Abstract
Background
Molecular tools for detecting malaria-infected mosquitoes with improved practicality, sensitivity and specificity, and high-throughput are required. A common PCR technique used to detect mosquitoes infected with Plasmodium spp. is a nested PCR assay based on the 18s-rRNA gene. However, this technique has several technical limitations, is laborious and time consuming.
Methods
In this study, a PCR-based on the Plasmodium cytochrome oxidase I (COX-I) gene was compared with the 18s-rRNA nested PCR using serial dilutions (330–0.0012 pg) of DNA from Plasmodium vivax, Plasmodium falciparum and Plasmodium knowlesi and with DNA from 48 positive and negative Kenyan mosquitoes (previously detected by using both ELISA and PCR). This assay for Plasmodium spp. DNA detection using the fast COX-I PCR assay was then performed individually on 2122 field collected mosquitoes (from the Solomon Islands) in which DNA was extracted from head and thorax.
Results
The fast COX-I PCR assay took 1 h to run and consistently detected as low as to 0.043 pg of parasite DNA (equivalent to two parasites) in a single PCR, while analyses with the 18s-rRNA nested PCR required 4 h to complete with a consistent detection threshold of 1.5 pg of DNA. Both assays produced concordant results when applied to the 48 Kenyan control samples with known Plasmodium spp. infection status. The fast COX-I PCR identified 23/2122 Plasmodium-infected mosquitoes from the Solomon Islands.
Conclusions
This new COX-I PCR adapted for a single PCR reaction is a faster, simpler, cheaper, more sensitive technique amenable to high-throughput analyses for Plasmodium DNA detection in mosquitoes and is comparable to the 18s-rRNA nested PCR. The improved sensitivity seen with the fast COX-I PCR will improve the accuracy of mosquito infection rate determination.
Keywords: Malaria, Plasmodium, Diagnosis, Sporozoite, Anopheles, 18s-rRNA, Cytochrome oxidase I, Solomon Islands, Vectors, DNA barcoding
Background
As worldwide malaria transmission intensity has decreased significantly over the last decade [1, 2], larger numbers of mosquitoes are required for analysis to determine accurate infection rates [3]. The ability to detect Plasmodium spp. sporozoites in the salivary glands of Anopheles species is required for malaria studies. Detecting and characterizing infective mosquitoes is necessary for vector incrimination [4], the estimation of the entomological inoculation rate [5], and when looking at transmission blocking immunity [6].
Techniques used to detect sporozoites in mosquitoes include: (i) dissection of salivary glands and examination under the microscope [7]; (ii) immunoassays to detect circumsporozoite proteins, i.e. enzyme-linked immunosorbent assays CSP-ELISA [8, 9], and rapid dipstick Immuno-Chromatographic Assays (Vec-Test™ Malaria) [10]; and (iii) PCR based assays. All three techniques have limitations in terms of practicality, sensitivity and specificity [11–13].
PCR-based methods have demonstrated a higher sensitivity for Plasmodium DNA detection than other methods allowing the detection of less than 10 sporozoites per µL of source material, overcoming some limitations in sensitivity in other methods used [4, 11, 14–18]. Among the molecular sporozoite detection methods, nested PCR targeting the Plasmodium 18s-rRNA gene is the method most extensively used [14, 15, 19] and is consequently considered the “standard” PCR [20]. In this method, a nested PCR protocol is used to first identify the presence of DNA from the Plasmodium genus and then up to six additional PCRs are required to identify all Plasmodium species causing human malaria [21]. Other methods involving the same target gene have been developed using a single multiplex PCR assay [16] and Taqman real-time PCR [4]. Recently, a PCR–RFLP was designed to target the Plasmodium cytochrome b mitochondrial gene [22].
The success of any PCR strategy is strongly influenced by the quality and quantity of the template, expertise of operators, stability of reagents, and can be affected by debris and carry-over from host cells or traces of reagents/template used during DNA extraction and reactions in the multi-step process [11]. Current PCR techniques may not be amenable for high throughput analysis, due to their laborious and time-consuming nature when hundreds or thousands of specimens may need to be screened for Plasmodium spp. infection. A fast, simple, sensitive and high-throughput method is required to improve detection of malaria infected or infective (when analyses are limited to head and thorax) mosquitoes.
Here, a new, more sensitive, and faster high-throughput PCR assay based on the Plasmodium cytochrome oxidase I (COX-I) gene was developed, and compared to the 18s-rRNA nested PCR method for Plasmodium spp. DNA detection using known positive and negative infected mosquitoes. The primary goal of this study was to provide a new molecular diagnostic tool with improved detection of malaria infections in mosquitoes both in terms of sensitivity and throughput, which can be used for malaria entomological studies and to develop and evaluate intervention strategies toward malaria control and/or elimination.
