Abstract
The phage-shock protein (Psp) response is an extracytoplasmic response system that is vital for maintenance of the cytoplasmic membrane when the cell encounters stressful conditions. The paradigm of the Psp response has been established in Escherichia coli. The response has been shown to be important for survival during the stationary phase, maintenance of the proton motive force across membranes and implicated in virulence. In this study, we identified a putative PspA homologue in Burkholderia pseudomallei, annotated as BPSL2105. Similar to the induction of PspA in E. coli, the expression of B. pseudomallei BPSL2105 was induced by heat shock. Deletion of BPSL2105 resulted in a survival defect in the late stationary phase coincident with dramatic changes in the pH of the culture medium. The B. pseudomallei BPSL2105 deletion mutant also displayed reduced survival in macrophage infection – the first indication that the Psp response plays a role during intracellular pathogenesis in this species. The purified protein formed large oligomeric structures similar to those observed for the PspA protein of E. coli, and PspA homologues in Bacillus, cyanobacteria and higher plants, providing further evidence to support the identification of BPSL2105 as a PspA-like protein in B. pseudomallei.
Introduction
Burkholderia pseudomallei is a Gram-negative bacterium and the causative agent of the disease melioidosis (White, 2003). Melioidosis is endemic to regions of Southeast Asia and Northern Australia where the bacterium is widely distributed in the soil (Currie et al., 2008; Dance, 2000). It is a common cause of community-acquired bacteraemic pneumonia (Chaowagul et al., 1989; Currie et al., 2010), but the disease can manifest in different forms depending on the route of exposure (Cheng & Currie, 2005). Transmission via the aerosol route is thought to be a significant risk and, consequently, B. pseudomallei has been classified as a Category B biological threat agent by the Centers for Disease Control and Prevention in the USA.
B. pseudomallei is an extremely persistent bacterium, able to survive in a diverse range of environments. Studies have shown that it is able to withstand such conditions as low pH, high salt concentrations and high temperatures, and has been recovered from distilled water several years after initial inoculation (reviewed by Inglis & Sagripanti, 2006). It has a large genome, consisting of two chromosomes which encode many genes associated with adaptation to different environmental conditions and resistance to a range of niche-related stresses (Holden et al., 2004). This adaptability has allowed the primarily environmental bacterium to colonize a further niche – the mammalian host. As an intracellular pathogen, B. pseudomallei can persist inside both phagocytic and non-phagocytic cells, where it must resist a novel panoply of stresses (Allwood et al., 2011; Jones et al., 1996). It is thought that, in cases of latency, bacteria reside intracellularly in a non-replicating form (Gan, 2005), only causing symptoms of disease later in life when the host immune system has been compromised (Ngauy et al., 2005).
The phage-shock protein (Psp) response is an extracytoplasmic stress response in bacteria that functions to maintain cell membrane integrity during stress (Darwin, 2005). It was initially observed in Escherichia coli, with first reports describing a protein produced at a high concentration during filamentous phage infection, subsequently termed PspA (Brissette et al., 1990). The Psp response is induced by a number of different stresses, many of which have a detrimental effect on the proton motive force (PMF) and therefore the Psp response is thought to have an important physiological role in maintaining the PMF across the cytoplasmic membrane (Kleerebezem et al., 1996). The Psp response is thought to maintain the integrity of the cytoplasmic membrane in times of stress, and has been shown to be important in survival and virulence-related processes in several species of bacteria (Darwin, 2013; Darwin & Miller, 2001; Joly et al., 2010). Its importance for survival during stationary phase growth has been demonstrated previously in E. coli (Weiner & Model, 1994).
The Psp response has been most studied in E. coli and Yersinia enterocolitica, where it has been shown to involve the products of the pspABC operon, regulated by PspF, the activity of which is in turn regulated by PspA (Dworkin et al., 2000; Elderkin et al., 2002; Jovanovic et al., 1996; 1999). When the Psp response is induced, the concentration of PspA increases, whereupon it is recruited to the cytoplasmic membrane (Yamaguchi et al., 2010). In vitro, PspA is able to form large oligomeric rings (Hankamer et al., 2004), which are able to bind to membrane phospholipids and reduce proton leakage through the damaged membrane (Kobayashi et al., 2007). Although the Psp systems of E. coli and Y. enterocolitica are considered the paradigm, there are a number of bacteria which possess isolated PspA homologues but lack other members of the psp operon. For example, Streptomyces lividans and Bacillus subtilis both possess a PspA homologue which is upregulated by known Psp-inducing conditions (Vrancken et al., 2008; Wolf et al., 2010).
In this study, we identified a putative PspA homologue in B. pseudomallei which responds to similar stresses as the Psp response in E. coli. We demonstrated its importance for survival during stationary phase growth and for intracellular survival in a macrophage cell line. In addition, the B. pseudomallei PspA-like protein was expressed and analysed by transmission electron microscopy (TEM), revealing the presence of a higher-order PspA-like protein species which assembled into ring-shaped oligomers.
Methods
Bacterial strains, culture conditions and mutant construction
E. coli and B. pseudomallei were cultured in Luria–Bertani (LB) broth at 37 °C with agitation, unless otherwise stated. Bacterial strains used are listed in Table 1. Antibiotics (chloramphenicol and kanamycin) were used at 50 μg ml− 1 final concentration.
Table 1. Strains and plasmids used.
| Strain or plasmid | Characteristics | Reference or source |
|---|---|---|
| E. coli | ||
| BL21(DE3) | F− ompT hsdSB(rB− , mB− ) gal dcm BL21(DE3) | Invitrogen |
| DH5α | F− ϕ80lacZΔM15 Δ(lacZYA–argF) U169 recA1 endA1 hsdR17 (rK–, mK+) phoA supE44 λ– thi-1 gyrA96 relA1 | E. coli genetic stock centre |
| S17-λ pir | λ pir recA thi pro hsdR − RP4-2(Km : : Tn7 Tc : : Mu) TprSmr | Laboratory collection |
| B. pseudomallei | ||
| K96243 | Clinical isolate from Thailand, sequenced strain | Holden et al. (2004) |
| ΔpspA | K96243 derivative; unmarked deletion ΔBPSL2105 | This study |
| ΔpspA/pBHR4.pspA | K96243 derivative; unmarked deletion ΔBPSL2105; pBHR4 : : BPSL2105, Cmr | This study |
| Plasmids | ||
| pCR-Blunt II-TOPO | Cloning vector for blunt-end PCR products, Kmr | Invitrogen |
| pDM4 | pNQ705 derivative, oriR6K, mobRP4, sacBR, Cmr | Milton et al. (1996) |
| pDM4.ΔpspA | pDM4 derivative with BPSL2105 flanking regions for mutagenesis, Cmr | This study |
| pBHR4-groS-RFP | pBHR1 derivative, turboFP635, PgroES, rrnB, Cmr | Wand et al. (2011) |
| pBHR4.pspA | pBHR4-groS-RFP derivative with PBPSL2105, BPSL2105, Cmr | This study |
| pET28a | E. coli expression vector, T7 lac promoter, Kmr | Novagen |
| pET28-His6-pspA | pET28a derivative, N-terminal His6-tagged BPSL2105, Kmr | This study |
Primers used for DNA amplification are listed in Table 2. The B. pseudomallei unmarked deletion mutant was made by homologous recombination using the pDM4 suicide vector by the method outlined in Logue et al. (2009). The pDM4.ΔpspA plasmid was constructed by ligation of BglII-linearized pDM4 with the truncated BPSL2105. The truncated gene was produced by amplifying upstream and downstream flanking regions of BPSL2105 from B. pseudomallei K96243 genomic DNA using primers BPSL2105 LF F/BPSL2105 LF R (upstream) and BPSL2105 RF F/BPSL2105 RF R (downstream). NdeI and BglII restriction sites were incorporated into either end of the flanks to aid manipulation. The resulting flanks were cloned into pCR-Blunt II-TOPO (Invitrogen) followed by digestion with NdeI and BglII for ligation with the pDM4 suicide vector. To construct B. pseudomalleiΔpspA, the pDM4.ΔpspA plasmid was transformed into E. coli S17-λ pir cells before conjugation with B. pseudomallei K96243. The merodiploid strain was screened for chloramphenicol resistance before sucrose selection was carried out to select for the second recombination event. Unmarked BPSL2105 deletion mutants were verified by PCR and Southern blot.
