Abstract
Medical conditions accompanying obesity often require drug therapy, but whether and how obesity alters the expression of drug-metabolizing enzymes and thus drug pharmacokinetics is poorly defined. Previous studies have shown that high-fat diet (HFD) feeding and subsequent obesity in mice lead to altered expression of transcriptional regulators for cytochrome P450 CYP2D6, including hepatocyte nuclear factor 4α (HNF4α, a transcriptional activator of CYP2D6) and small heterodimer partner (SHP, a transcriptional repressor of CYP2D6). The objective of this study was to examine whether diet-induced obesity alters CYP2D6 expression by modulating HNF4α and SHP expression. Male CYP2D6-humanized transgenic (Tg-CYP2D6) mice were fed with HFD or matching control diet for 18 weeks. Hepatic mRNA expression of CYP2D6 decreased to a small extent in the HFD group (by 31%), but the differences in CYP2D6 protein and activity levels in hepatic S9 fractions were found insignificant between the groups. Although hepatic SHP expression did not differ between the groups, HNF4α mRNA and protein levels decreased by ∼30% in the HFD group. Among major mouse endogenous cytochrome P450 genes, Cyp1a2 and Cyp2c37 showed significant decreases in the HFD group, whereas Cyp2e1 expression did not differ between groups. Cyp2b10 and Cyp3a11 expression was higher in the HFD group, with corresponding 2.9-fold increases in hepatic CYP3A activities in HFD-fed mice. Together, these results suggest that obesity has minimal effects on CYP2D6-mediated drug metabolism, although it modulates the expression of mouse endogenous P450s in a gene-specific manner.
Introduction
Obesity is a prevalent medical condition associated with increased incidences of chronic comorbidities, such as diabetes, cardiovascular diseases, and nonalcoholic fatty liver diseases (Fujioka, 2015), which often require drug treatment. Yet, our understanding of whether and how obesity alters the rate and extent of drug disposition, especially hepatic drug metabolism, remains incomplete. Clinical reports suggest that obesity leads to altered hepatic drug metabolism in a metabolic pathway-specific manner; however, the directional changes for cytochrome P450 (P450)-mediated metabolism are for the most part inconclusive.
CYP2D6 is a major drug-metabolizing enzyme responsible for metabolizing ∼25% of marketed drugs. Previous studies have shown important roles for transcriptional regulation of CYP2D6 (Koh et al., 2014; Pan et al., 2015). Hepatocyte nuclear factor 4α (HNF4α) transactivates CYP2D6 promoter by binding to a proximal promoter region, whereas small heterodimer partner (SHP) represses CYP2D6 expression by inhibiting HNF4α transactivation of CYP2D6 promoter (Koh et al., 2014). Of interest, obesity leads to altered expression/activity of different transcriptional regulators of CYP2D6. Specifically, high-fat diet (HFD) feeding in mice has been shown to decrease HNF4α expression (Xu et al., 2015) and increase SHP protein levels (Miao et al., 2009). However, whether these changes affect CYP2D6 expression remains unclear. In this study, we examined whether HFD feeding modulates CYP2D6-mediated drug metabolism by using CYP2D6-humanized transgenic (Tg-CYP2D6) mice. Furthermore, we explored how endogenous mouse P450 gene expression is altered in HFD-fed mice to provide insights into the effects of obesity on hepatic drug metabolism in general.
Materials and Methods
Animals.
Tg-CYP2D6 mice were previously described (Koh et al., 2014). Male Tg-CYP2D6 mice (6–8 week-old; n = 7–9 per group) were fed with either Adjusted Calories Diet (HFD; TD.88137, Teklad; Envigo, Huntingdon, Cambridgeshire, United Kingdom) or a matching low-fat control diet (TD.08485, Teklad) for 18 weeks in specific pathogen-free environment with 12-hour light and dark cycles. The mice had access to the diet and water ad libitum. All procedures were approved by the Institutional Animal Care and Use Committee in the University of Illinois at Chicago.
RNA Isolation and Quantitative Real Time-PCR.
