Abstract
Glyoxalase is an evolutionary highly conserved pathway present in all organisms. Conventional glyoxalase pathway has two enzymes, glyoxalase I (GLYI) and glyoxalase II (GLYII) that act sequentially to detoxify a highly cytotoxic compound methylglyoxal (MG) to D-lactate with the help of reduced glutathione. Recently, proteins with DJ-1/PfpI domain have been reported to perform the same conversion in a single step without the help of any cofactor and thus termed as “unique glyoxalase III” enzyme. Genome-wide analysis of glyoxalase genes have been previously conducted in Arabidopsis, rice and Soybean plants, but no such study was performed for one of the agricultural important model legume species, Medicago truncatula. A comprehensive genome-wide analysis of Medicago identified a total of putative 29 GLYI, 14 GLYII genes, and 5 glyoxalase III (DJ-1) genes. All these identified genes and their corresponding proteins were analyzed in detail including their chromosomal distribution, gene duplication, phylogenetic relationship, and the presence of conserved domain(s). Expression of all these genes was analyzed in different tissues as well as under two devastating abiotic stresses- salinity and drought using publicly available transcript data. This study revealed that MtGLYI-4, MtGLYII-6, and MtDJ-1A are the constitutive members with a high level of expression at all 17 analyzed tissues; while MtGLYI-1, MtGLYI-11, MtGLYI-5, MtGLYI-7, and MtGLYII-13 showed tissue-specific expression. Moreover, most of the genes displayed similar pattern of expression in response to both salinity and drought stress, irrespective of stress duration and tissue type. MtGLYI-8, MtGLYI-11, MtGLYI-6, MtGLYI-16, MtGLYI-21, and MtGLYII-9 showed up-regulation, while MtGLYI-17 and MtGLYI-7/9 showed down-regulation in response to both stresses. Interestingly, MtGLYI-14/15 showed completely opposite pattern of expression in these two stresses. This study provides an initial basis about the physiological significance of glyoxalase genes in plant development and stress response of Medicago that could be explored further.
Keywords: glyoxalase, Medicago truncatula, transcript alteration, gene duplication, expression profiling, abiotic stress, salinity, drought
Introduction
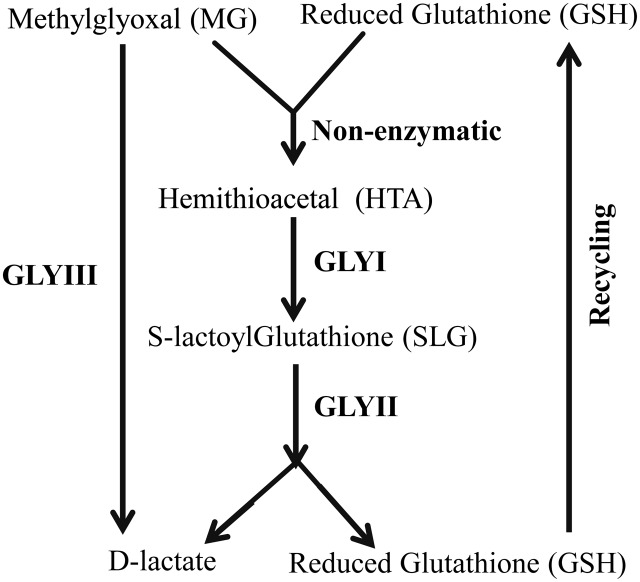
Glyoxalase pathway is one of the evolutionary highly conserved pathways which have been found in all era of organisms starting from lower prokaryotic Escherichia coli to higher eukaryotes Homo sapiens (Kaur et al., 2014a). Glyoxalase pathway detoxifies highly cytotoxic compound MG into its non-toxic form, D-lactate (Figure 1). Conventional glyoxalase pathway consists of two enzymes; GLYI and GLYII (Kaur et al., 2014c). MG and GSH generate a non-enzymatic adduct hemithioacetal (HTA). GLYI isomerizes HTA to S-lactyl-glutathione (SLG) which is further hydrolysed by GLYII into D-lactate and GSH (Figure 1). Recently, there are some reports of the presence of another unique glyoxalase member named Glyoxalase III (GLYIII) (Lee et al., 2012; Kwon et al., 2013; Ghosh et al., 2016). GLYIII could do the same conversion of MG into D-lactate in a single step (Figure 1).
FIGURE 1.
Schematic diagram of glyoxalase pathway. Conversion of MG into D-lactate could be performed in two different ways. First one, conventional glyoxalase pathway consists of two enzymes, GLYI and GLYII with an intermediate product of S-D-lactoylglutathione (SLG). The second and unique one is glyoxalase III (GLYIII) in a single step without the help of any cofactor.
Besides its proposed metabolic role of MG metabolism, glyoxalase enzymes have been found to be involved in various other cellular functions. Various complications of diabetes, such as nephropathy, retinopathy, neuropathy, and cardiovascular complications have been protected by glyoxalase system (Rabbani and Thornalley, 2011). Glyoxalase enzymes have also been reported to be involved in cell division and proliferation, and microtubule assembly of human (Thornalley, 1990). Thus, this pathway has been considered as “marker for cell growth and division.” Similarly, numerous studies suggested its potential role in stress tolerance pathways of plant (Kaur et al., 2014a). Transgenic plants over-expressing either GLYI or GLYII were found to provide significant tolerance against multiple abiotic stresses including salinity, drought, and heavy metal toxicity in various plant species (Singla-Pareek et al., 2003; Ghosh et al., 2014; Kaur et al., 2014a; Mustafiz et al., 2014). Moreover, overexpression of GLYI and GLYII genes together in transgenic tobacco and tomato plants confers extensive salt and heavy metal tolerance by maintaining the level of MG normal and decreasing oxidative stresses (Singla-Pareek et al., 2003, 2006; Alvarez Viveros et al., 2013). Thus MG and glyoxalases are considered as the potential biomarker for plant stress physiology (Kaur et al., 2014c).
Glyoxalase proteins have been extensively analyzed from various microorganisms and animal systems such as E. coli, H. sapiens, Saccharomyces cerevisiae (Kaur et al., 2013). In spite of great importance, very little study has been conducted on plant glyoxalases as compared to non-plant species. Plant glyoxalase activity has been first reported from Douglas fir needles by Smits and Johnson (1981). Presently, the presence of glyoxalase genes and corresponding enzyme activity has been studied from various plant species, such as rice, Arabidopsis, tomato, tobacco, soybean, wheat, Zea mays, sugarcane, Brassica etc. (Kaur et al., 2014a,c). The activity of plant GLYI has been reported to be dependent on either Ni2+ or Zn2+ (Jain et al., 2016). Plant GLYII enzymes possess various metals such as Fe2+, Zn2+, Mn2+ in their structure and these metal ions are essential for its optimum activity (Ghosh et al., 2014). However, the activity of GLYIII enzyme from E. coli has been reported to be metal independent (Subedi et al., 2011).
Most of the plant genes exist as a family due to the genome expansion and gene duplication events occurred during the evolution of plant (Guo, 2013). Availability of complete genome sequence databases of various plants species provides the opportunity to identify plants gene families. In silico genome-wide analyses of Arabidopsis thaliana, Glycine max, and Oryza sativa has been carried out to identify glyoxalase gene families (Mustafiz et al., 2011; Ghosh and Islam, 2016; Ghosh et al., 2016). There are 11 GLYI and 5 GLYII genes in Arabidopsis; 24 GLYI and 12 GLYII genes in G. max; and 11 potential GLYI and three GLYII genes in rice (Mustafiz et al., 2011; Ghosh and Islam, 2016). Expression profiling of all these genes has been performed at different developmental stages, tissues, and abiotic stresses using publicly available microarray database. It has been observed that AtGLYI-3, AtGLYII-2, and AtGLYII-5 expressed constitutively in all the tissues and developmental stages; while AtGLYI-7 and AtGLYII-2 are the most stress-inducible members (Mustafiz et al., 2011). In G. max, GmGLYI-7 and GmGLYII-8 showed maximum expression in all the developmental stages and tissues; while GmGLYI-6, GmGLYI-9, GmGLYI-20, GmGLYII-5, and GmGLYII-10 were highly abiotic stress responsive members (Ghosh and Islam, 2016). Similarly, OsGLYI-11, OsGLYII-2, and OsGLYII-3 expressed constitutively; while OsGLYI-11 and OsGLYII-3 were the most stress responsive members (Mustafiz et al., 2011).
In spite of having a handful genome sequence data deposited in the publicly available database, genome-wide analysis of glyoxalase gene families was not performed in Medicago truncatula. M. truncatula is considered as an excellent model plant due to its relatively small genome size and very high genetic transformation efficiency (Zhang et al., 2013). Moreover, this is a symbiotic legume plant that fixed 40–60 million tons of N2 per annum (Smil, 1999), is equivalent to US$7 to 10 billion (Graham and Vance, 2003). A detailed genome-wide identification of Medicago conventional (GLYI and GLYII) and unique glyoxalase (DJ-1) genes is presented along with their phylogenetic relationship, chromosomal distribution, gene and protein structure, genome duplication and transcript profiling. Results indicate the presence of 29 GLYI, 14 GLYII, and 5 DJ-1 genes in Medicago that codes for 35, 27, and 6 proteins, respectively. This is the largest glyoxalase protein family reported to date in any organism. Expression analysis of these genes based on publicly available microarray data indicates the differential regulation of glyoxalase family members depending on developmental and environmental cues. In particular, MtGLYI-8, MtGLYII-9, and MtDJ-1C are found to be the most up-regulated stress responsive members that might play an imperative role to minimize the accumulation of MG under stress. This study provides a strong base for further characterization and functional validation of glyoxalase genes from M. truncatula.
Materials and Methods
Identification and Nomenclature of Glyoxalase Genes in Medicago truncatula
Putative Medicago GLYI, GLYII and unique GLYIII (DJ-1) proteins were identified by BLASTP search at JCVI M. truncatula annotation database 4.0v11 (Young et al., 2011) with an e-value of 1 using previously reported Arabidopsis GLYI (GenBank: AEE28797.1), Brassica GLYII (GenBank: AAO26580.1) and rice DJ-1C (LOC_Os04g57590) protein sequence as query, respectively. Each of the identified sequences was used as secondary query subsequently until finding new members. Protein sequences of all BLAST hits were retrieved from the database and analyzed using the Hidden Markov Model (HMM) profile of Pfam2 with e-value of 1. The presence of glyoxalase domain (PF00903) or Metallo-beta-lactamase domain (PF00753) or DJ-1/PfpI domain (PF01965) confirmed the identity of the proteins as GLYI or GLYII or DJ-1, respectively. Identified proteins were nomenclature as prefix “Mt” for M. truncatula, followed by GLYI or GLYII or DJ-1. They have numbered arabically for GLYI and GLYII, or alphabetically for DJ-1 depending on their chromosomal position as mentioned in previous studies (Ghosh and Islam, 2016; Ghosh et al., 2016). Detailed information about the identified genes and corresponding proteins was retrieved using locus search option of M. truncatula annotation database 4.0v11 (Young et al., 2011). Sub-cellular localization of all these identified proteins was predicted using CELLO v.2.5: sub-cellular localization predictor3 (Yu et al., 2006), Wolf PSORT4 (Horton et al., 2007) and ChloroP5 (Emanuelsson et al., 1999) tools with default parameters (Tables 1–3).
Table 1.