Table 2. Primers used.
| Primer | Sequence (5′ → 3′) | Restriction site |
|---|---|---|
| BPSL2105 LF F | AGATCTTGAACGCGTGCATGGAATCG | BglII |
| BPSL2105 LF R | CATATGTTTGATCGTGCGCGAAATAG | NdeI |
| BPSL2105 RF F | CATATGGACCGCCTCGAAGCGCTGAA | NdeI |
| BPSL2105 RF R | AGATCTCGAGCATGCCGCCCGAGGTC | BglII |
| BPSL2105 prom F | CGATCGGCGCTGAACGCGTGCATGGA | PvuI |
| BPSL2105 comp R | GGATCCTTACTGCGCCGGCGTGTTCA | BamHI |
| RT PspA F | CGCGCACGATCAAAGGTCTG | – |
| RT PspA R | GCCGCTGTTCGTATCGGCTG | – |
| 16S rRNA F | GATGACGGTACCGGAAGAATAAGC | – |
| 16S rRNA R | CCATGTCAAGGGTAGGTAAGGTTT | – |
| PspA F | GCCGCCGGATCCATGTCGCTTTTCGACTCTATTTC | BamHI |
| PspA R | CGGCGGGAGCTCTTACTGCGCCGGCGTGTTCAG | SacI |
| BPSL2105-6 F | GTCGGTCGACAAGCTCAAAG | – |
| BPSL2105-6 R | GGATCGGTGAAGCGCAGTTG | – |
Restriction sites are underlined.
To construct pBHR4.pspA, the BPSL2105 gene and promoter were amplified from B. pseudomallei genomic DNA using primers BPSL2105 prom F/BPSL2105 comp R. This introduced PvuI and BamHI restriction sites onto either end of the fragment. The PCR product was cloned into pCR-Blunt II-TOPO before digestion and ligation with PvuI/BamHI-digested pBHR4-groS-RFP (Wand et al., 2011). The pBHR4.pspA plasmid was transferred by conjugation into B. pseudomalleiΔpspA. The complemented strain B. pseudomalleiΔpspA/pBHR4.pspA was then selected for by plating onto LB agar containing 50 μg chloramphenicol ml− 1 and the presence of the plasmid confirmed by colony PCR.
To investigate bacterial survival, overnight cultures were diluted in 100 ml LB broth and grown to the stationary phase by continuous incubation at 37 °C with agitation. The cultures were titrated for viable cells daily by plating on LB agar with or without appropriate antibiotics. pH was measured using a Hanna Piccolo Plus pH meter (Sigma).
RNA isolation and reverse transcription (RT)-PCR
Overnight cultures of B. pseudomallei K96243 were diluted to OD590 0.1 and grown at 37 °C for 6 h until the mid-exponential phase was reached. Cultures were divided into 1 ml aliquots and incubated at 37 °C and 50 °C. RNA samples were collected at selected time points for up to 30 min by addition of 2 ml RNAprotect (Qiagen). RNA was recovered using a RNeasy Mini kit (Qiagen) as instructed by the manufacturer. This resulted in RNA at a concentration of 100–400 ng μl− 1, quantified using a NanoDrop 1000 spectrophotometer. Residual DNA was removed by treating the RNA with TURBO DNA-free DNase (Ambion). During this step the RNA was standardized to a concentration of 125 ng μl− 1. Following this, the samples were reverse transcribed using Enhanced Avian Reverse Transcriptase (Sigma) according to the manufacturer's instructions. The resulting cDNA was amplified by PCR using Herculase II fusion DNA polymerase (Agilent Technologies) in a standard PCR. For each PCR, the appropriate controls with water and RNA in the absence of reverse transcriptase were included to ensure that amplifications were of cDNA and not contaminating genomic DNA. Transcripts of BPSL2105 were amplified using RT PspA F/RT PspA R primers, and 16S rRNA was amplified as a positive control using 16S rRNA F/16S rRNA R primers. PCR was also performed using primers (BPSL2105-6 F/BPSL2105-6 R) complementary to sequences overlapping both BPSL2105 and BPSL2106 to determine whether the genes were co-transcribed.
Intracellular survival assays
J774A.1 cells (ECACC) were seeded in a 24-well plate in Dulbecco's modified Eagle's medium (DMEM) supplemented with 1 % l-glutamine and 10 % FCS at a concentration of 4 × 105 cells ml− 1. The cells were incubated at 37 °C with 5 % CO2 overnight until they had reached a density of 1 × 106 cells ml− 1. A stationary phase culture was diluted in Leibovitz L-15 medium supplemented with 10 % FCS to OD590 0.35–0.4, equivalent to ∼1 × 108 c.f.u. ml− 1. This was serially diluted to 1 × 107 c.f.u. ml− 1. The DMEM was removed from the J774A.1 cells and replaced with L-15 media containing 1 × 107 c.f.u. ml− 1 bacteria. The cells were incubated at 37 °C for 30 min, and then the media was removed and the cells washed three times with PBS. To kill any extracellular bacteria, 1 ml L-15 supplemented with 1 mg kanamycin ml− 1 was added to each well and incubated at 37 °C. After 1 h, the antibiotic medium was removed and replaced with 1 ml L-15 supplemented with 250 μg kanamycin ml− 1, and incubated at 37 °C for 24 h. At selected time points the antibiotic medium was removed and the cells lysed by addition of 1 ml water. The lysate was serially diluted and cultured on LB agar to enumerate viable bacteria.
MIC determination
MIC determinations were carried out according to the method of Lambert & Pearson (2000) with some modifications. Briefly, stock solutions of H2O2, HCl, NaOH, NaCl, lysozyme and deferoxamine were prepared in LB broth at a concentration of 256 μg ml− 1. A 100 μl aliquot of the stock solution was added to 100 μl LB broth in the first column of a 96-well plate and a twofold dilution carried out across the plate. The bacterial inoculum was prepared by growing a stationary phase culture of B. pseudomallei, adjusting to OD590 0.35–0.40 in LB broth and then serially diluting to a concentration of 1 × 106 c.f.u. ml− 1. A 100 μl aliquot of this culture was added to each compound dilution that had been dispensed into the test wells. This provided a final compound dilution range of 64–0.03 μg ml− 1. NaOH was tested at a concentration of 0.5–1024 μg ml− 1. The 96-well plates were incubated at 37 °C for 18 h before the optical density was recorded for each well.
Expression and purification of a His6-tagged PspA-like protein
BPSL2105 was amplified by PCR using the primers PspA F/PspA R and cloned into the corresponding sites in the pET28a vector (Novagen). The resulting pET28-His6-pspA plasmid encoded N-terminal His6-tagged BPSL2105 (His6-PspA), verified by DNA sequencing (Eurofins MWG), with a molecular mass of 27.876 kDa. His6-PspA was expressed in E. coli BL21(DE3) as described previously (Elderkin et al., 2002), with or without 1.1 % CHAPS. Briefly, cultures were grown in LB broth to OD600 ∼0.6, following which protein expression was induced by addition of 1 mM IPTG. The induced cultures were incubated overnight at 18 °C before harvesting the cells by centrifugation. To extract the protein, the cells were resuspended in lysis buffer (100 mM Tris/HCl, pH 7.5, 50 mM NaCl, 75 mM NaSCN) and sonicated. The insoluble and soluble fractions were separated by centrifugation and the soluble fraction directly purified by Ni2+-affinity chromatography, according to the manufacturer's instructions. Size exclusion chromatography was performed using a Superdex 200 gel filtration column (GE Healthcare) calibrated with the following molecular mass standards: blue dextran (2000 kDa), β-amylase (200 kDa), alcohol dehydrogenase (150 kDa), albumin (66 kDa), carbonic anhydrase (29 kDa) and cytochrome c (12.4 kDa). Gel filtration buffer (20 mM Tris/HCl, pH 7.5, 200 mM NaCl, 75 mM NaSCN) was passed through the column at a flow rate of 1 ml min− 1. The 1 ml fractions were collected between 35 and 55 ml. The yield after gel filtration was 7.6 mg when purified from a 500 ml culture. For purification under denaturing conditions, the cells were resuspended in denaturing lysis buffer (20 mM Tris/HCl, pH 7.5, 500 mM NaCl, 1 mM β-mercaptoethanol and 6 M guanidine hydrochloride). Purification was carried out by Ni2+-affinity chromatography, according to the manufacturer's instructions, using a wash and elution buffer containing 20 mM Tris/HCl, pH 7.5, 500 mM NaCl, 1 mM β-mercaptoethanol, 8 M urea and 10/500 mM imidazole. Protein mass was confirmed by MS.
