ABSTRACT
Tubulointerstitial fibrosis (TIF) is recognized as a final phenotypic manifestation in the transition from chronic kidney disease (CKD) to end-stage renal disease (ESRD). Here we show that conditional inactivation of Sav1 in the mouse renal epithelium resulted in upregulated expression of profibrotic genes and TIF. Loss of Sav1 induced Stat3 activation and a senescence-associated secretory phenotype (SASP) that coincided with the development of tubulointerstitial fibrosis. Treatment of mice with the YAP inhibitor verteporfin (VP) inhibited activation of genes associated with senescence, SASPs, and activation of Stat3 as well as impeded the development of fibrosis. Collectively, our studies offer novel insights into molecular events that are linked to fibrosis development from Sav1 loss and implicate VP as a potential pharmacological inhibitor to treat patients at risk for developing CKD and TIF.
KEYWORDS: fibrosis, mouse, renal, senescence
INTRODUCTION
Chronic kidney disease (CKD) is currently increasing in prevalence in the United States. The National Kidney Foundation predicts that 26 million Americans have CKD and that millions of others are at risk. Tubulointerstitial fibrosis (TIF) occurs in patients with CKD and is characterized by myofibroblast activation, loss of capillary networks, and an accumulation of extracellular matrix and inflammatory cells (1–3). In fibrotic kidneys, the cortical interstitium is generally expanded, with an increased propensity to accumulate collagen (1–3). Epithelial cell death also occurs, resulting in reduced expression of epithelial markers and an increase in the expression of mesenchymal markers (4–6).
Although it is well recognized that TIF is a final manifestation in the transition from CKD to end-stage renal disease (ESRD), the mechanisms underlying the etiology of renal fibrosis are poorly understood, and as a result, progress toward finding an effective treatment has been hindered. Potential therapeutic targets for treatment of CKD have been examined through in vitro and in vivo studies in the last decade, but the findings from these studies have yet to be translated to viable treatment options for patients. Inhibition of the renin-angiotensin system (RAS), for example, has been shown to reverse CKD in animal models; however, subsequent work with RAS inhibitors or blockers in humans has failed to show similar efficacy (7–10). Additional studies have begun to examine targeting of nuclear transcription factors in pathways regulating lipid metabolism and inflammation, although most candidate pharmacological inhibitors of interest remain in preclinical stages (11). At present, dialysis or renal transplantation is still the only solution for patients with CKD.
Work in the last decade has begun to recognize the importance of genetic alterations in facilitating renal fibrotic development. Genome-wide association studies have identified loci associated with increased susceptibility to renal dysfunction in humans (12, 13). Additionally, mouse models and genome-wide transcriptome analyses of kidneys from patients with CKD and fibrosis have facilitated the identification of a number of signaling pathways associated with renal fibrosis, including the Notch, Wnt, and Hedgehog pathways (3). The further identification of signaling pathways associated with fibrosis increases the potential opportunities for targeted therapy to thwart fibrosis.
SALVADOR I (SAV1) is a WW domain-containing protein originally identified as one of the core components of the HIPPO signaling pathway. The HIPPO pathway is a conserved protein kinase pathway originally identified as a regulator of organ size (14–16). A number of cell type-dependent functions have been ascribed to HIPPO signaling, including regulation of stem/progenitor cells, cell proliferation, apoptosis, cell polarity, and cell-cell contact inhibition (17–21). At the center of the HIPPO pathway is the tumor suppressor gene Hippo, or its mammalian counterpart, mammalian STE-20 kinase 1 and 2 (MstI/2 or STK3/4). STK3/4 phosphorylates the scaffold protein SAV1, and together they phosphorylate and activate two other core components: LATS1/2 and its associated protein, MOB1. LATS1/2 in turn phosphorylates the main effector molecule of the HIPPO pathway, YAP, resulting in its inactivation and cytoplasmic sequestration by the 14-3-3 protein. Loss of HIPPO pathway function from SAV1 loss or loss of any of the core components has been suggested to result in YAP nuclear translocation and its association with transcription factors to constitutively activate growth-promoting genes involved in cancer. Specific knockout of Sav1 in the livers of mice has been shown to result in hepatomegaly and multifocal hepatic carcinomas (22). YAP amplification or constitutive overexpression has also been observed in a number of cancers (14, 21, 23–26).
Components of the HIPPO pathway have also been implicated in tissue fibrosis. Yap has been suggested to promote pulmonary fibrosis through its effects on mechanosignaling (27) and has been implicated in liver fibrosis by its role in hepatic stellate cell activation (28). Loss of Lats1/2 or abnormal accumulation of Yap has also been suggested to be important for kidney fibrosis after ischemic injury (29). Finally, most recently, Sav1 loss was also implicated in kidney fibrosis, although the role for SAV1 in CKD had not yet been documented prior to that study (30).