List of putative Glyoxalase I gene family members of Medicago truncatula.
| Gene name | Locus name | Transcripts | Coordinate (5′–3′) | Transcript length (bp) | CDS (bp) | Protein | Localization | ||
|---|---|---|---|---|---|---|---|---|---|
| Length (aa) | MW (kDa) | pI | |||||||
| MtGLYI-1 | Medtr1g022325 | Medtr1g022325.1 | 7056631–7055156 | 1683 | 1287 | 428 | 47.38 | 5.66 | Cha,b, Cya,b |
| MtGLYI-2 | Medtr1g115170 | Medtr1g115170.1 | 51845699–51846145 | – | 447 | 148 | 16.76 | 4.69 | Cya,b |
| MtGLYI-3 | Medtr2g005880 | Medtr2g005880.1 | 372509–376355 | 4086 | 546 | 181 | 20.60 | 5.26 | ExCa, Cya, Chb |
| MtGLYI-4 | Medtr2g023500 | Medtr2g023500.1 | 8303233–8298020 | 5211 | 864 | 287 | 32.10 | 5.26 | Cya,b |
| MtGLYI-5 | Medtr2g103460 | Medtr2g103460.1 | 44546518–44544702 | 2214 | 516 | 171 | 19.16 | 5.93 | Cya,b |
| MtGLYI-6 | Medtr2g103490 | Medtr2g103490.1 | 44556161–44555344 | 815 | 516 | 171 | 19.42 | 5.04 | Cya,b, Nub |
| MtGLYI-7 | Medtr3g110185 | Medtr3g110185.1 | 51187554–51183426 | 4129 | 2466 | 821 | 92.26 | 8.06 | Nua, Cyb |
| MtGLYI-8 | Medtr4g011020 | Medtr4g011020.1 | 2571851–2573439 | 1992 | 528 | 175 | 20.01 | 5.87 | Cya,b, Nub |
| MtGLYI-9 | Medtr4g039890 | Medtr4g039890.1 | 14172120–14173530 | 1411 | 744 | 247 | 27.56 | 7.33 | Cya,b |
| MtGLYI-10 | Medtr4g057685 | Medtr4g057685.1 | 21216977–21211083 | 8202 | 705 | 234 | 26.69 | 9.49 | Mta, Chb,c |
| Medtr4g057685.2 | 21216757–21211083 | 8202 | 561 | 186 | 20.95 | 5.33 | Cya, Nub | ||
| MtGLYI-11 | Medtr4g063500 | Medtr4g063500.1 | 23518703–23522474 | 4097 | 426 | 141 | 15.88 | 6.24 | Cya,b |
| Medtr4g063500.2 | 23518703–23522474 | 4097 | 387 | 128 | 14.40 | 6.52 | Cya,b | ||
| MtGLYI-12 | Medtr4g114080 | Medtr4g114080.1 | 46953345–46952651 | 1028 | 507 | 168 | 19.23 | 5.13 | ExCa, Nua, Chb |
| MtGLYI-13 | Medtr4g125860 | Medtr4g125860.1 | 52258825–52261268 | 3392 | 582 | 193 | 21.65 | 7.32 | Mta,b, Chb,c |
| MtGLYI-14 | Medtr4g132260 | Medtr4g132260.1 | 55265824–55262670 | 3155 | 846 | 281 | 31.47 | 4.73 | Cya,b |
| MtGLYI-15 | Medtr4g132270 | Medtr4g132270.1 | 55272349–55269373 | 2977 | 915 | 304 | 33.95 | 4.98 | Cya,b |
| Medtr4g132270.2 | 55271711–55269373 | 2977 | 846 | 281 | 31.55 | 4.88 | Cya,b | ||
| MtGLYI-16 | Medtr5g006360 | Medtr5g006360.1 | 881390–880412 | 1234 | 423 | 140 | 15.86 | 6.35 | Cya, Nub |
| MtGLYI-17 | Medtr5g006370 | Medtr5g006370.1 | 883839–883046 | 1856 | 420 | 139 | 15.66 | 5.48 | Nua, Mta, Cya, Chb |
| Medtr5g006370.2 | 883585–883046 | 1856 | 336 | 111 | 12.53 | 5.11 | Mta, Cya,b | ||
| Medtr5g006370.3 | 883839–883162 | 1856 | 390 | 129 | 14.59 | 6.3 | Nua, Mta, Cyb | ||
| MtGLYI-18 | Medtr5g009740 | Medtr5g009740.1 | 2433440–2432537 | 1209 | 351 | 116 | 12.82 | 7.57 | Mta, Chb |
| MtGLYI-19 | Medtr5g021610 | Medtr5g021610.1 | 8378105–8376835 | 1446 | 624 | 207 | 23.91 | 4.7 | ExCa, Nua,b |
| MtGLYI-20 | Medtr5g090990 | Medtr5g090990.1 | 39642769–39640064 | 3139 | 1287 | 428 | 47.37 | 5.61 | Cha,b, Cya, Nub |
| MtGLYI-21 | Medtr5g091060 | Medtr5g091060.1 | 39679453–39682352 | 3349 | 1314 | 437 | 48.36 | 5.34 | Cha,b,c |
| MtGLYI-22 | Medtr6g087120 | Medtr6g087120.1 | 33502501–33497925 | 5130 | 1047 | 348 | 39.15 | 7.96 | Mta, Chb,c |
| MtGLYI-23 | Medtr8g076160 | Medtr8g076160.1 | 32249509–32248727 | 1060 | 582 | 193 | 21.94 | 5.04 | ExCa,b, Cya |
| MtGLYI-24 | Medtr8g102980 | Medtr8g102980.1 | 43369583–43365194 | 4842 | 1044 | 347 | 38.82 | 7.03 | Cha,b,c |
| Medtr8g102980.2 | 43369583–43366886 | 4842 | 1029 | 342 | 38.18 | 7.86 | Cha,b,c | ||
| MtGLYI-25 | Medtr0003s0630 | Medtr0003s0630.1 | 351938–354470 | 2533 | 603 | 200 | 23.18 | 4.86 | Nua, Cya,b |
| MtGLYI-26 | Medtr0430s0010 | Medtr0430s0010.1 | 826–1190 | 365 | 258 | 85 | 9.63 | 9.30 | Cya |
| MtGLYI-27 | Medtr1239s0010 | Medtr1239s0010.1 | 34–1801 | 1768 | 1681 | 560 | 62.66 | 6.24 | Cya,b |
| MtGLYI-28 | Medtr1275s0010 | Medtr1275s0010.1 | 9–1946 | 1938 | 1548 | 515 | 55.66 | 8.22 | Mta, Cyb |
| MtGLYI-29 | Medtr1759s0010 | Medtr1759s0010.1 | 211–1341 | 1128 | 987 | 328 | 35.34 | 9.62 | Cha, ExCb |
bp base pair; MW, molecular weight; kDa, kilo Dalton; Ch, chloroplast; Cy, cytosol; Mt, mitochondria; ExC, extra cellular; Nu, nuclear. aLocalization prediction by CELLO v.2.5 (http://cello.life.nctu.edu.tw/). bLocalization prediction by pSORT (http://www.genscript.com/wolf-psort.html). cChloroplast localization signal confirmed by ChloroP (http://www.cbs.dtu.dk/services/ChloroP/).
Table 3.
List of putative Glyoxalase III gene family members of Medicago truncatula.
| Gene name | Locus name | Transcripts | Coordinate (5′–3′) | Transcript length (bp) | CDS (bp) | Protein | Localization | ||
|---|---|---|---|---|---|---|---|---|---|
| Length (bp) | MW (kDa) | pI | |||||||
| Mt DJ-1A | Medtr2g078060 | Medtr1g022325.1 | 32401642–32405412 | 3968 | 1287 | 428 | 46.87 | 7.25 | Cha,b,c |
| Mt DJ-1B | Medtr3g064093 | Medtr3g064093.1 | 28828715–28828368 | 510 | 279 | 92 | 10.43 | 6.33 | ExCa, Pma, Cyb |
| Mt DJ-1C | Medtr3g064115 | Medtr3g064115.1 | 28837989–28837425 | 626 | 285 | 94 | 10.25 | 6.77 | ExCa, Pma, Cya,b |
| Mt DJ-1D | Medtr3g064140 | Medtr3g064140.1 | 28843504–28840287 | 3588 | 1164 | 387 | 41.41 | 5.97 | Cya,b |
| MtDJ-1E | Medtr4g085900 | Medtr4g085900.1 | 33593600–33596516 | 3437 | 1356 | 451 | 48.20 | 9.42 | Mta, Cha,b,c |
| Medtr4g085900.2 | 33593600–33595499 | 3437 | 1014 | 337 | 36.25 | 6.95 | Pma, Mta,b, Cha,b,c | ||
Bp, base pair; MW, molecular weight; kDa, kilo Dalton; Ch, chloroplast; Cy, cytosol; Mt, mitochondria; ExC, extra cellular; Nu, nuclear; Pm, plasma membrane. aLocalization prediction by CELLO v.2.5 (http://cello.life.nctu.edu.tw/). bLocalization prediction by pSORT (http://www.genscript.com/wolf-psort.html). cChloroplast localization signal confirmed by ChloroP (http://www.cbs.dtu.dk/services/ChloroP/).
Chromosomal Localization and Gene Duplications
Chromosomal location for all the newly identified glyoxalase genes was pointed using genome browser of M. truncatula database 4.0v11 (Young et al., 2011) (Figure 2). Duplication events between the Medicago glyoxalase genes with that of Medicago, Arabidopsis, rice, and soybean were identified using plant genome duplication database6 analysis (Lee et al., 2013). The rate of synonymous (Ks) and non-synonymous substitution (Ka) was retrieved from plant genome duplication database, too (Table 4).
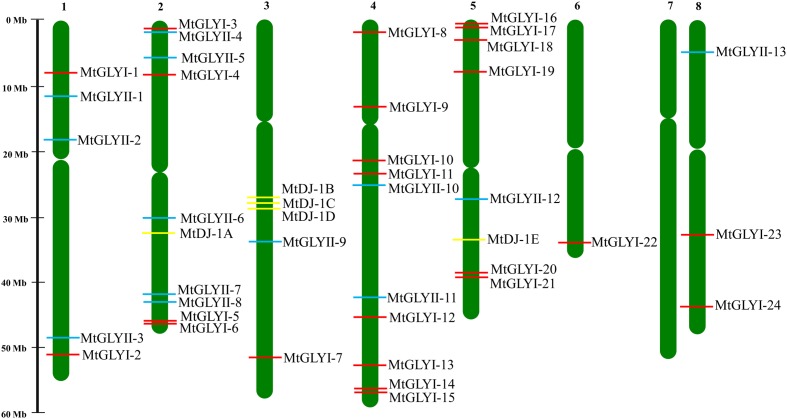
FIGURE 2.
Chromosomal distribution glyoxalase genes in Medicago truncatula. Presence and position of all identified conventional and unique glyoxalase genes were shown in seven different chromosomes. Chromosome numbers are indicated at the top of each bar. Chromosome 7 did not have any glyoxalase genes. The position of GLYI, GLYII, and DJ-1 genes are marked with red, cyan, and yellow color boxes. The name of each gene is mentioned just right side of the colored boxes.
Table 4.
List of glyoxalase gene duplication events between Medicago truncatula, Arabidopsis thaliana, Oryza sativa, and Glycine max.
| Orthologous genes | Paralogous genes | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Arabidopsis thaliana | Oryza sativa | Glycine max | Medicago truncatula | |||||||||||||
| Locus 1 | Locus 2 | Ka | Ks | Ka/Ks | Locus 2 | Ka | Ks | Ka/Ks | Locus 2 | Ka | Ks | Ka/Ks | Locus 2 | Ka | Ks | Ka/Ks |
| MtGLYI-4 | AtGLYI-3 | 0.13 | 1.61 | 0.08 | MtGLYI-14 | 0.14 | 0.51 | 0.27 | ||||||||
| MtGLYI-5 | AtGLYI-4 | 0.25 | 0 | - | OsGLYI-1 | 0.38 | 0 | - | MtGLYI-8 | 0.19 | 1.29 | 0.15 | ||||
| MtGLYI-7 | MtGLYI-22 | 0.29 | 2.15 | 0.13 | ||||||||||||
| MtGLYI-10 | AtGLYI-2 | 0.24 | 0 | - | GmGLYI-16 | 0.09 | 0.67 | 0.13 | ||||||||
| MtGLYI-11 | OsGLYI-3 | 0.21 | 0 | - | ||||||||||||
| MtGLYI-12 | AtGLYI-4 | 0.31 | 0 | - | OsGLYI-1 | 0.37 | 0 | - | MtGLYI-8 | 0.25 | 0 | - | ||||
| MtGLYI-13 | AtGLYI-11 | 0.24 | 3.39 | 0.07 | GmGLYI-23 | 0.09 | 0.36 | 0.25 | ||||||||
| MtGLYI-18 | AtGLYI-10 | 0.14 | 1.64 | 0.09 | ||||||||||||
| MtGLYI-19 | MtGLYI-23 | 0.12 | 0.92 | 0.13 | ||||||||||||
| MtGLYII-3 | GmGLYII-12 | 0.13 | 0.44 | 0.29 | ||||||||||||
| MtGLYII-6 | AtGLYII-2 | 0.18 | 0 | - | GmGLYII-6 | 0.11 | 0.56 | 0.19 | ||||||||
| MtGLYII-7 | AtGLYII-1 | 0.21 | 1.86 | 0.11 | OsGLYII-3 | 0.28 | 0 | - | GmGLYII-7 | 0.08 | 0.39 | 0.21 | MtGLYII-10 | 0.18 | 0.7 | 0.26 |
| MtGLYII-10 | AtGLYII-4 | 0.27 | 1.89 | 0.14 | OsGLYII-3 | 0.34 | 5.04 | 0.07 | GmGLYII-9 | 0.16 | 0.65 | 0.25 | ||||
| MtDJ-1A | AtDJ-1A | 0.29 | 1.64 | 0.17 | GmDJ-1E | 0.11 | 0.53 | 0.21 | ||||||||
| MtDJ-1B | GmDJ-1C | 0.48 | 0.80 | 0.6 | ||||||||||||
| MtDJ-1E | AtDJ-1C | 0.34 | 1.39 | 0.24 | GmDJ-1B | 0.13 | 0.65 | 0.19 | ||||||||
Multiple Sequence Alignment and Phylogenetic Analysis
Phylogenetic relationship among glyoxalase proteins from Arabidopsis, rice, soybean, and Medicago were investigated using MEGA 7.0 (Kumar et al., 2016) with the Neighbor-Joining method and 1000 bootstrap replicates (Figures 3, 8A). Multiple sequence alignment was performed using ClustalW (Larkin et al., 2007) (Figures 5, 7, 8C). Protein sequences used in the phylogenetic analysis are provided as Additional Files S1, S2, S3 GLYI, GLYII and DJ-1 proteins, respectively.