Methods
Plasmodium species reference strains and Plasmodium infected Anopheles
Plasmodium falciparum (HB3 strain) and Plasmodium vivax (Miami strain) specimens from culture (Dr. Michael Ferdig, University of Notre Dame; and BEI Resources [23] respectively) were used as reference strains. DNA from Plasmodium samples was extracted following directions in the E.Z.N.A. Blood DNA Mini Kit (Omega Bio-Tek, Norcross, GA). DNA of Plasmodium knowlesi (Malayan strain) was provided by Dr. John W. Barnwell, Centers for Disease Control and Prevention, USA. The concentration of the extracted DNA was determined using a Nanodrop 2000 (Thermo Scientific, Waltham, MA). For the validation of the new PCR, four sets of eight serial dilutions (using a 1:6 factor) for each DNA species was prepared (each by different operators), resulting in DNA concentrations of approximately 330 pg (dilution 1) to 0.0012 pg (dilution 8). Forty-eight DNA samples from Kenyan mosquitoes, infected (n = 24) and uninfected (n = 24) with Plasmodium spp. based on ELISA and nested-PCR (homogenized mosquito material was separated into two aliquots, to detect the CSP protein and Plasmodium DNA) [24, 25], were also included for PCR validation.
Mosquito preparation and dissection
Female adult Anopheles mosquitoes were captured by human landing catching (HLC) by consenting village residents in Western Province, Solomon Islands (n = 2122) and preserved in 70% ethanol (Burkot et al. pers. comm.). In the laboratory, the 70% ethanol was removed and replaced by 100% ethanol for 12 h at room temperature. The ethanol was decanted and the mosquitoes (in individual tubes) were dried at 37 °C for 15 min. Dried mosquitoes were dissected under the stereoscope. The head and thorax were separated from the abdomen using sterile toothpicks and placed in a 1.5 mL microfuge tube for further processing.
DNA extraction using a cetyltrimethylammonium bromide (CTAB)-based method
Dissected mosquito head and thorax were thoroughly ground for 20 min with a pulsating vortex mixer (VWR International, Radnor, PA) in 1.5 mL microfuge tubes containing two stainless steel beads of 3.2 mm (BioSpec Products, Inc. Bartlesville, OK) and 200 μL of 2% CTAB (Sigma-Aldrich, St Louis, MO). Samples were then incubated at 65 °C for 5 min. 200 μL of chloroform was added to each tube, the reagents were mixed, and then centrifuged at 12,000 rpm for 5 min. An isopropanol (200 µL) precipitation was performed on the transparent supernatant (at 12,000 rpm at 5 min). The centrifuged DNA pellet was washed with 70% ethanol (200 µL) and dried [26]. Each dried DNA sample was resuspended in 20 μL of PCR-grade water, gently shaken and incubated at 55 °C for 5 min. The concentration of DNA was determined using a Nanodrop 2000 and stored at −20 °C until further use.
18s-rRNA nested PCR
The sensitivity of the 18s-rRNA nested PCR [14, 19] (Table 1) to detect Plasmodium DNA was examined with serial dilutions of DNA from the reference Plasmodium strains and the 48 known Plasmodium positive and negative Kenyan Anopheles mosquitoes [24, 25] using the recombinant DNA polymerase (Invitrogen, Carlsbad, CA). One micro litre of DNA was used as template for nest-1 PCR and 1 µL of the resulting PCR product was used in nest-2 PCR reaction, both with a final volume of 10 μL (Table 1). Five micro litre of the nest-2 PCR product was loaded on a 1% agarose gel stained with SYBR®safe (Invitrogen, Carlsbad, CA) to confirm amplifications of the 235 bp product (Plasmodium positive) [14, 19].
Table 1.
PCR conditions for the 18s-rRNA nested-PCR and the new COX-I PCRs for Plasmodium sporozoite detection
| Diagnostic description | Reagents quantities and final concentration | Thermal profile |
|---|---|---|
| 18s-rRNA genus specific PCR nest-1 [19] |
1X PCR buffer, 80 μM dNTPmix, 0.8 mM MgCl2, 0.1 mM each primer (rPLU1–rPLU5), 0.25 U Taq polymerase | 94 °C for 4 min; 35 cycles of 94 °C for 30 s, 55 °C for 1 min, 72 °C for 1 min; and 72 °C for 4 min. Time: 120 min |
| 18s-rRNA genus specific PCR nest-2 [19] |
Same as nest-1 but using rPLU3–rPLU4 primers | Same as nest-1 but annealing temperature is 62 °C. Time: 120 min |
| Conventional COX-I PCR | 1X PCR buffera, 10 mM dNTPs, 0.4 mM each primer, 1.5 mM MgCl2 and 0.2 µL of recombinant Taq polymerase | 94 °C for 5 min; 40 cycles of 94 °C for 1 min, 62 °C for 1 min, 72 °C for 90 s; and 72 °C for 10 min. Time: 155 min |
| Fast COX-I PCR | 1X blood phusion buffera, 1 mM each primer, and 0.125 µL of blood phusion polymerase | 98 °C for 4 min; 70 cycles of 98 °C for 1 s, 69 °C for 5 s, 72 °C for 35 s; and 72 °C for 10 min. Time: 62 min |
aThe Blood Phusion buffer contains MgCl2 at a final concentration of 3 mM
Single step PCR for Plasmodium sporozoite detection based on the cytochrome oxidase I
The nucleotide sequences of human-Plasmodium species (P. falciparum, P. vivax, P. knowlesi, Plasmodium malariae, Plasmodium ovale wallikeri and Plasmodium ovale curtisi) cytochrome oxidase I (COX-I), contained in the mitochondrial genome, were downloaded from GeneBank [27] and aligned as described previously [28]. A set of primers, COX-IF (5′ AGAACGAACGCTTTTAACGCCTG 3′) and COX-IR (3′ ACTTAATGGTGGATATAAAGTCCATCCwGT 5′), were designed to amplify a polymorphic fragment in the COX-I gene (DNAstar Lasergene® 11 software, DNAstar Inc. Madison, WI). Two master-mixes were prepared, one using a recombinant DNA polymerase (Invitrogen, Carlsbad, CA) in 25 µL of PCR reaction (named the conventional COX-I PCR) (Table 1) and another prepared using the Blood Phusion polymerase (Thermo Scientific, Waltham, MA) with 15 μL of PCR reaction (named the fast COX-I PCR) (Table 1) using 2 µL of DNA template.