Western blotting
Proteins were separated by SDS-PAGE on a 12 % (w/v) acrylamide gel and transferred onto nitrocellulose transfer membranes (Protran; Whatman). After blocking for 1 h with 5 % milk powder in PBS and 0.05 % Tween, the blots were incubated with anti-His (27-4710-01; GE Healthcare) at 1 : 1000 dilution, followed by incubation with horseradish peroxidase-conjugated anti-mouse IgG (NA931; GE Healthcare) at 1 : 5000 dilution. Secondary antibody was detected using an enhanced chemiluminescence reagent (Western C; Bio-Rad).
TEM
PspA was negatively stained with 2 % (w/v) uranyl acetate on 50 % carbonyl Formvar grids and examined on a Hitachi H7000 transmission electron microscope operated at 80 kV with magnification ranging from × 50 000 to × 200 000. A protein-free control was used, whereby a negatively stained sample in the absence of any purified recombinant protein was imaged, to verify that the images observed were not due to staining or sample handling during image acquisition.
Statistical analysis
All data are reported as mean ± sem. Results were statistically analysed using a two-way ANOVA and Bonferroni post-tests. P < 0.05 was considered statistically significant.
Results
BPSL2105 from B. pseudomallei is a putative PspA homologue
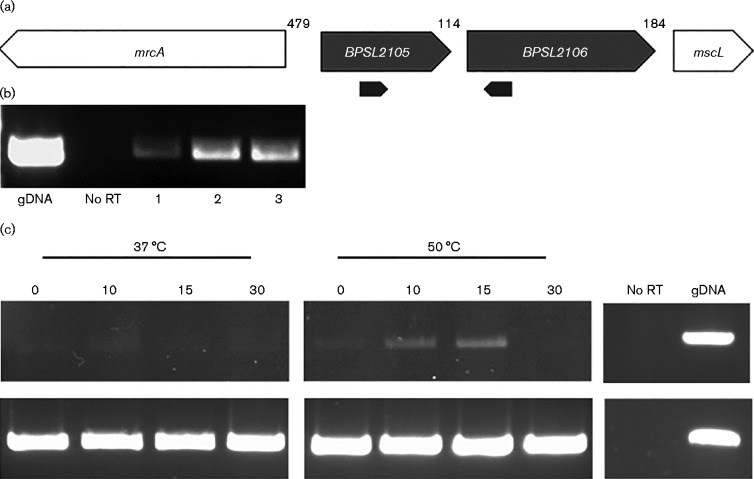
Several putative PspA homologues (annotated as PspA family proteins) from a number of Burkholderia species were compared with the B. pseudomallei K96243 sequenced genome. Although it is currently annotated as a hypothetical protein, BPSL2105 in B. pseudomallei K96243 has 99–100 % identity to many of these proteins, including proteins from both the less pathogenic Burkholderia thailandensis and the close relative Burkholderia mallei (Fig. S1, available in the online Supplementary Material; Cole et al., 2008; Gautier et al., 2008; Jovanovic et al., 2014a). The amino acid sequence of BPSL2105 was further compared with known PspAs in species such as E. coli and Y. enterocolitica using blast (http://blast.ncbi.nlm.nih.gov/Blast.cgi) (Fig. S1). This comparison showed low identity compared with the Enterobacteriaceae, e.g. BPSL2105 had 22 % identity to Y. enterocolitica PspA and 21 % overall identity to E. coli. Similarly, less well characterized PspA homologues in B. subtilis and S. lividans showed 28 and 31 % identity, respectively. However, PspAs in the Enterobacteriaceae are known to be coiled-coil proteins (Dworkin et al., 2000) and BPSL2105 is predicted to contain several α-helices as well as an amphipathic helix sequence in the initial 20 amino acids, comparable to PspA in E. coli (Jovanovic et al., 2014a) (see also Fig. S1). Despite this, no other members of the Psp regulon were found to be present in B. pseudomallei K96243. Instead, BPSL2105 is predicted to form an operon with BPSL2106, which is annotated as a putative membrane protein (Figs. 1a and S1). This gene order is observed in other close relatives of B. pseudomallei that possess PspA family proteins. RT-PCR was performed on RNA isolated from B. pseudomallei, verifying that BPSL2105 and BPSL2106 were co-transcribed (Fig. 1b).
Fig. 1.
BPSL2105 organization and expression. (a) Predicted operon of B. pseudomallei BPSL2105/2106. (b) Demonstration of co-transcription of BPSL2105 and BPSL2106 by RT-PCR. RNA samples were collected from three separate cultures (1, 2 and 3). Primers used during RT-PCR are represented by the black arrows. (c) Expression of BPSL2105 during extreme heat shock. RNA samples were collected at 0, 10, 15 and 30 min time points, purified and residual DNA removed. Following this, the samples were amplified using RT-PCR with primers amplifying a region either from BPSL2105 or from 16S rRNA genes. Controls were carried out using RNA in the absence of reverse transcriptase. gDNA, genomic DNA.
In order to determine whether BPSL2105 possessed PspA-like features, RT-PCR was carried out on mRNA isolated from B. pseudomallei grown under conditions known to cause upregulation of PspA. Previous studies have shown that heat shock at 50 °C causes a transient increase in the concentration of PspA in E. coli (Brissette et al., 1990). To investigate the effect of temperature on expression of BPSL2105 in B. pseudomallei, RNA was collected from cultures shocked at 50 °C and RT-PCR performed using primers RT PspA F/RT PspA R (Table 1) to amplify BPSL2105 mRNA. The PCR product was visualized using gel electrophoresis to provide a semiquantitative result. The results showed a transient increase in BPSL2105 expression at 50 °C compared with the incubation at 37 °C, with maximal induction at 10–15 min (Fig. 1c).
Another known inducer of E. coli PspA is hyperosmotic shock (Brissette et al., 1990). Whereas expression of PspA during heat shock is independent of the PspBC sensors, not present in B. pseudomallei, expression of PspA under high-salt conditions is partially dependent on these proteins (Weiner et al., 1991). RT-PCR was used to measure the relative level of expression of BPSL2105 in cultures exposed to 0.3 M NaCl compared with expression in unshocked samples. However, the results showed no increased expression of BPSL2105 compared with controls (data not shown).
The data from these studies provided preliminary evidence that BPSL2105 encodes a PspA-like protein in B. pseudomallei. BPSL2105 has a secondary structure comparable to the α-helical structure of E. coli PspA, which is essential for its regulatory and effector functions (Elderkin et al., 2005; Joly et al., 2009), and a key amphipathic helix, thought to play an important role in inner membrane binding and signal transduction in E. coli (Jovanovic et al., 2014a). In addition, BPSL2105 expression is increased in response to heat shock at 50 °C, a known Psp-inducing condition, further indicating that BPSL2105 is a PspA-like protein in this species.