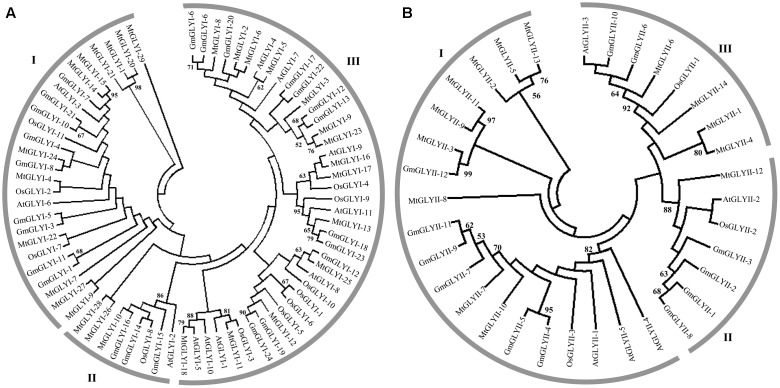
FIGURE 3.
Phylogenetic relationships of Medicago, Arabidopsis, rice, and soybean Glyoxalase proteins. Phylogenetic analysis of GLYI (A) and GLYII (B) proteins were performed separately using all the reported members from Medicago, Arabidopsis, rice, and soybean. The tree was constructed using full-length amino acid sequences based on a neighbor-joining method with 1000 bootstrap value.
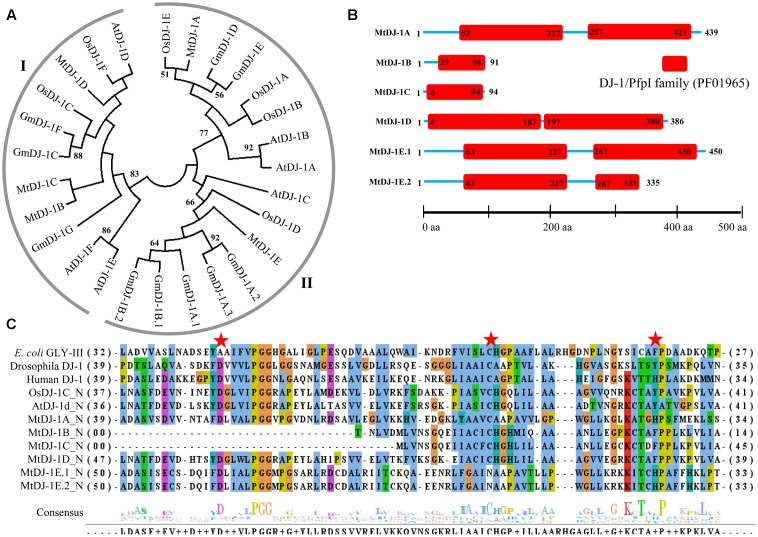
FIGURE 8.
Structural and sequence analysis of MtDJ-1 proteins. (A) Phylogenetic analysis of DJ-1 protein family members from Medicago, Arabidopsis, rice, and soybean was performed using their full-length amino acid sequences and neighbor-joining method with 1000 bootstrap value. (B) Protein structures of 6 MtDJ-1 proteins were analyzed using Pfam (http://www.sanger.ac.uk/Software/Pfam) for the presence of functional DJ-1/PfpI domain (PF01965). The exact position of each domain is indicated by the exact amino acid number inside the red box, while the amino acid number of full length of protein is indicated at each end of the line. The relative position of the domains could be interpreted by the scale given below. (C) Conserved DJ-1/PfpI domain (N-terminal one in case of two) of all MtDJ-1 proteins were aligned with that of previously reported well-characterized novel GLYIII proteins of E. coli, D. melanogaster, H. sapiens, rice (OsDJ-1C), and Arabidopsis (AtDJ-1d) using ClustalW program. Jalview multiple alignment editor programswere employed to generate the picture. Three conserved active side residues are marked with red stars.
FIGURE 5.
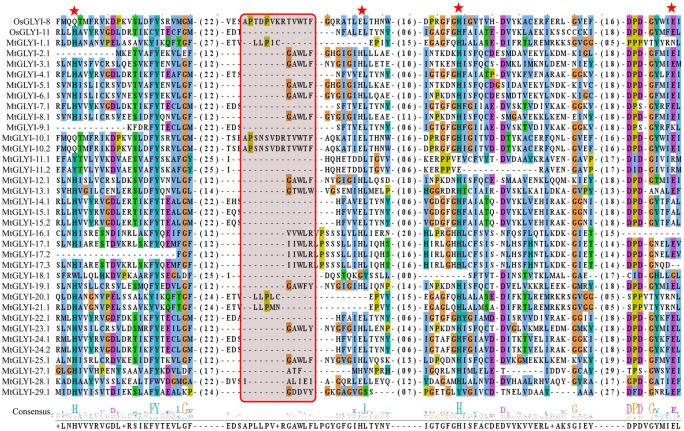
Sequence alignment of GLYI domains of all MtGLYI proteins along with that of OsGLYI-11.2 and OsGLYI-8. Conserved GLYI domains (first one in case of two domain) of all MtGLYI proteins were aligned with that of previously reported Ni2+-dependent OsGLYI-11.2 and Zn2+-dependent OsGLYI-8 using ClustalW program. The picture was generated using Jalview multiple alignment editor programs. Four conserved metal binding sites of GLYI proteins were represented as red stars, while region specific for Zn2+-dependent isoform was marked with a red shaded box.
FIGURE 7.
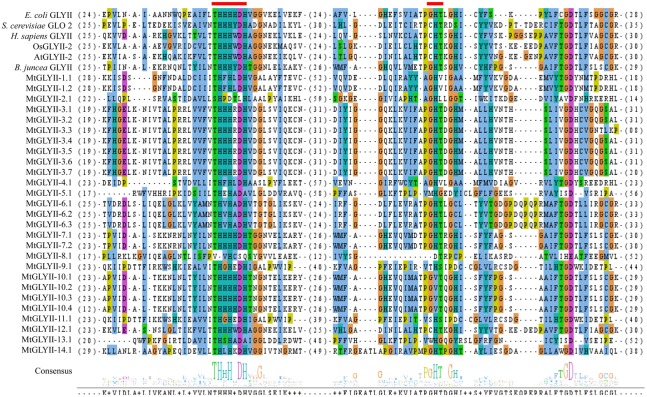
Sequence alignment of GLYII domains of all MtGLYII proteins along with that of other reported GLYII proteins from various species. Conserved GLYII domains of all MtGLYII proteins were aligned with that of previously reported GLYII proteins from E. coli, S. cerevisiae, H. sapiens, rice (OsGLYII-2), Arabidopsis (AtGLYII-2), and B. juncea using ClustalW program. The picture was generated using Jalview multiple alignment editor programs. Two conserved metal binding sites of GLYII proteins were indicated as red boxes.
Domain Architecture of MtGLYI, MtGLYII, and MtDJ-1 Proteins
All the predicted MtGLYI (35), MtGLYII (27), and MtDJ-1 (6) proteins were analyzed using Pfam2 (Finn et al., 2016) to confirm the presence of their respective conserved domains and their exact positions. The position of glyoxalase domain (PF00903) or Metallo-beta-lactamase domain (PF00753) or DJ-1/PfpI domain (PF01965) were identified for all GLYI or GLYII or DJ-1 proteins and the relative figure was generated (Figures 4, 6, 8B). The presence of some additional domains along with the conserved domain was marked in the corresponding figure at an approximate position.
FIGURE 4.
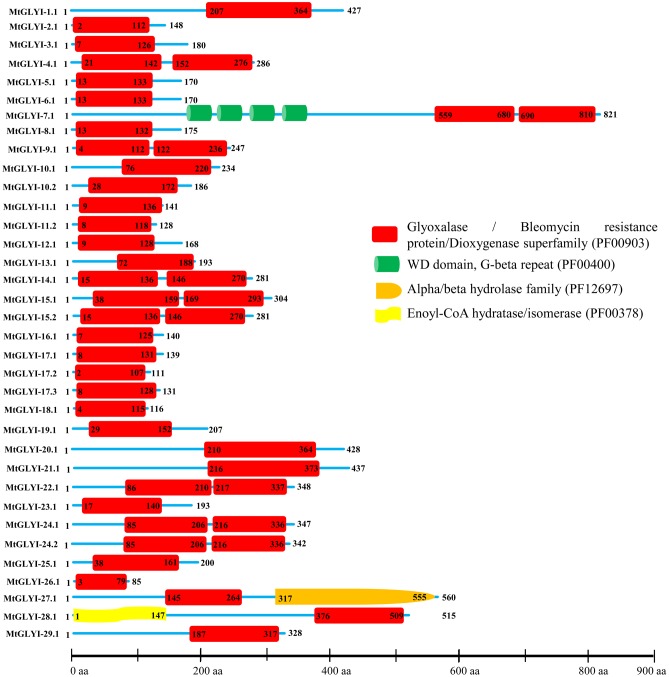
Schematic representation of MtGLYI proteins structure. Protein structures of 35 MtGLYI proteins are shown along with the names of all members on the left side of the figure. All MtGLYI proteins were analyzed using Pfam (http://www.sanger.ac.uk/Software/Pfam) for the presence of functional domain(s). All these proteins possess conserved glyoxalase domain (PF00903) along with some other domains such as WD domain, G-beta repeat (PF00400), Alpha/beta hydrolase family (PF12697) and Enoyl-CoA hydratase/isomerase (PF00378). Different domains are indicated by different colored artworks denoted at the right corner. Exact position and number of each domain are indicated by the amino acid number inside the box. The length of protein is also indicated by exact amino acid numbers and relative position of the domains could be interpreted by the scale given below.
FIGURE 6.
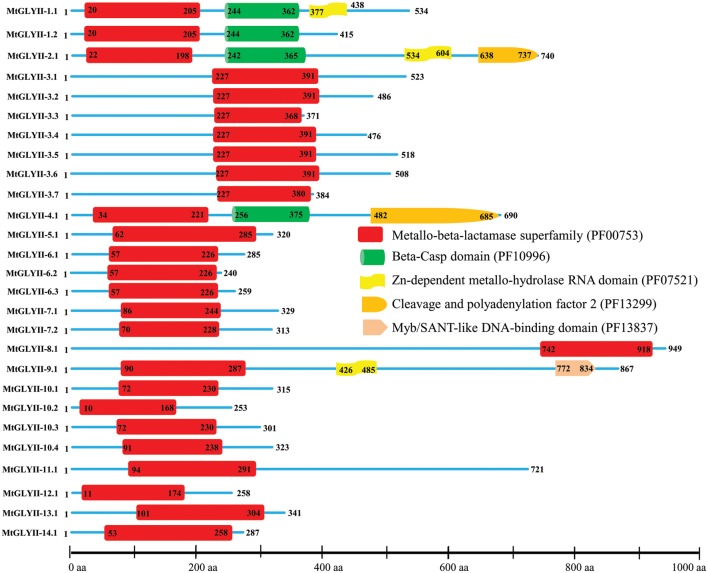
Schematic representation of MtGLYII proteins structure. Protein structures of 27 MtGLYII proteins are shown along with the names of all members on the left side of the figure. MtGLYII proteins were analyzed using Pfam (http://www.sanger.ac.uk/Software/Pfam) for the presence of functional domain(s). All these proteins possess conserved Metallo-beta-lactamase superfamily (PF00753) along with some other domains such as Beta-Casp domain (PF10996), Zn-dependent metallohydrolase RNA domain (PF07521), Cleavage and polyadenylation factor 2 (PF13299), and Myb/SANT-like DNA-binding domain (PF13837). Different domains are marked by different colored artworks denoted at the right side of the figure. The exact position of each domain or full-length proteins is indicated by the exact amino acid number inside or outside the box. The relative position of the domains could be interpreted by the scale given below.