The sensitivity of COX-I PCRs (conventional and fast) to detect Plasmodium DNA was evaluated using the DNA (serial dilutions) of reference strains and the 48 control samples from Kenya [24, 25]. Five micro litre of the PCR product was visualized on 1% agarose gel in order to confirm amplifications of the expected ~540 bp product (Plasmodium genus positive). Differences between the performance of the 18s-rRNA nested PCR and the COX-I PCRs were evaluated with the McNemar’s Chi Square test.
Fast COX-I PCR sequencing reaction
The PCR sequencing reactions were performed as previously described [28]. In brief, 8 μL of PCR product from the fast COX-I PCR was purified, and a sequencing-PCR performed with the COX-IF primer (Table 1). Samples were sequenced on an ABI 3730XL 96-capillary sequencer. Sequence analyses were performed using the DNASTAR Lasergene ® 11 software (DNAstar Inc. Madison, WI).
Results
Comparison of COX-I PCRs and 18s-rRNA nested-PCR using DNA (serial dilutions) of Plasmodium reference strains
The DNA extracted with the E.Z.N.A. Blood DNA Mini Kit from P. vivax and P. falciparum reference strains were 30 and 7 ng/µL (OD260:OD280 average of 1.93) respectively. The DNA of P. knowlesi was provided at 20 ng/µL. The performance of the fast COX-I PCRs (the conventional COX-I PCR results were not shown as they were similar to the fast COX-I PCR) and the 18s-rRNA nested PCR method for Plasmodium spp. detection were compared using eight serial dilutions (Fig. 1). For the fast COX-I PCR, consistent and successful amplifications (100% of positivity) were achieved for all parasite species down to dilution 6 (0.043 pg), followed by 25–75% of positivity for all parasites in dilution 7, and 25–50% of positivity of P. vivax and P. falciparum in the final dilution (0.0012 pg) (Fig. 1a).
Fig. 1.
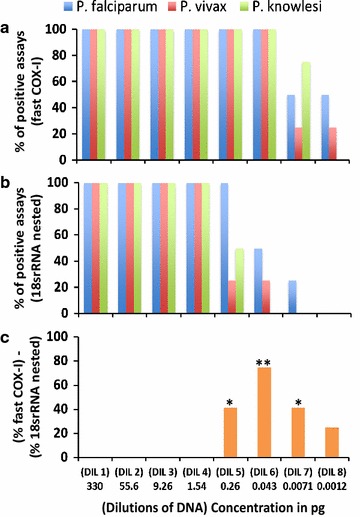
Plasmodium spp. detection using the fast COX-I PCR and the 18s-rRNA nested-PCR. a Percentage of successful Plasmodium DNA detection in four different assays using the fast COX-I PCR. b Percentage of successful Plasmodium DNA detection in four different assays using the 18s-rRNA nested PCR. c Percentage of Plasmodium DNA detected by COX-I PCR but not by the 18s-rRNA nested PCR based on 12 different assays; p value (McNemar’s Chi square test) = 0.036 (*) and 0.0038 (**). The predicted number of parasites based on Li et al. [32], in dilutions 1–8 were 15,348, 2586, 430, 71, 12, 2, 0.33 and 0.05 parasites respectively
In contrast, the 18s-rRNA nested PCR detected consistent amplifications of all DNA parasites down to dilution 4 (1.5 pg of DNA). The parasite positivity rate in dilutions 5, 6, and 7, was variable, while no PCR amplification was detected in the final dilution (Fig. 1b). Taking into account all the repetitions (12 repetitions using three operators for each dilution), both fast COX-I PCR and the 18s-rRNA nested PCRs detected parasites down to the fourth dilution (1.54 pg), however, for dilutions 5, 6, and 7, the percentage of Plasmodium DNA detected by fast COX-I was significantly higher when compared with the 18s-rRNA (P value ≤ 0.036) (Fig. 1c).
The expected bands of 235 bp (for the 18s-rRNA nested PCR) (Fig. 2a) and 540 bp (for the fast COX-I PCR) (Fig. 2b), were robust and no significant loss of intensity was seen between dilutions down to the last positive amplification. The conventional COX-I PCR showed bands with lower intensity and inconsistencies after dilution 4 (Fig. 2c) when compared to the fast COX-I PCR (Fig. 2b). Non-specific amplification was seen with the 18s-rRNA protocol but none with the COX-I PCRs (Fig. 2). Finally, PCR products (from the fast COX-I PCR) of P. vivax, P. falciparum, and P. knowlesi (all from dilution 6) were successfully sequenced and their identity confirmed (Table 2).
