTRs of mRNAs. Among these umiRNAs, miR-619-5p interacts with the largest number of target genes that have the greatest number of binding sites with complete complementarity of miR-619-5p and mRNAs. It is necessary to identify many miRNA binding sites in the mRNAs of these genes for the control of gene expression. Furthermore, it is important to control the expression of the corresponding gene complexes that are functionally associated with miRNAs. Therefore, we have studied a unique miR-619-5p that binds to the mRNAs of several hundred human and orthologous genes.
Methods
The nucleotide sequences of mRNAs of human genes (Homo sapience – Hsa) and orthologous genes (Bos mutus - The wild yak (Bmu), Callithrix jacchus – The common marmoset (Cja), Camelus dromedarius – Arabian camel (Cdr), Camelus ferus – The wild Bactrian camel (Cfe), Chlorocebus sabaeus – The green monkey (Csa), Colobus angolensis palliatus – The Angola colobus (Can), Equus caballus - The horse (Eca), Gorilla gorilla - The western gorilla (Ggo), Macaca fascicularis – The crab-eating macaque (Mfa), Macaca mulatta – The rhesus macaque (Mmu), Macaca nemestrina - Pig-tailed macaque (Mne), Mandrillus leucophaeus – The drill (Mle), Nomascus leucogenys - The northern white-cheeked gibbon (Nle), Ovis aries – The sheep (Oar), Pan paniscus - Bonobos (Ppa), Pan troglodytes – The common chimpanzee (Ptr), Papio anubis – The olive baboon (Pan), Pongo abelii - The Sumatran orangutan (Pab), Rhinopithecus roxellana – The golden snub-nosed monkey (Rro)) were downloaded from NCBI GenBank (http://www.ncbi.nlm.nih.gov) [15] in FASTA format using Lextractor002 script [11]. Nucleotide sequences of human mature miR-619-5p (GCUGGGAUUACAGGCAUGAGCC) were downloaded from the miRBase database (http://mirbase.org) [16]. The miR-619-5p binding sites in the 5’-untranslated regions (5’UTRs), the coding domain sequences (CDSs) and the 3’-untranslated regions (3’UTRs) of several genes were predicted using the MirTarget program [12]. This program defines the features of binding: a) the localization of miRNA binding sites in the 5’UTRs, the CDSs and the 3’UTRs of the mRNAs; b) the free energy of hybridization (∆G, kJ/mole). The ratio ΔG/ΔGm (%) was determined for each site (ΔGm equals the free energy of miRNA binding with its perfect complementary nucleotide sequence).
Results
The search of 2,750 human microRNAs (miRNAs) binding sites in 12,175 mRNAs of human genes using the MirTarget program has been completed. The mRNAs have different miRNA binding site origins, lengths, quantities, and properties. The list of miR-619-5p target genes and the positions of binding sites are outlined in Table 1. miR-619-5p is 22 nucleotides in length and is coded by an intron of the slingshot protein phosphatase 1 (SSH1) gene, which is located on chromosome 12 [17, 18]. mRNAs of 201 genes have complete complementary binding sites for miR-619-5p (ΔG/ΔGm = 100%). Therefore, the energy of interaction of miR-619-5p with mRNA of all the genes listed in the table is the same and equal to ΔG = −121 kJ/mole.
Table 1.
Positions of miR-619-5p binding sites and disease or function of target genes
| Gene | Site, nt | Disease or function | PMID | Gene | Site, nt | Disease or function | PMID |
|---|---|---|---|---|---|---|---|
| ACSL6 | 4639 | prostate cancer | 19064571 | MRPS25 | 1609 | uncharacterized | 26302410 |
| ADAL | 2041 | proliferation | 23645737 | MSH3 | 4139 | carcinogenesis | 24934723 |
| ADAM17 | 3466 | breast cancer | 22967992 | NANOS1 | 3219 | retinoblastoma | 25100735 |
| AGMAT | 2207 | renal carcinoma | 14648699 | NCMAP | 2259 | uncharacterized | |
| AK1 | 1449 | hypertension | 23863634 | NDUFAF7 | 1697 | leukemia | 24292274 |
| AKT2 | 4571 | neuroblastoma | 23468863 | NDUFC2 | 1646 | colon cancer | 25804238 |
| ALDH3A2 | 2617 | detoxification | 9829906 | NLN | 4215 | Parkinson’s D. | 25378390 |
| ANKRD16 | 2165 | breast cancer | 20453838 | NRIP2 | 2075 | atopic asthma | 17075290 |
| AP5B1 | 4316 | differentiation | 15146197 | NSL1 | 3063 | kinetochore-protein | 16585270 |
| ARGFX | 2642 | development | 20565723 | NXPE3 | 7447 | hepatocarcinoma | 26883180 |
| ARHGEF39 | 1307 | tumorogenesis | 22327280 | OPTN | 2332 | glaucoma | 26302410 |
| ARL11 | 1033 | tumorogenesis | 18337727 | PAG1 | 8156 | prostatic cancer | 21092590 |
| ATCAY | 2991 | schizophrenia | 19165527 | PAQR5 | 4439 | ovarian cancer | 21761364 |
| ATP1A2 | 4410 | tumorogenesis | 23474907 | PARK2 | 3729 | Parkinson’s D. | 26860075 |
| BCL2L15 | 2650 | apoptosis | 16690252 | PBLD | 2077 | hepatocarcinoma | 26594798 |
| BPNT1 | 1128 | ovarian cancer | 20628624 | PCGF5 | 5089 | Alzheimer’s D. | 16385451 |
| C15orf40 | 523 | uncharacterized | PCSK5 | 8613 | tumorogenesis | 21094132 | |
