Abstract
Oprozomib is an oral proteasome inhibitor currently under investigation in patients with hematologic malignancies or solid tumors. Oprozomib elicits potent pharmacological actions by forming a covalent bond with the active site N-terminal threonine of the 20S proteasome. Oprozomib has a short half-life across preclinical species and in patients due to systemic clearance via metabolism. Potential for drug-drug interactions (DDIs) could alter the exposure of this potent therapeutic; therefore, a thorough investigation of pathways responsible for metabolism is required. In the present study, the major drug-metabolizing enzyme responsible for oprozomib metabolism was identified in vitro. A diol of oprozomib was found to be the predominant metabolite in human hepatocytes, which formed via direct epoxide hydrolysis. Using recombinant epoxide hydrolases (EHs) and selective EH inhibitors in liver microsomes, microsomal EH (mEH) but not soluble EH (sEH) was found to be responsible for oprozomib diol formation. Coincubation with 2-nonylsulfanyl-propionamide, a selective mEH inhibitor, resulted in a significant decrease in oprozomib disappearance (>80%) with concurrent complete blockage of diol formation in human hepatocytes. On the contrary, a selective sEH inhibitor did not affect oprozomib metabolism. Pretreatment of hepatocytes with the pan-cytochrome P450 (P450) inhibitor 1-aminobenzotriazole resulted in a modest reduction (∼20%) of oprozomib metabolism. These findings indicated that mEH plays a predominant role in oprozomib metabolism. Further studies may be warranted to determine whether drugs that are mEH inhibitors cause clinically significant DDIs with oprozomib. On the other hand, pharmacokinetics of oprozomib is unlikely to be affected by coadministered P450 and sEH inhibitors and/or inducers.
Introduction
The proteasome is a multicatalytic enzyme complex and key to the ubiquitin-dependent pathway for intracellular protein degradation (Kisselev and Goldberg, 2001; Ciechanover, 2005). The highly regulated ubiquitin proteasome system affects a wide variety of cellular processes and is critical for cellular homeostasis. Inhibition of proteasome leads to accumulation of misfolded proteins and induction of cell cycle arrest and apoptosis. Transformed cells, particularly multiple myeloma (MM) cells, possess elevated levels of proteasome activity. Consequently, these actively proliferating malignant cells are more sensitive to proteasome inhibitors than normal cells (Kumatori et al., 1990; Loda et al., 1997; Li and Dou, 2000; Almond and Cohen, 2002). Proteasome inhibition has emerged as an effective therapeutic strategy for the treatment of MM and some lymphomas. Bortezomib (Velcade), the first-generation proteasome inhibitor, was approved in 2003 (Bross et al., 2004; Chauhan et al., 2005; O’Connor et al., 2005). Carfilzomib (Kyprolis), a peptide epoxyketone (Fig. 1), represents a new generation of proteasome inhibitors. It selectively binds to the N-terminal threonine of the proteasome via two covalent bonds to form a morpholino ring (Harshbarger et al., 2015), and has demonstrated favorable clinical safety and efficacy profiles compared with bortezomib (Kuhn et al., 2007). Carfilzomib has been approved for use as a single agent and in combination with lenalidomide and dexamethasone in patients with relapsed and/or refractory MM (Thompson, 2013). Notably, carfilzomib in combination with dexamethasone has doubled progression-free survival in patients with relapsed and/or refractory MM compared with bortezomib and dexamethasone in the phase 3 ENDEAVOR clinical trial (Dimopoulos et al., 2016). The introduction of proteasome inhibitors and immunomodulatory drugs has revolutionized MM treatment with the current overall survival of MM patients increased by 2- to 3-fold (Anderson, 2013).
Fig. 1.
Structures of carfilzomib and oprozomib. The numbers 1, 2, and 3 indicate three possible cleavage sites of oprozomib by peptidases. PR-025 is the cleavage product containing an epoxyketone at site 1 of oprozomib.
The success of bortezomib and carfilzomib has inspired efforts to develop oral proteasome inhibitors with improved flexibility in dose administration, specifically oral administration, for improved patient compliance and convenience. Ixazomib (Ninlaro) and oprozomib, representing peptide boronate and peptide epoxyketone, respectively, are two orally bioavailable proteasome inhibitors under clinical investigation (Rajan and Kumar, 2016). Ixazomib in combination with lenalidomide and dexamethasone administrated once weekly demonstrated efficacy in a phase 3 study (Moreau et al., 2016). Oprozomib (Fig. 1) is being evaluated in phase 1/2 clinical trials in patients with hematologic malignancy or solid tumors (Infante et al., 2016). Similar to carfilzomib, oprozomib inhibits primarily the chymotrypsin-like activity of the constitutive and immunoproteasomes via covalent modification of the N-terminal threonine (Zhou et al., 2009). It has shown a promising clinical activity in patients with relapsed and/or refractory MM either as a single agent or in combination with pomalidomide and dexamethasone (Prada et al., 2015; Shah et al., 2015; Vij et al., 2015).
Oprozomib is a tripeptide analog (Fig. 1) of carfilzomib that was developed to improve absorption. It was believed that smaller peptides were likely to be more effectively delivered across the small intestine (Hamman et al., 2005; Zhou et al., 2009). When dosed via oral gavage to Balb/C mice at 30 mg/kg, more than 80% proteasome inhibition was observed in blood, liver, and adrenal gland 1 hour post-administration (Zhou et al., 2009). Furthermore, oprozomib demonstrated improved metabolic stability in vitro across multiple species compared with carfilzomib. Nonetheless, oprozomib still displayed a high systemic clearance and short half-life (Fang et al., 2015). This is not surprising because both the peptide backbone and the epoxyketone warhead could be susceptible to metabolism (Yang et al., 2011; Wang et al., 2013). The rapid clearance of oprozomib was primarily mediated by metabolism, supported by the observation that the parent compound was a minor component in urine and bile samples from rats, as well as urine samples from human (in-house unpublished data). In addition, oprozomib has shown a relatively large interpatient pharmacokinetic (PK) variability (Infante et al., 2016).
The objective of the present study was to characterize the major metabolic pathways and key enzymes responsible for oprozomib metabolism in vitro using cryopreserved human hepatocytes, recombinant enzymes, and liver cellular fractions in the presence and absence of selective enzyme inhibitors. Deciphering the role of various enzymes responsible for oprozomib elimination is important in drug-drug interaction (DDI) risk assessment and mitigation of exaggerated pharmacology of this potent therapeutic. Particularly, it is important for cytochrome P450 (P450)–mediated DDIs since P450 isoforms are responsible for the primary metabolism of the majority of small molecule drugs on the market (Wienkers and Heath, 2005). Defining the primary metabolic routes could also help shed light on the observed oprozomib PK variabilities in patients.
Materials and Methods
Materials.
All chemicals and reagents were obtained from Sigma (St. Louis, MO) and liquid chromatography (LC) solvents were obtained from Fisher Scientific (Pittsburgh, PA). Oprozomib and authentic metabolite standards were synthesized and characterized at Onyx Pharmaceuticals, an Amgen subsidiary (South San Francisco, CA). Deuterated oprozomib (d5-oprozomib, five deuterium on the benzyl ring) was obtained from PepTech Corp. (Bedford, MA). A selective microsomal epoxide hydrolase (EH) inhibitor, 2-nonylsulfanyl-propionamide (NSPA) was previously synthesized (Morisseau et al., 2008), and a selective soluble EH (sEH) inhibitor 1-trifluoromethoxyphenyl-3-(1-propionylpiperidin-4-yl) urea (TPPU) was previously characterized (Rose et al., 2010). The structures of these two inhibitors are shown in Supplemental Fig. 1. Human recombinant microsomal EH (mEH, purity 80%) and sEH (purity 95%) were prepared and characterized as described previously (Morisseau et al., 2000, 2011). Supersomes containing cDNA-expressed human P450s, P450 reductase, and cytochrome b5 were purchased from BD Bioscience (San Jose, CA). Pooled human cryopreserved hepatocytes (20 donors, mixed gender) and media were purchased from BioreclamationIVT (Baltimore, MD). Liver subcellular fractions, including male or female liver microsomes, and cytosols (10 donors per gender, age: 21–58) were purchased from Sekisui Xenotech (Kansas City, KS). Pooled, gender-mixed liver microsomes were prepared by mixing an equal volume of male (10 donors) and female (10 donors) liver microsomes.
Metabolic Profiling of Oprozomib in Human Hepatocytes.