Fig. 2.
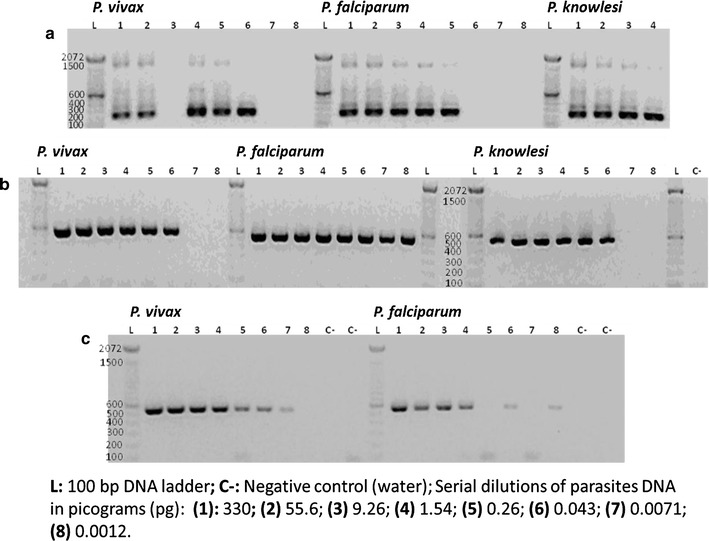
Electrophoresis gels of PCR products obtained from the 18s-rRNA nested-PCR and COX-I PCRs using serial dilutions of parasite DNAs. a PCR products (235 bp) from nest 2 PCR using the 18s-rRNA nested PCR. Dilution 3 for P. vivax did not amplified; PCR products for dilutions 5–8 for P. knowlesi were negatives and not included in this figure. b PCR products (540 bp) from the fast COX-I PCR. The PCR product bands have consistent size and intensity through all the positive dilutions. c PCR products (540 bp) from the conventional COX-I PCR. This PCR was not tested in P. knowlesi. Bands for P. falciparum did not amplify consistently
Table 2.
Summary of sequenced DNA samples (positive controls, Kenya and Solomon Islands) based on Plasmodium COX-I gene
| Sample | Species IDa | E valueb | Coveragec (%) | Identityd | [GenBank identifier]e |
|---|---|---|---|---|---|
| PCR product from dilution 6 (0.043 pg of DNA) | |||||
| P. falciparum | P. falciparum | 0 | 100 | 99.7% | [KM065500.1] |
| P. vivax | P. vivax | 1.3e–84 | 100 | 98.4% | [KF668441.1] |
| P. knowlesi | P. knowlesi | 8e–180 | 100 | 100% | [AB444108.1] |
| Known infective mosquitoes from the field (Kenya Highlands) | |||||
| Mosquitoes (n = 21) | P. falciparum | 0 | 100 | 99.7% | [KM065500.1] |
| Mosquitoes (n = 3) | P. ovale s.l. | 1.21e–151 | 100 | 97.5% | [KF018660.1] |
| Positive mosquitoes from Western Province, Solomon Islands | |||||
| Mosquitoes (n = 17) | P. falciparum | 0 | 100 | 99.7% | [KM065500.1] |
| Mosquitoes (n = 5) | P. vivax | 0 | 99 | 99.6 | [KF668441.1] |
| Mosquito (n = 1) | P. ovale wallikeri | 7.1e–110 | 100 | 92.2% | [HQ712053.1] |
aBest BLASTed hit in GeneBank
bProbability of observing the result by chance
cPercentage coverage of entire sequence against best hit
dPercentage of similarity against best hit
eGeneBank ID of the best hit
Sensitivity and specificity between 18s-rRNA nested PCR and fast COX-I PCR for Plasmodium detection
DNA samples (n = 48) obtained from known infected and uninfected mosquitoes from Kenya [24, 25] were used to validate the new COX-I PCR assays. All known infected samples (n = 24) were positive for Plasmodium species using the 18s-rRNA nested PCR and the fast COX-I PCR (100% sensitivity), while 22/24 of the samples were positive with the conventional COX-I PCR assay (92% sensitivity). All 24 uninfected samples were negative when using the three techniques (100% specificity). The PCR products from the fast COX-I PCR were sequenced and database comparisons demonstrated that 21 samples were P. falciparum and 3 samples were P. ovale s.l. (Table 2).
Presence of Plasmodium infective mosquitoes from the Solomon Islands
The DNA (head and thorax) of 2122 wild-caught anopheline mosquitoes from the Solomon Islands, was extracted using a CTAB-based method and screened for the presence of Plasmodium DNA using the fast COX-I PCR. An average of 47.89 μg/μL (range 10.6–150.2) of DNA was obtained, with an average OD260:OD280 of 2.01 (range 1.7–2.3). Twenty-three samples were positive for Plasmodium DNA in Anopheles farauti mosquitoes. Seventeen were positive for P. falciparum, five for P. vivax, and one for P. ovale wallikeri (Table 2).