| C17orf75 | 2895 | uncharacterized | PDAP1 | 1926 | proliferation | 23555679 | |
| C17orf75 | 3672 | PDCD4 | 3221 | tumorogenesis | 26871813 | ||
| C21orf58 | 2668 | uncharacterized | 11707072 | PEX2 | 3056 | cerebellar ataxia | 21392394 |
| C4orf19 | 2068 | uncharacterized | PGPEP1 | 1476 | liver cirrhosis | 25687677 | |
| C6orf170 | 4113 | uncharacterized | 20159594 | PIK3R2 | 3345 | tumorogenesis | 26677064 |
| C8orf44 | 336** | uncharacterized | PNPLA1 | 1991 | childhood obesity | 19390624 | |
| C8orf44 | 1626 | PODNL1 | 1876 | uncharacterized | 12477932 | ||
| C9orf85 | 871 | uncharacterized | POFUT1 | 4679 | hepatocarcinoma | 27003260 | |
| CACNB2 | 4301 | hypertension | 25966706 | POLH | 5550 | ovarian cancer | 25831546 |
| CACNG8 | 3218 | cardiomyopathy | 26710323 | PPM1K | 2192 | diabetes mellitus | 23446828 |
| CACNG8 | 5006 | PPP1R12B | 5156 | childhood asthma | 23640410 | ||
| CACNG8 | 7535 | PRRG4 | 998 | Parkinson’s D | 19772629 | ||
| CALHM1 | 2896 | Alzheimer’s D. | 26944452 | PSMB2 | 2925 | proteolysis | 21660142 |
| CCBE1 | 3321 | ovarian cancer | 19935792 | PTCD3 | 4116 | osteosarcoma | 19427859 |
| CCDC114 | 261* | dyskinesia | 23506398 | PTK6 | 2233 | tumorogenesis | 27311570 |
| CD109 | 6841 | bladder cancer | 20946523 | QRFPR | 1949 | metabolic S. | 16648250 |
| CD36 | 4042 | atherosclerosis | 16515687 | RAB11FIP1 | 4928 | cell transport | 26790954 |
| CD68 | 1398 | carcinomas | 21113139 | RAB3IP | 3975 | tumorogenesis | 12007189 |
| CDAN1 | 4296 | erythropoiesis | 19336738 | 7022 | |||
| CDHR3 | 4878 | asthma | 25848009 | RAB7L1 | 1693 | Parkinson’s D. | 26914237 |
| CEP68 | 4394 | cervical cancer | 17570516 | RBBP9 | 1818 | tumorogenesis | 21933118 |
| CHST5 | 2946 | colon carcinoma | 12107080 | RGS3 | 205* | cardiovascular D. | 24375609 |
| CHST6 | 2979 | dystrophy | 20539220 | RPS6KA6 | 7136 | tumorogenesis | 26732474 |
| CHST6 | 3876 | SCN11A | 5871 | neurophaty | 25791876 | ||
| CIAO1 | 2416 | tumorogenesis | 9556563 | SEPT11 | 4033 | hepatocarcinoma | 20419844 |
| CIAO1 | 3814 | SEPT14 | 1575 | Parkinson’s D | 27115672 | ||
| CLEC19A | 1747 | lectin | 12975309 | SGTB | 3142 | lymphopoesis | 2158125 |
| CLTC | 7006 | pancreatic cancer | 23228632 | SH3GLB1 | 4856 | prostate cancer | 27748942 |
| CORO2A | 2227 | colon cancer | 23490283 | SLC15A2 | 4333 | hepatocarcinoma | 25965825 |
| COX18 | 1264 | tumorogenesis | 20819778 | SLC17A5 | 2389 | cardiovascular D | 27872510 |
| CPM | 2698 | renal carcinoma | 23172796 | SLC26A2 | 5066 | colorectal cancer | 23840040 |
| CPM | 4996 | SLC26A4 | 4210 | hearing loss | 27729126 | ||
| CPT2 | 2557 | sudden death | 21641254 | SLC28A2 | 2196 | chronic hepatitis C | 23195617 |
| CYB5RL | 3426 | transcription | 16344560 | SLC7A11 | 6304 | tumorogenesis | 26729415 |
| CYP20A1 | 2539 | tumorogenesis | 15191668 | SLC7A14 | 8487 | breast cancer | 20379614 |
| CYP20A1 | 4709 | SNX22 | 902 | liver-disease | 21988832 | ||
| CYP27C1 | 3823 | self-rated health | 20707712 | SOWAHC | 3417 | retrotransposon | 22234889 |
| CYP2W1 | 2176 | colorectal cancer | 22993331 | SPATA13 | 5020 | colorectal cancer | 17599059 |
| DAP3 | 1842 | breast cancer | 22287761 | SPATA5 | 5648 | microcephaly | 26299366 |
| DCAF10 | 3305 | lung cancer | 28336923 | SPATS2 | 3332 | breast cancer | 20379614 |
| DCAF10 | 4559 | SPN | 5287 | tumorogenesis | 25551301 | ||
| DCLRE1C | 2966 | Omenn syndrome | 25981738 | STAC2 | 2241 | inherited ataxias | 16713569 |
| DDOST | 1782 | hyperglycemia | 22305527 | SYNJ2BP | 1298 | breast cancer | 19349195 |
| DHODH | 1709 | melanoma | 21430780 | SYNJ2BP | 4175 | ||
| DHRS9 | 1281* | tumorogenesis | 26254099 | TCEB1 | 1964 | tumorogenesis | 23083832 |
| DNAL1 | 4925 | dyskinesia | 15845866 | TIGD6 | 3439 | uncharacterized | |
| DSCR6 | 1706 | Down syndrome | 10814524 | TMEM156 | 1593 | uncharacterized | |
| ERBB3 | 5104 | tumorogenesis | 26689995 | TMEM19 | 3510 | uncharacterized | |
| FADS6 | 1777 | liver disease | 21988832, | TMEM213 | 875 | uncharacterized | |
| FAM161A | 2785 | retinal disease | 25749990 | TMEM214 | 1190 | uncharacterized | |
| FAM227A | 4981 | cancer | 26759717 | TMEM50B | 1026 | uncharacterized | |
| FAM84B | 3626 | tumorogenesis | 25980316 | TMEM56 | 1243 | nicotine dependence | 20379614 |
| FBLIM1 | 2126 | breast cancer, | 23645746 | TMF1 | 4736 | prostate cancer | 19330832 |
| FBXL22 | 1411 | cardiomyopathy | 24324551 | TMOD2 | 7816 | bladder cancer | 15095301 |
| FBXO27 | 1535 | leukemia | 126433 | TNFRSF10A | 1621 | cancer | 27780136 |
| FGD4 | 7619 | cancer | 22589722 | TNFRSF10D | 1532 | cancer | 26542757 |
| FKBP14 | 1515 | ovarian cancer | 27931282 | TOP3A | 3814 | leukaemia | 22050635 |
| FKBP14 | 2129 | TPRG1L | 1754 | uncharacterized | |||
| FKBP5 | 7114 | schizophrenia | 25522420 | TRIM72 | 1885 | ischemia | 26790476 |