Pooled cryopreserved human hepatocytes were thawed at 37°C, centrifuged at 65g in 45 ml thawing medium, and resuspended in maintenance medium. Cell viability and density were measured by a trypan blue exclusion assay. For metabolic profiling, suspensions (1.0 ml) containing hepatocytes (0.5 million cells/ml) were incubated with 10 µM of oprozomib for 1 hour. Reactions were conducted in a 37°C cell culture incubator with 100% relative humidity, 95% air, and 5% CO2. Final incubation contained 0.2% of dimethylsulfoxide (DMSO). Reactions were terminated by addition of an equal volume of acetonitrile with 0.1% formic acid (FA). Control incubation was prepared as follows: a mixture containing hepatocytes only was incubated for 1 hour, quenched with acetonitrile with 0.1% FA, and then spiked with 10 µM of oprozomib. Supernatants (600 µl) were concentrated under a nitrogen stream and then reconstituted in 100 µl of 40% acetonitrile/water (0.1% FA). Metabolite identification was conducted using a Thermo Oribtrap Q Exactive high-resolution mass spectrometer equipped with a Thermo Accela LC system from Thermo Fisher Scientific (Waltham, MA). Metabolite structures were assigned based on the accurate mass obtained from full scan mode at a resolution of 70,000 and tandem mass spectrometry (MS/MS) fragmentation pattern from data-dependent acquisition at a resolution of 17,500. For the quantification of oprozomib diol (PR-176) and (S)-2-amino-1-((R)-2-methyloxiran-2-yl)-3-phenylpropan-1-one (PR-025) diol formation in human hepatocytes, cells (0.25 ml, 0.5 million cells/ml) were incubated with 2 µM of oprozomib in 12-well, collagen-coated plates (Thermo Scientific). At each time point (0, 10, 20, 30, and 60 minutes), reactions were terminated by the addition of an equal volume of acetonitrile containing 100 ng/ml internal standard (IS) d5-oprozomib. After centrifugation, supernatants were collected and analyzed for oprozomib and the major metabolites by LC-MS/MS as described subsequently.
Epoxide Hydrolysis by Recombinant Human mEH and sEH In Vitro.
Recombinant human EHs were kept at −80°C until use. Briefly, incubations (0.1 ml) were conducted in 0.1 M Tris-HCl buffer (pH 9.0 for mEH; pH 7.5 for sEH) containing 0.1 mg/ml fatty acid-free bovine serum albumin (BSA) (Morisseau et al., 2000, 2011). Enzyme activities of mEH and sEH (5 µg/ml) toward epoxide hydrolysis were confirmed using their corresponding probe substrates (50 µM) cis-stilbene oxide (cis-SO) and trans-stilbene oxide (trans-SO), respectively. To determine whether oprozomib is a substrate for recombinant mEH and/or sEH, enzymes were prewarmed for 5 minutes at 37°C before the addition of oprozomib (2 µM) to initiate the reaction. The enzyme concentration used was from 1 to 10 µg/ml for mEH and up to 100 μg/ml for sEH. After 0, 10, 20, or 30 minutes as indicated, reactions were terminated by addition of 2 volumes of acetonitrile containing 0.1% FA and supernatants were collected for LC-MS/MS analysis.
Kinetics of Oprozomib Epoxide Hydrolysis in Recombinant mEH and Human Liver Microsomes (HLMs).
Reaction kinetic parameters (Vmax and Km) were determined using recombinant mEH (2 µg/ml, calculated based on 80% purity) and oprozomib (10–600 µM) in 0.1 M Tris-HCl buffer (pH 9.0) containing 0.1 mg/ml BSA. Kinetic studies in male and female HLMs were conducted in 0.1 M potassium phosphate buffer (pH 7.4) containing 0.1 mg/ml BSA. The final microsomal protein concentration was optimized to be 0.5 mg/ml and oprozomib was 10–600 µM. The final DMSO concentration was 0.5%. Oprozomib was added to initiate the reaction after prewarming enzymes for 5 minutes. Incubations (0.2 ml) were conducted at 37°C for 30 minutes and terminated by addition of 2 volumes of acetonitrile containing 100 ng/ml of IS d5-oprozomib. For kinetic studies of cis-SO, incubations (0.4 ml) were optimized and performed using 4 μg/ml recombinant mEH or 0.1 mg/ml microsomal proteins and cis-SO (1–200 μM) at 37°C for 10 minutes. The reaction was terminated by addition of one volume acetonitrile containing 50 μM trans-SO as IS. Supernatants were collected by centrifugation and analyzed by either ultra-performance LC-MS/MS or ultra-performance LC linked with a photodiode array (PDA) detector as described subsequently. Each set of data was fit to a simple Michaelis-Menten kinetics model using nonlinear regression data analysis (GraphPad Prism version 6; GraphPad Software Inc., La Jolla, CA). Each experimental reaction condition was conducted in triplicate.
NSPA was used as the selective mEH inhibitor to inhibit mEH enzyme activities in HLMs (Morisseau et al., 2008). Briefly, prewarmed pooled HLMs (0.5 mg/ml) were incubated with oprozomib (10 μM) in the absence or presence of NSPA (0.02–100 μM) in 0.1 M potassium phosphate buffer (pH 7.4, 0.1 mg/ml BSA) at 37°C. After 15 minutes, reactions were quenched with 2 volumes of acetonitrile containing 0.1% FA and supernatants were isolated for quantitative analysis. The inhibitory effects of NSPA on cis-SO (50 μM) hydrolysis were also determined under similar conditions except for a lower microsomal protein concentration (0.1 mg/ml) and shorter incubation time (10 minutes). The IC50 values were estimated using nonlinear regression data analysis (GraphPad Prism version 6).
Inhibition of Oprozomib Epoxide Hydrolysis in Human Hepatocytes in the Presence of EH Inhibitors.
Pooled cryopreserved human hepatocytes were thawed and resuspended in hepatocyte maintenance medium, and cell viability and density were measured. Incubation mixtures (0.1 ml) contained human hepatocytes (0.5 million cells/ml), oprozomib (2 μM), or cis-SO (50 μM), and inhibitors NSPA or TPPU (10 μM). The final solvent DMSO concentration was 0.2%. At each time point (0, 2, 5, 10, 20, 40, and 60 minutes), reactions were terminated by the addition of 2 volumes of acetonitrile containing either IS d5-oprozomib or trans-SO. After centrifugation, supernatants (150 μl) were collected and analyzed using LC-MS/MS or LC-PDA detection as described subsequently.
Inhibition of P450-Mediated Oprozomib Metabolism in Human Hepatocytes by 1-Aminobenzotriazole (1-ABT).
The role of P450s on oprozomib metabolism in human hepatocytes was investigated using a pan-P450 inhibitor 1-ABT. Pooled cryopreserved human hepatocytes were thawed at 37°C, centrifuged, and resuspended in hepatocyte maintenance medium. Incubations (0.25 ml) containing hepatocytes (0.5 million cells/ml) were preincubated with 0.5 mM 1-ABT or solvent (0.1% DMSO) for 30 minutes. Oprozomib was added at a final concentration of 2 µM to initiate the reactions. Reactions were conducted in a 37°C cell culture incubator with 100% relative humidity, 95% air, 5% CO2. At each time point (0, 2, 5, 10, 20, 40, and 60 minutes), reactions were terminated by the addition of an equal volume of acetonitrile containing IS d5-oprozomib. After centrifugation, supernatants were collected for the quantification of oprozomib disappearance and PR-176 formation by LC-MS/MS.
Metabolic Stability of Oprozomib in HLMs and Recombinant P450s.
Metabolic stability of oprozomib in pooled HLMs (mixed gender) was conducted in the presence and absence of cofactor NADPH. The value of intrinsic clearance was determined by the rate of parent compound disappearance (Obach, 1999). Briefly, oprozomib (1 µM) was mixed with liver microsomes (0.25 mg/ml) with or without 1 mM NADPH in 0.1 M potassium phosphate buffer (pH 7.4) containing 3.3 mM of MgCl2. The incubation mixtures were kept at 37°C for 1, 5, 10, 20, 30, and 40 minutes. The disappearance of oprozomib was monitored using a LC-MS/MS method described subsequently. In addition, a preliminary in vitro P450 phenotyping study was conducted by incubating oprozomib (1 µM) with six major recombinant P450 isoforms (125 pmol/ml) 1A2, 2D6, 3A4, 2C8, 2C9, and 2C19 individually, in the presence of 1 mM NADPH at 37°C for 0, 15, 30, and 60 minutes. The percentage of oprozomib remaining after incubation was calculated by comparing the peak response with that from the zero-minute incubation.
Inhibition of P450 Activities by Oprozomib.