Discussion
The excess of host DNA may interfere with the performance of Plasmodium PCR diagnosis. The DNA from P. falciparum (HB3 strain) was pure as was obtained from long-term culture, while DNA from the P. vivax (Miami strain) and P. knowlesi (Malayan strain) was a mix of parasite and primate hosts DNA (as there is not a long-term in vitro culture available for these species). The presence of host DNA may explain the lower sensitivity of the 18s-rRNA for P. vivax and P. knowlesi in dilution 5 and 6 (Fig. 1a), however this limitations was overcome by the fast-COX-I PCR.
The fast COX-I PCR consistently detected down to 0.043 pg of DNA (dilution 6), equivalent to two parasites [29], which is >460-fold more sensitive for Plasmodium DNA detection than other PCR techniques (Table 3) [4, 18, 22, 30]. This may be explained by both the higher number of the COX-I gene copies (up to 150) while 18s-rRNA has only eight or fewer [31], the well-designed COX-I primers and the use of the Blood Phusion polymerase, a proofreading polymerase with a processivity-enhanced domain [32] that performs in the presence of strong PCR inhibitors, including collagen and melanin, compounds of the insect cuticle [33]. An infected mosquito can carry several thousand down to seven sporozoites of Plasmodium spp. in their salivary glands [34] suggesting that the fast COX-I PCR is sufficient for identifying infective mosquitoes.
Table 3.
Summary of other PCR techniques for Plasmodium sporozoite detection
| Molecular sporozoite detection approach [ref] | DNA extraction | Plasmodium species | Cycling time in min | DNA limit of detection |
|---|---|---|---|---|
| 18s-rRNA nested PCR protocol [4, 15] | Livak or DNAzol methods | P. vivax, P. falciparum, P. ovale, P. malariae | 294 | 0.2 ng–0.2 pg |
| 18s-rRNA single PCR [4, 15] | P. vivax, P. falciparum, P. ovale, P. malariae | 205 | 2 ng–4 pg | |
| 18s-rRNA Taqman assay [4] | P. falciparum, P. ovale, P. malariae, P. vivax | 47 | 0.2 pg | |
| 18s-rRNA single PCR Tassanakajon [4, 18] | P. falciparum | 60* | 2 pg | |
| Cytochrome B single PCR [22] | IsoQuick nucleic acid extraction kit | P. vivax, P. falciparum | 96 | 0.2 pg |
| DHFR-TS nested [30] | Chelex | P. falciparum | >294 | 4–40 pg |
| Fast COX-I single PCR [this manuscript] | CTAB | P. vivax, P. falciparum, P. ovale s.l., P. knowlesi, P. ovale wallikeri | 62 | 0.043 pg |
min minutes, ng nanograms, pg picograms
* The original paper from Tassanakajon et al. [18] did not include times for denaturation and final extension
The cycling time for the fast COX-I PCR, is completed in an hour, a shorter time than the other techniques (Table 3). This will enable the processing of larger quantities of samples in shorter periods of time reducing processing time and costs. The PCR cost of processing 2122 DNA samples for Plasmodium spp. using the 18s-rRNA nested PCR or the conventional COX-I PCR is ~892 USD, while for the fast COX-I PCR is ~552 USD (Table 4). The fast COX-I PCR minimizes the risk of contamination and amplification of non-specific bands—the two primary technical limitations in nested PCR strategies or when DNA was derived from mosquitoes stored in ethanol or isopropanol [4, 35]. This will be particularly important when looking at vector incrimination or large numbers of mosquito samples where infection rates might be low such as with secondary vectors or vectors with low vectorial capacity.
Table 4.
Summary of cost analysis for the 18s-rRNA nested and COX-I PCRs for Plasmodium spp. detection
| PCR technique | Required PCR reagents | Estimated cost of the PCR kit | µL of polymerase used per reaction | Cost of PCR diagnosis per sample | Cost of 2122 reactions for Plasmodium detection |
|---|---|---|---|---|---|
| 18s-rRNA nested PCR | Taq polymerase kit (Invitrogen) | ~210 USD | 0.1 µL for nest-1 and 0.1 µL nest-2 | ~0.42 USD | ~892 USD |
| Conventional COX-I single PCR | Taq polymerase kit (Invitrogen) | ~210 USD | 0.2 µL in a single reaction | ~0.42 USD | ~892 USD |
| Fast COX-I single PCR | Blood Phusion polymerase kit (Thermo) | ~418 USD | 0.125 µL in a single reaction | ~0.26 USD | ~552 USD |
For a set of 24 known Plasmodium positive mosquitoes [24, 25], all PCRs were positive with the fast COX-I PCR, which confirms that the new PCR is able to detect Plasmodium DNA in samples from the field. The conventional PCR, which uses a recombinant DNA polymerase and the same primers (Table 1), did not amplify 2/24 of the positive samples. This may be explained by low parasite DNA quality or quantity and/or presence of PCR inhibitors in the samples. In either case, the fast COX-I PCR was able to overcome these limitations and identified these samples as positives (P. falciparum).