In the current study, we generated a mouse model with conditional knockout of Sav1 in the renal epithelium. We observed that Sav1 inactivation resulted in genetic and phenotypic alterations suggestive of renal fibrosis that were further accelerated upon induction of acute kidney injury (AKI) by use of aristolochic acid (AA). Induction of fibrosis by AA and Sav1 loss was associated with an induction of senescence and a senescence-associated secretory phenotype (SASP) that correlated with Stat3 activation. Inhibition of TIF with verteporfin (VP), a Yap inhibitor, resulted in downregulation of genes associated with senescence and SASP and in inhibition of Stat3 activation. Collectively, these results support a role for Sav1 loss in regulating interstitial fibrosis and suggest that events associated with senescence and Stat3 activation are the mechanisms underlying renal fibrosis induced by Sav1 loss.
RESULTS
Conditional inactivation of Sav1 in mouse renal tubule cells results in phenotypic changes resembling kidney fibrosis.
To determine the phenotypes associated with conditional deletion of Sav1 in murine renal proximal tubules in vivo, we crossed Ksp-CreER transgenic mice to Sav1floxed/floxed (Sav1f/f) mice to generate Ksp-CreER/Sav1fl/fl mice (referred to as KS mice from this point on). Ksp-cadherin is a tissue-specific member of the cadherin family that is expressed exclusively in the epithelial cells of the adult kidney and the developing genitourinary tract (31). Ksp-CreER transgenic mice express a tamoxifen-inducible Cre recombinase under the control of a 1.4-kb Ksp-cadherin promoter (31). Sav1f/f mice harbor loxP sites flanking exon 3 of Sav1 (22). Conditional deletion of Sav1 in the renal tubule epithelial cells of KS mice can therefore be achieved by administering tamoxifen by oral gavage (Fig. 1a).
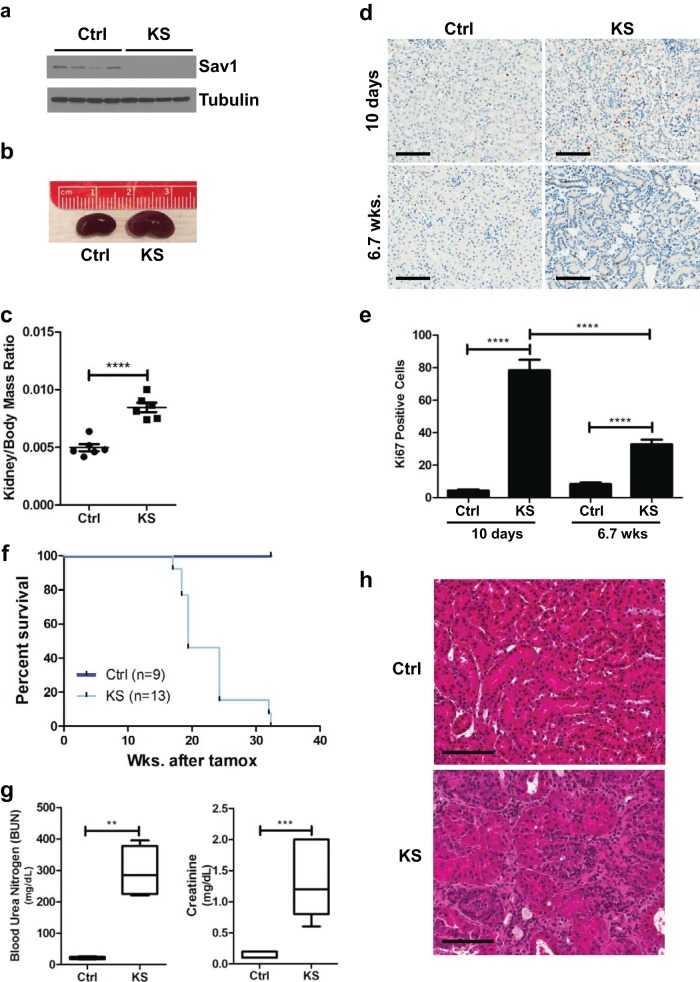
FIG 1.