Deletion of BPSL2105 in B. pseudomallei results in a growth defect in the late stationary phase
A deletion mutant was constructed in BPSL2105 using the pDM4 suicide vector and sacB counter-selection (Logue et al., 2009). This mutant, B. pseudomalleiΔpspA, was evaluated for phenotypes known to induce PspA in other bacteria. The stationary phase of growth has been shown to be an important inducer of the Psp response in E. coli where PspA is rapidly accumulated in the cell after one day in the stationary phase (Weiner & Model, 1994). Further to this, growth of an E. coli ΔpspABC strain shows a sharp decline in viability after day 9 at the stationary phase compared with the WT.
B. pseudomalleiΔpspA was grown in LB broth at 37 °C with aeration for the duration of the experiment. Both B. pseudomallei WT and ΔpspA mutant maintained a density of ∼5 × 109 c.f.u. ml− 1 during the first 6 days (144 h). From day 7, B. pseudomalleiΔpspA began to decline in viability compared with the WT (Fig. 2a). The WT phenotype was restored in ΔpspA complemented with the plasmid expressing BPSL2105. The pH of the cultures was monitored daily and was found to have increased from 7.5 to ∼8.5 over 192 h (8 days) in the B. pseudomalleiΔpspA culture, whereas the WT and complemented strain were able to maintain the pH between 6.5 and 7 over the course of the experiment (Fig. 2b).
Fig. 2.
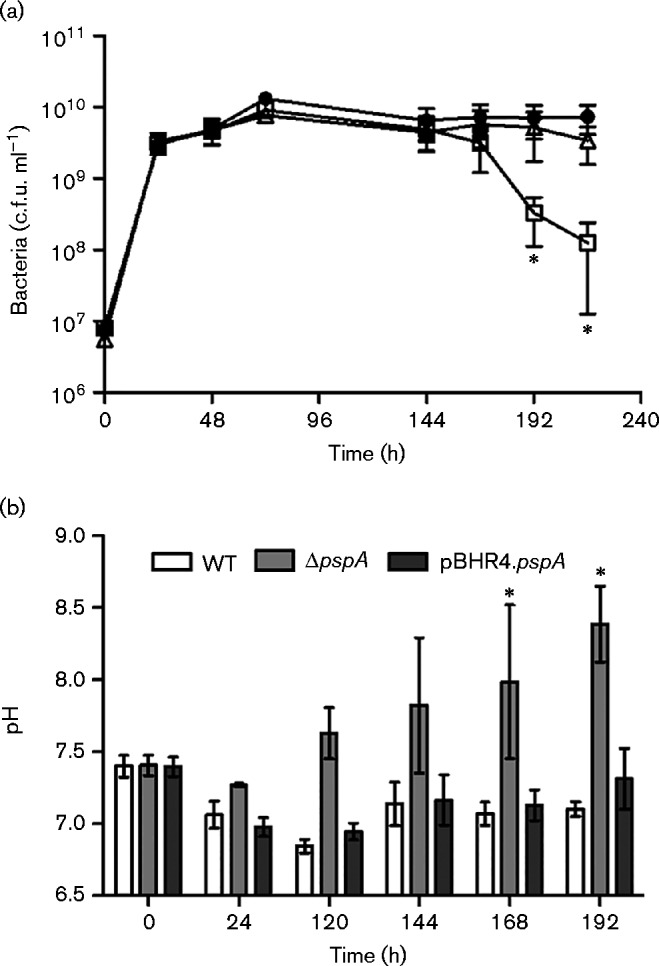
BPSL2105 contributes to late stationary phase survival. (a) Growth of B. pseudomallei over 216 h in 100 ml LB broth cultures. Overnight cultures of WT B. pseudomallei K96243 (•), ΔpspA (▪) and ΔpspA complemented with pBHR4.pspA (Δ) were diluted in 100 ml LB broth to OD590 0.1 and grown to the stationary phase by continuous incubation at 37 °C with agitation. Survival was measured by plating on LB agar with or without appropriate antibiotics. (b) pH of B. pseudomallei cultures measured over 192 h. Values are the mean ± sem from three independent experiments; *P < 0.05.
B. pseudomalleiΔpspA is more susceptible to macrophage killing during the late stationary phase
The Psp response is implicated in intracellular pathogenesis as psp genes are upregulated during macrophage infection in bacteria such as Salmonella enterica, Shigella flexneri and M. tuberculosis (Datta et al., 2015, Eriksson et al., 2003; Lucchini et al., 2005). B. pseudomallei is an intracellular pathogen and the ability to multiply within macrophages is essential for virulence (Pilatz et al., 2006; Stevens et al., 2003, 2004). In order to investigate the ability of B. pseudomalleiΔpspA to survive intracellularly, an infection assay was performed using J774A.1 murine macrophages. B. pseudomallei strains were grown for 6 days in 100 ml LB broth and used to infect a J774A.1 macrophage cell line at m.o.i. 10. The number of viable intracellular bacteria was measured by lysing the macrophages with water and culturing the bacteria. The WT bacteria were able to replicate intracellularly, but B. pseudomalleiΔpspA showed a reduction in the number of intracellular bacteria at 24 h (Fig. 3). Complementation of B. pseudomalleiΔpspA with the plasmid expressing BPSL2105 restored intracellular growth to WT levels. This result was divergent to that seen when macrophages were infected with exponential phase B. pseudomalleiΔpspA, where there was no difference in intracellular survival compared with the WT strains at 24 h post-infection (data not shown).
Fig. 3.
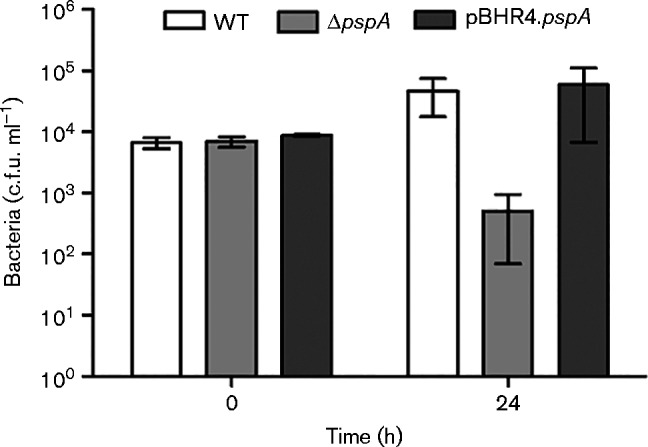
Intracellular survival of stationary phase B. pseudomallei in J774A.1 macrophages. Macrophage cells were infected with 1 × 107 c.f.u. ml− 1 bacteria for 30 min, followed by incubation with 1 mg kanamycin ml− 1 for 1 h to kill any extracellular bacteria. The cells were maintained in the presence of 250 μg kanamycin ml− 1 and periodically lysed in order to enumerate the number of intracellular bacteria. Values are the mean ± sem from at least three independent experiments; *P < 0.05.
B. pseudomallei-infected macrophages are prone to lysis by 24 h from high intracellular numbers of bacteria. This causes the internalized bacteria to be released into the kanamycin-containing media, killing any extracellular bacteria. Lactate dehydrogenase release was measured to verify that the reduction in intracellular bacteria at 24 h was not caused by increased cytotoxicity of the mutant. The lactate dehydrogenase levels in the media were similar for cells infected with B. pseudomallei WT, ΔpspA and the complemented strain (data not shown).
Macrophages infected with B. pseudomalleiΔpspA showed a decrease in intracellular bacteria to those of the original inoculum, indicating killing rather than just growth inhibition. Macrophages have a variety of mechanisms for killing phagocytosed bacteria. In an attempt to determine which of these mechanisms the mutant was more susceptible to, the bacteria were exposed to a range of conditions. Bacteria were grown to the stationary phase by incubation in 100 ml LB broth at 37 °C for 6 days. The level of growth of the stationary phase bacteria was measured under a range of conditions, including H2O2, NaCl, HCl, NaOH, lysozyme and an iron chelator, deferoxamine. Overall there was no difference between the susceptibility of either WT or the ΔpspA mutant to osmotic stress, pH, low iron or lysozyme. Similarly, both strains were highly susceptible to oxidative stress, with a MIC of 0.125–0.25 μg H2O2 ml− 1 (Fig. S2).