Total RNAs were isolated from mouse liver tissues using Trizol (Life Technologies/Thermo Fisher Scientific, Sunnyvale, CA) and used as template for cDNA synthesis using High-Capacity cDNA Reverse Transcription Kit (Life Technologies/Thermo Fisher Scientific). Quantitative real time-PCR (qRT-PCR) was performed using StepOnePlus Real-Time PCR System (Thermo Fisher Scientific). Predesigned gene expression assays and primers were listed in supplemental data (Supplemental Table 1). The relative mRNA expression level was determined after normalizing the expression levels by those of mouse Gapdh (2–ΔΔCt method).
Western Blot.
Mouse liver lysates were resolved using 4–20% Mini-PROTEAN TGX Precast Protein Gels (Bio-Rad, Hercules, CA). CYP2D6 (cat. no. 458246, BD Gentest; BD, Franklin Lakes, NJ); HNF4α (K9218; R&D Systems, Minneapolis, MN); SHP (sc-30169; Santa Cruz Biotechnology, Dallas, TX); and β-actin (A1978; Sigma-Aldrich, St. Louis, MO) primary antibodies were used. SuperSignal West Pico Chemiluminescent Substrate (34080; ThermoFisher, Rockford, IL) was used for detection. The images were recorded and analyzed using FluorChem E system and AlphaView software (ProteinSimple, San Jose, CA).
Measurement of CYP2D6 and CYP3A Activities.
Liver tissues (∼200 mg) were accurately weighed for S9 preparation. The samples were homogenized on ice using tissue homogenizer (PowerGen 125; Fisher Scientific) in 0.05 M Tris-HCl (pH 7.0) buffer containing 150 mM KCl, 2 mM EDTA, and protease inhibitor cocktail (P8340; Sigma-Aldrich). The homogenate was centrifuged at 9000g for 20 minutes at 4°C. The supernatant was collected and volume measured. Protein content in S9 fraction was measured by BCA protein assay (Pierce Biotechnology, Rockford, IL). Total S9 protein amount per liver = (S9 protein concentration × S9 volume / liver weight used for S9 preparation) × total liver weight. CYP2D6 activity was measured as previously described (Koh et al., 2014). Briefly, for the measurement of CYP2D6 activity, S9 fractions (∼3 mg/ml) were incubated with debrisoquine (200 μM) for 15 minutes, and the reaction was stopped by adding 1 vol of ice-cold acetonitrile with internal standard (paraxanthine, 800 ng/ml). The concentration of 4-hydroxydebrisoquine was measured by liquid chromatography–tandem mass spectroscopy (LC-MS/MS) (Agilent 6410 Triple Quadrupole LC/MS) using electrospray ion source (ESI) in positive ion mode, detecting [M+H]+ ions. Multiple reaction monitoring (MRM) data acquisition was employed: m/z = 192.3/132.2 for 4-hydroxydebrisoquine and m/z = 181.1/124.1 for the internal standard. For CYP3A activity, mouse liver S9 (∼0.6 mg/ml) was incubated with triazolam (23 μM) for 5 minutes and the reaction was stopped by adding 1 vol of ice-cold acetonitrile containing internal standard (1-hydroxytriazolam-D4, 40 ng/ml). The concentrations of 1-hydroxytriazolam were measured by LC-MS/MS (Agilent 1200 HPLC with AB Sciex QTRAP 5500 System) using ESI source in positive ion mode, detecting [M+H]+ ions. MRM for 1-hydroxytriazolam and 1-hydroxytriazolam-D4 were m/z = 359.4/176.3 and m/z = 363.2/335.0, respectively.
Statistical Analysis.
All data were presented as mean ± S.D. Statistical analyses for qRT-PCR and Western blot results were performed using Student’s t test between control and HFD group using Prism 6.0 (Graphpad Software, Inc., La Jolla, CA). For correlation study, Pearson’s correlation test and linear regression was used as appropriate.
Results and Discussion
Diet-Induced Obesity Model in Tg-CYP2D6 Mice.