| FXN | 3288 | metabolic disease | 26717909 | TRPM7 | 8079 | neuroblastoma | 27402209 |
| GDPD1 | 1559 | phosphodiesterase | 18991142 | TRPM7 | 8221 | carcinoma | 26779625 |
| GEMIN8 | 2172 | neuropathy | 16434402 | TXNDC15 | 2460 | thrombosis | 21642008 |
| GGT6 | 1956 | ovarian cancer | 25356737 | TYW5 | 3692 | schizophrenia | 23974872 |
| GK5 | 3808 | glioblastoma | 25936394 | UACA | 6120 | lung cancer | 22407486 |
| GK5 | 6355 | glioblastoma | 25936394 | UACA | 6120 | thyroid diseases | 15358194 |
| GLB1L | 2224 | phosphatase | 21382349 | UBIAD1 | 2881 | cancer | 23759948 |
| GOLGA3 | 7240 | immune disease | 17711851 | UBXN2A | 1665 | colon cancer | 24625977 |
| GP2 | 1877 | crohn disease | 22891285 | UPK1B | 1513 | cancer | 16354592, |
| GPR65 | 3309 | tumorogenesis | 24152439 | UQCRB | 1269 | colorectal cancer | 22545919 |
| GPR65 | 3309 | immune diseases | 15665078 | USP29 | 2* | protease | 10958632 |
| GPR82 | 2664 | uncharacterized | VHL | 3764 | tumorogenesis | 27460078 | |
| GPRIN2 | 6676 | schizophrenia | 27244233 | VHL | 3898 | ||
| GTPBP10 | 1873 | prostate cancer | 27409348 | VWA2 | 3366 | colon cancer | 15580307 |
| H6PD | 5754 | tumorogenesis | 15221007 | WDR73 | 1736 | microcephaly | 25466283 |
| HM13 | 1745 | glioblastoma | 28198167 | XIAP | 5681 | ovarian cancer | 26779627 |
| IFIT3 | 1864 | pancreatic cancer | 25650658 | YAE1D1 | 1548 | oral cancer | 23318452 |
| ISY1 | 686** | uncharacterized | ZBTB24 | 4842 | hepatocarcinoma | 27730394 | |
| IYD | 1658 | hypothyroidism. | 18765512 | ZC3H12D | 2812 | Acute lung injury | 26059755 |
| KIAA1456 | 2536 | colorectal cancer | 24743840 | ZDHHC20 | 3390 | tumorogenesis | 20334580 |
| KIF11 | 3598 | tumorogenesis | 28011472 | ZFP30 | 3463 | hypertension | 19851296 |
| KLHL23 | 2570 | tumorogenesis | 23676014 | ZNF114 | 1827 | transcription factor | 8467795 |
| KPNA1 | 5711 | breast cancer | 26052702 | ZNF197 | 3446 | thyroid cancer | 12682018 |
| KREMEN1 | 2199 | schizophrenia | 20153141 | ZNF320 | 5534 | glioblastoma | 11536051 |
| KREMEN1 | 2792 | schizophrenia | 20153141 | ZNF429 | 2081** | transcription factor | 7865130 |
| LAX1 | 2057 | uncharacterized | ZNF445 | 8820 | transcription factor | 16368201 | |
| LILRA6 | 2201 | tumorogenesis | 26769854 | ZNF461 | 3087 | transcription factor | 15004467 |
| LIMD1 | 5735 | breast cancer | 27656835 | ZNF549 | 3736 | transcription factor | 16344560 |
| LIMS1 | 3931 | cancer | 27590440 | ZNF557 | 4791 | transcription factor | 15851553 |
| LMOD3 | 3224 | myopathy | 25250574 | ZNF626 | 4620 | liver diseases | 18255255 |
| LMOD3 | 3993 | Alzheimer’s D | 22881374 | ZNF667 | 3240 | transcription factor | 17397802 |
| METTL6 | 1188 | breast cancer | 25151356 | ZNF716 | 2799 | cardiovascular D | 24376456 |
| MR1 | 3664 | hepatocarcinoma | 26823810 | ZNF780B | 5415 | transcription factor | 15057824 |
| MREG | 1540 | pulmonary D | 20463177 | ZNF84 | 4920 | transcription factor | 11856868 |
| ZNF841 | 3422 | transcription factor | 24280104 |
Notes: * - 5’UTR, **- CDS; others – 3’UTR, D - disease
The mRNAs of 201 human genes have complete complementary binding sites of miR-619-5p in the 3’UTR (214 sites), CDS (3 sites), and 5’UTR (4 sites). The mRNAs of 27 genes have four binding sites, seven genes have five binding sites, and CATAD1, ICA1L, GK5, POLH, and PRR11 genes have six miR-619-5p binding sites. The mRNAs of OPA3 and CYP20A1 genes have eight and ten binding sites, respectively. All of these sites are located in the 3’UTRs of mRNAs.
The target genes of the miR-619-5p carry out one or more different functions and are involved in the development of various diseases (Table 1).
The mRNAs of the C17orf75, C8orf44, CIAO1, CPM, CYP20A1, DCAF10, FKBP14, RAB3IP, SYNJ2BP, VHL genes have two complete complementary binding sites for miR-619-5p, and the mRNA of the CACNG8 gene has three such binding sites. This indicates a stronger dependence of the expression of these genes on miR-619-5p.
One of the methods to establish the credibility of the presence of miRNA binding site in the mRNA is to verify this site in the mRNAs of orthologous genes. In finding the miRNA binding sites raises the question of the level of reliability of the found sites. One effective way to establish the credibility of the binding sites is to establish binding sites in the orthologous genes and the identification of orthologous miRNA. Location of binding site in the protein coding region facilitates its conservation in evolution, especially if the corresponding oligopeptide plays an important role in the function of the protein. miR-619-5p binding sites with complete complementarity (ΔG/ΔGm is 100%) to the mRNAs of the four genes are located in the 5’UTRs (Table 2).
Table 2.