B. pseudomallei BPSL2105 assembles into multimeric complexes
We have recently demonstrated that E. coli PspA assembles into higher-order, multimeric complexes (Male et al., 2014). We therefore sought to establish whether B. pseudomallei BPSL2105 forms similar structures. His6-tagged BPSL2105 (His6-PspA) was recombinantly expressed and purified by Ni2+-affinity chromatography before being imaged by TEM. The purified His6-PspA protein, purified in the absence of the detergent CHAPS, was analysed by gel filtration, resulting in two peaks that eluted at 49.5 and 83.0–109.0 ml (Fig. 4a). The wider peak between 83.0 and 109.0 ml may correspond to dimeric (81.9 ml) and monomeric (90.3 ml) PspA species, estimated by molecular mass standards (Fig. S3) and studying the elution of a denatured form of the protein at 88.1 ml (Fig. 4a).
Fig. 4.
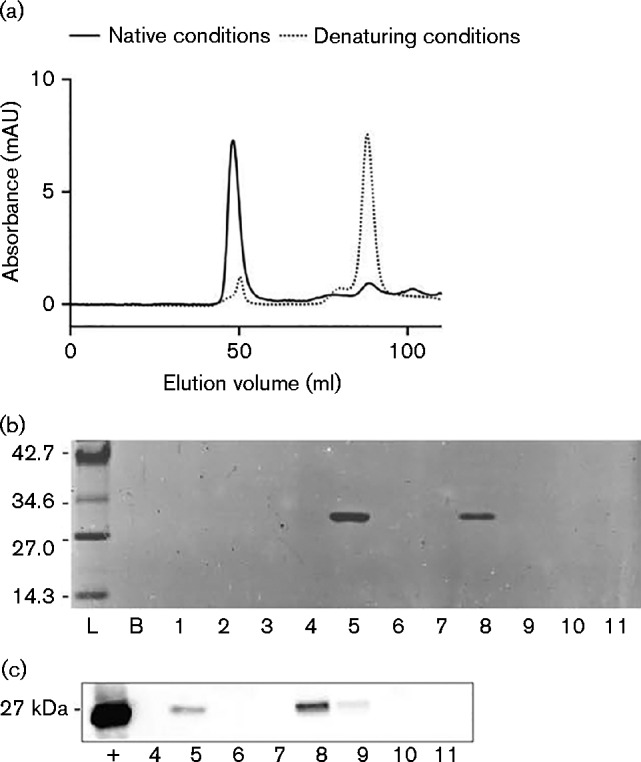
Determining the oligomeric state of purified His6-PspA protein. (a) Size exclusion chromatography trace of His6-PspA showing a void volume at 49.5 ml containing the aggregated and higher-order PspA protein species, and a collection of peaks between 83.0 and 109.0 ml containing putative dimeric and monomeric His6-PspA species. Included is an overlay of His6-PspA protein purified under denaturing conditions. AU, absorbance units. (b) SDS-PAGE of His6-PspA purified by gel filtration. Lanes represent pooled fractions from 0–10 (lane 1), 10–20 (lane 2), 20–30 (lane 3), 30–40 (lane 4), 40–50 (lane 5), 50–65 (lane 6), 65–80 (lane 7), 80–90 (lane 8), 90–100 (lane 9), 100–110 (lane 10) and 110–120 ml (lane 11). Values in kDa. (c) Western blot confirming the presence of His6-PspA protein. Fractions are the same as those outlined in (b);+, His6-PspA positive control.
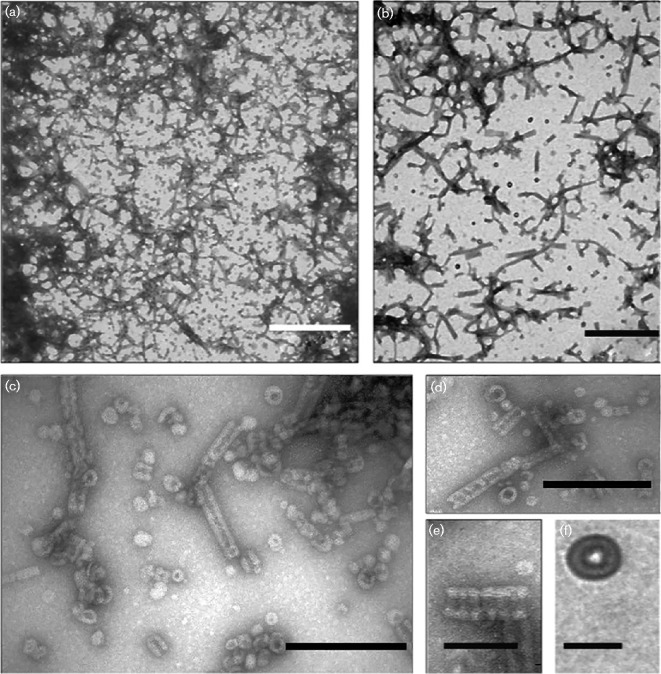
The presence of His6-PspA in both peaks and its absence elsewhere was confirmed by SDS-PAGE and Western blot (Fig. 4b, c). The peak at 49.5 ml (void volume fraction) showed the presence of His6-PspA with expected molecular mass (Fig. S3) and corresponding to a species with a molecular mass >200 kDa. In other bacteria, the high-molecular-mass species of PspA and its homologues have been shown to include the putative 36-meric ring and other higher-order PspA complexes (Fuhrmann et al., 2009; Hankamer et al. 2004). To investigate the higher-order species formed by B. pseudomallei PspA, the His6-PspA eluted at 49.5 ml was negatively stained with uranyl acetate and imaged by TEM. Three different species were observed: ring-shaped complexes, rod-shaped species of varying lengths and aggregated PspA forming large mesh structures (Fig. 5). No such structures were observed in PspA-free controls.
Fig. 5.
Transmission electron micrographs of negatively stained PspA protein complexes. (a, b) Mixtures of rings, rod-like complexes and mesh-like structures are readily visible. Bar, 500 nm. (c, d) Both the ring structures and rod-like complexes are visible in this field of view. Bar, 200 nm. (e) Close-up of a rod-like structure, clearly showing the indentations and striations that indicate stacking of ring-like structures. This is also an example of the tapered end observed occasionally. Bar, 100 nm. (f) The putative 36-mer, ring-like PspA structure. Bar, 40 nm.
The first type of PspA structure observed was a ring-shaped complex (Fig. 5c, d, f), similar to those previously observed in E. coli (Hankamer et al. 2004). The outer diameter of the B. pseudomallei PspA ring was ∼40 nm, slightly larger than the E. coli PspA 36-mer (Hankamer et al. 2004) and more similar to the size of the clathrin-like species previously observed in E. coli (30–40 nm) (Standar et al., 2008). Despite the difference in size, B. pseudomallei PspA appeared to form ring-like structures, visible as a ring of weak contrast with a black, stain-filled region in the middle, indicating the presence of a central hole with a diameter of ∼10–12 nm (Fig. 5c, d, f).
The second species observed was analogous to the rod-like complexes formed by the PspA homologue Vipp1 in the cyanobacterium Synechocystis (Fuhrmann et al., 2009). Slight variation in the diameter between the B. pseudomallei PspA rod-like complexes was observed, with a mean diameter between 40 and 45 nm (Fig. 5c, d). Along each rod-like complex, striations and indentations were visible, even at lower magnification, similar to those seen in the Vipp1 complexes (Fuhrmann et al., 2009). These characteristics were uniform, suggestive of ring stacking and were ∼15 nm apart (Fig. 5e). Some rod-like structures observed showed several structural anomalies, such as curved shapes and tapered ends (Fig. 5d, e), suggesting that the rings were not necessarily stacked directly on top of one another during formation of the rods. The ring-like structures were observed in supercomplexes in the form of a mesh-like structure created by interactions between the rod-like species (Fig. 5a, b). These large complexes appeared to be ordered structures, unlike the aggregated protein observed in ∼5–10 % of the sample (Fig. S4).