The relative potential of oprozomib to inhibit six major P450 isoforms was assessed in pooled HLMs (mixed gender) with and without 30-minute preincubation. The probe substrates used were phenacetin (CYP1A2), amodiaquine (CYP2C8), diclofenac (CYP2C9), S-mephenytoin (CYP2C19), dextromethorphan (CYP2D6), midazolam, and testosterone (CYP3A4/5). The incubation mixtures (0.2 ml) contained oprozomib at various concentrations (0–30 μM), a probe substrate, microsomal proteins, and 1.5 mM NADPH in 0.1 M potassium phosphate buffer (pH 7.4) with 3 mM MgCl2 for 15 minutes. To examine time-dependent inhibition, oprozomib was preincubated with liver microsomes for 30 minutes in the presence of NADPH, followed by the addition of probe substrates. The concentrations of P450 probe substrates were approximately equal to their reported Km values. The microsomal protein concentration was 0.05 or 0.1 mg/ml. The low protein concentrations were chosen to minimize possible binding of oprozomib to microsomal proteins or lipids, as well as reduce oprozomib metabolism via epoxide hydrolysis.
Quantification of Oprozomib and Its Metabolites Using LC-MS/MS.
Quantification of oprozomib and its major metabolites was performed using either an AB Sciex QTrap 4500 or 5500 mass spectrometer equipped with an electrospray ionization source (Redwood City, CA). Chromatographic separation was achieved using a Waters (Milford, MA) XBridge BEH C18 column (3.5 μ, 2.1 mm × 50 mm) and water (A)/acetonitrile (B) containing 0.1% FA as mobile phases at a flow rate of 0.5 ml/min. The mobile phase started at 10% B for 0.5 minute, linearly increased to 65% B within 1 minute, held for 0.3 minute before increasing to 95% B within 0.2 minute, held at 95% B for 1.5 minutes, and returned to 10% B within 0.2 minute. Multiple reactions monitoring of the transitions m/z 533.0 > 126.0, 551.3 > 325.2, 206.0 > 120, 224.0 > 150.0, and 538.2 > 126.1 was used to detect oprozomib, PR-176, PR-025, PR-025 diol, and IS d5-oprozomib, respectively. Calibration curves were generated by plotting the peak area ratio for each compound and IS versus the corresponding concentrations of synthetic metabolite standards, and fitting with a linear regression equation.
Quantification of cis-SO, trans-SO, R,R-Hydroxybenzoin, and meso-Hydroxybenzoin Using LC-PDA Detection.
Formation of R,R-hydroxybenzoin and meso-hydroxybenzoin from cis-SO and trans-SO hydrolysis was used for measuring mEH and sEH activities in vitro, respectively. Quantification of these two diol products was performed using a Shimadzu LC-20AD system coupled with a PDA detector (Columbia, MD). Chromatographic separation was achieved using a Waters XBridge BEH C18 column (2.1 mm × 100 mm, 3.5 μm) and water (A)/acetonitrile (B) containing 0.1% FA as mobile phases at a flow rate of 0.3 ml/min. The mobile phase started at 10% B for 0.5 minute, linearly increased to 95% B at 5 minutes, and held for 2 minutes before returning to initial 10% B within 0.5 minute. The total running time was 9 minutes. Quantification of the diol metabolites was calculated by fitting their peak area ratio (metabolites/IS) to the calibration curves that were generated by using authentic metabolite standards R,R-hydroxybenzoin and meso-hydroxybenzoin, respectively.
Statistical Analysis.
GraphPad Prism version 6 was used for the statistical analysis. Data represent mean ± S.D. The mean values of the measured values of each treatment group were compared using Student’s t test. Means were considered statistically different from one another if P < 0.05.
Results
Oprozomib Metabolism in Human Hepatocytes.
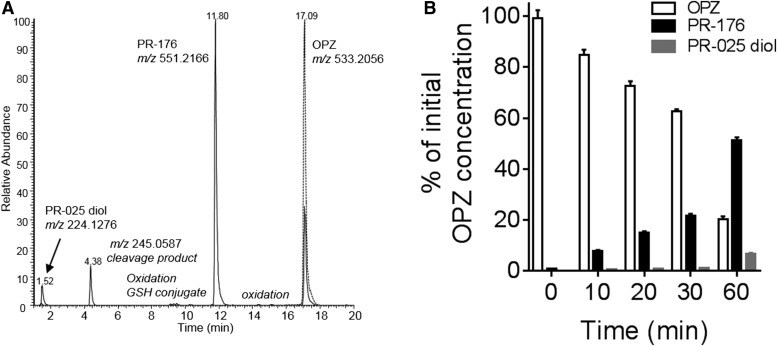
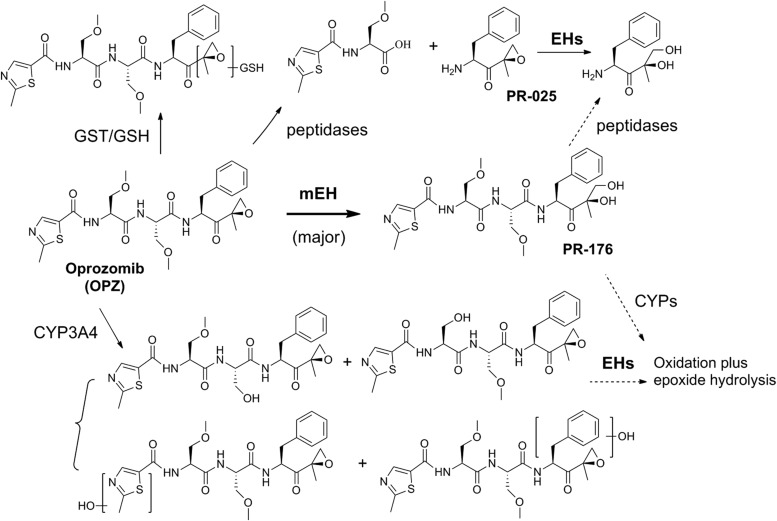
Incubation in cryopreserved human hepatocytes with oprozomib for 60 minutes led to the formation of several metabolites, which were detected by LC-MS/MS (Fig. 2). A diol of oprozomib (PR-176) was formed from direct epoxide hydrolysis (Fig. 2A). This appeared as the predominant metabolite based upon both mass spectrometer ion signals and ultraviolet absorbance. A peptide bond cleavage product with m/z at 245.0587 (cleavage on bond 2) was also observed, albeit at relatively low levels. Another low abundance metabolite with m/z at 224.1276 (diol of PR-025) resulted from peptide cleavage (cleavage on bond 1) and epoxide hydrolysis was detected. In addition, multiple trace metabolites, including from hydroxylation and/or de-methylation, direct glutathione (GSH) conjugation, and the combination of oxidation and epoxide hydrolysis or GSH conjugation, were also observed.
Fig. 2.
In vitro metabolism of oprozomib in human hepatocytes. (A) Representative ion chromatograms of oprozomib metabolites formed in human hepatocytes. Dash line: control sample, cell suspensions spiked with oprozomib; solid line: cell suspensions after 1-hour incubation with oprozomib. (B) Quantitative oprozomib (OPZ) disappearance and metabolite formation in human hepatocytes. At each time point, the levels of oprozomib, PR-176, and PR-025 diol in cell suspensions were quantified and then normalized as the percentage of initial oprozomib concentration at zero minutes. Data represent mean ± S.D. from three replicate incubations. PR-176 was the predominant metabolite formed via epoxide hydrolysis within 1-hour incubation.
Quantification of oprozomib disappearance and its major metabolite formation in hepatocyte incubation was performed using synthesized standards. As shown in Fig. 2B, PR-176 was readily formed after incubation with oprozomib (2 µM). After 60-minute incubation, nearly 55% of total oprozomib was recovered as the diol, about 7% was recovered as the diol of PR-025, and 20% remained as oprozomib. These quantitative results suggest that epoxide hydrolysis is the major metabolic pathway for oprozomib in human hepatocytes. Other minor pathways, such as peptide cleavage, oxidation, and GSH conjugation, combined to contribute to the remaining less than 20% of oprozomib from the initial incubation. It is worth mentioning that in rat hepatocyte incubations, two oprozomib conjugates via direct GSH conjugation were identified as major metabolites, in addition to PR-176 (Supplemental Fig. 2).
Oprozomib Hydrolysis by Recombinant Human mEH but Not sEH.