Variation of positions and nucleotide sequences of miR-619-5p binding sites in the 5’UTRs of mRNAs of mammal genes
| Species | Gene | Position of site, nt |
Nucleotide sequence |
|---|---|---|---|
| Hsa | CCDC114 | 261 | GCAUGCUGGCUCAUGCCUGUAAUCCCAGCACUUUGG |
| Hsa | DHRS9 | 1281 | GCGCGGUGGCUCAUGCCUGUAAUCCCAGCACUUUGG |
| Hsa | RGS3 | 205 | GCGCAGUGGCUCAUGCCUGUAAUCCCAGCACUUUGG |
| Ptr | RGS3 | 1 | GCGCAGUGGCUCAUGCCUGUAAUCCCAGCACUUUGG |
| Nle | RGS3 | 205 | GCACGGUGGCUCAUGCCUGUAAUCCCAGCACUUUGG |
| Hsa | USP29 | 2 | CUGGCCAGGCUCAUGCCUGUAAUCCCAGCACUUUGG |
| Pab | USP29 | 52 | CUGGCCAGGCUCAUGCCUGUAAUCCCAGCACUUUGG |
| Nle | USP29 | 52 | CUGGCCAGGCUCAUGCCUGUAAUCCCAGCACUUUGG |
| Mle | USP29 | 47 | CUGGCCAGGCUCAUGCCUGUAAUCCCAGCACUUUGG |
| Can | USP29 | 98 | CUGGCCAGGCUCAUGCCUGUAAUCCCAGCAUUUUGG |
| Ggo | USP29 | 100 | CUGGCCAGGCUCAUGCCUGUAAUUCCAGCACUUUGG |
| Rro | USP29 | 52 | CUGGCCAGGCUCAUGCCUGUAAUCGCAGCACUUUGG |
Before the 5’ end and after the 3’ end of miR-619-5p binding site, nucleotides are not homologous. The mRNAs of RGS3 and USP29 orthologous genes have binding sites in H. sapiens, N. leucogenys, P. abelii, M. leucophaeus, C. angolensis palliatus, G. gorilla, and R. roxellana.
miR-619-5p has two binding sites in the 5’UTRs of mRNAs of ANAPC16, CYB5D2, and PRR5 and three binding sites in the mRNA of DNASE1.
mRNAs of some genes have binding sites for miR-619-5p within their 5’UTRs and 3’UTRs or CDSs and 3’UTRs. For example, ATAD3C, C14orf182, and CYB5RL have miR-619-5p binding sites in the 5’UTRs and 3’UTRs, and C8orf44, ISY1, and ZNF714 have miR-619-5p binding sites in the CDSs and 3’UTRs.
The nucleotide sequences of miR-619-5p binding sites are located in the CDSs of the C8orf44, C8H8orf44, ISY1, ZNF429, and ZNF714 genes and encode the following oligopeptides (Table 3). C8H8orf44, C8orf44, and ISY1 genes encode the WLMPVIP oligopeptide, which is also present in the orthologous proteins of P. abelii, P. anubis, P. paniscus, and P. troglodytes. The mRNA of transcription factor ZNF429 and ZNF429 genes binding sites are encoded the AHACNP oligopeptide in the another reading frame. The first two oligopeptides are encoded in one open reading frame (ORF) and the amino acid sequences are highly conserved. The homologous oligonucleotide of the miR-619-5p binding site in the mRNA of ZNF714 gene codes for an oligopeptide in a different ORF.
Table 3.
Variation of amino acid sequences coding in miR-619-5p binding sites in the mRNAs of orthologous genes
| Species | Gene | Amino acid sequence |
|---|---|---|
| Hsa | C8orf44 | HWKGRARWLMPVIPALWEAKA |
| Hsa | C8H8orf44 | HWKGRARWLMPVIPALWEAKA |
| Pab | C8H8orf44 | HWKGWARWLTPVIPALWEAKA |
| Pan | C8H8orf44 | HWKGRARWLMPAIPALWEAKX |
| Ppa | C8H8orf44 | HWKGRAQWLTPVIPALWEAKA |
| Ptr | C8H8orf44 | HWKGRAQWLTPVIPALWEAKA |
| Hsa | ISY1 | EKERQVRWLMPVIPALWEAEA |
| Hsa | ZNF714 | KIQQGMVAHACNPNTLRGLGE |
| Ggo | ZNF714 | KIQQGMVAHACNPNTLRGLGE |
| Ptr | ZNF714 | KIQQGMVAHACNPNTXRGLGE |
| Ppa | ZNF714 | KIQQGMVAHACNPNTLRGLGE |
| Hsa | ZNF429 | IHRMGVVAHACNPSTLGGRGG |
| Mfa | ZNF429 | IHRLGVVAHACNPSTLGGRGG |
| Mmu | ZNF429 | IHRLGVVAHACNPSTLGGRGG |
| Mne | ZNF429 | IHRLGVVAHACNPSTLGGRGG |
The presence of miR-619-5p binding sites in the CDSs of five genes with different functions and the evolutionary conservation of these sites signify the role of miRNA in the regulation of the expression of these genes. The nucleotide sequences of specific regions of mRNAs of C8H8orf44, C8orf44, ISY1, ZNF429, and ZNF714 genes that contain miR-619-5p binding sites in the CDSs are homologous among themselves and to the binding sites located in the 5’UTRs and 3’UTRs.
The miRNA binding sites in the coding region, as opposed to the 3’UTR and 5’UTR, clearly demonstrate the relationship between miRNA and mRNA by their conserved amino acid sequences in orthologous proteins. miRNA binding site can be translated by two open reading frames that encode WLTPVIPA and AHACNPS oligopeptides. In the third reading frame, the miR-619-5p binding site has a stop codon. However, in the genes studied, no such sequence was found. In the absence of complete complementarity between miR-619-5p and its binding site, miR-619-5p uses a site containing the corresponding mutation in the CDS for the regulation of gene expression. Thus, a single miRNA binding site in the mRNA of various genes may correspond to three different oligopeptides. Generally, one out of these three oligopeptides is present in the proteins encoded by the orthologous genes.
ISY1 orthologous genes in H. sapiens, P. troglodytes, and N. leucogenys encode a protein containing QVRWLMPVIPALWEAEAGGSQA oligopeptide sequence (Table 4).
Table 4.