As the B. pseudomallei PspA higher-order structures observed above were purified in the absence of the detergent CHAPS, this soluble version of the protein may not be functional. As an additional control, the membrane-associated form of this protein was therefore purified with the addition of 1 % CHAPS to an extraction buffer and eluted fractions (Fig. S5). The purified protein had similar properties to the protein purified in the absence of CHAPS. We also observed similar higher-order species forming for this protein as for PspA purified in the absence of CHAPS (Fig. S6), demonstrating that these higher-order structures are not an artefact of the purification conditions or indicative of inactive protein.
Discussion
The Psp response is a poorly understood stress response system, expression of which is induced by changing conditions in the extracellular environment (Darwin, 2005; Joly et al., 2010). Its main function appears to be maintaining the integrity of the cytoplasmic membrane by generating a network of PspA complexes that are recruited to the cytoplasmic membrane during induction of the response (Yamaguchi et al., 2010). Loss of a functional Psp response results in the dissipation of the PMF under certain inner membrane stress conditions (Kleerebezem et al., 1996). This leads to defects in functions such as metal iron transport and biofilm formation (Beloin et al., 2004; Karlinsey et al., 2010).
The Psp response has been best characterized in the Enterobacteriaceae and few studies have been reported for other bacteria. We sought to determine whether B. pseudomallei, an opportunistic pathogen of medical importance, possessed a Psp response and whether it played a similar role in survival as reported for the enterobacterial paradigm. Bioinformatic analysis was carried out to identify whether Psp homologues were present in B. pseudomallei. Only a putative PspA homologue, BPSL2105, was identified, which showed low identity when compared with Y. enterocolitica or E. coli PspA amino acid sequences. However, the similarity in secondary structure to known PspAs from the enterobacteria supports the identification of BPSL2105 as a PspA homologue. PspA and its homologues are coiled-coil proteins containing four α-helical domains (Joly et al., 2009). In E. coli, interactions between the N-terminal amphipathic helices are important for intra- and intermolecular signalling, which allows the protein to switch between its role as negative regulator and membrane-associated effector of stress response (Jovanovic et al., 2014a). Although the primary sequence of this initial helix is not conserved in BPSL2105, it may nonetheless contribute to the ability to form similar protein interactions under conditions of stress.
The complete Psp regulon appears to be absent in B. pseudomallei; however, the presence of a predicted membrane protein that is co-transcribed with BPSL2105 bears resemblance to other species with only a single PspA homologue (Vrancken et al., 2008; Wolf et al., 2010). It is common with this response to find this arrangement as many of the psp genes appear to be dispensable (Darwin, 2005). For example, several Gram-positive bacteria, such as B. subtilis and S. lividans, have been reported to possess Psp-like responses without the accompanying regulatory function. The arrangement in B. pseudomallei could be similar to that reported for these species, which both contain a single PspA homologue in an operon with a predicted membrane protein, but lack a full psp operon (Vrancken et al., 2008; Wolf et al., 2010). Despite these differences to the accepted paradigm in enterobacteria, the response to heat stress in B. pseudomallei showed comparable induction of BPSL2105 to that of E. coli PspA, indicating a Psp-like response in this species. Conversely, there was no apparent change in the level of BPSL2105 expression in response to hyper osmotic shock, despite the proximity of the mscL gene encoding a mechanosensitive channel, involved in the cell's response to osmotic pressure changes. In E. coli, the Psp response to hyperosmotic shock is partially dependent on the regulatory proteins PspB and PspC (Weiner et al., 1991). The absence of these proteins in B. pseudomallei may account for the lack of BPSL2105 induction under similar conditions.
A PspA homologue is known to be expressed during the stationary phase in B. pseudomallei (Wongtrakoongate et al., 2007), and it has been shown that PspA is important for stationary phase survival in E. coli as its loss results in a severe loss of fitness during this phase of growth (Weiner & Model, 1994). We have shown that the loss of BPSL2105 resulted in reduced viability of B. pseudomallei after several days of prolonged growth. As with the E. coli mutant, B. pseudomalleiΔpspA cultures showed an increase in pH over the course of the experiment, demonstrating a reduction in the ability of the bacteria to maintain the extracellular pH. Although not shown in this study, this lack of control strongly suggests dissipation of the PMF, which demonstrates a strong link to other PspA homologues which function to maintain the PMF in response to stress (Kleerebezem et al., 1996). Whether the decrease in viability of the B. pseudomalleiΔpspA is linked to the change in its ability to maintain a tolerable extracellular pH has not been proven, but the S. lividans ΔpspA mutant showed a severe decrease in viability compared with the WT strain under alkaline conditions, indicating that this may be possible (Vrancken et al., 2008). It has been proposed that the stationary phase is the normal state of affairs for bacteria, which rarely encounter conditions suitable for exponential growth in natural niches (Kolter et al., 1993), indicating that BPSL2105 may play a more significant role under environmental survival conditions.
In B. pseudomallei, a PspA homologue has previously been shown to be downregulated at the stationary phase in an rpoE mutant, along with a number of other proteins important for responding to stress (Thongboonkerd et al., 2007). This resulted in lower tolerance to osmotic and oxidative stress, and also in reduced viability in mammalian phagocytes. In order to identify a more specific role for PspA under these conditions, an intracellular survival assay was carried out using a mouse macrophage cell line. This demonstrated that B. pseudomalleiΔpspA was less able to survive when phagocytosed in the stationary phase and, as such, the loss of PspA at the stationary phase may be indicative of a possible role in vivo, where bacteria are more likely to be maintained at the stationary phase stage of growth. B. pseudomalleiΔpspA was able to survive initial uptake by macrophage cells but subsequently the number of intracellular bacteria declined. In addition to the Psp response, other extracytoplasmic stress responses are known to have an important role in intracellular survival and resisting the diverse stresses encountered within phagocytic cells (reviewed by Rowley et al., 2006). The survival of B. pseudomalleiΔpspA exposed to selected stresses in vitro to mimic macrophage killing mechanisms failed to identify the specific cause for the reduced intracellular survival of the mutant: there was no difference in survival between the WT and mutant strains under any of the conditions tested. This may indicate that it may not necessarily be a single mechanism killing the mutant. The Psp response may therefore have an important role in this complex and harsh environment, which has been implicated in macrophage infection in S. enterica, S. flexneri and M. tuberculosis (Datta et al., 2015; Eriksson et al., 2003; Lucchini et al., 2005).
PspA is hypothesized to stabilize the cytoplasmic membrane in times of stress by forming large structures that associate with the membrane to prevent leakage, particularly of protons, to maintain the PMF essential for many key bacterial processes (Kobayashi et al., 2007). Previous studies have observed the formation of 36-mers by E. coli PspA in vitro (Hankamer et al., 2004), supported by further data showing that PspA forms up to 36-mers in vivo (Jovanovic et al., 2014b). Similarly, in B. subtilis the Lia system contains a PspA homologue, LiaH, which has been shown to form large oligomeric rings (Wolf et al., 2010). The ability to form large oligomeric rings is also observed for the Psp homologue Vipp1 in cyanobacteria (Fuhrmann et al., 2009). It has been demonstrated that the E. coli PspA structures interact with the phospholipids when in an oligomeric form (Kobayashi et al., 2007). This study has shown that, similar to PspA of E. coli and B. subtilis, B. pseudomallei BPSL2105 assembles into large multimeric complexes. The α-helical domains in other PspA and PspA-like proteins have been shown to be vital for complex formation (Aseeva et al., 2004; Joly et al. 2009), and this may be the case in B. pseudomallei where the secondary structure of BPSL2105 resembles these proteins. It may be postulated that these large complexes are the physiologically relevant form of PspA, functioning as scaffolds to maintain membrane integrity in the face of membrane disruption caused by stress.