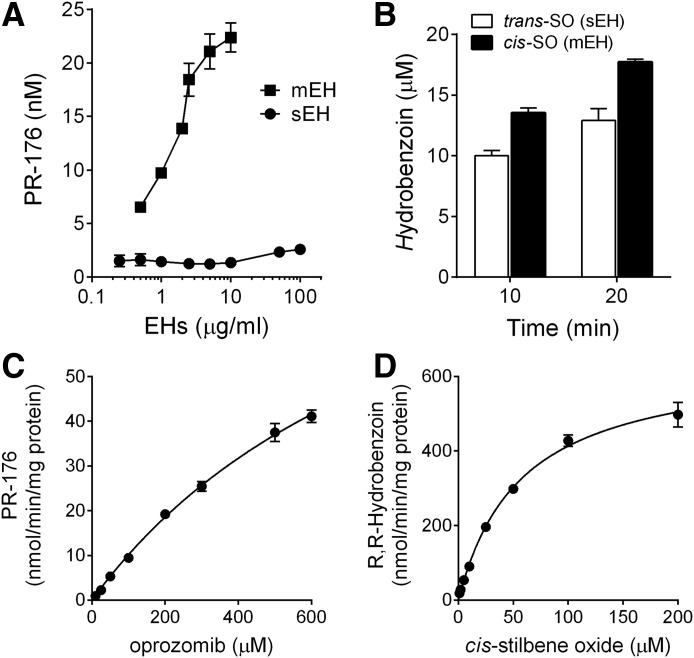
Since epoxide hydrolysis was identified to be the major metabolic pathway, recombinant human mEH and sEH were used to investigate the capacity of the different EH isoforms to hydrolyze oprozomib epoxide. Formation of PR-176 increased linearly with both incubation time (up to 30 minutes, data not shown) and mEH concentration (0.5–10 μg/ml) as shown in Fig. 3A. In contrast, no significant formation of PR-176 was observed when oprozomib was incubated with recombinant sEH for 30 minutes, at enzyme concentrations up to 100 μg/ml. In a parallel experiment, both mEH and sEH can readily convert their corresponding probe substrates, cis-SO or trans-SO, respectively, in a time-dependent manner (Fig. 3B).
Fig. 3.
Epoxide hydrolysis by recombinant microsomal and soluble EHs. (A) Epoxide hydrolysis of oprozomib by recombinant human mEH and sEH. Formation of PR-176 was quantified after incubation of oprozomib (2 µM) with various concentrations of enzymes for 30 minutes. A pilot study has shown a linear formation of PR-176 within 30 minutes under the current conditions. (B) Epoxide hydrolysis of cis-SO and trans-SO (50 µM) by mEH and sEH (5 µg/ml), respectively. Significant diol product formation was observed after incubation for 10 or 20 minutes, confirming the hydrolysis activities of EHs. (C) Kinetics of the formation of PR-176 using recombinant mEH. A range of oprozomib concentrations were incubated with mEH (2 µg/ml) at 37°C for 30 minutes. (D) Kinetics of cis-SO hydrolysis using recombinant mEH. A series of cis-SO concentrations were incubated with mEH (4 μg/ml) at 37°C for 10 minutes. Data represent mean ± S.D. from three replicate incubations. The Km and Vmax values were estimated by fitting the curve into the Michaelis-Menten equation.
The kinetic properties of oprozomib epoxide hydrolysis were subsequently evaluated using recombinant mEH. The highest oprozomib concentration (up to 600 µM) was chosen based on its aqueous solubility in the phosphate buffer system. An optimized protein concentration (2 μg/ml) and incubation time (30 minutes) were applied in the study to ensure the linearity over the course of the reaction with sufficient amounts of PR-176 formation for accurate quantification. As shown in Fig. 3C, formation of PR-176 increased with oprozomib concentration up to 600 μM without reaching a plateau. Fitting the data into the Michaelis-Menten equation estimated the values of Km and Vmax of oprozomib at 974 ± 112 μM and 109 ± 8.6 nmol/min/mg proteins, respectively. While the values of Km and Vmax could not be more accurately determined under the current assay conditions, the estimation suggests a relatively high Km value for oprozomib epoxide hydrolysis with recombinant mEH. As a control, the values of Km and Vmax of cis-SO were determined to be 56.9 ± 3.8 μM and 648 ± 17 nmol/min/mg proteins, respectively, which were comparable to previously reported values (Morisseau et al., 2011).
Inhibition of Oprozomib Epoxide Hydrolysis by a Selective mEH Inhibitor in HLMs.
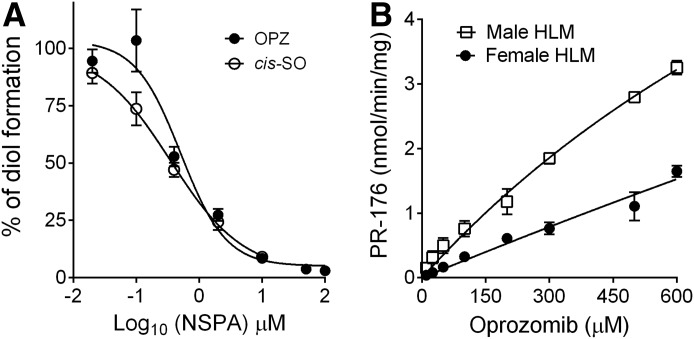
NSPA is a selective mEH inhibitor that has been previously characterized with recombinant rat and human mEH for its potency (Morisseau et al., 2008, 2011). Here, we evaluated its potency in inhibiting hydrolysis of cis-SO and oprozomib in HLMs. As seen in Fig. 4A, NSPA significantly inhibited cis-SO hydrolysis with a mean IC50 value of 0.35 μM, and oprozomib diol formation with a mean IC50 value of 0.51 μM in HLMs. The kinetic properties of oprozomib diol formation in both male and female HLMs were also determined (Fig. 4B). Similar to what was observed in the kinetic study with recombinant mEH (Fig. 3), formation of PR-176 essentially increased linearly with oprozomib concentrations without reaching a plateau. As a result, the values of Km and Vmax could not be accurately estimated using nonlinear regression analysis. Considering the Km values were likely over 5-fold higher than 50 µM, the apparent ratios of Vmax/Km values were roughly calculated from the slopes of the PR-176 formation rate over a low oprozomib concentration range (10–50 μM). As a result, the mean Vmax/Km values were estimated as 8.43 and 3.20 μl/min/mg protein in male and female HLMs, respectively. In addition, the mean values of Vmax/Km toward cis-SO and oprozomib epoxide hydrolysis in liver microsomes from rats and dogs were also estimated (Supplemental Table 1). As a control, the estimated mean Vmax/Km values for cis-SO were comparable to values reported in the literature (Kitteringham et al., 1996).
Fig. 4.
Inhibition and kinetics of PR-176 formation in HLMs. (A) Inhibition of mEH activity by NSPA in HLMs. Pooled HLMs (0.5 mg/ml) were incubated with oprozomib (10 μM) or cis-SO (50 µM) in the presence of various concentrations of NSPA at 37°C for 15 minutes as indicated. The IC50 values were estimated using nonlinear regression data analysis. (B) Kinetics of PR-176 formation in male and female HLMs was conducted in 0.1 M potassium phosphate buffer (pH 7.4, 0.1 mg/ml BSA). Incubations were performed at 37°C for 30 minutes. Data were fitted into the Michaelis-Menten equation for calculation of the Km and Vmax values. Data represent mean ± S.D. from triplicate incubations.
Effect of the mEH Inhibitor NSPA on Oprozomib Metabolism in Human Hepatocytes.
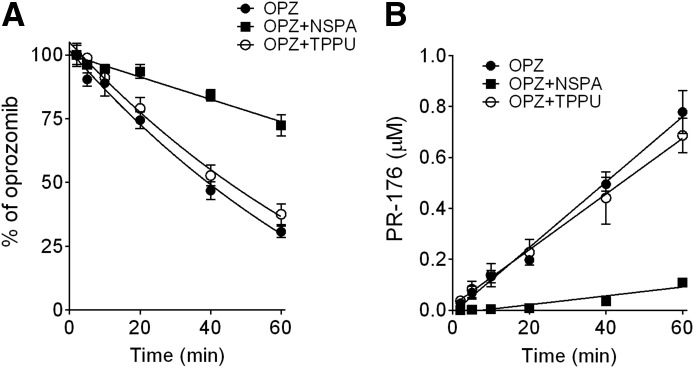
Both NSPA and TPPU were used to further determine the importance of mEH and sEH, respectively, on oprozomib metabolism in human hepatocytes (Fig. 5). The metabolic stability of the two inhibitors, TPPU and NSPA, in human hepatocytes was first determined. TPPU was stable in human hepatocytes over a 1-hour incubation, whereas NSPA was labile with a half-time value of ∼15 minutes. Thus, a concentration of NSPA (10 μM) 20-fold higher than the measured in vitro IC50 values on mEH activity was used in the human hepatocyte study. The inhibitory effect of NSPA in human hepatocytes was first confirmed using cis-SO as the probe substrate, where over 80% inhibition of cis-SO hydration was observed within 10-minute incubation; however, inhibition was reduced to 60% with a longer incubation time (up to 20 minutes; data not shown). The rate of oprozomib metabolism in human hepatocytes was significantly inhibited (∼80%) by NSPA cotreatment (9.7 ± 0.9 μl/min/106 cells) compared with the vehicle (DMSO) control (40.7 ± 1.4 μl/min/106 cells). Consistently, formation of PR-176 was also significantly decreased (>90% inhibition) by NSPA treatment up to the 60-minute incubation (Fig. 5B). In contrast, the sEH inhibitor TPPU did not significantly inhibit oprozomib metabolism (35.0 ± 1.5 μl/min/106 cells) and PR-176 formation (Fig. 5).