The fast COX-I PCR was successfully tested in different anopheline species with different human malaria parasites. The Anopheles mosquitoes tested included Anopheles farauti, Anopheles hinesorum, Anopheles lungae, and Anopheles solomonis from the Solomon Islands, and Anopheles funestus, Anopheles coustani, Anopheles maculipalpis, Anopheles theileri, and Anopheles leesoni amongst others from Kenya, suggesting that this PCR can be used across vector species. This PCR-sequencing approach functioned across human Plasmodium species including P. knowlesi. The COX-I primers had 100% of identity and 100% coverage with at least 26 different Plasmodium species including parasites from lizards, birds, rodents and non-human primates, which may be relevant in assessing malaria transmission in particular settings (e.g. forest border areas). At the core of this technique (DNA barcoding), the use of COX-I relies in the use of a set of primers that recognize a flanking conserved region for Plasmodium spp. surrounding an internal variable region that allows species identification by sequencing of the amplified fragment.
Conclusion
The fast COX-I PCR designed for Plasmodium species sporozoite detection is more sensitive, less expensive, and faster than other PCR strategies utilized at present. This functionally better diagnostic may be utilized in both research, intervention strategies and monitoring studies towards identifying infected and infective mosquitoes.
Authors’ contributions
DFE: conceived and designed the COX-I PCRs, performed laboratory work (dissections, DNA extractions, molecular diagnostic PCRs and sequencing experiments), data analyses and writing of the manuscript. NAD, VM, JD, HX, XY and JN: participated in laboratory work, data analyses and critically reviewed the manuscript. HB, AA, HR and RC: participated in field mosquito collection. TLR, TRB and JS: coordinated field work, participated in field mosquito collection and critically reviewed the manuscript. FHC: participates in supervision of the study and critically review the manuscript. NFL: coordinated field-work, participated in field mosquitoes collection, supervision of the study, data analyses and critically reviewed the manuscript. All authors read and approved the final manuscript.
Acknowledgements
The authors are very grateful to the communities of Western Province in Solomon Islands and the community of Bigege in the Western Highlands of Kenya for their assistance and cooperation. In particular, Danyal Odabasi and Jance Oscar in the Solomon Islands as well as the Kenya Medical Research Institute/US Centers for Disease Control and Prevention, Kisumu. Technical assistance from Ngor Majak Anyieth, Heather Boldt, Brooke Brown, Daniel Olivieri, Daniel Pichler, Keith Loh, Emily Kruse, Thomas Rieth and Emma Troth in the University of Notre Dame molecular laboratory is acknowledged.
Competing interests
The authors declare that they have not competing interests.
Availability of data and materials
The datasets used and/or analyzed during the current study available from the corresponding author on reasonable request.
Ethical statement
Ethical approval for the collection of mosquitoes with HLC was obtained from the National Health Research & Ethics Committee, Solomon Islands (HRC13/14 and HRC14/16), the James Cook University Human Research Ethics Committee, Australia (H4914 and H4915), the University of Notre Dame Institutional Review Board (FWA 00002462) and University Hospitals Case Medical Centre Institutional Review Board for Human Investigation, USA (05-11-11).
Funding
This work was primarily sponsored by Gates Foundation Malaria Transmission Consortium (MTC) Grant ID 45114. In addition, the National Institute of Allergy and Infectious Diseases of the National Institutes of Health for the International Center of Excellence in Malaria Research in the Southwest Pacific (subaward to James Cook University; award number U19AI08986) is gratefully acknowledged for support to collect the Solomon Islands samples. The content is solely the responsibility of the authors and does not necessarily represent the official views of the funders.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
- CSP-ELISA
circumsporozoite protein-enzyme-linked immunosorbent assay
- COX-I
cytochrome oxidase I
- 18s-rRNA
18 small subunit ribosomal RNA
- BEI Resources
the biological and emerging infections resources program
- min
minute
- sec
second
- µL
microlitre
- pg
picogram
- ng
nanogram
Contributor Information
Diego F. Echeverry, Email: decheve1@nd.edu
Nicholas A. Deason, Email: Nicholas.deason@nih.gov
Victoria Makuru, Email: victoria.makuru.1@nd.edu.
Jenna Davidson, Email: jenna.r.davidson.60@nd.edu.
Honglin Xiao, Email: hxiao@nd.edu.
Julie Niedbalski, Email: niedbalski.14@nd.edu.
Xiaoyu Yu, Email: xyu4@nd.edu.
Jennifer C. Stevenson, Email: jennyc.stevenson@macharesearch.org
Hugo Bugoro, Email: bugorohugo@yahoo.com.
Allan Aparaimo, Email: apairamo@gmail.com.
Hedrick Reuben, Email: hedrick2010@gmail.com.
Robert Cooper, Email: bob_cooper5@gmail.com.