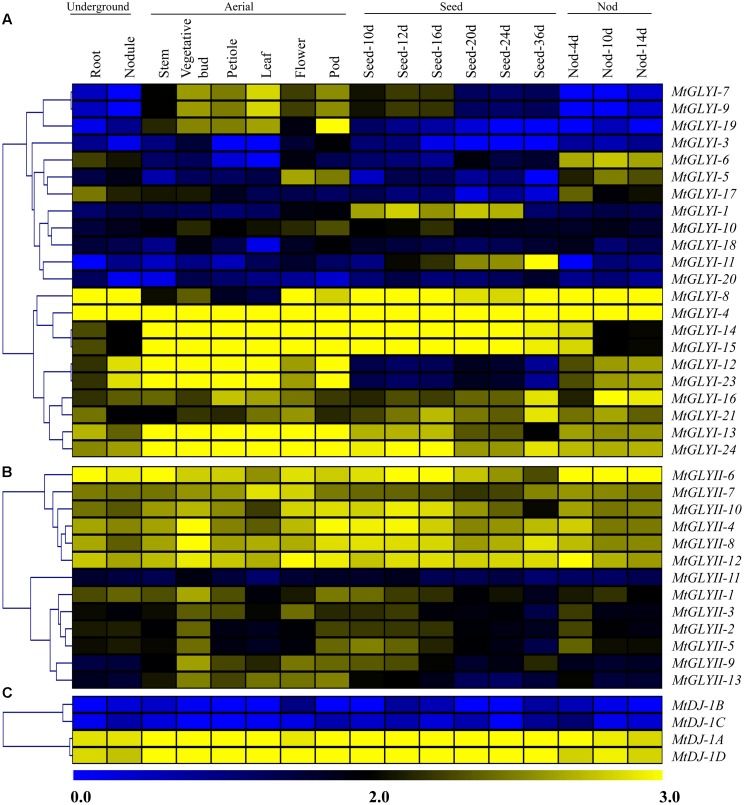
Expression Profiling of Glyoxalase Genes at Different Tissues of M. truncatula
Genome-wide microarray data of conventional and unique glyoxalase genes were obtained from the M. truncatula gene expression Atlas (MtGEA) Project database7 (Benedito et al., 2008). The corresponding probesets for all MtGLYI, MtGLYII, and MtDJ-1 genes were identified using NetAffx Analysis Center online Probe Match tool8 with default parameters based on respective CDS sequence as query. Genes with more than one probe, probe with the highest expression value was considered for analysis; and single probe for many genes was considered as the same transcriptional profile. Normalized transcript data was obtained for 17 different tissues, including underground tissues-root and nodule; aerial tissues- stem, vegetative bud, petiole, leaf, flower, pod; seed development (seed of 10, 12, 16, 20, 24, and 36 days); and nod development (nod of 4, 10, and 14 days). These normalized expression data was used to generate heatmap (Figure 9) and hierarchically clustering based on the Manhattan correlation with average linkage using the Institute for Genomic Research MeV software package9 (Saeed et al., 2006).
FIGURE 9.
Expression profiling of Medicago glyoxalase genes with hierarchical clustering at different developmental stages/tissues. Genome-wide microarray data of conventional GLYI (A) and GLYII (B); and novel glyoxalase III (C) genes were obtained from the Medicago truncatula gene expression Atlas (MtGEA) Project database (https://mtgea.noble.org/v2/). Normalized transcript data was obtained for 17 different tissues, including underground tissues-root and nodule; aerial tissues- stem, vegetative bud, petiole, leaf, flower, pod; seed development (seed of 10, 12, 16, 20, 24, and 36 days); and nod development (nod of 4, 10, and 14 days). These normalized expression data was used to generate heatmap with hierarchical clustering based on the Manhattan correlation with average linkage using MeV software package. Color scale below heat map shows the level of expression; yellow indicates high transcript abundance while blue indicates a low level of transcript abundance.
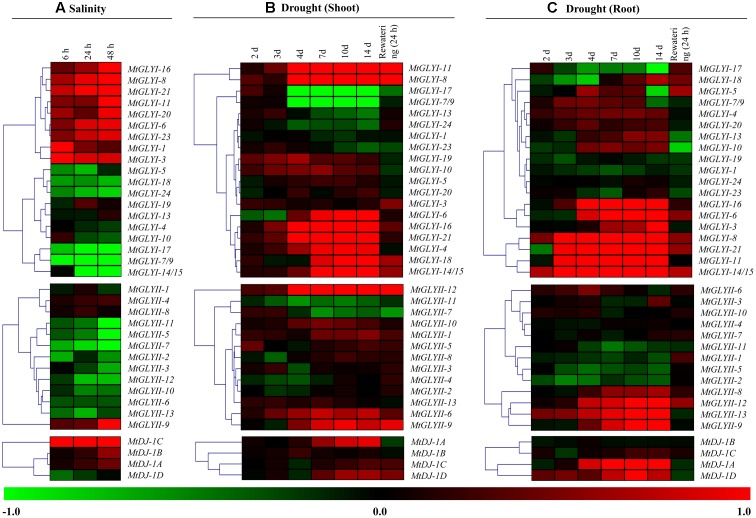
Microarray-Based Expression Analysis of Medicago glyoxalase Genes in Response to Salinity and Drought
Normalized and curated perturbation expression data of Medicago glyoxalase genes were retrieved from Genevestigator Affymetrix Medicago genome array10 (Hruz et al., 2008; Zimmermann et al., 2008) using the already identified corresponding probe. The log2 fold change ratio was retrieved using Genevestigator with default parameters using experiments id: MT-00011: Effects of salt stress on M. truncatula seedlings (GSE14029)11 and MT-00013: Global reprogramming of transcription and metabolism in M. truncatula during progressive drought and after rewatering (E-MTAB-2681) (Zhang et al., 2014). Detailed informations about the gene expression experimental design is provided as Additional File S4. These experiments were performed with minimum three replicates and fold change values were calculated in comparison to the untreated control 0 h seedlings in case of salinity and an untreated 2 days control sample of respective tissue in the case of drought. Generation of heat maps (Figure 10) and hierarchical clustering with Manhattan distance metric method was performed using Institute for Genomic Research MeV software package9 (Saeed et al., 2006).
FIGURE 10.
Expression analyses of Medicago glyoxalase genes in response to two devastating abiotic stresses. Normalized and curated perturbation expression data of the Medicago glyoxalase genes were retrieved from publicly available Genevestigator Affymetrix Medicago genome array (https://www.genevestigator.com/gv/plant.jsp) using the already identified corresponding probesets of experiments with id: MT-00011 and MT-00013 (Details provided in the “Materials and Methods” section). Relative expression data of all available MtGLYI, MtGLYII, and MtDJ-1 were combined and analyzed in response to different duration of salinity (A); and drought with shoot tissue (B) and root tissue (C). Fold change of expression data was calculated by comparing with the corresponding mock samples. Heat maps with hierarchical clustering were generated using Manhattan distance metric method of MeV software package. Color scale below heat map shows the type of transcript alteration; red indicates up-regulation and green indicate down-regulation of the corresponding transcript.
Results
Identification of Glyoxalase Proteins in Medicago truncatula
Glyoxalase proteins have been well characterized from various plant as well as animal species. The genome-wide analysis identified a total of 35 unique Medicago GLYI proteins encoded by 29 GLYI genes. These genes were found to be located on different chromosomes of Medicago (Figure 2). They were nomenclature as MtGLYI-1 to MtGLYI-29 according to the order of their chromosomal location (Table 1). The number of Medicago GLYI genes was found to be greater than previously reported Arabidopsis (10), rice (11), and soybean (24) GLYI genes (Mustafiz et al., 2011; Ghosh and Islam, 2016). Similarly, a total of 27 Medicago GLYII proteins encoded by 14 GLYII genes were identified. Both the number of GLYII genes and proteins were found to be greater than previously reported Arabidopsis (5 genes and 9 proteins), rice (3 genes and 4 proteins), and soybean (12 genes and 23 proteins) (Mustafiz et al., 2011; Ghosh and Islam, 2016). They were symbolized as MtGLYII-1 to MtGLYII-14 like MtGLYI genes (Table 2).
Table 2.
List of putative Glyoxalase II gene family members of Medicago truncatula.
| Gene name | Locus name | Transcripts | Coordinate (5′–3′) | Transcript length (bp) | CDS (bp) | Protein | Localization | ||
|---|---|---|---|---|---|---|---|---|---|
| Length (aa) | MW (kDa) | pI | |||||||
| MtGLYII-1 | Medtr1g032500 | Medtr1g032500.1 | 11561502–11550336 | 11903 | 1605 | 534 | 60.13 | 6.73 | Cya,b |
| Medtr1g032500.2 | 11561502–11551704 | 11903 | 1248 | 415 | 47.02 | 7.58 | Cya,b | ||
| MtGLYII-2 | Medtr1g050492 | Medtr1g050492.1 | 19478035–19486897 | 8863 | 2223 | 740 | 82.00 | 4.82 | Cya,b, Chb |
| MtGLYII-3 | Medtr1g110300 | Medtr1g110300.1 | 49751952–49742911 | 9300 | 1572 | 523 | 58.66 | 6.9 | Cya, Chb |
| Medtr1g110300.2 | 49751952–49743319 | 9300 | 1461 | 486 | 54.32 | 6.03 | Cya, Chb | ||
| Medtr1g110300.3 | 49751952–49745653 | 9300 | 1116 | 371 | 41.53 | 5.84 | Cya, Chb | ||
| Medtr1g110300.4 | 49751952–49745288 | 9300 | 1431 | 476 | 53.22 | 6.2 | Cya, Chb | ||
| Medtr1g110300.5 | 49751952–49743319 | 9300 | 1557 | 518 | 58.30 | 6.12 | Cya, Chb | ||
| Medtr1g110300.6 | 49751952–49745288 | 9300 | 1527 | 508 | 57.20 | 6.28 | Cya, Chb | ||
| Medtr1g110300.7 | 49751952–49745802 | 9300 | 1155 | 384 | 42.64 | 5.62 | Cya, Chb | ||
| MtGLYII-4 | Medtr2g006180 | Medtr2g006180.1 | 489282–485609 | 4028 | 2073 | 690 | 76.96 | 6.37 | Cya,b |
| MtGLYII-5 | Medtr2g018660 | Medtr2g018660.1 | 5892053–5896603 | 5924 | 963 | 320 | 35.87 | 6.18 | Nua, Cyb |
| MtGLYII-6 | Medtr2g072190 | Medtr2g072190.1 | 30330754–30334767 | 4591 | 858 | 285 | 31.43 | 8.42 | Mta,b |
| Medtr2g072190.2 | 30330754–30333959 | 4591 | 723 | 240 | 26.47 | 8.93 | Mta,b, Chb,c | ||
| Medtr2g072190.3 | 30330754–30334011 | 4591 | 780 | 259 | 28.77 | 7.72 | Mta,b, Chb,c | ||
| MtGLYII-7 | Medtr2g099090 | Medtr2g099090.1 | 42458819–42455183 | 4198 | 990 | 329 | 36.10 | 8.65 | Mta, Chb,c |
| Medtr2g099090.2 | 42458690–42455183 | 4198 | 942 | 313 | 34.51 | 8.57 | Mta, Chb,c | ||
| MtGLYII-8 | Medtr2g101390 | Medtr2g101390.1 | 43590697–43596569 | 6147 | 2853 | 950 | 104.98 | 6.29 | Nua, Cya, Mta, Chb,c |
| MtGLYII-9 | Medtr3g089020 | Medtr3g089020.1 | 33966445–33957865 | 9309 | 2604 | 867 | 96.36 | 8.11 | Cha,b,c |
| MtGLYII-10 | Medtr4g068100 | Medtr4g068100.1 | 25474066–25470040 | 4584 | 948 | 315 | 34.99 | 6.78 | Mta, Nub |
| Medtr4g068100.2 | 25472955–25470040 | 4584 | 762 | 253 | 28.03 | 5.55 | Cya,b | ||
| Medtr4g068100.3 | 25474066–25470489 | 4584 | 906 | 301 | 33.49 | 6.78 | Mta, Pma, ExCa, Nub | ||
| Medtr4g068100.4 | 25474066–25470040 | 4584 | 972 | 323 | 35.86 | 8.14 | Mta, Nub | ||
| MtGLYII-11 | Medtr4g103770 | Medtr4g103770.1 | 42917141–42924712 | 7724 | 2166 | 721 | 80.73 | 9.83 | Mta, Chb,c |
| MtGLYII-12 | Medtr5g068440 | Medtr5g068440.1 | 28969883–28974477 | 5037 | 777 | 258 | 28.53 | 5.34 | Cya,b |
| MtGLYII-13 | Medtr8g017270 | Medtr8g017270.1 | 5810801–5804905 | 6295 | 1026 | 341 | 38.00 | 7.68 | Cha,b,c, Mta, Pma |
| MtGLYII-14 | Medtr2166s0010 | Medtr2166s0010.1 | 62–925 | 861 | 864 | 287 | 31.26 | 6.6 | ExCa, Cya, Chb |
Bp, base pair; MW, molecular weight; kDa, kilo Dalton; Ch, chloroplast; Cy, cytosol; Mt, mitochondria; ExC, extra cellular; Nu, nuclear. aLocalization prediction by CELLO v.2.5 (http://cello.life.nctu.edu.tw/). bLocalization prediction by pSORT (http://www.genscript.com/wolf-psort.html). cChloroplast localization signal confirmed by ChloroP (http://www.cbs.dtu.dk/services/ChloroP/).