Fig. 5.
Inhibition of epoxide hydrolysis of oprozomib in human hepatocytes by the mEH inhibitor NSPA, but minimal inhibition by the sEH inhibitor TPPU. (A) Percentage of oprozomib remaining in human hepatocytes. (B) Formation of PR-176 in human hepatocytes. Cryopreserved human hepatocytes were incubated with oprozomib (2 μM) in the presence of NSPA, TPPU, or vehicle (0.1% DMSO) for various incubation times. Both oprozomib disappearance and PR-176 formation were quantified using LC-MS/MS. Data represent mean ± S.D. from triplicate incubations. The percentage of oprozomib was calculated using the remaining oprozomib concentration divided by zero-minute oprozomib concentration.
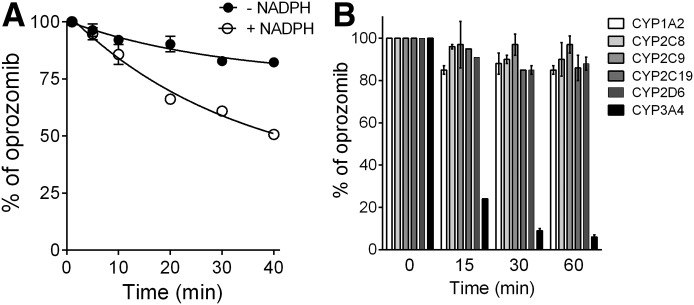
Metabolic Stability of Oprozomib in HLMs in the Presence and Absence of NADPH.
Metabolic identification and quantitation results (Figs. 2 and 5) in human hepatocytes have suggested a minor role of P450s on oprozomib metabolism. HLM contains high hepatic mEH activity, and is also a common in vitro system for assessment of P450-mediated metabolism. Therefore, microsomes were expected to be a relevant in vitro system to study the metabolism of oprozomib. Interestingly, in pooled HLMs, the presence of NADPH significantly increased the intrinsic clearance of oprozomib by 3.5-fold compared with that in the absence of NADPH (70.7 ± 2.9 versus 20.2 ± 1.8 µl/min/mg protein) (Fig. 6A). These results indicated a significant role of oxidation pathways over epoxide hydrolysis for oprozomib metabolism in HLMs, which is not consistent with what was observed in human hepatocytes (Fig. 2).
Fig. 6.
Metabolism of oprozomib by HLMs and recombinant P450s. (A) Percentage of oprozomib remaining in pooled HLMs in the presence or absence of cofactor NADPH. Initial oprozomib concentration was 1 µM. (B) P450 phenotyping on oprozomib oxidation using six major P450 isoforms. Oprozomib was incubated with six recombinant P450 isoforms for various incubation times, individually. The loss of oprozomib was quantified using LC-MS/MS assays. Data represent mean ± S.D. from triplicate incubations.
Oprozomib Showed Time-Dependent Inhibition of CYP3A4 in HLMs.
In vitro inhibition potential of oprozomib on major P450 isoforms (CYP1A2, 2C8, 2C9, 2C19, 2D6, and 3A4/5) was also evaluated using pooled HLMs (Table 1). Oprozomib did not inhibit the activities of CYP1A2, 2C8, 2C9, 2C19, and 2D6 at concentrations up to 30 µM with or without 30-minute preincubation. Oprozomib (30 µM) reduced the activity of CYP3A4/5 by 45% and 33% as measured by testosterone 6β-hydroxylation and midazolam 1′-hydroxylation assays, respectively, without preincubation. The inhibitory effect was further enhanced with 30-minute preincubation, where the IC50 values decreased to 12 and 5.5 µM with midazolam and testosterone as substrates, respectively (Table 1). This time-dependent inhibition was NADPH-dependent and resistant to dilution (data not shown). The ratios of the maximum inactivation rate constant (kinact) to the concentration of the inactivator that gives half the maximum rate of inactivation (KI) were about 2.0 and 1.4 ml/min/µmol, respectively, with midazolam and testosterone as marker substrates. In addition, a preliminary P450 phenotyping study using recombinant P450 isoforms showed oprozomib was a substrate of CYP3A4 (Fig. 6B).
TABLE 1.
In vitro evaluation of oprozomib as an inhibitor of six human P450 enzymes
Results representing the average data (i.e., percentage of control activity) obtained from duplicate samples from each test article concentration were used to calculate the IC50 values. The IC50 values were calculated using GraphPad Prism (version 6). The highest concentration of oprozomib tested in the assays was 30 µM.
| Enzyme |
P450 Reaction |
Inhibition of Human P450s by Oprozomib | |||
|---|---|---|---|---|---|
| Direct (0-Minute Preincubation) |
Time Dependent (30-Minute Preincubation) |
||||
| IC50 |
Inhibition at 30 µMa |
IC50 |
Inhibition at 30 µMa |
||
| µM | % | µM | % | ||
| CYP1A2 | Phenacetin O-deethylation | >30 | 21 | >30 | NAb |
| CYP2C8 | Amodiaquine N-dealkylation | >30 | 20 | >30 | 13 |
| CYP2C9 | Diclofenac 4′-hydroxylation | >30 | 6.3 | >30 | 24 |
| CYP2C19 | S-Mephenytoin 4′-hydroxylation | >30 | 16 | >30 | 7.2 |
| CYP2D6 | Dextromethorphan O-demethylation | >30 | 0.2 | >30 | 3.7 |
| CYP3A4/5 | Testosterone 6β-hydroxylation | >30 | 45 | 5.5 | 89 |
| CYP3A4/5 | Midazolam 1′-hydroxylation | >30 | 33 | 12 | 68 |
NA, not applicable.
The inhibition (%) was calculated using the data for the highest concentration of test article evaluated and the following formula: inhibition (%) = 100% − % activity remaining of solvent control.
No value was obtained since the rates at the highest concentration of oprozomib evaluated (30 µM) were higher than the control rates.
Effect of the pan-P450 Inhibitor 1-ABT on Oprozomib Metabolism in Human Hepatocytes.
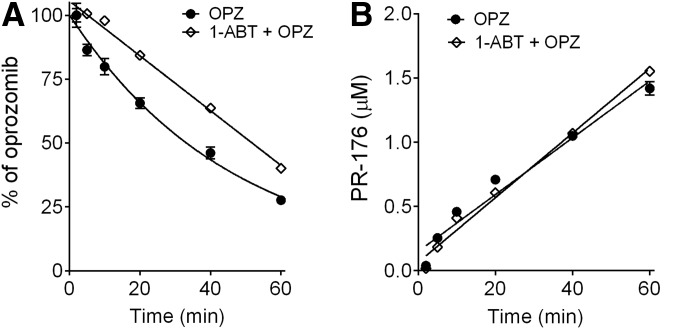
In this experiment, the rates of oprozomib metabolism in human hepatocytes in the absence and presence of pan-P450 inhibitor 1-ABT were compared. After preincubation with 1-ABT (0.5 mM) to inactivate P450 enzymes (in control experiments, 1-ABT reduced testosterone oxidation by 90%), oprozomib disappearance was modestly reduced by ∼20% (clearance with 1-ABT: 31.6 ± 1.4 μl/min/106 cells; without 1-ABT: 41.8 ± 1.2 μl/min/106 cells) (Fig. 7A). As expected, 1-ABT did not affect the formation of PR-176 (Fig. 7B). In addition, pretreatment of hepatocytes with 0.5 mM 1-ABT completely blocked the formation of oprozomib metabolites from direct P450-mediated oxidation, as well as secondary products through oxidation and epoxide hydrolysis, as shown in Fig. 2.
Fig. 7.