Thomas R. Burkot, Email: tom.burkot@jcu.edu.au
Tanya L. Russell, Email: tanya.russell@jcu.edu.au
Frank H. Collins, Email: frank@nd.edu
Neil F. Lobo, Email: nlobo@nd.edu
References
- 1.WHO . World malaria report 2016. Geneva: World Health Organization; 2016. [Google Scholar]
- 2.O’Meara WP, Mangeni JN, Steketee R, Greenwood B. Changes in the burden of malaria in sub-Saharan Africa. Lancet Infect Dis. 2010;10:545–555. doi: 10.1016/S1473-3099(10)70096-7. [DOI] [PubMed] [Google Scholar]
- 3.Hay SI, Smith DL, Snow RW. Measuring malaria endemicity from intense to interrupted transmission. Lancet Infect Dis. 2008;8:369–378. doi: 10.1016/S1473-3099(08)70069-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bass C, Nikou D, Blagborough AM, Vontas J, Sinder RE, Williamson MS, et al. PCR-based detection of Plasmodium in Anopheles mosquitoes: a comparison of a new high-throughput assay with existing methods. Malar J. 2008;7:177. doi: 10.1186/1475-2875-7-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Himeidan YE, Elzaki MM, Kweka EJ, Ibrahim M, Elhassan IM. Pattern of malaria transmission along the Rahad River basin Eastern Sudan. Parasit Vectors. 2011;4:109. doi: 10.1186/1756-3305-4-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gonçalves D, Hunziker P. Transmission-blocking strategies: the roadmap from laboratory bench to the community. Malar J. 2016;15:95. doi: 10.1186/s12936-016-1163-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beier JC, Perkins PV, Koros JK, Onyango FK, Gargan TP, Wirtz RA, et al. Malaria sporozoite detection by dissection and ELISA to assess infectivity of afrotropical Anopheles (Diptera: Culicidae) J Med Entomol. 1990;27:377–384. doi: 10.1093/jmedent/27.3.377. [DOI] [PubMed] [Google Scholar]
- 8.Wirtz RA, Zavala F, Charoenvit Y, Campbell GH, Burkot TR, Schneider I, et al. Comparative testing of monoclonal antibodies against Plasmodium falciparum sporozoites for ELISA development. Bull World Health Org. 1987;65:39–45. [PMC free article] [PubMed] [Google Scholar]
- 9.Burkot TR, Graves PM, Cattan JA, Wirtz RA, Gibson FD. The efficiency of sporozoite transmission in the human malarias, Plasmodium falciparum and P. vivax. Bull World Health Org. 1987;65:375–380. [PMC free article] [PubMed] [Google Scholar]
- 10.Appawu MA, Bosompem KM, Dadzie S, Mckakpo US, Anim-Baidoo I, Dykstra E, et al. Detection of malaria sporozoites by standard ELISA and VecTestTM dipstick assay in field-collected anopheline mosquitoes from a malaria endemic site in Ghana. Trop Med Int Health. 2003;8:1012–1017. doi: 10.1046/j.1360-2276.2003.00127.x. [DOI] [PubMed] [Google Scholar]
- 11.Arez AP, Lopes D, Pinto J, Franco AS, Snounou G, do Rosario VE. Plasmodium sp.: optimal protocols for PCR detection of low parasite numbers from mosquito (Anopheles sp.) samples. Exp Parasitol. 2000;94:269–272. doi: 10.1006/expr.2000.4496. [DOI] [PubMed] [Google Scholar]
- 12.Aonuma H, Suzuki M, Iseki H, Perera N, Nelson B, Igarashi I, et al. Rapid identification of Plasmodium-carrying mosquitoes using loop-mediated isothermal amplification. Biochem Biophys Res Commun. 2008;376:671–676. doi: 10.1016/j.bbrc.2008.09.061. [DOI] [PubMed] [Google Scholar]
- 13.Swain S, Mohanty A, Mahapatra N, Parida SK, Marai NS, Tripathy HK, et al. The development and evaluation of a single step multiplex PCR for simultaneous detection of Anopheles annularis group mosquitoes, human host preference and Plasmodium falciparum sporozoite presence. Trans R Soc Trop Med Hyg. 2009;103:1146–1152. doi: 10.1016/j.trstmh.2009.03.022. [DOI] [PubMed] [Google Scholar]
- 14.Snounou G, Singh B. Nested PCR analysis of Plasmodium parasites. Methods Mol Med. 2002;72:189–203. doi: 10.1385/1-59259-271-6:189. [DOI] [PubMed] [Google Scholar]
- 15.Snounou G, Viriyakosol S, Jarra W, Thaithong S, Brown KN. Identification of the four human malaria parasite species in field samples by the polymerase chain reaction and detection of a high prevalence of mixed infections. Mol Biochem Parasitol. 1993;58:283–292. doi: 10.1016/0166-6851(93)90050-8. [DOI] [PubMed] [Google Scholar]
- 16.Padley D, Moody AH, Chiodini PL, Saldanha J. Use of a rapid, single-round, multiplex PCR to detect malarial parasites and identify the species present. Ann Trop Med Parasitol. 2003;97:131–137. doi: 10.1179/000349803125002977. [DOI] [PubMed] [Google Scholar]
- 17.Bassene H, Kengne P, Ndiath MO, Sokhna C, Dupressoir T, Fontenille D, et al. Comparison of PCR, ELISA-CSP and direct microscopic observation methods for the detection of Plasmodium falciparum sporozoites in Anopheles gambiae M in Senegal. Bull Soc Pathol Exot. 2009;102:233–237. [PubMed] [Google Scholar]