In the same way, genome-wide searching of unique GLYIII proteins in Medicago identified six DJ-1 proteins encoded by five genes. They were named as MtDJ-1A to MtDJ-1E like previously reported members from other species (Ghosh et al., 2016) (Table 3). Although the number of genes was almost similar, but the number of MtDJ-1 proteins was less than previously reported Arabidopsis (6 genes and 11 proteins), rice (6 genes and 12 proteins) (Ghosh et al., 2016) and soybean (7 genes and 11 proteins) (unpublished data) counterparts. In all cases of MtGLYI, MtGLYII, and MtDJ-1 families, proteins number was found to be greater than that of genes (Tables 1–3), that indicate the presence of alternate splicing event in Medicago glyoxalase transcripts.
Detailed Analysis of Newly Identified MtGLYI, MtGLYII, and MtDJ-1 Members
All the newly identified glyoxalase members were analyzed further in terms of the length of transcripts, coding DNA sequence (CDS), and polypeptide; molecular weight, isoelectric point and sub-cellular localization of the corresponding protein. CDS length of MtGLYI members was found to be fluctuating from 258 bp (MtGLYI-26) to 2466 bp (MtGLYI-7) with an average of 1533 bp. Thus, the largest member of MtGLYI protein family is MtGLYI-7 with a polypeptide length of 821 aa and molecular weight of 92.26 kDa. Subsequently, the smallest member is MtGLYI-26 with 85 aa in length and 9.63 kDa in molecular weight (Table 1). MtGLYI protein family members showed an average length of 510 aa and predicted molecular weight of 57.04 kDa. Isoelectric point (pI) is an important parameter of protein that determined the net charge of a protein under certain physiological conditions. Members of MtGLYI family showed the broad difference in their pI-value ranging from 4.69 (MtGLYI-2) to 9.62 (MtGLYI-29). Out of 35 MtGLYI proteins, 24 of them showed a pI-value less than 7.0 (neutral pH) and the rest 11 showed more than 7.0. Thus both positively and negatively charged MtGLYI proteins co-exist in the system under particular physiological condition. MtGLYI proteins were predicted to be localized at cytosol, chloroplast, mitochondria, nucleus, and extracellular spaces. The majority of MtGLYI family members are found to be localized in the cytosol. Chloroplast localization of MtGLYI-21, MtGLYI-24.1, and MtGLYI-24.2 were confirmed by all three tools namely CELLO, Wolf PSORT, and ChloroP. Localization signal for mitochondria, nucleus or extra-cellular matrix were found to be predicted by either CELLO or Wolf PSORT (Table 1).
The length of MtGLYII CDS varied from 723 bp (MtGLYII-6.2) to 2853 bp (GmGLYII-8.1) with an average of 2579 bp (Table 2). The largest member (MtGLYII-8.1) has a polypeptide length of 950 aa with a molecular weight of 104.98 kDa; whereas MtGLYII-6.2, the smallest member has polypeptide length and molecular weight of 240 aa and 26.47 kDa, respectively (Table 2). In terms of pI, MtGLYII-2 and MtGLYII-6.2 showed the most acidic (4.82) and basic (8.93) value, respectively. Like MtGLYI members, the majority of MtGLYII proteins (17 out of 27) showed acidic pI-value, while rest 10 MtGLYII members showed basic pI-value (Table 2). Similar to MtGLYI, most of the MtGLYII proteins showed cytosolic localization. Chloroplast localization of MtGLYII-9.1 and MtGLYII-13.1 was confirmed by all three tools-CELLO, Wolf PSORT, and ChloroP; and mitochondrial localization of MtGLYII-6.1 was confirmed by both CELLO and Wolf pSORT. Nucleus and extracellular localization of few members were predicted by either CELLO or Wolf pSORT.
Like conventional glyoxalase enzymes (GLYI + GLYII), different physio-chemical properties of unique glyoxalase III proteins were analyzed. The length of MtDJ-1 CDS was varied from 279 bp (MtDJ-1B) to 1356 bp (MtDJ-1E.1) with an average of 898 bp (Table 3). Consequently, the smallest MtDJ-1 member (MtDJ-1B) is 91 aa in length with a molecular weight of 10.43 kDa; while the largest one is MtDJ-1E.1 with a length of 450 aa and molecular weight of 48.20 kDa (Table 3). Isoelectric value (pI) varies from 5.69 (MtDJ-1D) to 9.42 (MtDJ-1E.1); with four acidic and two basic members at neutral pH. Chloroplast localization of MtDJ-1A, MtDJ-1E.1, and MtDJ-1E.2 was confirmed by all three tools-CELLO, Wolf PSORTand ChloroP; followed by cytosolic localization of MtDJ-1C and MtDJ-1D, and mitochondrial localization of MtDJ-1E.2 was confirmed by both CELLO and Wolf pSORT tools. Localization of extracellular matrix and plasma membrane of few members were only predicted by either CELLO or Wolf pSORT tool (Table 3).
Chromosomal Distribution and Gene Duplication
All glyoxalase genes were found to be distributed randomly in seven different chromosomes of Medicago out of total eight chromosomes (Figure 2). A detailed chromosome map was constructed to exactly point out the newly identified MtGLYI, MtGLYII, and MtDJ-1 genes on different Medicago chromosomes. Five of MtGLYI (MtGLYI-25 to MtGLYI-29) and one of the MtGLYII genes (MtGLYII-14) were present in scaffold region and thus unable to point out them in chromosomes. Rest 24 MtGLYI genes were found to be unevenly distributed in seven Medicago chromosomes. Chromosome 4 contains a maximum number of eight GLYI genes, followed by chromosome 5 with six and chromosome 2 with four members. However, chromosomes 1 and 8 have two GLYI genes each, and only one GLYI gene each is present in chromosomes 3 and 6 (Figure 2). Similarly, 13 MtGLYII genes were found to be located on six different chromosomes (Figure 2). The gene density per chromosome is highly uneven, where chromosome 2 contains a maximum of five GLYII genes, followed by three genes in chromosome 1 and two genes in chromosome 4. However, chromosomes 3, 5, and 8 have only one GLYII gene each (Figure 2). Out of 5 MtDJ-1 genes, maximum three genes present in chromosome 3 and one member each in chromosomes 2 and 5. All these genes were found to be distributed in a highly uneven fashion in terms of chromosome number and position. Chromosome 4 was found to be the highly dense with 10 genes in total; while chromosomes 2, 3, and 5 contain at least one member of each gene family (Figure 2).
Gene duplication and divergence are two most important phenomenon of plant evolution. Two major angiosperms (eudicots and monocots) are calculated to diverge from 125–140 Mya to 170–235 Mya ago (Lee et al., 2013). This largely results in subsequent chromosomal duplication and gain/loss of genes. To identify glyoxalase gene duplication among four plants (Arabidopsis, Rice, Soybean, and Medicago) genome, plant genome duplication database12 were analyzed (Lee et al., 2013) and the data is presented as Table 4. In this study, five MtGLYI and one MtGLYII sister-gene pairs have been observed (Table 4). The gene pairs are MtGLYI-4/MtGLYI-14, MtGLYI-5/MtGLYI-8, MtGLYI-7/MtGLYI-22, MtGLYI-12/MtGLYI-8, MtGLYI-19/MtGLYI-23, and MtGLYII-7/MtGLYII-10. Moreover, orthologous of MtGLY genes were found in three other well-characterized plant genomes of Arabidopsis, Soybean, and Rice. Non-synonymous to synonymous substitutions (Ka/Ks) ratio indicates the evolutionary history and direction of selection (Li et al., 1981; Chen et al., 2014). A pair of genes with Ka/Ks < 1 indicates purifying selection, Ka/Ks = 1 implies neutral drifting, and lastly Ka/Ks > 1 implies positive or Darwinian selection (Juretic et al., 2005; Chen et al., 2014). Ka/Ks ratio of all gene pairs was found to be below 1 indicating the influence of purifying selection in the evolution of glyoxalase genes among Arabidopsis, rice, soybean, and Medicago (Table 4).
Phylogenetic Analysis of Conventional Glyoxalase Genes among Four Well-Characterized Plant Species
Glyoxalase genes from Arabidopsis, rice, soybean, and Medicago were analyzed further for their sequence similarities and evolutionary divergence through phylogenetic analysis. Three major classes GLYI family members were identified in the phylogenetic tree (Figure 3A). Based on various previous studies, these classes may belong to Ni2+-dependent GLYI, Zn2+-dependent GLYI, and functionally diverse inactive GLYI. Clade-I had the largest GLYI members from different plant species, while clade-II had the lowest number of members only from Arabidopsis and rice genome (Figure 3A). Clade-I contains three rice GLYI members OsGLYI-2, OsGLYI-7, and OsGLYI-11; and two members of Arabidopsis GLYI family AtGLYI-3 and AtGLYI-6 those have been considered as Ni2+-dependent GLYI enzyme (Kaur et al., 2013; Mustafiz et al., 2014). Thus, members of this clade are supposed to show Ni2+-dependent GLYI activity. Similarly, clade-II has previously reported Zn2+-dependent rice (OsGLYI-8) and Arabidopsis (AtGLYI-2) member which indicated to be Zn2+-dependent GLYI clusters. This confirmed the dominance of Ni2+-dependent GLYI over Zn2+-dependent isoform in plant species (Figure 3A) which had been recently proved in Arabidopsis, too (Jain et al., 2016). Members of clade-III are considered as functionally diverged GLYI members as it contains OsGLYI-10, which did not possess conventional GLYI activity (unpublished data).
Similarly, phylogenetic analysis of GLYII proteins could be divided into three major classes (I–III) (Figure 3B). One of rice GLYII protein (OsGLYII-1) and one Arabidopsis GLYII protein (AtGLYII-3) was previously reported to possess sulfur dioxygenase (SDO) activity (Kaur et al., 2014b). Both of them were present in the clade-III, representing the functional divergence members of GLYII family. As AtGLYII-2 was found to be localized in mitochondria (Marasinghe et al., 2005), other members of clade II (one from rice, OsGLYII-2), four proteins from soybean and one from Medicago, MtGLYII-12) could be mitochondrial localized. Lastly, Clade-I might contain all the active and cytoplasmic localized GLYII members from four plant species.
Structural and Sequence Analysis of MtGLYI Proteins
The presence of conserved glyoxalase domain (PF00903) in all the predicted 35 MtGLYI proteins was analyzed using Pfam. Analyses revealed that all MtGLYI protein has at least one GLYI domain, while some of them have two. Out of 35, nine MtGLYI proteins contained two consecutive GLYI domains (Figure 4). Single GLYI protein with two domains had been reported from G. max (Ghosh and Islam, 2016), S. cerevisiae (Frickel et al., 2001), O. sativa (Mustafiz et al., 2014), and Plasmodium falciparum (Deponte et al., 2007). Apart from conserved GLYI domain, some proteins have addition domains, such as WD domain (PF00400), Alpha/beta hydrolase family (PF12697) and Enoyl-CoA hydratase/isomerase (PF00378). The role of these domains in association with GLYI proteins is still not clear. GLYI domain (only N-terminal one in a case of two domain containing members) sequences of all MtGLYI proteins were aligned with that of Ni2+-dependent OsGLYI-11.2 and Zn2+-dependent OsGLYI-8 (Kaur et al., 2013) proteins to predict on their metal ion dependency and catalytic activity (Figure 5). Enzymatic activity of GLYI proteins has been found to be dependent on metal ions (Kaur et al., 2013). This dependency could be easily predicted based on the size of GLYI domain; domain length of ∼120 aa showed Ni2+-dependence and length of 142 aa showed Zn2+-dependence (Kaur et al., 2013). According to the alignment, Zn2+-dependent OsGLYI-8 has an extra stretch of 12 aa (Figure 5, marked with rectangle), that is completely absent in Ni2+-dependent OsGLYI-11.2 proteins. Proteins with this stretch of amino acids such as MtGLYI-10.1 and MtGLYI-10.2 are found to be Zn2+ dependent, while proteins like MtGLYI-4, MtGLYI-7, MtGLYI-9, MtGLYI-11, MtGLYI-14, MtGLYI-15, MtGLYI-18, MtGLYI-22, and MtGLYI-24 might be Ni2+ dependent. Some of the proteins possess partial regions, thus difficult to predict (Figure 5).