Amino acid sequences coding in miR-619-5p binding sites in the mRNA of ISY1 gene of orthologous genes
| Species | Amino acid sequence |
|---|---|
| Hsa | PGVRELFEKERQVRWLMPVIPALWEAEAGGSQALPPPRKTRAELMKA |
| Ptr | PGVRELFEKERQVRWLMPVIPALWEAEAGGSQALPPPRKTRAELMKA |
| Nle | PGVRELFEKERQARWLTPVIPALWEAEAGGSQALPPPRKTRAELMKA |
| Hsa* | PGVRELFEKEP----------------------LPPPRKTRAELMKA |
| Bmu | PGVRELFEKEP----------------------LPPPRKTRAELMKA |
| Cdr | PGVRELFEKEP----------------------LPPPRKTRAELMKA |
| Cfa | PGVRELFEKEP----------------------LPPPRKTRAELMKA |
| Cja | PGVRELFEKEP----------------------LPPPRKTRAELMKA |
| Eca | PGVRELFEKEP----------------------LPPPRKTRAELMKA |
| Ggg | PGVRELFEKEP----------------------LPPPRKTRAELMKA |
| Mmu | PGVRELFEKEP----------------------LPPPRKTRAELMKA |
| Nle | PGVRELFEKEP----------------------LPPPRKTRAELMKA |
| Oar | PGVRELFEKEP----------------------LPPPRKTRAELMKA |
| Pab | PGVRELFEKEP----------------------LPPPRKTRAELMKA |
| Ppa | PGVRELFEKEP----------------------LPPPRKTRAELMKA |
| Rro | PGVRELFEKEP----------------------LPPPRKTRAELMKA |
* RAB43 - human ISY1 paralog gene
However, the RAB43 gene, which is paralogous to human ISY1, lacks the nucleotide sequence encoding the QVRWLMPVIPALWEAEAGGSQA oligopeptide. Additionally, ISY1 gene in the genomes of other animals also lacks the nucleotide sequence encoding this oligopeptide (Table 4).
Nucleotide sequences of miR-619-5p binding sites in the mRNAs of ADAM17, ALDH3A2, and ARL11 orthologous genes are shown in Table 5.
Table 5.
Variation of nucleotide sequences of miR-619-5p binding sites in the 3’UTR of mRNAs of ADAM17, ALDH3A2, and ARL11 of orthologs
| Species | Gene | Position, nt | Nucleotide sequence |
|---|---|---|---|
| Hsa | ADAM17 | 3466 | TGGGAGTGGTGGCTCATGCCTGTAATCCCAGCACTTGGAGAGG |
| Cat | ADAM17 | 3485 | GGGGCGCAGTGGCTCATGCCTGTAATCCCAGCACTTTGGGAGG |
| Mmul | ADAM17 | 3491 | GGGGCGCGGTGGCTCATGCCTGTAATCCCAGCACTTTGGGAGG |
| Mne | ADAM17 | 3438 | GGGGCGCGGTGGCTCATGCCTGTAATCCCAGCACTTTGGGAGG |
| Ptr | ADAM17 | 3449 | TGGGAGTGGTGGCTCATGCCTGTAATCCCAGCACTTGGAGAGG |
| Rro | ADAM17 | 3425 | GGGGCGCGGTGGCTCATGCCTGTAATCCCAGCACTTTGGGAGG |
| Hsa | ALDH3A2 | 2617 | CGGGCGTGGTGGCTCATGCCTGTAATCCCAGCACTTTGGGAGG |
| Cja | ALDH3A2 | 3444 | CGGGCGTGGTGGCTCATGCCTGTAATCCCAGCACTTTAGGAGG |
| Ggo | ALDH3A2 | 2712 | CGGGCGTGGTGGCTCATGCCTGTAATCCCAGCACTTTGGGAGG |
| Mmul | ALDH3A2 | 2509 | CGGACATGGTGGCTCATGCCTGTAATCCCAGCACTTTGGGAGG |
| Mne | ALDH3A2 | 2504 | CGGACATGGTGGCTCATGCCTGTAATCCCAGCACTTTGGGAGG |
| Nle | ALDH3A2 | 2714 | TGGTCATGGTGGCTCATGCCTGTAATCCCAGCACTTTGGGAGG |
| Pab | ALDH3A2 | 2297 | TGGGCATGGTGGCTCATGCCTGTAATCCCAGCACTTTGGGAGG |
| Ppa | ALDH3A2 | 2715 | CGGGCATGGTGGCTCATGTCTGTAATCCCAGCACTTTGGGAGG |
| Ptr | ALDH3A2 | 2711 | CGGGCATGGTGGCTCATGTCTGTAATCCCAGCACTTTGGGAGG |
| Rro | ALDH3A2 | 2727 | CGGACGTGGTGGCTCATGCCTGTAATCCCAGCACTTTGGGAGG |
| Hsa | ARL11 | 1033 | TTGGCCCGGTGGCTCATGCCTGTAATCCCAGCACTGTGGGAGA |
| Cat | ARL11 | 1642 | CAGATGCAGTGGCTCATGCCTGTAATCCCAGCACTTTGGGTGG |
| Mfa | ARL11 | 1698 | CAGATGCAGTGGCTCATGCCTGTAATCCCAGCACTTTGGGTGG |
| Mmul | ARL11 | 1747 | CAGATGCAGTGGCTCATGCCTGTAATCCCAGCACTTTGGGTGG |
| Mne | ARL11 | 1024 | TTGGCACGGTGGCTCATGCCTGTAATCCCAGCACTTTGGGAGA |
| Mne | ARL11 | 1471 | CAGATGCAGTGGCTCATGCCTGTAATCCCAGCACTTTGGGTGG |
| Ptr | ARL11 | 1353 | CGGGCATGGTGGCTCATGTCTGTAATCCCAGCACTTTGGGAGG |
| Rro | ARL11 | 1254 | CAGGTGCAGTGGCTCATGCCTGTAATCCCAGCACTTTGGGCGG |
These orthologous genes are characterized by highly conserved nucleotide sequence GGCTCATGCCTGTAATCCCAGC of miR-619-5p binding sites. This shows that the interaction of miR-619-5p with mRNAs of these genes is conserved during evolution. Some of the human miR-619-5p target genes and their corresponding orthologous genes have two miR-619-5p binding sites in their mRNAs.
Table 6 shows the nucleotide sequences of two miR-619-5p binding sites in the 3’UTR of mRNAs of ERBB3, FBLIM1, and FKBP14 orthologous genes.
Table 6.