Tg-CYP2D6 mice have been used as an in vivo model for studying transcriptional regulation of CYP2D6 (Koh et al., 2014; Pan et al., 2015). To evaluate the effect of obesity on CYP2D6 expression in vivo, we first established a diet-induced obesity model in Tg-CYP2D6 mice by chronic HFD feeding. Male Tg-CYP2D6 mice were fed with either HFD or a matching control diet for 18 weeks ad libitum. Mice in the HFD group gained more weight than those from control diet group since week 8 (Supplemental Fig. 1A). The livers from HFD-fed mice were pale and enlarged, suggesting hepatic steatosis (Supplemental Fig. 1B). The mouse liver weight was 1.68 ± 0.38 and 3.62 ± 1.02 g in control and HFD groups, respectively (n = 6–8 per group, p < 0.001). Liver-to-body weight ratio was also increased significantly in HFD mice (Supplemental Fig. 1C). Although the S9 protein yield per gram of liver was 25% lower in HFD-fed mice, the total protein amount in S9 fractions per liver was ∼60% higher in the group (Supplemental Fig. 1D), suggesting increased numbers and/or sizes of parenchymal cells upon HFD. In humans, the liver volume (determined by using magnetic resonance imaging) in obese-to-morbidly obese subjects was 1.25- to 1.5-fold larger (Lewis et al., 2006; Bian et al., 2014) than that in subjects with normal body weight (Johnson et al., 2005). Of note, the extent of increases in liver size is smaller in humans than in mice, and the relative contribution between accumulating fats and the expansion of parenchymal cells to increased liver size in humans is unclear.
CYP2D6 Expression Did Not Differ between HFD and Control Mice.
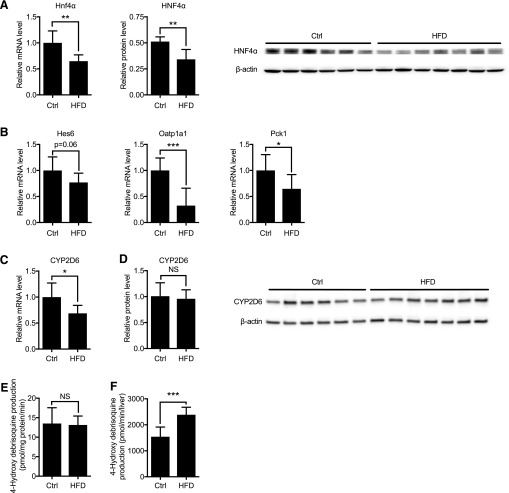
Previous reports showed that HFD decreases HNF4α expression by inducing microRNA-34a expression (Xu et al., 2015) and that HFD feeding increases SHP protein levels by enhancing SHP protein stability (Miao et al., 2009). To validate if expression of these transcription regulators is altered in obese Tg-CYP2D6 mice, hepatic mRNA and protein levels of HNF4α and SHP were examined in HFD-fed Tg-CYP2D6 mice. HNF4α mRNA and protein levels decreased significantly (35% and 33%, respectively) in the HFD group of Tg-CYP2D6 mice (Fig. 1A). This was accompanied by decreased expression of known HNF4α target genes in the liver [i.e., hes family basic helix-loop-helix (bHLH) transcription factor 6 (Hes6), organic anion–transporting polypeptides 1a1 (Oatp1a1), and phosphoenolpyruvate carboxykinase 1 (Pck1)] (Hayhurst et al., 2001; Martinez-Jimenez et al., 2010; Fang et al., 2012) (Fig. 1B). Contrary to the previous report, SHP mRNA and protein levels did not differ between HFD and control groups (Supplemental Fig. 2). Differences in mouse study design (i.e., CV57 versus Tg-CYP2D6 with C57/BL background) may explain the apparent discrepancy. Also, although a matching low-fat control diet was used in this study, the diet used for the control group was not specified in the previous study. Differences in diet and strain have been shown to result in differential composition of gut microbiota in mice (Ussar et al., 2015). Accumulating evidence indicates that gut microbiota modulates multiple aspects of physiology, including the composition of bile acids and subsequent intestinal farnesoid X receptor (FXR) signaling (Wahlstrom et al., 2016). The products of intestinal FXR signaling, such as fibroblast growth factor 15, can alter SHP protein stability in the liver (Miao et al., 2009).
Fig. 1.