In conclusion, B. pseudomallei BPSL2105 encodes a PspA-like protein that is induced in response to extreme heat shock, similar to the PspA protein of E. coli. It is important for survival during stationary phase and during infection of macrophages. The purified protein is able to assemble into high-order oligomers. This behaviour may aid in the stabilization of the B. pseudomallei cytoplasmic membrane during induction of the Psp-like response as a result of stressful conditions. Recently, it has been proposed that in E. coli PspA is potentially targeted to areas of the inner membrane associated with peptidoglycan biosynthesis machinery (Jovanovic et al., 2014b). In light of this, the presence of a gene encoding a penicillin-binding protein, mrcA, directly upstream of BPSL2105 may be significant. Future studies are needed to elucidate the importance of this and the function of downstream genes, such as BPSL2106, in the B. pseudomallei Psp response.
Supplementary Data
Supplementary Data
Abbreviations:
- PMF
proton motive force
- Psp
phage-shock protein
- RT
reverse transcription
- TEM
transmission electron microscopy
References
- Allwood E.M., Devenish R.J., Prescott M., Adler B., Boyce J.D. (2011). Strategies for intracellular survival of Burkholderia pseudomallei Front Microbiol 2170. 10.3389/fmicb.2011.00170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aseeva E., Ossenbühl F., Eichacker L.A., Wanner G., Soll J., Vothknecht U.C. (2004). Complex formation of Vipp1 depends on its α-helical PspA-like domain J Biol Chem 27935535–35541 10.1074/jbc.M401750200. [DOI] [PubMed] [Google Scholar]
- Beloin C., Valle J., Latour-Lambert P., Faure P., Kzreminski M., Balestrino D., Haagensen J.A.J., Molin S., Prensier G., other authors (2004). Global impact of mature biofilm lifestyle on Escherichia coli K-12 gene expression Mol Microbiol 51659–674 10.1046/j.1365-2958.2003.03865.x. [DOI] [PubMed] [Google Scholar]
- Brissette J.L., Russel M., Weiner L., Model P. (1990). Phage shock protein, a stress protein of Escherichia coli Proc Natl Acad Sci U S A 87862–866 10.1073/pnas.87.3.862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaowagul W., White N.J., Dance D.A.B., Wattanagoon Y., Naigowit P., Davis T.M.E., Looareesuwan S., Pitakwatchara N. (1989). Melioidosis: a major cause of community-acquired septicemia in northeastern Thailand J Infect Dis 159890–899 10.1093/infdis/159.5.890. [DOI] [PubMed] [Google Scholar]
- Cheng A.C., Currie B.J. (2005). Melioidosis: epidemiology, pathophysiology, and management Clin Microbiol Rev 18383–416 10.1128/CMR.18.2.383-416.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole C., Barber J.D., Barton G.J. (2008). The Jpred 3 secondary structure prediction server Nucleic Acids Res 36 (Suppl. 2), W197–W201 10.1093/nar/gkn238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Currie B.J., Dance D.A., Cheng A.C. (2008). The global distribution of Burkholderia pseudomallei and melioidosis: an update Trans R Soc Trop Med Hyg 102 (Suppl. 1), S1–S4 10.1016/S0035-9203(08)70002-6. [DOI] [PubMed] [Google Scholar]
- Currie B.J., Ward L., Cheng A.C. (2010). The epidemiology and clinical spectrum of melioidosis: 540 cases from the 20 year Darwin prospective study PLoS Negl Trop Dis 4e900. 10.1371/journal.pntd.0000900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dance D.A. (2000). Melioidosis as an emerging global problem Acta Trop 74115–119 10.1016/S0001-706X(99)00059-5. [DOI] [PubMed] [Google Scholar]
- Darwin A.J. (2005). The phage-shock-protein response Mol Microbiol 57621–628 10.1111/j.1365-2958.2005.04694.x. [DOI] [PubMed] [Google Scholar]
- Darwin A.J. (2013). Stress relief during host infection: the phage shock protein response supports bacterial virulence in various ways PLoS Pathog 9e1003388. 10.1371/journal.ppat.1003388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darwin A.J., Miller V.L. (2001). The psp locus of Yersinia enterocolitica is required for virulence and for growth in vitro when the Ysc type III secretion system is produced Mol Microbiol 39429–445 10.1046/j.1365-2958.2001.02235.x. [DOI] [PubMed] [Google Scholar]
- Datta P., Ravi J., Guerrini V., Chauhan R., Neiditch M.B., Shell S.S., Fortune S.M., Hancioglu B., Igoshin O.A., Gennaro M.L. (2015). The Psp system of Mycobacterium tuberculosis integrates envelope stress-sensing and envelope-preserving functions Mol Microbiol 97408–422 10.1111/mmi.13037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dworkin J., Jovanovic G., Model P. (2000). The PspA protein of Escherichia coli is a negative regulator of σ54-dependent transcription J Bacteriol 182311–319 10.1128/JB.182.2.311-319.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elderkin S., Jones S., Schumacher J., Studholme D., Buck M. (2002). Mechanism of action of the Escherichia coli phage shock protein PspA in repression of the AAA family transcription factor PspF J Mol Biol 32023–37 10.1016/S0022-2836(02)00404-7. [DOI] [PubMed] [Google Scholar]
- Elderkin S., Bordes P., Jones S., Rappas M., Buck M. (2005). Molecular determinants for PspA-mediated repression of the AAA transcriptional activator PspF J Bacteriol 1873238–3248 10.1128/JB.187.9.3238-3248.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson S., Lucchini S., Thompson A., Rhen M., Hinton J.C.D. (2003). Unravelling the biology of macrophage infection by gene expression profiling of intracellular Salmonella enterica Mol Microbiol 47103–118 10.1046/j.1365-2958.2003.03313.x. [DOI] [PubMed] [Google Scholar]
- Fuhrmann E., Bultema J.B., Kahmann U., Rupprecht E., Boekema E.J., Schneider D. (2009). The vesicle-inducing protein 1 from Synechocystis sp. PCC 6803 organizes into diverse higher-ordered ring structures Mol Biol Cell 204620–4628 10.1091/mbc.E09-04-0319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan Y.H. (2005). Interaction between Burkholderia pseudomallei and the host immune response: sleeping with the enemy? J Infect Dis 1921845–1850 10.1086/497382. [DOI] [PubMed] [Google Scholar]
- Gautier R., Douguet D., Antonny B., Drin G. (2008). heliquest: a web server to screen sequences with specific α-helical properties Bioinformatics 242101–2102 10.1093/bioinformatics/btn392. [DOI] [PubMed] [Google Scholar]
- Hankamer B.D., Elderkin S.L., Buck M., Nield J. (2004). Organization of the AAA+ adaptor protein PspA is an oligomeric ring J Biol Chem 2798862–8866 10.1074/jbc.M307889200. [DOI] [PubMed] [Google Scholar]
- Holden M.T.G., Titball R.W., Peacock S.J., Cerdeño-Tárraga A.M., Atkins T., Crossman L.C., Pitt T., Churcher C., Mungall K., other authors (2004). Genomic plasticity of the causative agent of melioidosis, Burkholderia pseudomallei Proc Natl Acad Sci U S A 10114240–14245 10.1073/pnas.0403302101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inglis T.J.J., Sagripanti J.L. (2006). Environmental factors that affect the survival and persistence of Burkholderia pseudomallei Appl Environ Microbiol 726865–6875 10.1128/AEM.01036-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joly N., Burrows P.C., Engl C., Jovanovic G., Buck M. (2009). A lower-order oligomer form of phage shock protein A (PspA) stably associates with the hexameric AAA+ transcription activator protein PspF for negative regulation J Mol Biol 394764–775 10.1016/j.jmb.2009.09.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joly N., Engl C., Jovanovic G., Huvet M., Toni T., Sheng X., Stumpf M.P.H., Buck M. (2010). Managing membrane stress: the phage shock protein (Psp) response, from molecular mechanisms to physiology FEMS Microbiol Rev 34797–827 10.1111/j.1574-6976.2010.00240.x. [DOI] [PubMed] [Google Scholar]