Inhibition of P450 activities for oprozomib in human hepatocytes by 1-ABT. (A) Percentage of oprozomib remaining in human hepatocytes. (B) Formation of PR-176 in human hepatocytes. Cryopreserved human hepatocytes were incubated with oprozomib (2 μM) after preincubation with 1-ABT (0.5 mM) or vehicle (0.1% DMSO) for 30 minutes. Both oprozomib disappearance and PR-176 formation after various incubation times were quantified using LC-MS/MS. Data represent mean ± S.D. from triplicate incubations. The percentage of oprozomib was calculated using the remaining oprozomib concentration divided by zero-minute oprozomib concentration.
Discussion
Peptide epoxyketone drugs represent a new class of proteasome inhibitors that are highly efficacious for the treatment of patients with MM. Since these inhibitors act via covalent modification, they induce prolonged pharmacodynamic effects in spite of a short duration of systemic exposure (Yang et al., 2011; Wang et al., 2013). Proteasome activity recovery primarily relies on the rate of de novo proteasome synthesis, with a half-life of about 20–72 hours across a variety of tissues in preclinical species, except for blood after dosing with oprozomib (Zhou et al., 2009; Dou and Zonder, 2014). Different from the first-generation epoxyketone proteasome inhibitor carfilzomib, which is administered intravenously due to the lack of oral bioavailability, oprozomib demonstrated improved metabolic stability and was orally bioavailable (Zhou et al., 2009). However, oprozomib still displayed high systemic clearance, a short half-life, and relatively high PK variability in preclinical species and patients (Fang et al., 2015; Infante et al., 2016).
To understand the parameters governing the elimination of oprozomib, we conducted a number of in vitro metabolism studies. As summarized in Scheme 1, the in vitro metabolism data showed that hydrolysis of oprozomib epoxide to form the diol PR-176 by EH is the major metabolic pathway in human hepatocytes, with peptide cleavage by peptidases, oxidation by P450s, and direct GSH conjugation representing minor alternative metabolic pathways. Consistent with the results in human hepatocytes, PR-176 was also found to be a major circulating metabolite in dogs, the non-rodent toxicity species, as well as human plasma samples from a first-in-human clinical study (in-house unpublished data). Interestingly, GSH conjugation, an alternative pathway to detoxify xenobiotic epoxides, was minor in human hepatocytes. Although cryopreservation may reduce the in vitro GSH-conjugating capacity in both human and rat hepatocytes (Sohlenius-Sternbeck and Schmidt, 2005), two GSH conjugates of oprozomib were identified as major metabolites in addition to diol in cryopreserved rat hepatocytes. In addition, multiple GSH conjugates were observed for clozapine, a positive control for GSH adduct formation in both rat and human hepatocytes (Supplemental Fig. 3). Consistent with the in vitro observation, no detectable GSH conjugates were observed in human plasma and urine samples; on the other hand, GSH conjugates were observed in vivo in rat plasma samples (in-house unpublished data). These data indicate GSH conjugation plays a minor role in oprozomib metabolism in humans.
Scheme 1.
Proposed major metabolic pathways of oprozomib in human hepatocytes. Oprozomib is predominantly metabolized by mEH in human hepatocytes, whereas P450s, peptidases, and glutathione S-transferase (GST)/GSH play minor roles. Oxidative and cleavage products containing an epoxyketone could be further metabolized via epoxide hydrolysis.
The predominant role of EHs on oprozomib metabolism could have implications on potential DDIs and therefore may influence development strategy. Several EHs are found in mammals; among them, two major isoforms, mEH and sEH, have been extensively characterized (Fretland and Omiecinski, 2000; Kodani and Hammock, 2015). Both isoforms are expressed ubiquitously throughout the mammalian body with the highest expression generally in the liver (Gill and Hammock, 1980; Pacifici et al., 1988; Coller et al., 2001). Collectively, data from recombinant EHs (Fig. 3), liver microsomes (Fig. 4), and hepatocytes in the presence or absence of selective inhibitors (Fig. 5) indicate that the epoxide ring opening of oprozomib was catalyzed by mEH, but not sEH. Therefore, oprozomib PK is unlikely to be affected by co-administration with sEH inhibitors. On the other hand, oprozomib is primarily metabolized by mEH. Some amide-containing drugs such as valpromide or progabide have been shown to inhibit in vivo mEH activity (Fretland and Omiecinski, 2000; Kodani and Hammock, 2015). Thus, further studies may be warranted to determine whether drugs that are mEH inhibitors cause clinically significant DDIs with oprozomib. Moreover, mEH is involved in the metabolism of numerous xenobiotics, including 1,3-butadiene, naphthalene, carbamazepine, and phenytoin (Decker et al., 2009). It is possible that oprozomib acts as a mEH inhibitor that affects the clearance of coadministered drugs, which are mainly metabolized by mEH. However, a preliminary in vitro study in HLMs showed that oprozomib does not significantly inhibit cis-SO epoxide hydrolysis at a concentration up to 100 µM (data not shown).
The high variability of oprozomib PK observed in preclinical species and patients could be attributed to variability of absorption and/or metabolism by mEH. Expression of mEH in ex vivo tissues was shown to vary considerably among individuals (up to 8-fold) (Hassett et al., 1997), and that expression is affected by age, sex, diseases, and environmental factors (Václavíková et al., 2015). In addition, two human polymorphisms have been identified in the coding region of the mEH gene: exon 3 (a Tys113His mutation), resulting in a 50% decrease in enzyme activity in vitro, and exon 4 (a His139Arg mutation), resulting in a 25% increase in enzyme activity in vitro (Pinarbasi et al., 2010). However, the variant alleles appeared to exert only a modest impact on mEH enzymatic activities (within 2-fold) in vivo (Kroetz et al., 1993; Hosagrahara et al., 2004). Based on this information and the high systemic clearance of oprozomib, interindividual difference in mEH activity may contribute to the high interindividual PK variability of oprozomib in patients (Infante et al., 2016), but it is unlikely to be the main factor. Variable absorption, particularly associated with a narrow absorption window, may be another key factor in oprozomib PK variability (Teague et al., 2013).
In contrast to the major role of mEH, a minor role of P450 on oprozomib metabolism was observed in human hepatocytes (Fig. 7). Using the pan-P450 inhibitor 1-ABT, a modest decrease in oprozomib disappearance (<20%) was observed. This corroborates the result that oxidative products are minor metabolites in human hepatocytes (Fig. 2), as well as in plasma samples from patients (in-house unpublished data). Thus, although oprozomib is a substrate of CYP3A4, coadministration of oprozomib with other drugs that are P450 modulators would be unlikely to significantly alter the PK of oprozomib. In addition, oprozomib did display time-dependent inhibition of CYP3A4/5 in HLMs (Fig. 6); however, the inhibitory effect was not manifested in vivo in a clinical DDI study where oprozomib was administered orally and coadministered with midazolam (Tsimberidou et al., 2016).
Choosing relevant in vitro systems is critical in understanding the metabolism and associated DDI potentials for oprozomib and other peptide epoxyketone inhibitors. When studying oprozomib metabolism in HLMs (Fig. 6), a higher level of P450-mediated metabolism was observed in contrast to the results observed in human hepatocytes (Fig. 7). Moreover, the high levels of oxidative metabolites in HLMs were also not consistent with the in vivo human metabolite profiling results (in-house unpublished data). The distinct observation of oprozomib metabolism in HLMs versus human hepatocytes might be due to significant loss of mEH activity during microsomal subcellular fractionation. In hepatocytes, besides the endoplasmic reticulum membrane, mEH is also present on the plasma membrane facing the extracellular medium (Levy, 1996), thus a significant portion of the hepatocyte mEH activity might be lost when preparing microsomes. On the other hand, P450 activities are concentrated in the endoplasmic reticulum membrane, which could explain why HLM incubation may bias metabolism toward P450 oxidation.
Furthermore, it is worth discussing the limitations of using hepatocytes in evaluation of the metabolism and extrapolation of the in vivo clearance of peptide epoxyketone analogs. Hepatocytes provided consistent metabolic profiles for oprozomib with those from in vivo samples. However, the rates of oprozomib disappearance and diol formation from in vitro hepatocyte incubation would underpredict the in vivo clearance of oprozomib, which was measured higher than hepatic blood flow in preclinical species (Fang et al., 2015). There are several possible reasons. First, the rate of diol formation obtained from in vitro kinetic incubation may underestimate the efficiency of mEH. For EH-mediated hydrolysis, the epoxide is rapidly trapped as a covalent hydroxyl alkyl enzyme intermediate. The rate of diol formation is limited by the slow hydrolysis of the intermediate (Armstrong, 1999). Second, the activity of epoxide hydrolases is generally not well characterized in commercial preparations. The effects of reagents and in vitro preparation procedure on epoxide hydrolase activity have not been systematically studied to the best of our knowledge. Third, extrahepatic metabolism also plays an important role in the elimination of this series of compounds because EHs and peptidases are ubiquitously expressed throughout the body. Therefore, the rate of metabolism in hepatocytes could be used simply as a tool to rank order analogs for prioritizing in vivo studies instead of extrapolating clearance during lead optimization.