Expression and activity of HNF4α and CYP2D6 in HFD-fed Tg-CYP2D6 mice. Liver tissues were collected from Tg-CYP2D6 mice after 18 weeks of HFD or matching control (Ctrl) diet feeding. (A) Relative Hnf4α mRNA levels (left) determined by qRT-PCR were normalized by that of control group. Relative HNF4α protein levels (right) were normalized against β-actin band intensities. (B) Hepatic mRNA levels of representative HNF4α target genes were determined by qRT-PCR and normalized by that of control group. (C) Hepatic mRNA level of CYP2D6 was normalized by that of control group. (D) CYP2D6 protein level in hepatic S9 fraction (right) was determined using Western blot normalized against β-actin band intensities. (E) CYP2D6 activity in mouse liver S9 fraction was measured using debrisoquine as a substrate. CYP2D6 activity is presented as 4-hydroxydebrisoquine formation rate per mg S9 protein. (F) Total CYP2D6 activity per liver was the product of CYP2D6 activity per mg S9 protein and total S9 protein level per liver. N = 6–8 per group for qRT-PCR and activity assay. Data shown are mean ± S.D. NS, not significant, *p < 0.05, **p < 0.01, ***p < 0.001.
Importantly, hepatic mRNA levels of CYP2D6 decreased to a small extent in the HFD group (Fig. 1C). However, hepatic CYP2D6 protein and activity levels did not differ between HFD and control groups (Fig. 1, D and E). The total CYP2D6 activity per liver was even higher in HFD-fed mice owing to the greater S9 protein amounts (Fig. 1F). Of interest, results from currently available clinical data suggest that the metabolic rates of CYP2D6 substrate drugs are similar in obese and nonobese subjects. For example, Cheymol et al. (1995) reported no differences in the clearance of dexfenfluramine (a CYP2D6 substrate of hepatic extraction ratio ∼0.3) between obese and nonobese subjects after intravenous infusion. It appears plausible that the obesity-induced increase in liver size observed in humans does not involve the expansion of parenchymal cells, unlike in mice.
Obesity Had Differential Effects on Mouse Endogenous P450s.
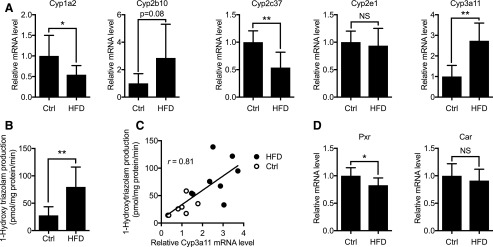
Considering that HNF4α is a global regulator of hepatic genes, including those encoding P450s (Jover et al., 2009), we examined the mRNA expression of representative mouse endogenous P450s. qRT-PCR results showed pathway-dependent changes in their expression in HFD-fed mice (Fig. 2A and Supplemental Fig. 3). Cyp1a2 and Cyp2c37 expression decreased significantly in HFD-fed mice, whereas Cyp2e1 expression did not differ between the groups. As for mouse endogenous Cyp2ds, expression of Cyp2d26 (the most abundant hepatic Cyp2d isoform) did not change upon HFD feeding, whereas other isoforms (Cyp2d10, Cyp2d22, and Cyp2d40) decreased significantly. Of note, Cyp3a11 expression increased significantly in HFD-fed mice (Fig. 2A). Although this is consistent with previous reports (Kim et al., 2004; Kudo et al., 2009; Geng et al., 2015), others have reported decreased hepatic Cyp3a11 expression or activity upon HFD feeding in mice (Yoshinari et al., 2006; Ghose et al., 2011; Wahlang et al., 2014). Differences in diet composition and housing condition may have contributed to the discrepancy. For instance, feeding of mice with HFD containing saturated fat (as in our HFD) induced Cyp3a11 expression, but HFD containing unsaturated fat decreased it (Geng et al., 2015). Also, mice housed in conventional conditions have been reported to exhibit low-grade inflammation in the liver on HFD feeding, although mice in a specific pathogen-free environment do not (Muller et al., 2016). Hepatic inflammation is known to cause CYP3A downregulation in humans and mice by downregulating the expression and function of transcription factors for CYP3A4 (Aitken et al., 2006). Indeed, the hepatic expression of inflammation markers (e.g., IL1β, IL6, or TNFα) was increased in HFD-fed mice in the studies that observed decreased Cyp3a11 expression (Yoshinari et al., 2006; Ghose et al., 2011; Wahlang et al., 2014). Of note, hepatic expression of IL1β, IL6, and TNFα did not differ between HFD and control groups in our study (Supplemental Fig. 4).
Fig. 2.