Variation of nucleotide sequences of two miR-619-5p binding sites in the 3’UTR of mRNAs of ERBB3, FBLIM1, and FKBP14 of orthologs
| Species | Gene | Position, nt | Nucleotide sequence |
|---|---|---|---|
| Hsa | ERBB3 | 4950 | CGGGCATGGTGGCTCATGCCTGTAATCTCAGCACTTTGGGAG |
| Hsa | ERBB3 | 5104 | TGGGTGCAGTGGCTCATGCCTGTAATCCCAGCCAGCACTTTG |
| Csa | ERBB3 | 4989 | CGGGCATGGTGGCTCATGCCTGTAATCCTAGCACTTTGGGAG |
| Csa | ERBB3 | 5149 | TGGGCGCTGTGGCTCATGCCTGCAATCCCAGCACTTTGGGAG |
| Mfa | ERBB3 | 5114 | TGGGCATGGTGGCTCATGCCTGTAATCCCAGCACTTTGGGAG |
| Mfa | ERBB3 | 5269 | TGGGCGCTGTGGCTCATGCCTGCAATCCCAGCCCTTTGGGAG |
| Mmu | ERBB3 | 5114 | TGGGCATGGTGGCTCATGCCTGTAATCCCAGCACTTTGGGAG |
| Mmu | ERBB3 | 5269 | TGGGCGCTGTGGCTCATGCCTGCAATCCCAGCCCTTTGGGAG |
| Mne | ERBB3 | 5112 | CGGGCATGGTGGCTCATGCCTGTAATCCCAGCACTTTGGGAG |
| Mne | ERBB3 | 5267 | TGGGCGCTGTGGCTCATGCCTGCAATCCCAGCCCTTTGGGAG |
| Pan | ERBB3 | 5106 | CGGGCATGGTGGCTCATGCCTGTAATCCCAGCACTTTGGGAG |
| Pan | ERBB3 | 5274 | TGGGCGCTGTGGCTCATGCCTGCAGTCCCAGCACTTTGGGAG |
| Ptr | ERBB3 | 5105 | CGGGCATGGTGGCTCATGCCTGTAATCTCAGCACTTTGGGAG |
| Ptr | ERBB3 | 5243 | TGGGTGCAGTGGCTCATGCCTGTAATCCCAGCCAGCACTTTG |
| Mne | FBLIM1 | 1938 | TGGGCGTGGTGGCTCATGCCTGTAATCCCTGCACTTTGGGAG |
| Mne | FBLIM1 | 5267 | TGGGCGCTGTGGCTCATGCCTGCAATCCCAGCCCTTTGGGAG |
| Pab | FKBP14 | 1514 | CAGGCACGGTGGCTCACGCCTGTAATCCCAGCACTTCGGGAG |
| Pab | FKBP14 | 2128 | TGGGTGTGGTGGCTCATGCCTGTAATCCCAGCACTTTGGGGG |
Notes: The black type indicates the binding site of miR-619-5p
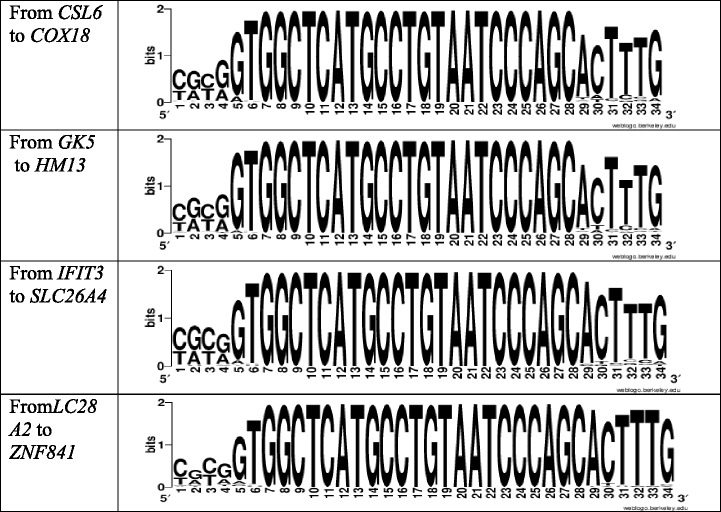
Table 7 shows the degree of conservation of miR-619-5p binding sites in the 201 mRNAs of target genes. All mRNAs with complete complementarity to miR-619-5p binding sites (ΔG/ΔGm is 100%) were divided into four groups, and the frequency of occurrence of nucleotides was determined in each group. The results suggest that miR-619-5p binding sites are highly conserved. The binding site GGCTCATGCCTGTAATCCCAGC does not change and in each of the four gene groups the observed variability of nucleotides on the right and left is high.
Table 7.
Variation of nucleotide sequences of mRNA region with miR-619-5p binding sites (See Additional file 1, 2, 3 and 4)
Discussion
Here we have identified many miRNAs binding sites in the mRNAs of 201 human genes which indicates that umiRNAs act as coordinators of gene expression by participating in many biological processes. Previous studies have shown the influences of miRNAs on the expression of genes that encode the transcription factors [19, 20] and on the expression of proteins that participate in the cellular cycle [3, 21–23], apoptosis [4, 24–26], and stress responses [27]. It was shown the role of the mir-619-5p in the regulation of different pathological processes [28]. It was investigated the correlations between the expression of MALAT1 and miR-619-5p, in addition to the association between the clinicopathological features and survival outcomes of patients with stage II and III colorectal cancer tumors [28]. It was observed, that hsa-miR-619-5p and hsa-miR-1184 microRNA expression significantly increased in prostatic cancer. MicroRNA-gene-net analysis indicated that miR-619-5p and other some miRNAs had the most important and extensive regulatory function for Qi-stagnation syndromes and Qi-deficiency syndromes in coronary heart disease [29].
One or several umiRNAs regulating the expression of hundreds of genes can create a system of interconnected processes in cells and organisms. Such role of these umiRNAs is possible because they circulate in the blood and have access to nearly all cells of an organism [30–32]. Our results provide the basis for studying the systemic roles of unique and normal miRNAs in the regulation of gene expression in human cells. The expression of many target genes is regulated by umiRNAs does not allow individual mRNAs of target genes to be expressed in more degree than others. The greater expression of one mRNA, the larger number of umiRNAs bind to this mRNA. This allows one umiRNA to maintain a certain balance of expression of the corresponding target genes. If umiRNA expression changes, such system is vulnerable. This will cause the development of pathology in the cell, tissue or body.
Conclusions
The majority of miR-619-5p binding sites are located in the 3’UTRs of mRNAs of target genes. Some genes have miRNA binding sites in the 5’UTRs of mRNAs. It is necessary to maintain nucleotide sequences of the binding site of umiRNA in the CDSs of several genes. Different genes have binding sites for miRNAs that are read in different open reading frames. Therefore, identical nucleotide sequences encode different amino acids in different proteins. In encoded proteins, these sites encode conservative oligopeptides. The binding sites of miR-619-5p in 3’UTRs, 5’UTRs and CDSs are conservative in the orthologous mammalian genes.