- Jones A.L., Beveridge T.J., Woods D.E. (1996). Intracellular survival of Burkholderia pseudomallei Infect Immun 64782–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovanovic G., Weiner L., Model P. (1996). Identification, nucleotide sequence, and characterization of PspF, the transcriptional activator of the Escherichia coli stress-induced psp operon J Bacteriol 1781936–1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovanovic G., Rakonjac J., Model P. (1999). in vivo and in vitro activities of the Escherichia coli σ54 transcription activator, PspF, and its DNA-binding mutant, PspFΔHTH J Mol Biol 285469–483 10.1006/jmbi.1998.2263. [DOI] [PubMed] [Google Scholar]
- Jovanovic G., Mehta P., McDonald C., Davidson A.C., Uzdavinys P., Ying L., Buck M. (2014a). The N-terminal amphipathic helices determine regulatory and effector functions of phage shock protein A (PspA) in Escherichia coli J Mol Biol 4261498–1511 10.1016/j.jmb.2013.12.016. [DOI] [PubMed] [Google Scholar]
- Jovanovic G., Mehta P., Ying L., Buck M. (2014b). Anionic lipids and the cytoskeletal proteins MreB and RodZ define the spatio-temporal distribution and function of membrane stress controller PspA in Escherichia coli Microbiology 1602374–2386 10.1099/mic.0.078527-0. [DOI] [PubMed] [Google Scholar]
- Karlinsey J.E., Maguire M.E., Becker L.A., Crouch M.-L.V., Fang F.C. (2010). The phage shock protein PspA facilitates divalent metal transport and is required for virulence of Salmonella enterica sv. Typhimurium Mol Microbiol 78669–685 10.1111/j.1365-2958.2010.07357.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleerebezem M., Crielaard W., Tommassen J. (1996). Involvement of stress protein PspA (phage shock protein A) of Escherichia coli in maintenance of the protonmotive force under stress conditions EMBO J 15162–171. [PMC free article] [PubMed] [Google Scholar]
- Kobayashi R., Suzuki T., Yoshida M. (2007). Escherichia coli phage-shock protein A (PspA) binds to membrane phospholipids and repairs proton leakage of the damaged membranes Mol Microbiol 66100–109 10.1111/j.1365-2958.2007.05893.x. [DOI] [PubMed] [Google Scholar]
- Kolter R., Siegele D.A., Tormo A. (1993). The stationary phase of the bacterial life cycle Annu Rev Microbiol 47855–874 10.1146/annurev.mi.47.100193.004231. [DOI] [PubMed] [Google Scholar]
- Lambert R.J.W., Pearson J. (2000). Susceptibility testing: accurate and reproducible minimum inhibitory concentration (MIC) and non-inhibitory concentration (NIC) values J Appl Microbiol 88784–790 10.1046/j.1365-2672.2000.01017.x. [DOI] [PubMed] [Google Scholar]
- Logue C.A., Peak I.R.A., Beacham I.R. (2009). Facile construction of unmarked deletion mutants in Burkholderia pseudomallei using sacB counter-selection in sucrose-resistant and sucrose-sensitive isolates J Microbiol Methods 76320–323 10.1016/j.mimet.2008.12.007. [DOI] [PubMed] [Google Scholar]
- Lucchini S., Liu H., Jin Q., Hinton J.C.D., Yu J. (2005). Transcriptional adaptation of Shigella flexneri during infection of macrophages and epithelial cells: insights into the strategies of a cytosolic bacterial pathogen Infect Immun 7388–102 10.1128/IAI.73.1.88-102.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Male A.L., Oyston P.C.F., Tavassoli A. (2014). Self-assembly of Escherichia coli phage shock protein A Adv Microbiol 4353–359 10.4236/aim.2014.47042. [DOI] [Google Scholar]
- Milton D.L., O'Toole R., Horstedt P., Wolf-Watz H. (1996). Flagellin A is essential for the virulence of Vibrio anguillarum J Bacteriol 1781310–1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngauy V., Lemeshev Y., Sadkowski L., Crawford G. (2005). Cutaneous melioidosis in a man who was taken as a prisoner of war by the Japanese during World War II J Clin Microbiol 43970–972 10.1128/JCM.43.2.970-972.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilatz S., Breitbach K., Hein N., Fehlhaber B., Schulze J., Brenneke B., Eberl L., Steinmetz I. (2006). Identification of Burkholderia pseudomallei genes required for the intracellular life cycle and in vivo virulence Infect Immun 743576–3586 10.1128/IAI.01262-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowley G., Spector M., Kormanec J., Roberts M. (2006). Pushing the envelope: extracytoplasmic stress responses in bacterial pathogens Nat Rev Microbiol 4383–394 10.1038/nrmicro1394. [DOI] [PubMed] [Google Scholar]
- Standar K., Mehner D., Osadnik H., Berthelmann F., Hause G., Lünsdorf H., Brüser T. (2008). PspA can form large scaffolds in Escherichia coli FEBS Lett 5823585–3589 10.1016/j.febslet.2008.09.002. [DOI] [PubMed] [Google Scholar]
- Stevens M.P., Friebel A., Taylor L.A., Wood M.W., Brown P.J., Hardt W.D., Galyov E.E. (2003). Burkholderia pseudomallei type III secreted protein, BopE, facilitates bacterial invasion of epithelial cells and exhibits guanine nucleotide exchange factor activity J Bacteriol 1854992–4996 10.1128/JB.185.16.4992-4996.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens M.P., Haque A., Atkins T., Hill J., Wood M.W., Easton A., Nelson M., Underwood-Fowler C., Titball R.W., other authors (2004). Attenuated virulence and protective efficacy of a Burkholderia pseudomallei bsa type III secretion mutant in murine models of melioidosis Microbiology 1502669–2676 10.1099/mic.0.27146-0. [DOI] [PubMed] [Google Scholar]
- Thongboonkerd V., Vanaporn M., Songtawee N., Kanlaya R., Sinchaikul S., Chen S.T., Easton A., Chu K., Bancroft G.J., Korbsrisate S. (2007). Altered proteome in Burkholderia pseudomallei_rpoE operon knockout mutant: insights into mechanisms of rpoE operon in stress tolerance, survival, and virulence J Proteome Res 61334–1341 10.1021/pr060457t. [DOI] [PubMed] [Google Scholar]
- Vrancken K., Van Mellaert L., Anné J. (2008). Characterization of the Streptomyces lividans PspA response J Bacteriol 1903475–3481 10.1128/JB.01966-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wand M.E., Müller C.M., Titball R.W., Michell S.L. (2011). Macrophage and Galleria mellonella infection models reflect the virulence of naturally occurring isolates of B. pseudomallei, B. thailandensis and B. oklahomensis BMC Microbiol 1111. 10.1186/1471-2180-11-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner L., Model P. (1994). Role of an Escherichia coli stress-response operon in stationary-phase survival Proc Natl Acad Sci U S A 912191–2195 10.1073/pnas.91.6.2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner L., Brissette J.L., Model P. (1991). Stress-induced expression of the Escherichia coli phage shock protein operon is dependent on sigma 54 and modulated by positive and negative feedback mechanisms Genes Dev 51912–1923 10.1101/gad.5.10.1912. [DOI] [PubMed] [Google Scholar]
- White N.J. (2003). Melioidosis Lancet 3611715–1722 10.1016/S0140-6736(03)13374-0. [DOI] [PubMed] [Google Scholar]
- Wolf D., Kalamorz F., Wecke T., Juszczak A., Mäder U., Homuth G., Jordan S., Kirstein J., Hoppert M., other authors (2010). In-depth profiling of the LiaR response of Bacillus subtilis J Bacteriol 1924680–4693 10.1128/JB.00543-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wongtrakoongate P., Mongkoldhumrongkul N., Chaijan S., Kamchonwongpaisan S., Tungpradabkul S. (2007). Comparative proteomic profiles and the potential markers between Burkholderia pseudomallei and Burkholderia thailandensis Mol Cell Probes 2181–91 10.1016/j.mcp.2006.08.006. [DOI] [PubMed] [Google Scholar]
- Yamaguchi S., Gueguen E., Horstman N.K., Darwin A.J. (2010). Membrane association of PspA depends on activation of the phage-shock-protein response in Yersinia enterocolitica Mol Microbiol 78429–443 10.1111/j.1365-2958.2010.07344.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data