Mouse endogenous P450 expression upon HFD feeding in Tg-CYP2D6 mice. Liver tissues were collected from mice after 18 weeks of HFD or matching control (Ctrl) diet feeding. (A) Hepatic mRNA levels of drug-metabolizing P450s were measured using qRT-PCR and normalized to those of the control group. (B) Mouse CYP3A activity in liver S9 fraction was measured using triazolam as a substrate. CYP3A activity is presented as 1-hydroxytriazolam formation rate. (C) Mouse CYP3A activity correlated with Cyp3a11 relative mRNA level (Pearson correlation efficiency r = 0.81, p < 0.001). (D) Hepatic mRNA levels of Pxr and Car were determined using qRT-PCR and normalized by those of the control group. N = 7–8 per group. Data shown are mean ± S.D. NS, not significant, *p < 0.05, **p < 0.01.
Considering that Cyp3a11 is a mouse homolog of CYP3A4, the most important drug-metabolizing enzyme in humans, the changes in Cyp3a11 expression were further verified at the enzyme activity level using triazolam as a probe substrate. Hepatic CYP3A activity in mouse S9 fraction was significantly higher in mice fed with HFD (Fig. 2B). CYP3A activity correlated well with Cyp3a11 mRNA levels (r = 0.81) (Fig. 2C), suggesting that increased CYP3A enzyme activity in HFD mice is in part attributable to enhanced Cyp3a11 expression. Pregnane X receptor (PXR) and constitutive androstane receptor (CAR) are ligand-activated nuclear receptors and major transcriptional activators of Cyp3a11 expression in mice (Li et al., 2016). qRT-PCR results showed that mRNA expression of neither Pxr nor Car increased in HFD-fed mice (Fig. 2D), suggesting that altered expression of these transcription factors is not responsible for Cyp3a11 induction in HFD-fed mice. Whether HFD feeding modulates (e.g., increases) the hepatic levels of the endogenous ligands of PXR or CAR (e.g., bilirubin, bile acids, and steroid hormones) (Bjorkholm et al., 2009) remains unclear. Of interest, HFD feeding was shown to cause adrenal cortical hyperplasia in mice, leading to higher plasma levels of corticosteroids (i.e., PXR activators) (Swierczynska et al., 2015).
Clinical evidence is conflicting on whether obesity affects CYP3A4-mediated drug metabolism in humans. A population pharmacokinetic analysis of midazolam [a CYP3A4 substrate with hepatic extraction ratio 0.3–0.5 (Trouvin et al., 1988)] in morbidly obese patients showed no change in systemic clearance but decreased oral clearance of midazolam compared with that in healthy volunteers (Brill et al., 2014). On the other hand, the systemic clearance of triazolam (a CYP3A4 substrate of intermediate hepatic extraction ratio) after intravenous dose did not differ between obese subjects and those with normal weight (Derry et al., 1995). In human liver tissues, CYP3A4 protein levels in liver biopsies collected from obese patients (during laparoscopic gastric bypass or biliopancreatic diversion surgery) were negatively correlated with patients body mass index (Ulvestad et al., 2013), indicating repressive effects of obesity on CYP3A4 expression. Considering that morbid obesity is often accompanied by comorbidity (e.g., diabetes) that incurs chronic hepatic inflammation, decreased CYP3A4 expression in the liver tissues may result in part from underlying inflammation in obese subjects. Definitive evidence on how increased body weight impacts hepatic CYP3A4 expression in the absence of inflammation in humans is currently lacking.
To summarize, results from our diet-induced obesity model in Tg-CYP2D6 mice and comparison against clinical data suggest that obesity will likely have minimal effects on CYP2D6-mediated drug metabolism. Together, results from this study provide insight into how obesity may impact hepatic drug disposition.
Abbreviations
- HFD
high-fat diet
- HNF4α
hepatocyte nuclear factor 4α
- P450
cytochrome P450
- PXR
pregnane X receptor
- qRT-PCR
quantitative real time-PCR
- SHP
small heterodimer partner
- Tg-CYP2D6
CYP2D6-humanized transgenic mouse
Authorship Contributions
Participated in research design: Ning, Jeong.
Conducted experiments: Ning.
Performed data analysis: Ning, Jeong.
Wrote or contributed to the writing of the manuscript: Ning, Jeong.