- 18.Tassanakajon A, Boonsaeng V, Wilairat P, Panyim S. Polymerase chain reaction detection of Plasmodium falciparum in mosquitoes. Trans R Soc Trop Med Hyg. 1993;87:273–275. doi: 10.1016/0035-9203(93)90124-9. [DOI] [PubMed] [Google Scholar]
- 19.Singh B, Bobogare A, Cox-Singh J, Snounou G, Abdullah MS, Rahman HA. A genus- and species-specific nested polymerase chain reaction malaria detection assay for epidemiologic studies. Am J Trop Med Hyg. 1999;60:687–692. doi: 10.4269/ajtmh.1999.60.687. [DOI] [PubMed] [Google Scholar]
- 20.Alonso P, Barnwell J, Bell D, Hanson K, Mendis K, Moonen B, et al. A research agenda for malaria eradication: diagnoses and diagnostics. PLoS Med. 2011;8:e1000396. doi: 10.1371/journal.pmed.1000406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fuehrer HP, Noedl H. Recent advances in detection of Plasmodium ovale: implications of separation into the two species Plasmodium ovale wallikeri and Plasmodium ovale curtisi. J Clin Microbiol. 2014;52:387–391. doi: 10.1128/JCM.02760-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hasan AU, Suguri S, Sattabongkot J, Fujimoto C, Amakawa M, Harada M, et al. Implementation of a novel PCR based method for detecting malaria parasites from naturally infected mosquitoes in Papua New Guinea. Malar J. 2009;8:182. doi: 10.1186/1475-2875-8-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Molestina RE. BEI Resources: a biological resource center for parasitologists. Trends Parasitol. 2010;26:559–560. doi: 10.1016/j.pt.2010.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.St Stevenson J, Laurent B, Lobo NF, Cooke MK, Kahindi SC, Oriango RM, et al. Novel vectors of malaria parasites in the western highlands of Kenya. Emerg Infect Dis. 2012;18:1547–1549. doi: 10.3201/eid1809.120283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.St Laurent B, Cooke M, Krishnankutty SM, Asih P, Mueller JD, Kahindi S, et al. Molecular characterization reveals diverse and unknown malaria vectors in the Western Kenyan Highlands. Am J Trop Med Hyg. 2016;94:327–335. doi: 10.4269/ajtmh.15-0562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sambrook J, Russell D. Molecular cloning: a laboratory manual. 3. New York: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 27.Benson DA, Clark K, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. GenBank. Nucleic Acids Res. 2015;43:D30–D35. doi: 10.1093/nar/gku1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Echeverry DF, Deason NA, Davidson J, Makuru V, Xiao H, Niedbalski J, et al. Human malaria diagnosis using a single-step direct-PCR based on the Plasmodium cytochrome oxidase III gene. Malar J. 2016;15:128. doi: 10.1186/s12936-016-1185-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li F, Niu C, Ye B. Nested polymerase chain reaction in detection of Plasmodium vivax sporozoites in mosquitoes. Chin Med J. 2001;114:654–658. [PubMed] [Google Scholar]
- 30.Harada M, Ishikawa H, Matsuoka H, Ishii A, Suguri S. Estimation of the sporozoite rate of malaria vectors using the polymerase chain reaction and a mathematical model. Acta Med Okayama. 2000;54:165–171. doi: 10.18926/AMO/32275. [DOI] [PubMed] [Google Scholar]
- 31.Isozumi R, Fukui M, Kaneko A, Chan CW, Kawamoto F, Kimura M. Improved detection of malaria cases in island settings of Vanuatu and Kenya by PCR that targets the Plasmodium mitochondrial cytochrome c oxidase III (cox3) gene. Parasitol Int. 2015;64:304–308. doi: 10.1016/j.parint.2014.09.006. [DOI] [PubMed] [Google Scholar]
- 32.Thermo-Fisher scientific. User guide: Phusion Blood Direct PCR Kit. Waltham. 2015.
- 33.Radstrom P, Knutsson R, Wolffs P, Lovenklev M, Lofstrom C. Pre-PCR processing—strategies to generate PCR-compatible samples. Mol Biotechnol. 2004;26:133–146. doi: 10.1385/MB:26:2:133. [DOI] [PubMed] [Google Scholar]
- 34.Beier JC, Davis JR, Vaughan JA, Noden BH, Beier MS. Quantitation of Plasmodium falciparum sporozoites transmitted in vitro by experimentally infected Anopheles gambiae and Anopheles stephensi. Am J Trop Med Hyg. 1991;44:564–570. doi: 10.4269/ajtmh.1991.44.564. [DOI] [PubMed] [Google Scholar]
- 35.Harrison GF, Foley DH, Rueda LM, Melanson VR, Wilkerson RC, Long LS, et al. Plasmodium-specific molecular assays produce uninterpretable results and non-Plasmodium spp. sequences in field-collected Anopheles vectors. Am J Trop Med Hyg. 2013;89:1117–1121. doi: 10.4269/ajtmh.12-0581. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study available from the corresponding author on reasonable request.