The active site of GLYI proteins had a conserved motif of H/QEH/QE irrespective of their metal ion dependency. These four residues were found to be conserved in both OsGLYI-8 and OsGLYI-11.2 proteins, and marked with red stars (Figure 5). Among these four residues, the position of first glutamate residue showed maximum divergence in GmGLYI proteins. MtGLYI-11.1 and MtGLYI-11.2 proteins showed variation in all the four conserved active side residues. Proteins with the presence of all these four highly conserved residues are expected to have functional GLYI enzyme activity as per suggestion of previous studies (Mustafiz et al., 2014; Ghosh and Islam, 2016). Eight MtGLYI proteins such as MtGLYI-4.1, MtGLYI-7.1, MtGLYI-10.1, MtGLYI-10.2, MtGLYI-22.1, MtGLYI-24.1, MtGLYI-24.2, and MtGLYI-28.1 possess all four residues and expected to have function GLYI enzyme activity.
Sequence Analysis of MtGLYII Proteins for Conserved Domain and Active Sites
Pfam analysis confirmed the presence of conserved Metallo-beta-lactamase domain (PF00753) in all MtGLYII proteins either alone or with some other domains (Figure 6). Out of total 27 proteins, 22 MtGLYII proteins had only Metallo-beta-lactamase domain, while rest 5 contained additional domain such as Beta-Casp domain (PF10996), Zn-dependent metallo-hydrolase RNA domain (PF07521), Cleavage and polyadenylation factor 2 (PF13299), Myb/SANT-like DNA-binding domain (PF13837) (Figure 6). The significance of these additional domains is not clear in relation to GLYII enzyme activity. Similar to GLYI, GLYII proteins contained two well-conserved domain; one is metal binding motif (THXHXDH) and another one is active site motif (C/GHT) (Ghosh et al., 2014). The presence of both these motifs is crucial for a functional GLYII enzyme. Therefore, sequences of all MtGLYII proteins were aligned with that of E. coli, yeast, human, rice, Arabidopsis and Brassica GLYII proteins (Figure 7). The position of both these motifs was marked with red boxes (Figure 7). Out of 27 identified MtGLYII proteins, only two (MtGLYII-2.1 and MtGLYII-8.1) do not possess the conserved metal binding residues, but 13 of them do not have the active site motif (Figure 7). MtGLYII-1.1, MtGLYII-2.1, MtGLYII-2.2, MtGLYII-4.1, MtGLYII-5.1, MtGLYII-8.1, MtGLYII-9.1, MtGLYII-10.1-4, MtGLYII-11.1, and MtGLYII-13.1 proteins are either missing metal binding or active side residues, thus might not possess functional GLYII enzyme activity.
Analysis of Unique Glyoxalase III Proteins in Medicago
The presence of unique glyoxalase III proteins has been reported in many microorganisms and plants. GLYIII activity was found in the proteins with DJ-1/PfpI domain. Like conventional glyoxalase enzymes, DJ-1 proteins from Arabidopsis, rice, soybean, and Medicago were analyzed using Mega 5.2 tool (Figure 8A). There were two major clades in the tree, named- clades I and II. Among them, AtDJ-1D and OsDJ-1C were experimentally proven to have substantial GLYIII enzymatic activity (Kwon et al., 2013; Ghosh et al., 2016). Proteins present in clade-I along with these two might have significant GLYIII activity; while clade-II members could have less activity.
All previously reported rice and Arabidopsis DJ-1 proteins were found to contain two DJ-1/PfpI domains, while that of E. coli, Drosophila, and human were found to contain only single domain (Ghosh et al., 2016). DJ-1/PfpI domain has an average size of around 140 to 150 amino acids in all species except E. coli. Sequence analysis of MtDJ-1 proteins revealed that most of them have two DJ-1/PfpI domain, too (Figure 8B). Only MtDJ-1B and MtDJ-1C has single DJ-1/PfpI domain with a size of 69 and 88 amino acids each. These two might have partial DJ-1/PfpI domain. However, the domains of other MtDJ-1 proteins are connected by a linker of 40 amino acids (MtDJ-1A and MtDJ-1E) or 10 amino acids (MtDJ-1D). The N-terminal DJ-1/PfpI domains of all MtDJ-1 proteins were aligned with that of E. coli, Drosophila, human DJ-1 proteins; and AtDJ-1D and OsDJ-1C proteins (Figure 8C). All reported DJ-1 proteins with GLYIII contain a conserved catalytic triad of aspartate/glutamate, cysteine, and tyrosine/histidine residues (Ghosh et al., 2016). All these three residues were marked with red stars (Figure 8C). Among them, the central cysteine residue is the key for GLYIII enzymatic activity due to it’s directly involvement in catalysis (Lin et al., 2012). All MtDJ-1 proteins possessed this highly conserved cysteine residues, except MtDJ-1E (Figure 8C). The absence of this important residue in MtDJ-1E protein might lead to complete lack of GLYIII activity as proven by previously for AtDJ-1C (Kwon et al., 2013). However, the absence of any of other two conserved residues led to partial loss of GLY III activity in case of AtDJ-1e and AtDJ-1f (Kwon et al., 2013). Thus, MtDJ-1A should be the most efficient GLYIII enzyme of Medicago with all three conserved residues, followed by MtDJ-1D (Figure 8C).
Expression Profiling of Glyoxalase Genes in M. truncatula
To investigate the expression patterns of Medicago glyoxalase genes, expression data for a diverse set of 17 Medicago tissues was retrieved and analyzed. All these tissues could be broadly divided into four classes; such as underground, aerial, seed and nod; and represented as heat maps (Figures 9A–C). Different members of MtGLY gene families showed different tissue specificity. Among all MtGLYI genes, MtGLYI-4 showed the highest level of constitutive expression in all these analyzed tissues, followed by MtGLYI-24, MtGLYI-13, MtGLYI-21, and MtGLYI-16. MtGLYI-8 also maintained a high level of expression except for some aerial tissues; while MtGLYI-14 and MtGLYI-15 showed low expression only in underground and nod tissues. There are some clusters of genes that showed expression in particular type of tissues only. MtGLYI-7, MtGLYI-9 and MtGLYI-19 showed medium to high-level expression in aerial tissues only, while MtGLYI-5, MtGLYI-6 and MtGLYI-17 showed a similar pattern of expression only in underground and nod tissues. The presence of seed-specific GLYI transcripts has been previously reported in rice, Arabidopsis, and soybean (Mustafiz et al., 2014; Ghosh and Islam, 2016). Similarly, MtGLYI-1 and MtGLYI-11 has medium to high level of expression in different seed maturation stages and low expression in other tissues (Figure 9A). However, some members maintained the high level of expression in all tissues except for seed only, such as MtGLYI-12 and MtGLYI-23. This indicates toward the presence of a complex regulatory network for MtGLYI gene expression.
On the other hand, two distinct clades were observed in case of MtGLYII and MtDJ-1 gene expression data (Figures 9B,C). One set of genes maintained high level of constitutive expression in all tissues, while the other set of genes had very low level of expression. Out of 13 analyzed MtGLYII genes, 6 genes such as MtGLYII-4, MtGLYII-6, MtGLYII-7, MtGLYII-8, MtGLYII-10, and MtGLYII-12 showed medium to high-level expression in all the tissues with very few exceptions (Figure 9B). Among others, MtGLYII-11 showed the lowest level of expression in all the tissues, while MtGLYII-1, MtGLYII-3, MtGLYII-9, and MtGLYII-13 genes showed significant expression in the aerial tissues only. In the case of MtDJ-1 family members; MtDJ-1A and MtDJ-1D showed a high level of expression in all these tissues, while MtDJ-1B and MtDJ-1C showed the lowest level (Figure 9C). As compared to MtGLYI family members, MtGLYII and MtDJ-1 family members showed less complexity and tissue specificity.
Expression of MtGLYI, MtGLYII, and MtDJ-1 Genes in Response to Salinity and Drought
To investigate the physiological role of Medicago glyoxalase genes in response to abiotic stress, corresponding transcript expression was analyzed under salinity and drought stresses. In response to salt stress (3 days old Jemalong A17 seedlings were treated with 180 mM NaCl) (Li et al., 2009), log2 fold change ratio was analyzed at 6, 24, and 48 h of stress as compared to their 0 h expression level. A cluster of MtGLYI genes-MtGLYI-16, MtGLYI-8, MtGLYI-21, MtGLYI-11, MtGLYI-20, MtGLYI-6, MtGLYI-23, MtGLYI-1, and MtGLYI-3 showed early up-regulation (6 h) with further enhancement with stress duration (48 h). However, another set of MtGLYI genes-MtGLYI-17, MtGLYI-7/9, MtGLYI-14/15, MtGLYI-5, MtGLYI-18, and MtGLYI-24 showed strong down-regulation at all three-time points (Figure 10A). Rest members-MtGLYI-4, MtGLYI-10, MtGLYI-13, and MtGLYI-19 showed slight alteration as compared to the untreated control sample. On contrary, only one MtGLYII member-MtGLYII-9 showed strong up-regulation, followed by MtGLYII-4 and MtGLYII-8 (Figure 10A). Similarly, MtDJ-1C showed stable up-regulation from 6 to 48 h of salt stress, followed by MtDJ-1B and MtDJ-1A (Figure 10A). Rest of MtGLYII and MtDJ-1 members showed a different degree of down-regulation.
A complex differential expression pattern was observed for all three gene families in response to drought stress at both shoot (Figure 10B) and root (Figure 10C). Most of the genes showed less sensitivity during early days of stress (2 and 3 days). From day-4 and onward, some MtGLYI genes (MtGLYI-11, MtGLYI-8, MtGLYI-6, MtGLYI-16, MtGLYI-21, MtGLYI-4, MtGLYI-18, and MtGLYI-14/15) showed strong up-regulation; while two other members (MtGLYI-17 and MtGLYI-7/9) showed drastic down-regulation. In case of MtGLYII; MtGLYII-12, MtGLYII-6, and MtGLYII-9 showed a similar pattern of up-regulation from 4 to 14 days of drought stress. All these up/down-regulated genes maintain this pattern after 24 h of rewatering, too. Similarly, drought stress causes severe alteration of MtGLYI, MtGLYII, and MtDJ-1 genes at root tissues (Figure 10C). MtGLYI-16, MtGLYI-6, MtGLYI-3, MtGLYI-8, MtGLYI-21, MtGLYI-11, and MtGLYI-14/15 genes showed sharp up-regulation from day-3 to onward at root tissue, and come down to normal level after rewatering (Figure 10C). Likewise, MtGLYII-12, MtGLYII-13, MtGLYII-9, and MtDJ-1A, MtDJ-1D maintained up-regulated transcripts among MtGLYII and MtDJ-1 families from very early to late drought stress. As root has a direct contact with soil water, re-watering brings back the expression level of most glyoxalase genes into their ground level at root tissue (Figure 10C). All these expression data indicated the diverse role of different conventional and unique glyoxalase members in different stress modulation pathways of Medicago plant.