Footnotes
This work was supported by the National Institutes of Health [Grants R01HD065532 and R01GM112746]. This research was supported in part by a Graduate Student Fellowship Award from the American Association of Pharmaceutical Scientists Foundation.
 This article has supplemental material available at dmd.aspetjournals.org.
This article has supplemental material available at dmd.aspetjournals.org.
References
- Aitken AE, Richardson TA, Morgan ET. (2006) Regulation of drug-metabolizing enzymes and transporters in inflammation. Annu Rev Pharmacol Toxicol 46:123–149. [DOI] [PubMed] [Google Scholar]
- Bian H, Hakkarainen A, Lundbom N, Yki-Järvinen H. (2014) Effects of dietary interventions on liver volume in humans. Obesity (Silver Spring) 22:989–995. [DOI] [PubMed] [Google Scholar]
- Björkholm B, Bok CM, Lundin A, Rafter J, Hibberd ML, Pettersson S. (2009) Intestinal microbiota regulate xenobiotic metabolism in the liver. PLoS One 4:e6958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brill MJ, van Rongen A, Houwink AP, Burggraaf J, van Ramshorst B, Wiezer RJ, van Dongen EP, Knibbe CA. (2014) Midazolam pharmacokinetics in morbidly obese patients following semi-simultaneous oral and intravenous administration: a comparison with healthy volunteers. Clin Pharmacokinet 53:931–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheymol G, Weissenburger J, Poirier JM, Gellee C. (1995) The pharmacokinetics of dexfenfluramine in obese and non-obese subjects. Br J Clin Pharmacol 39:684–687. [PMC free article] [PubMed] [Google Scholar]
- Derry CL, Kroboth PD, Pittenger AL, Kroboth FJ, Corey SE, Smith RB. (1995) Pharmacokinetics and pharmacodynamics of triazolam after two intermittent doses in obese and normal-weight men. J Clin Psychopharmacol 15:197–205. [DOI] [PubMed] [Google Scholar]
- Fang B, Mane-Padros D, Bolotin E, Jiang T, Sladek FM. (2012) Identification of a binding motif specific to HNF4 by comparative analysis of multiple nuclear receptors. Nucleic Acids Res 40:5343–5356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka K. (2015) Current and emerging medications for overweight or obesity in people with comorbidities. Diabetes Obes Metab 17:1021–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng T, Xia L, Russo S, Kamara D, Cowart LA. (2015) Prosteatotic genes are associated with unsaturated fat suppression of saturated fat-induced hepatic steatosis in C57BL/6 mice. Nutr Res 35:812–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghose R, Omoluabi O, Gandhi A, Shah P, Strohacker K, Carpenter KC, McFarlin B, Guo T. (2011) Role of high-fat diet in regulation of gene expression of drug metabolizing enzymes and transporters. Life Sci 89:57–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayhurst GP, Lee YH, Lambert G, Ward JM, Gonzalez FJ. (2001) Hepatocyte nuclear factor 4alpha (nuclear receptor 2A1) is essential for maintenance of hepatic gene expression and lipid homeostasis. Mol Cell Biol 21:1393–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson TN, Tucker GT, Tanner MS, Rostami-Hodjegan A. (2005) Changes in liver volume from birth to adulthood: a meta-analysis. Liver Transpl 11:1481–1493. [DOI] [PubMed] [Google Scholar]
- Jover R, Moya M, Gómez-Lechón MJ. (2009) Transcriptional regulation of cytochrome p450 genes by the nuclear receptor hepatocyte nuclear factor 4-alpha. Curr Drug Metab 10:508–519. [DOI] [PubMed] [Google Scholar]
- Kim S, Sohn I, Ahn JI, Lee KH, Lee YS, Lee YS. (2004) Hepatic gene expression profiles in a long-term high-fat diet-induced obesity mouse model. Gene 340:99–109. [DOI] [PubMed] [Google Scholar]