In summary, in vitro characterization of oprozomib metabolism revealed that epoxide hydrolysis mediated by mEH, rather than sEH, is the major metabolic pathway for oprozomib metabolism, consistent with findings from in vivo samples. Oxidative metabolism plays a minor role in oprozomib elimination. These data suggest P450 and sEH activity modulators when coadministered with oprozomib would unlikely significantly affect oprozomib PK. On the other hand, the results in the present study indicate that mEH inhibitors administered concomitantly with oprozomib may affect oprozomib PK. Further studies may be warranted to determine whether drugs that are mEH inhibitors cause clinically significant DDIs with oprozomib. Hepatocytes serve as a good in vitro system to assess the metabolic profiles of peptide epoxyketone inhibitors but generally underestimate the rate of in vivo clearance of oprozomib. Metabolic findings using liver microsomes should be interpreted with caution since the microsomal system may be biased toward P450-mediated metabolism and may underestimate non-P450 metabolism for peptide epoxyketones.
Acknowledgments
We thank to Dr. Larry Wienkers, Dr. Ji Ma, Dr. Brooke Rock, and Dr. Lixia Jin from the Department of Pharmacokinetics and Drug Metabolism, Amgen Inc., for reviewing the manuscript and helpful suggestions on study design and data analysis.
Abbreviations
- 1-ABT
1-aminobenzotriazole
- BSA
bovine serum albumin
- cis-SO
cis-stilbene oxide
- DDI
drug-drug interaction
- DMSO
dimethylsulfoxide
- EH
epoxide hydrolase
- FA
formic acid
- GSH
glutathione
- HLM
human liver microsome
- IS
internal standard
- LC
liquid chromatography
- mEH
microsomal epoxide hydrolase
- MM
multiple myeloma
- MS/MS
tandem mass spectrometry
- NSPA
2-nonylsulfanyl-propionamide
- P450
cytochrome P450
- PDA
photodiode array
- PK
pharmacokinetic
- PR-025
(S)-2-amino-1-((R)-2-methyloxiran-2-yl)-3-phenylpropan-1-one
- PR-176
oprozomib diol
- sEH
soluble epoxide hydrolase
- TPPU
1-trifluoromethoxyphenyl-3-(1-propionylpiperidin-4-yl) urea
- trans-SO
trans-stilbene oxide
Authorship Contributions
Participated in research design: Zhi. Wang, Fang, Zhe. Wang.
Conducted experiments: Zhi. Wang, Fang, Teague, Wong.
Contributed new reagents or analytic tools: Morisseau, Hammock.
Performed data analysis: Zhi. Wang, Fang, Teague, Wong, Morisseau, Rock, Zhe. Wang.
Wrote or contributed to the writing of the manuscript: Zhi. Wang, Fang, Morisseau, Hammock, Rock, Zhe. Wang.
Footnotes
The work was supported in part by the National Institutes of Health National Institute of Environmental Health Sciences [Grant R01 ES002710].
 This article has supplemental material available at dmd.aspetjournals.org.
This article has supplemental material available at dmd.aspetjournals.org.
References
- Almond JB, Cohen GM. (2002) The proteasome: a novel target for cancer chemotherapy. Leukemia 16:433–443. [DOI] [PubMed] [Google Scholar]
- Anderson KC. (2013) Therapeutic advances in relapsed or refractory multiple myeloma. J Natl Compr Canc Netw 11 (Suppl 5):676–679. [DOI] [PubMed] [Google Scholar]
- Armstrong RN. (1999) Kinetic and chemical mechanism of epoxide hydrolase. Drug Metab Rev 31:71–86. [DOI] [PubMed] [Google Scholar]
- Bross PF, Kane R, Farrell AT, Abraham S, Benson K, Brower ME, Bradley S, Gobburu JV, Goheer A, Lee SL, et al. (2004) Approval summary for bortezomib for injection in the treatment of multiple myeloma. Clin Cancer Res 10:3954–3964. [DOI] [PubMed] [Google Scholar]
- Chauhan D, Hideshima T, Mitsiades C, Richardson P, Anderson KC. (2005) Proteasome inhibitor therapy in multiple myeloma. Mol Cancer Ther 4:686–692. [DOI] [PubMed] [Google Scholar]
- Ciechanover A. (2005) Intracellular protein degradation: from a vague idea, through the lysosome and the ubiquitin-proteasome system, and onto human diseases and drug targeting (Nobel lecture). Angew Chem Int Ed Engl 44:5944–5967. [DOI] [PubMed] [Google Scholar]
- Coller JK, Fritz P, Zanger UM, Siegle I, Eichelbaum M, Kroemer HK, Mürdter TE. (2001) Distribution of microsomal epoxide hydrolase in humans: an immunohistochemical study in normal tissues, and benign and malignant tumours. Histochem J 33:329–336. [DOI] [PubMed] [Google Scholar]
- Decker M, Arand M, Cronin A. (2009) Mammalian epoxide hydrolases in xenobiotic metabolism and signalling. Arch Toxicol 83:297–318. [DOI] [PubMed] [Google Scholar]
- Dimopoulos MA, Moreau P, Palumbo A, Joshua D, Pour L, Hájek R, Facon T, Ludwig H, Oriol A, Goldschmidt H, et al. ENDEAVOR Investigators (2016) Carfilzomib and dexamethasone versus bortezomib and dexamethasone for patients with relapsed or refractory multiple myeloma (ENDEAVOR): a randomised, phase 3, open-label, multicentre study. Lancet Oncol 17:27–38. [DOI] [PubMed] [Google Scholar]
- Dou QP, Zonder JA. (2014) Overview of proteasome inhibitor-based anti-cancer therapies: perspective on bortezomib and second generation proteasome inhibitors versus future generation inhibitors of ubiquitin-proteasome system. Curr Cancer Drug Targets 14:517–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y, Wang Z, Zhang T, Teague J, and Wang Z (2015) Contribution of epoxide hydrolase and cytochrome P450 (CYP) enzymes on oprozomib disposition. AAPS 2015-001707.