Discussion
Previous studies have shown that glyoxalase proteins play important role in the development and abiotic stress modulation of a plant (Kaur et al., 2014a,c). Genome-wide identification of glyoxalase gene family has been carried out in Arabidopsis, rice, and soybean (Mustafiz et al., 2011; Ghosh and Islam, 2016). The present study of genome-wide analysis of glyoxalase genes in M. truncatula identified a total of 29 GLYI, 14 GLYII, and 5 DJ-1 genes. The number of glyoxalase genes in M. truncatula was found to be the maximum as compared to the previous reports; Arabidopsis (10 GLYI, 5 GLYII, and 6 DJ-1 genes) (Mustafiz et al., 2011; Kwon et al., 2013), rice (11 GLYI, 3 GLYII, and 6 DJ-1 genes) (Mustafiz et al., 2011; Ghosh et al., 2016) and soybean (24 GLYI, 12 GLYII, and 7 DJ-1 genes) (Ghosh and Islam, 2016) (unpublished data). The number of glyoxalase genes in four different plant species could not be correlated with their respective genome size. The genome size of Medicago is 375 Mb (Young et al., 2011); soybean genome size is 1.1 Gb (Schmutz et al., 2010); size of rice genome is 466 Mb (Yu et al., 2002) and that of Arabidopsis is 125 Mb (Bennett et al., 2003). In spite of one third genome size as compared to soybean, another legume plant Medicago has a greater number of conventional glyoxalase genes and an almost similar number of unique glyoxalase genes (Tables 1–3). However, a similar level of both conventional and unique glyoxalase genes is present in Arabidopsis and rice even though rice has four times bigger genome size than Arabidopsis. Plants tend to duplicate its important genes to generate novel isoform or increase number to adopt with different adverse conditions and developmental process (Gu et al., 2003; Chen et al., 2014). In the present analysis, a total of six duplicated glyoxalase gene pairs were observed in Medicago (Table 4). Moreover, orthologs of Medicago glyoxalase genes were found in Arabidopsis, rice, and soybean too. Maximum 11 orthologs pairs were found with Arabidopsis, followed by nine with soybean and five with rice (Table 4).
One of the possible reasons behind the greater number and gene duplication event of plant glyoxalase genes as compared to their animal counterpart is the functional divergence of genes (Gu, 2003). Different types of subfunctionalization or neofunctionalization have arrived due to the functional divergence (Gu, 2003). In the present study, out of 35 MtGLYI proteins several of them did not possess all four conserved functional metal binding site and might be non-functional GLYI enzyme (Figure 5). Glutathione S-transferase activity has been observed for one of the earlier predicted soybean GLYI (accession no. X68819) (Skipsey et al., 2000), while sulfur dioxygenase-like activity observed for Arabidopsis (AtGlx2-5) and rice (OsGLYII-1) GLYII proteins (Kaur et al., 2014b). Moreover, GLYI proteins could be broadly divided into two classes based on their metal activation; Zn2+ and non-Zn2+ (mainly Ni2+/Co2+) (Jain et al., 2016). This metal ion specificity could be easily predicted based on their GLYI domain’s amino acid sequence and length (Suttisansanee et al., 2011; Kaur et al., 2013). Based on these criteria, maximum MtGLYI proteins were predicted be Ni2+/Co2+-activated (Figure 5) similar to previously predicted functional GLYI of soybean (16 out of 20) (Ghosh and Islam, 2016), rice (3 out of 4 active OsGLYIs) (Kaur et al., 2013) and Arabidopsis (2 out of 3 functional AtGLYIs) (Jain et al., 2016). Coexistence of both these metal activated form has been reported recently in model plant A. thaliana (Jain et al., 2016), and earlier in a eubacterial species, Pseudomonas aeruginosa (Sukdeo and Honek, 2007).
Glyoxalase transcripts were found to be altered in response to certain tissue or developmental stages specificity in the previous studies on Arabidopsis, rice, and soybean (Mustafiz et al., 2011; Ghosh and Islam, 2016). Transcript abundance analysis of glyoxalase genes at different developmental stages revealed the constitutive expression pattern of AtGLYI2, AtGLYI3, AtGLYI6, AtGLYI9, AtGLYI11, AtGLYII2, and AtGLYII5 of Arabidopsis; and OsGLYI2, OsGLYI4, OsGLYI6, OsGLYI7, OsGLYI8, OsGLYI11, OsGLYII2, and OsGLYII3 at all the stages (Mustafiz et al., 2011). Similar pattern of constitute expression was observed for GmGLYI7, GmGLYI8, GmGLYI21, and GmGLYII8 transcripts of soybean at the developmental stages (Ghosh and Islam, 2016). However, some other glyoxalase members such as AtGLYI8, OsGLYI3, and OsGLYI10 showed highly tissue specific (seed) expression (Mustafiz et al., 2011). Expression profile of all the conventional glyoxalase (MtGLYI and MtGLYII) and novel glyoxalase (MtDJ-1) genes was analyzed at different developmental stages (Figure 9). Medicago glyoxalase transcripts showed a similar pattern of tissue preferences. Out of all members; MtGLYI-4, MtGLYII-6, MtDJ-1A, and MtDJ-1D showed the constitutive high level of expression at all the analyzed stages. Members of all three gene families (MtGLYI, MtGLYII, and MtDJ-1) showed distinct two clusters according to their transcript abundance in underground, aerial, seed, and pod tissues (Figure 9). One cluster of genes maintained high-level expression in all analyzed tissues, while the other cluster showed a low level of expression. Meanwhile, some other members such as MtGLYI-1, MtGLYI-6, MtGLYI-7, MtGLYI-9, MtGLYI-13, MtGLYII-1, and MtGLYII-13 showed preference toward certain stages/tissue types, indicated their imperative role in the developmental transition/regulation. Genes with low transcript abundance in the developmental stages/tissues might have other stress or cellular or metabolic regulation.
Various abiotic stresses cause increased in vitro accumulation of MG in plant tissues (Kaur et al., 2014c). The role of MG detoxification glyoxalase machinery had been well characterized in literature that provided tolerance against multiple abiotic stresses (Graham and Vance, 2003; Yu et al., 2006). Recently, manipulation of rice glyoxalase pathway has been reported to provide tolerance against three major abiotic stresses such as salinity, drought and heat; and to reduce sheath blight fungus (Rhizoctonia solani) induced damage (Gupta et al., 2017). The possible mechanisms behind these observed tolerance against diverse abiotic and biotic stresses might involve improved MG detoxification, reduced oxidative damage, protected ultra-structure of organelles (chloroplast and mitochondria), and maintained photosynthetic machinery (Gupta et al., 2017). Similar pattern of stress tolerance was observed when the activity of complete glyoxalase pathway was enhanced in transgenic tobacco (Singla-Pareek et al., 2003) and tomato plant (Alvarez Viveros et al., 2013). However, over-expression of either GLYI or GLYII gene also provides tolerance against various abiotic stresses (Singla-Pareek et al., 2008; Ghosh et al., 2014; Mustafiz et al., 2014; Kaur et al., 2017). Thus, the stress tolerance potential of glyoxalase members have been well proved by various studies.
Different glyoxalase members of Arabidopsis and rice such as AtGLYI4, AtGLYI7, OsGLYI6, and OsGLYI11 were previously reported for their highly stress inducible expression pattern (Mustafiz et al., 2011). Similarly, several soybean glyoxalase members such as GmGLYI6, GmGLYI9, GmGLYI20, GmGLYII5, and GmGLYII10 were reported for their highly abiotic stress responsive expression (Ghosh and Islam, 2016). Moreover, few other soybean glyoxalase members such as GmGLYI-6/9, GmGLYI-20, GmGLYII-6, and GmGLYII-10 showed biotic stress specific modulation in response to various pathogenic infection (Ghosh, 2016). Stress specific transcript alteration is a highly complex process and vary depending on the duration and type of stress; and genotype of plant. Expression pattern of all Medicago glyoxalase genes was further analyzed in response to two devastating abiotic stresses-salinity and drought (Figure 10). Different members of Medicago glyoxalase gene families responded differently in response to the duration of stress (salinity and drought) and types of tissue analyzed (drought) (Figure 10). MtGLYI-8, MtGLYI-21, MtGLYII-9, and MtDJ-1C found to be the highly up-regulated member in response to salinity; while MtGLYI-8, MtGLYI-11, MtGLYI-21, MtGLYII-9, MtGLYII-13, and MtDJ-1A are the drought specific up-regulated members (Figure 10). Thus MtGLYI-8, MtGLYI-21, and MtGLYII-9 found to be the highly up-regulated abiotic stress responsive member that could be characterized further. Overall the present study provides basic information about Medicago glyoxalase members that will facilitate the identification of appropriate candidate gene(s) for generating abiotic stress resistant crop plants.
Conclusion
In summary, a comprehensive in silico genome-wide analysis of Medicago glyoxalase gene families (MtGLYI, MtGLYII, and MtDJ-1) were performed. A total of 29 MtGLYI, 14 MtGLYII and 5 unique GLYIII (MtDJ-1) genes were identified from M. truncatula genome sequence that code for 35 GLYI, 27 GLYII, and 6 DJ-1 proteins, respectively. This is the largest identified glyoxalase gene family till date in any organism. All the identified members were further investigated for their classification, evolution, metal dependency, putative enzyme activity, and tissue/developmental-specific expression. Transcript abundance data of Medicago glyoxalase genes showed their major participation in the regulation of Medicago development. Specifically, the present study identified few Medicago glyoxalase members could be explored further their stress tolerance potential. In future, functional analyses of these genes will enable them as an ideal candidate for transgenic applications.
Author Contributions
AG designed and performed the experiments, analyzed the data and wrote the manuscript.
Conflict of Interest Statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
AG acknowledges Shahjalal University of Science and Technology, Sylhet, Bangladesh for providing the logistic support and Department of Biochemistry and Molecular Biology of the same University for providing the laboratory space. The support of Genevestigator team is acknowledged, too.
Abbreviations
- aa
amino acid
- bp
base pair
- d
day
- GLYI
glyoxalase I
- GLYII
glyoxalase II
- GSH
reduced glutathione
- h
hour
- MG
methylglyoxal
Footnotes
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.00836/full#supplementary-material
FILE S1 | Sequences of glyoxalase I proteins from various species used for phylogenetic analysis of MtGLYI proteins.
FILE S2 | Sequences of glyoxalase II proteins from various species used for phylogenetic analysis of MtGLYII proteins.
FILE S3 | Sequences of novel glyoxalase III (DJ-1) proteins from various species used for phylogenetic analysis of MtDJ-1 proteins.
FILE S4 | Details information about the gene expression experiments.