Additional files
Variation of nucleotide sequences of mRNA region with miR-619-5p binding sites of genes from CSL6 to COX18 (Conservative binding sites are in bold) (PDF 218 kb)
Variation of nucleotide sequences of mRNA region with miR-619-5p binding sites of genes from GK5 to HM13 (Conservative binding sites are in bold) (PDF 106 kb)
Variation of nucleotide sequences of mRNA region with miR-619-5p binding sites of genes from IFIT3 to SLC26A4 (Conservative binding sites are in bold) (PDF 139 kb)
Variation of nucleotide sequences of mRNA region with miR-619-5p binding sites of genes from LC28A2 to ZNF841 (Conservative binding sites are in bold). The data given in the Additional files 1, 2, 3 and 4 demonstrate the variability of the nucleotides before and after the binding sites of miR-619-5p, which is shown in the Weblogo schemes in the table 8. (PDF 151 kb)
Acknowledgements
We thank Leducq foundation and the European Union project «Muscle Stress Relief». Also we thank PhD Berillo O. for her help in the collection of data.
Funding
This study was supported by a grant (N0491/ГФ4) from the Ministry of Education and Science, Kazakhstan Republic, SRI of Biology and Biotechnology Problems, al-Farabi Kazakh National University, and Institute for Anesthesiology and Intensive Operative Care Medical Faculty Mannheim, Mannheim, Germany.
Availability of data and materials
The data sets supporting the results of this article are included within the article and its additional files and publicly available.
Authors’ contributions
SA, RN and AI conceived of the study and drafted the manuscript. SA, RN, AI, SL, AP, IP and AA made substantial contributions to acquisition of data, to interpretation and modification of the data. All authors involved in drafting the manuscript, read and approved the final version of the manuscript.
Competing interests
The authors declares that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
- CDSs
Coding domain sequences
- miRNAs
Micrornas
- ORF
Open reading frame
- Umirna
Unique miRNA
Footnotes
Electronic supplementary material
The online version of this article (doi:10.1186/s12864-017-3811-6) contains supplementary material, which is available to authorized users.
Contributor Information
Shara Atambayeva, Email: atambayevashara@gmail.com.
Raigul Niyazova, Email: Raigul.Nyiyazova@kaznu.kz.
Anatoliy Ivashchenko, Email: a.iavashchenko@gmail.com.
Anna Pyrkova, Email: Anna.Pyrkova@kaznu.kz.
Ilya Pinsky, Email: ilya.pinskyi@mail.ru.
Aigul Akimniyazova, Email: akimniyazova@gmail.com.
Siegfried Labeit, Email: labeit@embl.de.
References
- 1.Doxakis E. Principles of miRNA-Target Regulation in Metazoan Models. Int J Mol Sci. 2013;14:16280–16302. doi: 10.3390/ijms140816280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tang G. siRNA and miRNA: an insight into RISCs. Trends Biochem Sci. 2005;30:106–114. doi: 10.1016/j.tibs.2004.12.007. [DOI] [PubMed] [Google Scholar]
- 3.Luo Q, Li X, Li J, Kong X, Zhang J, Chen L, Huang Y, Fang L. MiR-15a is underexpressed and inhibits the cell cycle by targeting CCNE1 in breast cancer. Int J Oncol. 2013;43:1212–1218. doi: 10.3892/ijo.2013.2034. [DOI] [PubMed] [Google Scholar]
- 4.Li X, Chen YT, Josson S, Mukhopadhyay NK, Kim J, Freeman MR, Huang WC. MicroRNA-185 and 342 inhibit tumorigenicity and induce apoptosis through blockade of the SREBP metabolic pathway in prostate cancer cells. PLoS One. 2013;8:e70987. doi: 10.1371/journal.pone.0070987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Qian NS, Liu WH, Lv WP, Xiang X, Su M, Raut V, Chen YL, Dong JH. Upregulated MicroRNA-92b regulates the differentiation and proliferation of EpCAM-positive fetal liver cells by targeting C/EBPß. PLoS One. 2013;8:e68004. doi: 10.1371/journal.pone.0068004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Poethig RS. Vegetative phase change and shoot maturation in plants. Curr Top Dev Biol. 2013;105:125–152. doi: 10.1016/B978-0-12-396968-2.00005-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ling YH, Ding JP, Zhang XD, Wang LJ, Zhang YH, Li YS, Zhang ZJ, Zhang XR. Characterization of microRNAs from goat (Capra hircus) by Solexa deep-sequencing technology. Genet Mol Res. 2013;12:1951–1961. doi: 10.4238/2013.June.13.4. [DOI] [PubMed] [Google Scholar]
- 8.Knyazev EN, Fomicheva KA, Mikhailenko DS, et al. Plasma Levels of hsa-miR-619-5p and hsa-miR-1184 Differ in Prostatic Benign Hyperplasia and Cancer. Bull Exp Biol Med. 2016;161:108–11. doi: 10.1007/s10517-016-3357-7. [DOI] [PubMed] [Google Scholar]
- 9.Hou J, Wang J, Lin C, Fu J, Ren J, Li L, Guo H, Han X, Liu J. Circulating MicroRNA Profiles Differ between Qi-Stagnation and Qi-Deficiency in Coronary Heart Disease Patients with Blood Stasis Syndrome. Evid Based Complement Alternat Med. 2014;2014:926962. doi: 10.1155/2014/926962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Swaminathan S, Suzuki K, Seddiki N, Kaplan W, Cowley MJ, Hood CL, et al. Differential regulation of the Let-7 family of microRNAs in CD4+ T cells alters IL-10 expression. J Immunol. 2012;188:6238–6246. doi: 10.4049/jimmunol.1101196. [DOI] [PubMed] [Google Scholar]
- 11.Ivashchenko A, Berillo O, Pyrkova A, Niyazova R, Atambayeva S. MiR-3960 binding sites with mRNA of human genes. Bioinformation. 2014;10(7):423–7. doi: 10.6026/97320630010423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ivashchenko A, Berillo O, Pyrkova A, Niyazova R, Atambayeva S. The properties of binding sites of miR-619-5p, miR-5095, miR-5096 and miR-5585-3p in the mRNAs of human genes. Biomed Res Int. 2014;2014:720715. doi: 10.1155/2014/720715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ivashchenko A, Berillo O, Pyrkova A, Niyazova R. Binding Sites of miR-1273 Family on the mRNA of Target Genes. Biomed Res Int. 2014;2014:620530. doi: 10.1155/2014/620530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ivashchenko A, Berillo O, Pyrkova A, Niyazova R, Atambayeva S. The binding sites of unique miRNAs in the human mRNAs. J Biotechnol. 2014;185:125–37. [Google Scholar]