- Fretland AJ, Omiecinski CJ. (2000) Epoxide hydrolases: biochemistry and molecular biology. Chem Biol Interact 129:41–59. [DOI] [PubMed] [Google Scholar]
- Gill SS, Hammock BD. (1980) Distribution and properties of a mammalian soluble epoxide hydrase. Biochem Pharmacol 29:389–395. [DOI] [PubMed] [Google Scholar]
- Hamman JH, Enslin GM, Kotzé AF. (2005) Oral delivery of peptide drugs: barriers and developments. BioDrugs 19:165–177. [DOI] [PubMed] [Google Scholar]
- Harshbarger W, Miller C, Diedrich C, Sacchettini J. (2015) Crystal structure of the human 20S proteasome in complex with carfilzomib. Structure 23:418–424. [DOI] [PubMed] [Google Scholar]
- Hassett C, Lin J, Carty CL, Laurenzana EM, Omiecinski CJ. (1997) Human hepatic microsomal epoxide hydrolase: comparative analysis of polymorphic expression. Arch Biochem Biophys 337:275–283. [DOI] [PubMed] [Google Scholar]
- Hosagrahara VP, Rettie AE, Hassett C, Omiecinski CJ. (2004) Functional analysis of human microsomal epoxide hydrolase genetic variants. Chem Biol Interact 150:149–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Infante JR, Mendelson DS, Burris HA, 3rd, Bendell JC, Tolcher AW, Gordon MS, Gillenwater HH, Arastu-Kapur S, Wong HL, Papadopoulos KP. (2016) A first-in-human dose-escalation study of the oral proteasome inhibitor oprozomib in patients with advanced solid tumors. Invest New Drugs 34:216–224. [DOI] [PubMed] [Google Scholar]
- Kisselev AF, Goldberg AL. (2001) Proteasome inhibitors: from research tools to drug candidates. Chem Biol 8:739–758. [DOI] [PubMed] [Google Scholar]
- Kitteringham NR, Davis C, Howard N, Pirmohamed M, Park BK. (1996) Interindividual and interspecies variation in hepatic microsomal epoxide hydrolase activity: studies with cis-stilbene oxide, carbamazepine 10, 11-epoxide and naphthalene. J Pharmacol Exp Ther 278:1018–1027. [PubMed] [Google Scholar]
- Kodani SD, Hammock BD. (2015) The 2014 Bernard B. Brodie award lecture-epoxide hydrolases: drug metabolism to therapeutics for chronic pain. Drug Metab Dispos 43:788–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroetz DL, Loiseau P, Guyot M, Levy RH. (1993) In vivo and in vitro correlation of microsomal epoxide hydrolase inhibition by progabide. Clin Pharmacol Ther 54:485–497. [DOI] [PubMed] [Google Scholar]
- Kuhn DJ, Chen Q, Voorhees PM, Strader JS, Shenk KD, Sun CM, Demo SD, Bennett MK, van Leeuwen FW, Chanan-Khan AA, et al. (2007) Potent activity of carfilzomib, a novel, irreversible inhibitor of the ubiquitin-proteasome pathway, against preclinical models of multiple myeloma. Blood 110:3281–3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumatori A, Tanaka K, Inamura N, Sone S, Ogura T, Matsumoto T, Tachikawa T, Shin S, Ichihara A. (1990) Abnormally high expression of proteasomes in human leukemic cells. Proc Natl Acad Sci USA 87:7071–7075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy D. (1996) Membrane proteins which exhibit multiple topological orientations. Essays Biochem 31:49–60. [PubMed] [Google Scholar]
- Li B, Dou QP. (2000) Bax degradation by the ubiquitin/proteasome-dependent pathway: involvement in tumor survival and progression. Proc Natl Acad Sci USA 97:3850–3855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loda M, Cukor B, Tam SW, Lavin P, Fiorentino M, Draetta GF, Jessup JM, Pagano M. (1997) Increased proteasome-dependent degradation of the cyclin-dependent kinase inhibitor p27 in aggressive colorectal carcinomas. Nat Med 3:231–234. [DOI] [PubMed] [Google Scholar]
- Moreau P, Masszi T, Grzasko N, Bahlis NJ, Hansson M, Pour L, Sandhu I, Ganly P, Baker BW, Jackson SR, et al. TOURMALINE-MM1 Study Group (2016) Oral ixazomib, lenalidomide, and dexamethasone for multiple myeloma. N Engl J Med 374:1621–1634. [DOI] [PubMed] [Google Scholar]
- Morisseau C, Beetham JK, Pinot F, Debernard S, Newman JW, Hammock BD. (2000) Cress and potato soluble epoxide hydrolases: purification, biochemical characterization, and comparison to mammalian enzymes. Arch Biochem Biophys 378:321–332. [DOI] [PubMed] [Google Scholar]
- Morisseau C, Bernay M, Escaich A, Sanborn JR, Lango J, Hammock BD. (2011) Development of fluorescent substrates for microsomal epoxide hydrolase and application to inhibition studies. Anal Biochem 414:154–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morisseau C, Newman JW, Wheelock CE, Hill Iii T, Morin D, Buckpitt AR, Hammock BD. (2008) Development of metabolically stable inhibitors of Mammalian microsomal epoxide hydrolase. Chem Res Toxicol 21:951–957. [DOI] [PubMed] [Google Scholar]
- O’Connor OA, Wright J, Moskowitz C, Muzzy J, MacGregor-Cortelli B, Stubblefield M, Straus D, Portlock C, Hamlin P, Choi E, et al. (2005) Phase II clinical experience with the novel proteasome inhibitor bortezomib in patients with indolent non-Hodgkin’s lymphoma and mantle cell lymphoma. J Clin Oncol 23:676–684. [DOI] [PubMed] [Google Scholar]
- Obach RS. (1999) Prediction of human clearance of twenty-nine drugs from hepatic microsomal intrinsic clearance data: An examination of in vitro half-life approach and nonspecific binding to microsomes. Drug Metab Dispos 27:1350–1359. [PubMed] [Google Scholar]
- Pacifici GM, Temellini A, Giuliani L, Rane A, Thomas H, Oesch F. (1988) Cytosolic epoxide hydrolase in humans: development and tissue distribution. Arch Toxicol 62:254–257. [DOI] [PubMed] [Google Scholar]
- Pinarbasi H, Silig Y, Pinarbasi E. (2010) Microsomal epoxide hydrolase polymorphisms. Mol Med Rep 3:723–727. [DOI] [PubMed] [Google Scholar]
- Prada CP, Shain KH, Voorhees P, Gabrail N, Abidi M, Zonder J, Boccia R, Richardson PG, Neuman L, Wong H, et al. (2015) Oprozomib (OPZ) and dexamethasone (DEX) in patients (PTs) with relapsed and/or refractory multiple myeloma (RRMM): Updated results from dose escalation in a phase 1b/2, multicenter, open-label study. Clin Lymphoma Myeloma Leuk 15:e273. [Google Scholar]
- Rajan AM, Kumar S. (2016) New investigational drugs with single-agent activity in multiple myeloma. Blood Cancer J 6:e451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose TE, Morisseau C, Liu JY, Inceoglu B, Jones PD, Sanborn JR, Hammock BD. (2010) 1-Aryl-3-(1-acylpiperidin-4-yl)urea inhibitors of human and murine soluble epoxide hydrolase: structure-activity relationships, pharmacokinetics, and reduction of inflammatory pain. J Med Chem 53:7067–7075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah J, Niesvizky R, Stadtmauer E, Rifkin RM, Berenson J, Berdeja JG, Sharman JP, Lyons R, Klippel Z, Wong H, et al. (2015) Oprozomib, pomalidomide, and dexamethasone (OPomd) in patients (Pts) with relapsed and/or refractory multiple myeloma (RRMM): initial results of a phase 1b study. Blood 126:378. [Google Scholar]
- Sohlenius-Sternbeck AK, Schmidt S. (2005) Impaired glutathione-conjugating capacity by cryopreserved human and rat hepatocytes. Xenobiotica 35:727–736. [DOI] [PubMed] [Google Scholar]
- Teague J, Wang Z, Jones J, Jiang J, Muchamuel T, Phizackerley K, Wong H, and Kirk C (2013) Understanding the absorption characteristics of the oral (PO) proteasome inhibitor oprozomib in the gastrointestinal (GI) tract. AAPS 2013-000358.
- Thompson JL. (2013) Carfilzomib: a second-generation proteasome inhibitor for the treatment of relapsed and refractory multiple myeloma. Ann Pharmacother 47:56–62. [DOI] [PubMed] [Google Scholar]
- Tsimberidou AM, Ou Y, Xu Y, Wang Z, Harvey RD, Mita A, Sharma S, Papadopoulos K, Wang D, Pinchasik D, Demirhan E, et al. (2016) A phase I study of oprozomib to assess drug-drug interaction with midazolam in patients with advanced malignancies. Clin Pharmacol Ther 99: S100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Václavíková R, Hughes DJ, Souček P. (2015) Microsomal epoxide hydrolase 1 (EPHX1): Gene, structure, function, and role in human disease. Gene 571:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vij R, Savona M, Siegel D, Kaufman JL, Badros A, Ghobrial I, Paner A, Jagannath S, Jakubowiak A, Mikhael J, et al. (2015) Updated results from a multicenter, open-label, dose-escalation phase 1b/2 study of single-agent oprozomib (OPZ) in patients (PTs) with hematologic malignancies, including multiple myeloma (MM). Haematologica 100:251. [Google Scholar]
- Wang Z, Yang J, Kirk C, Fang Y, Alsina M, Badros A, Papadopoulos K, Wong A, Woo T, Bomba D, et al. (2013) Clinical pharmacokinetics, metabolism, and drug-drug interaction of carfilzomib. Drug Metab Dispos 41:230–237. [DOI] [PubMed] [Google Scholar]
- Wienkers LC, Heath TG. (2005) Predicting in vivo drug interactions from in vitro drug discovery data. Nat Rev Drug Discov 4:825–833. [DOI] [PubMed] [Google Scholar]
- Yang J, Wang Z, Fang Y, Jiang J, Zhao F, Wong H, Bennett MK, Molineaux CJ, Kirk CJ. (2011) Pharmacokinetics, pharmacodynamics, metabolism, distribution, and excretion of carfilzomib in rats. Drug Metab Dispos 39:1873–1882. [DOI] [PubMed] [Google Scholar]
- Zhou HJ, Aujay MA, Bennett MK, Dajee M, Demo SD, Fang Y, Ho MN, Jiang J, Kirk CJ, Laidig GJ, et al. (2009) Design and synthesis of an orally bioavailable and selective peptide epoxyketone proteasome inhibitor (PR-047). J Med Chem 52:3028–3038. [DOI] [PubMed] [Google Scholar]