- Koh KH, Pan X, Shen HW, Arnold SL, Yu AM, Gonzalez FJ, Isoherranen N, Jeong H. (2014) Altered expression of small heterodimer partner governs cytochrome P450 (CYP) 2D6 induction during pregnancy in CYP2D6-humanized mice. J Biol Chem 289:3105–3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudo T, Shimada T, Toda T, Igeta S, Suzuki W, Ikarashi N, Ochiai W, Ito K, Aburada M, and Sugiyama K (2009) Altered expression of CYP in TSOD mice: a model of type 2 diabetes and obesity. Xenobiotica 39:889–902. [DOI] [PubMed] [Google Scholar]
- Lewis MC, Phillips ML, Slavotinek JP, Kow L, Thompson CH, Toouli J. (2006) Change in liver size and fat content after treatment with Optifast very low calorie diet. Obes Surg 16:697–701. [DOI] [PubMed] [Google Scholar]
- Li CY, Renaud HJ, Klaassen CD, Cui JY. (2016) Age-specific regulation of drug-processing genes in mouse liver by ligands of xenobiotic-sensing transcription factors. Drug Metab Dispos 44:1038–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Jimenez CP, Kyrmizi I, Cardot P, Gonzalez FJ, Talianidis I. (2010) Hepatocyte nuclear factor 4alpha coordinates a transcription factor network regulating hepatic fatty acid metabolism. Mol Cell Biol 30:565–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao J, Xiao Z, Kanamaluru D, Min G, Yau PM, Veenstra TD, Ellis E, Strom S, Suino-Powell K, Xu HE, et al. (2009) Bile acid signaling pathways increase stability of small heterodimer partner (SHP) by inhibiting ubiquitin-proteasomal degradation. Genes Dev 23:986–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller VM, Zietek T, Rohm F, Fiamoncini J, Lagkouvardos I, Haller D, Clavel T, Daniel H. (2016) Gut barrier impairment by high-fat diet in mice depends on housing conditions. Mol Nutr Food Res 60:897–908. [DOI] [PubMed] [Google Scholar]
- Pan X, Lee YK, Jeong H. (2015) Farnesoid X receptor agonist represses cytochrome P450 2D6 expression by upregulating small heterodimer partner. Drug Metab Dispos 43:1002–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swierczynska MM, Mateska I, Peitzsch M, Bornstein SR, Chavakis T, Eisenhofer G, Lamounier-Zepter V, Eaton S. (2015) Changes in morphology and function of adrenal cortex in mice fed a high-fat diet. Int J Obes 39:321–330. [DOI] [PubMed] [Google Scholar]
- Trouvin JH, Farinotti R, Haberer JP, Servin F, Chauvin M, Duvaldestin P. (1988) Pharmacokinetics of midazolam in anaesthetized cirrhotic patients. Br J Anaesth 60:762–767. [DOI] [PubMed] [Google Scholar]
- Ulvestad M, Skottheim IB, Jakobsen GS, Bremer S, Molden E, Asberg A, Hjelmesæth J, Andersson TB, Sandbu R, Christensen H. (2013) Impact of OATP1B1, MDR1, and CYP3A4 expression in liver and intestine on interpatient pharmacokinetic variability of atorvastatin in obese subjects. Clin Pharmacol Ther 93:275–282. [DOI] [PubMed] [Google Scholar]
- Ussar S, Griffin NW, Bezy O, Fujisaka S, Vienberg S, Softic S, Deng L, Bry L, Gordon JI, Kahn CR. (2015) Interactions between gut microbiota, host genetics and diet modulate the predisposition to obesity and metabolic syndrome. Cell Metab 22:516–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahlang B, Song M, Beier JI, Cameron Falkner K, Al-Eryani L, Clair HB, Prough RA, Osborne TS, Malarkey DE, Christopher States J, and Cave MC (2014) Evaluation of Aroclor 1260 exposure in a mouse model of diet-induced obesity and non-alcoholic fatty liver disease. Toxicol Appl Pharmacol 279:380–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahlström A, Sayin SI, Marschall HU, Bäckhed F. (2016) Intestinal crosstalk between bile acids and microbiota and its impact on host metabolism. Cell Metab 24:41–50. [DOI] [PubMed] [Google Scholar]
- Xu Y, Zalzala M, Xu J, Li Y, Yin L, Zhang Y. (2015) A metabolic stress-inducible miR-34a-HNF4α pathway regulates lipid and lipoprotein metabolism. Nat Commun 6:7466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshinari K, Takagi S, Yoshimasa T, Sugatani J, Miwa M. (2006) Hepatic CYP3A expression is attenuated in obese mice fed a high-fat diet. Pharm Res 23:1188–1200. [DOI] [PubMed] [Google Scholar]