- 15.National Center for Biotechnology Information. http://www.ncbi.nlm.nih.gov
- 16.Griffiths-Jones S, Grocock R, van Dongen S, Bateman A, Enright A. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:140–144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cummins JM, He Y, Leary RJ, Pagliarini R, Diaz LA, Jr, Sjoblom T, Barad O, Bentwich Z, Szafranska AE, Labourier E, Raymond CK, Roberts BS, Juhl H, Kinzler KW, Vogelstein B, Velculescu VE. The colorectal microRNAome. Proc Natl Acad Sci U S A. 2006;103:3687–3692. doi: 10.1073/pnas.0511155103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ple H, Landry P, Benham A, Coarfa C, Gunaratne PH, Provost P. The repertoire and features of human platelet microRNAs. PLoS One. 2012;7:e50746. doi: 10.1371/journal.pone.0050746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cui Q, Yu Z, Pan Y, Purisima EO, Wang E. MicroRNAs preferentially target the genes with high transcriptional regulation complexity. Biochem Biophys Res Commun. 2007;352(3):733–8. doi: 10.1016/j.bbrc.2006.11.080. [DOI] [PubMed] [Google Scholar]
- 20.Yan L, Kang M, Qin Z, Zhang W, Li Y, Ou H. An intronic miRNA regulates expression of the human endothelial nitric oxide synthase gene and proliferation of endothelial cells by a mechanism related to the transcription factor SP-1. PLoS One. 2013;8:e70658. doi: 10.1371/journal.pone.0070658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang P, Zou F, Zhang X, Li H, Dulak A, Tomko RJ, Jr, Lazo JS, Wang Z, Zhang L, Yu J. microRNA-21 negatively regulates Cdc25A and cell cycle progression in colon cancer cells. Cancer Res. 2013;69:8157–8165. doi: 10.1158/0008-5472.CAN-09-1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cui X, Witalison EE, Chumanevich AP, Chumanevich AA, Poudyal D, et al. The induction of microRNA-16 in colon cancer cells by protein arginine deiminase inhibition causes a p53-dependent cell cycle arrest. PLoS One. 2013;8:e53791. doi: 10.1371/journal.pone.0053791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Y, Zheng X, Zhang Z, Zhou J, Zhao G, et al. MicroRNA-149 inhibits proliferation and cell cycle progression through the targeting of ZBTB2 in human gastric cancer. PLoS One. 2012;7:e41693. doi: 10.1371/journal.pone.0041693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Y, Lee CG. MicroRNA and cancer focus on apoptosis. J Cell Mol Med. 2009;13:12–23. doi: 10.1111/j.1582-4934.2008.00510.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li C, Hashimi SM, Good DA, Cao S, Duan W, Plummer PN, Mellick AS, Wei MQ. Apoptosis and microRNA aberrations in cancer. Clin Exp Pharmacol Physiol. 2012;39:739–746. doi: 10.1111/j.1440-1681.2012.05700.x. [DOI] [PubMed] [Google Scholar]
- 26.Lima RT, Busacca S, Almeida GM, Gaudino G, Fennell DA, Vasconcelos MH. MicroRNA regulation of core apoptosis pathways in cancer. Eur J Cancer. 2011;47:163–174. doi: 10.1016/j.ejca.2010.11.005. [DOI] [PubMed] [Google Scholar]
- 27.Cawley K, Logue SE, Gorman AM, Zeng Q, Patterson J, Gupta S, Samali A. Disruption of microRNA biogenesis confers resistance to ER stress-induced cell death upstream of the mitochondrion. PLoS One. 2013;8:e73870. doi: 10.1371/journal.pone.0073870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qiu G, Zhang X, Zhang S, Liu P, Wu W, Zhang J, Dai S. Dysregulation of MALAT1 and miR-619-5p as a prognostic indicator in advanced colorectal carcinoma. Oncol Lett. 2016;12(6):5036–5042. doi: 10.3892/ol.2016.5312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hou J, Wang J, Lin C, Fu J, Ren J, Li L, Guo H, Han X, Liu J. Circulating MicroRNA Profiles Differ between Qi-Stagnation and Qi-Deficiency in Coronary Heart Disease Patients with Blood Stasis Syndrome. Hindawi Publishing Corporation Evidence-Based Complementary and Alternative Medicine. 2014;2014:1–9. doi: 10.1155/2014/926962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kumar S, Keerthana R, Pazhanimuthu A, Perumal P. Overexpression of circulating miRNA-21 and miRNA-146a in plasma samples of breast cancer patients. Indian J Biochem Biophys. 2013;50:210–214. [PubMed] [Google Scholar]
- 31.Smith-Vikos T, Slack FJ. MicroRNAs circulate around Alzheimer’s disease. Genome Biol. 2013;14(7):125. doi: 10.1186/gb4116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reshmi G, Chandra SS, Babu VJ, Babu PS, Santhi WS, Ramachandran S, Lakshmi S, Nair AS, Pillai MR. Identification and analysis of novel microRNAs from fragile sites of human cervical cancer: computational and experimental approach. Genomics. 2011;97:333–340. doi: 10.1016/j.ygeno.2011.02.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Variation of nucleotide sequences of mRNA region with miR-619-5p binding sites of genes from CSL6 to COX18 (Conservative binding sites are in bold) (PDF 218 kb)
Variation of nucleotide sequences of mRNA region with miR-619-5p binding sites of genes from GK5 to HM13 (Conservative binding sites are in bold) (PDF 106 kb)
Variation of nucleotide sequences of mRNA region with miR-619-5p binding sites of genes from IFIT3 to SLC26A4 (Conservative binding sites are in bold) (PDF 139 kb)
Variation of nucleotide sequences of mRNA region with miR-619-5p binding sites of genes from LC28A2 to ZNF841 (Conservative binding sites are in bold). The data given in the Additional files 1, 2, 3 and 4 demonstrate the variability of the nucleotides before and after the binding sites of miR-619-5p, which is shown in the Weblogo schemes in the table 8. (PDF 151 kb)
Data Availability Statement
The data sets supporting the results of this article are included within the article and its additional files and publicly available.