References
- Alvarez Viveros M. F., Inostroza-Blancheteau C., Timmermann T., Gonzalez M., Arce-Johnson P. (2013). Overexpression of GlyI and GlyII genes in transgenic tomato (Solanum lycopersicum Mill.) plants confers salt tolerance by decreasing oxidative stress. Mol. Biol. Rep. 40 3281–3290. 10.1007/s11033-012-2403-4 [DOI] [PubMed] [Google Scholar]
- Benedito V. A., Torres-Jerez I., Murray J. D., Andriankaja A., Allen S., Kakar K., et al. (2008). A gene expression atlas of the model legume Medicago truncatula. Plant J. 55 504–513. 10.1111/j.1365-313X.2008.03519.x [DOI] [PubMed] [Google Scholar]
- Bennett M. D., Leitch I. J., Price H. J., Johnston J. S. (2003). Comparisons with Caenorhabditis (approximately 100 Mb) and Drosophila (approximately 175 Mb) using flow cytometry show genome size in Arabidopsis to be approximately 157 Mb and thus approximately 25% larger than the Arabidopsis genome initiative estimate of approximately 125 Mb. Ann. Bot. 91 547–557. 10.1093/aob/mcg057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Chen Z., Zhao H., Zhao Y., Cheng B., Xiang Y. (2014). Genome-wide analysis of soybean HD-Zip gene family and expression profiling under salinity and drought treatments. PLoS ONE 9:e87156 10.1371/journal.pone.0087156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deponte M., Sturm N., Mittler S., Harner M., Mack H., Becker K. (2007). Allosteric coupling of two different functional active sites in monomeric Plasmodium falciparum glyoxalase I. J. Biol. Chem. 282 28419–28430. 10.1074/jbc.M703271200 [DOI] [PubMed] [Google Scholar]
- Emanuelsson O., Nielsen H., von Heijne G. (1999). ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci. 8 978–984. 10.1110/ps.8.5.978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn R. D., Coggill P., Eberhardt R. Y., Eddy S. R., Mistry J., Mitchell A. L., et al. (2016). The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 44 D279–D285. 10.1093/nar/gkv1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frickel E. M., Jemth P., Widersten M., Mannervik B. (2001). Yeast glyoxalase I is a monomeric enzyme with two active sites. J. Biol. Chem. 276 1845–1849. 10.1074/jbc.M005760200 [DOI] [PubMed] [Google Scholar]
- Ghosh A. (2016). Expression analysis of soybean (Glycine max) glyoxalase genes indicates their biotic stress specific alteration. Res. Rev. J. Bot. Sci. 5 1–5. [Google Scholar]
- Ghosh A., Islam T. (2016). Genome-wide analysis and expression profiling of glyoxalase gene families in soybean (Glycine max) indicate their development and abiotic stress specific response. BMC Plant Biol. 16:87 10.1186/s12870-016-0773-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh A., Kushwaha H. R., Hasan M. R., Pareek A., Sopory S. K., Singla-Pareek S. L. (2016). Presence of unique glyoxalase III proteins in plants indicates the existence of shorter route for methylglyoxal detoxification. Sci. Rep. 6:18358 10.1038/srep18358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh A., Pareek A., Sopory S. K., Singla-Pareek S. L. (2014). A glutathione responsive rice glyoxalase II, OsGLYII-2, functions in salinity adaptation by maintaining better photosynthesis efficiency and anti-oxidant pool. Plant J. 80 93–105. 10.1111/tpj.12621 [DOI] [PubMed] [Google Scholar]
- Graham P. H., Vance C. P. (2003). Legumes: importance and constraints to greater use. Plant Physiol. 131 872–877. 10.1104/pp.017004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu X. (2003). Functional divergence in protein (family) sequence evolution. Genetica 118 133–141. 10.1023/A:1024197424306 [DOI] [PubMed] [Google Scholar]
- Gu Z., Steinmetz L. M., Gu X., Scharfe C., Davis R. W., Li W. H. (2003). Role of duplicate genes in genetic robustness against null mutations. Nature 421 63–66. 10.1038/nature01198 [DOI] [PubMed] [Google Scholar]
- Guo Y. L. (2013). Gene family evolution in green plants with emphasis on the origination and evolution of Arabidopsis thaliana genes. Plant J. 73 941–951. 10.1111/tpj.12089 [DOI] [PubMed] [Google Scholar]
- Gupta B. K., Sahoo K. K., Ghosh A., Tripathi A. K., Anwar K., Das P., et al. (2017). Manipulation of glyoxalase pathway confers tolerance to multiple stresses in rice. Plant Cell Environ. 10.1111/pce.12968 [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Horton P., Park K. J., Obayashi T., Fujita N., Harada H., Adams-Collier C. J., et al. (2007). WoLF PSORT: protein localization predictor. Nucleic Acids Res. 35 W585–W587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hruz T., Laule O., Szabo G., Wessendorp F., Bleuler S., Oertle L., et al. (2008). Genevestigator v3: a reference expression database for the meta-analysis of transcriptomes. Adv. Bioinform. 2008:420747 10.1155/2008/420747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain M., Batth R., Kumari S., Mustafiz A. (2016). Arabidopsis thaliana Contains both Ni2+ and Zn2+ dependent glyoxalase i enzymes and ectopic expression of the latter contributes more towards abiotic stress tolerance in E. coli. PLoS ONE 11:e0159348 10.1371/journal.pone.0159348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juretic N., Hoen D. R., Huynh M. L., Harrison P. M., Bureau T. E. (2005). The evolutionary fate of MULE-mediated duplications of host gene fragments in rice. Genome Res. 15 1292–1297. 10.1101/gr.4064205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur C., Ghosh A., Pareek A., Sopory S. K., Singla-Pareek S. L. (2014a). Glyoxalases and stress tolerance in plants. Biochem. Soc. Trans. 42 485–490. 10.1042/BST20130242 [DOI] [PubMed] [Google Scholar]
- Kaur C., Mustafiz A., Sarkar A. K., Ariyadasa T. U., Singla-Pareek S. L., Sopory S. K. (2014b). Expression of abiotic stress inducible ETHE1-like protein from rice is higher in roots and is regulated by calcium. Physiol. Plant. 152 1–16. 10.1111/ppl.12147 [DOI] [PubMed] [Google Scholar]
- Kaur C., Singla-Pareek S. L., Sopory S. K. (2014c). Glyoxalase and methylglyoxal as biomarkers for plant stress tolerance. Critic. Rev. Plant Sci. 33 429–456. 10.1007/s00709-010-0144-6 [DOI] [Google Scholar]
- Kaur C., Tripathi A. K., Nutan K. K., Sharma S., Ghosh A., Tripathi J. K., et al. (2017). A nuclear-localized rice glyoxalase I enzyme, OsGLYI-8, functions in the detoxification of methylglyoxal in the nucleus. Plant J. 89 565–576. 10.1111/tpj.13407 [DOI] [PubMed] [Google Scholar]
- Kaur C., Vishnoi A., Ariyadasa T. U., Bhattacharya A., Singla-Pareek S. L., Sopory S. K. (2013). Episodes of horizontal gene-transer and gene-fusion led to co-existence of different metal-ion specific glyoxalase I. Sci. Rep. 3:3076 10.1038/srep03076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Tamura K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for Bigger Datasets. Mol. Biol. Evol. 33 1870–1874. 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon K., Choi D., Hyun J., Jung H., Baek K., Park C. (2013). Novel glyoxalases from Arabidopsis thaliana. FEBS J. 280 3328–3339. 10.1111/febs.12321 [DOI] [PubMed] [Google Scholar]
- Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23 2947–2948. 10.1093/bioinformatics/btm404 [DOI] [PubMed] [Google Scholar]
- Lee J. Y., Song J., Kwon K., Jang S., Kim C., Baek K., et al. (2012). Human DJ-1 and its homologs are novel glyoxalases. Hum. Mol. Genet. 21 3215–3225. 10.1093/hmg/dds155 [DOI] [PubMed] [Google Scholar]
- Lee T. H., Tang H., Wang X., Paterson A. H. (2013). PGDD: a database of gene and genome duplication in plants. Nucleic Acids Res. 41 D1152–D1158. 10.1093/nar/gks1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D., Su Z., Dong J., Wang T. (2009). An expression database for roots of the model legume Medicago truncatula under salt stress. BMC Genomics 10:517 10.1186/1471-2164-10-517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W. H., Gojobori T., Nei M. (1981). Pseudogenes as a paradigm of neutral evolution. Nature 292 237–239. 10.1038/292237a0 [DOI] [PubMed] [Google Scholar]
- Lin J., Prahlad J., Wilson M. A. (2012). Conservation of oxidative protein stabilization in an insect homologue of parkinsonism-associated protein DJ-1. Biochemistry 51 3799–3807. 10.1021/bi3003296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marasinghe G. P., Sander I. M., Bennett B., Periyannan G., Yang K. W., Makaroff C. A., et al. (2005). Structural studies on a mitochondrial glyoxalase II. J. Biol. Chem. 280 40668–40675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustafiz A., Ghosh A., Tripathi A. K., Kaur C., Ganguly A. K., Bhavesh N. S., et al. (2014). A unique Ni -dependent and methylglyoxal-inducible rice glyoxalase I possesses a single active site and functions in abiotic stress response. Plant J. 78 951–963. 10.1111/tpj.12521 [DOI] [PubMed] [Google Scholar]
- Mustafiz A., Singh A. K., Pareek A., Sopory S. K., Singla-Pareek S. L. (2011). Genome-wide analysis of rice and Arabidopsis identifies two glyoxalase genes that are highly expressed in abiotic stresses. Funct. Integr. Genomics 11 293–305. 10.1007/s10142-010-0203-2 [DOI] [PubMed] [Google Scholar]
- Rabbani N., Thornalley P. J. (2011). Glyoxalase in diabetes, obesity and related disorders. Semin. Cell Dev. Biol. 22 309–317. 10.1016/j.semcdb.2011.02.015 [DOI] [PubMed] [Google Scholar]
- Saeed A. I., Bhagabati N. K., Braisted J. C., Liang W., Sharov V., Howe E. A., et al. (2006). TM4 microarray software suite. Methods Enzymol. 411 134–193. 10.1016/S0076-6879(06)11009-5 [DOI] [PubMed] [Google Scholar]
- Schmutz J., Cannon S. B., Schlueter J., Ma J., Mitros T., Nelson W., et al. (2010). Genome sequence of the palaeopolyploid soybean. Nature 463 178–183. 10.1038/nature08670 [DOI] [PubMed] [Google Scholar]
- Singla-Pareek S., Reddy M., Sopory S. (2003). Genetic engineering of the glyoxalase pathway in tobacco leads to enhanced salinity tolerance. Proc. Natl. Acad. Sci. U.S.A. 100 14672–14677. 10.1073/pnas.2034667100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singla-Pareek S., Yadav S., Pareek A., Reddy M., Sopory S. (2008). Enhancing salt tolerance in a crop plant by overexpression of glyoxalase II. Trans. Res. 17 171–180. 10.1007/s11248-007-9082-2 [DOI] [PubMed] [Google Scholar]
- Singla-Pareek S. L., Yadav S. K., Pareek A., Reddy M. K., Sopory S. K. (2006). Transgenic tobacco overexpressing glyoxalase pathway enzymes grow and set viable seeds in zinc-spiked soils. Plant Physiol. 140 613–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skipsey M., Andrews C. J., Townson J. K., Jepson I., Edwards R. (2000). Cloning and characterization of glyoxalase I from soybean. Arch. Biochem. Biophys. 374 261–268. 10.1006/abbi.1999.1596 [DOI] [PubMed] [Google Scholar]
- Smil V. (1999). Nitrogen in crop production: an account of global flows. Glob. Biogeochem. Cycles 13 647–662. 10.1029/1999GB900015 [DOI] [Google Scholar]
- Smits M. M., Johnson M. A. (1981). Methylgloxal: enzyme distributions relative to its presence in Douglas-fir needles and absence in Douglas-fir needle callus. Arch. Biochem. Biophys. 208 431–439. 10.1016/0003-9861(81)90529-4 [DOI] [PubMed] [Google Scholar]
- Subedi K. P., Choi D., Kim I., Min B., Park C. (2011). Hsp31 of Escherichia coli K-12 is glyoxalase III. Mol. Microbiol. 81 926–936. 10.1111/j.1365-2958.2011.07736.x [DOI] [PubMed] [Google Scholar]
- Sukdeo N., Honek J. F. (2007). Pseudomonas aeruginosa contains multiple glyoxalase I-encoding genes from both metal activation classes. Biochim. Biophys. Acta 1774 756–763. 10.1016/j.bbapap.2007.04.005 [DOI] [PubMed] [Google Scholar]
- Suttisansanee U., Lau K., Lagishetty S., Rao K. N., Swaminathan S., Sauder J. M., et al. (2011). Structural variation in bacterial glyoxalase I enzymes: investigation of the metalloenzyme glyoxalase I from Clostridium acetobutylicum. J. Biol. Chem. 286 38367–38374. 10.1074/jbc.M111.251603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornalley P. J. (1990). The glyoxalase system: new developments towards functional characterization of a metabolic pathway fundamental to biological life. Biochem. J. 269 1–11. 10.1042/bj2690001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young N. D., Debelle F., Oldroyd G. E., Geurts R., Cannon S. B., Udvardi M. K., et al. (2011). The Medicago genome provides insight into the evolution of rhizobial symbioses. Nature 480 520–524. 10.1038/nature10625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C. S., Chen Y. C., Lu C. H., Hwang J. K. (2006). Prediction of protein subcellular localization. Proteins 64 643–651. 10.1002/prot.21018 [DOI] [PubMed] [Google Scholar]
- Yu J., Hu S., Wang J., Wong G. K., Li S., Liu B., et al. (2002). A draft sequence of the rice genome (Oryza sativa L. ssp. indica). Science 296 79–92. 10.1126/science.1068037 [DOI] [PubMed] [Google Scholar]
- Zhang C., Zhang H., Zhao Y., Jiang H., Zhu S., Cheng B., et al. (2013). Genome-wide analysis of the CCCH zinc finger gene family in Medicago truncatula. Plant Cell Rep. 32 1543–1555. 10.1007/s00299-013-1466-6 [DOI] [PubMed] [Google Scholar]
- Zhang J. Y., Cruz D. E. C. M. H., Torres-Jerez I., Kang Y., Allen S. N., Huhman D. V., et al. (2014). Global reprogramming of transcription and metabolism in Medicago truncatula during progressive drought and after rewatering. Plant Cell Environ. 37 2553–2576. 10.1111/pce.12328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann P., Laule O., Schmitz J., Hruz T., Bleuler S., Gruissem W. (2008). Genevestigator transcriptome meta-analysis and biomarker search using rice and barley gene expression databases. Mol. Plant 1 851–857. 10.1093/mp/ssn048 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
FILE S1 | Sequences of glyoxalase I proteins from various species used for phylogenetic analysis of MtGLYI proteins.
FILE S2 | Sequences of glyoxalase II proteins from various species used for phylogenetic analysis of MtGLYII proteins.
FILE S3 | Sequences of novel glyoxalase III (DJ-1) proteins from various species used for phylogenetic analysis of MtDJ-1 proteins.
FILE S4 | Details information about the gene expression experiments.