Conditional knockout of Sav1 in mouse renal tubule cells results in decreased survival, renal failure, and histological abnormalities. (a) Immunoblot of whole-cell lysates isolated from the cortex tissue of KspCreER; Sav1f/f mice 0.8 week after treatment with vehicle (control [Ctrl]) or tamoxifen (KS), confirming the knockout of Sav1 expression. (b) Representative photomicrographs showing an enlarged kidney from a KS mouse compared to a kidney from an age-matched Ctrl mouse. Kidneys were taken from mice at 6 weeks post-tamoxifen treatment. (c) Comparison of kidney-to-body-mass ratios of KS mice (n = 5) and age-matched control mice (n = 6). Kidneys were taken from mice at 6 weeks post-tamoxifen treatment. (d) Representative images of Ki67-stained mouse kidney sections from mice 10 days and 6.7 weeks after treatment with vehicle or tamoxifen. (e) Bar graph showing percentages of Ki67-positive cells in the renal cortexes of Ctrl (n = 4) and KS (n = 4) mice 10 days and 6.7 weeks after treatment with vehicle or tamoxifen. Cells in three images (magnification, ×10) per mouse were counted and averaged. (f) Kaplan-Meier survival curves comparing cohorts of Ctrl (n = 9) and KS (n = 13) mice. (g) Blood urea nitrogen (BUN) and creatinine levels in blood plasmas from Ctrl (n = 6) and moribund KS (n = 5) mice. (h) Histological abnormalities shown in a representative H&E-stained section of renal tissue from a KS mouse (moribund) compared to one from an age-matched Ctrl mouse. All vehicle-treated mice were given sunflower oil by oral gavage. Error bars in all graphs represent standard deviations. **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001. Magnification, ×20 for all images; bars = 100 μm.
Prior work has shown that specific ablation of Sav1 in hepatocytes gives rise to hepatomegaly, with a 10% increase in liver size by 4 months of age (22). Similar to the phenotype observed in the liver, we observed that loss of Sav1 at 6 weeks post-tamoxifen treatment induced a significant increase in the kidney-to-body-mass ratio (Fig. 1b and c). Ki67 staining of kidney sections revealed an increase in Ki67 staining in the nuclei of renal tubule cells immediately post-tamoxifen treatment that persisted even at 6 weeks post-tamoxifen treatment (Fig. 1d and e). We observed more Ki67-positive cells at 10 days post-tamoxifen treatment than at 6 weeks post-tamoxifen treatment. We speculate that this spike in proliferation may have been a nonspecific consequence of initial Cre-ER activation, as we previously observed small increases in Ki67 staining immediately following tamoxifen treatment that subsided at later time points (data not shown). Collectively, these results suggest that an increase in cell proliferation may contribute to an overgrowth of kidneys in KS mice compared to those of age-matched controls.
KS mice had a decreased survival time (median = 19.4 weeks) compared to that of control mice (median not reached) (Fig. 1f). At the time of necropsy, significantly elevated levels of blood urea nitrogen and creatinine were found in the plasmas of KS mice compared to those of control mice, confirming renal failure as the likely cause of premature death (Fig. 1g). Histological examination of KS kidney sections by hematoxylin and eosin (H&E) staining revealed a number of abnormalities, including dilation of renal tubules and an expansion of the renal interstitium with increased interstitial cellularity (Fig. 1h).
Sav1 loss in the renal epithelium results in gene expression changes associated with renal fibrosis and senescence.
To begin to understand the consequences associated with Sav1 inactivation in renal tubules, we isolated RNAs from renal cortex tissues of tamoxifen-treated Ksp-CreER/Sav1fl/fl (KS) mice and vehicle-treated Ksp-CreER/Sav1fl/fl (control [Ctrl]) mice 6 weeks after treatment. Transcriptome sequencing (RNA-seq) analysis revealed 4,378 genes upregulated >1.5-fold and 3,362 genes downregulated >1.5-fold (with a t test false discovery rate [FDR] of <0.05). Examination of differentially regulated genes by Ingenuity Pathway Analysis (IPA) showed Tgf-β1 to be the top upstream regulator of genes activated 2-fold or more by Sav1 loss. Tgf-β1 is a cytokine that has long been recognized as a key mediator in the pathogenesis of renal fibrosis (32, 33). Further analysis of the RNA-seq data showed collagen genes and genes associated with fibrosis to be increased upon Sav1 depletion (Fig. 2a). To further confirm that loss of Sav1 induces activation of profibrotic genes, we performed real-time quantitative reverse transcription-PCR (qRT-PCR) on selected genes (34) at an earlier time point (3 weeks) after tamoxifen/vehicle treatment. Consistent with the results from the RNA-seq analysis, we observed significant activation of the profibrotic genes assessed by real-time qRT-PCR (Fig. 2b). Masson's trichrome staining and immunohistochemistry (IHC) revealed accumulations of collagen and vimentin, respectively, in KS kidneys compared to those of age-matched controls at 6 weeks posttreatment (Fig. 2c). An increase in staining for the T-cell marker CD3 also revealed that loss of Sav1 may trigger increased T-cell infiltration or expansion (Fig. 2c).
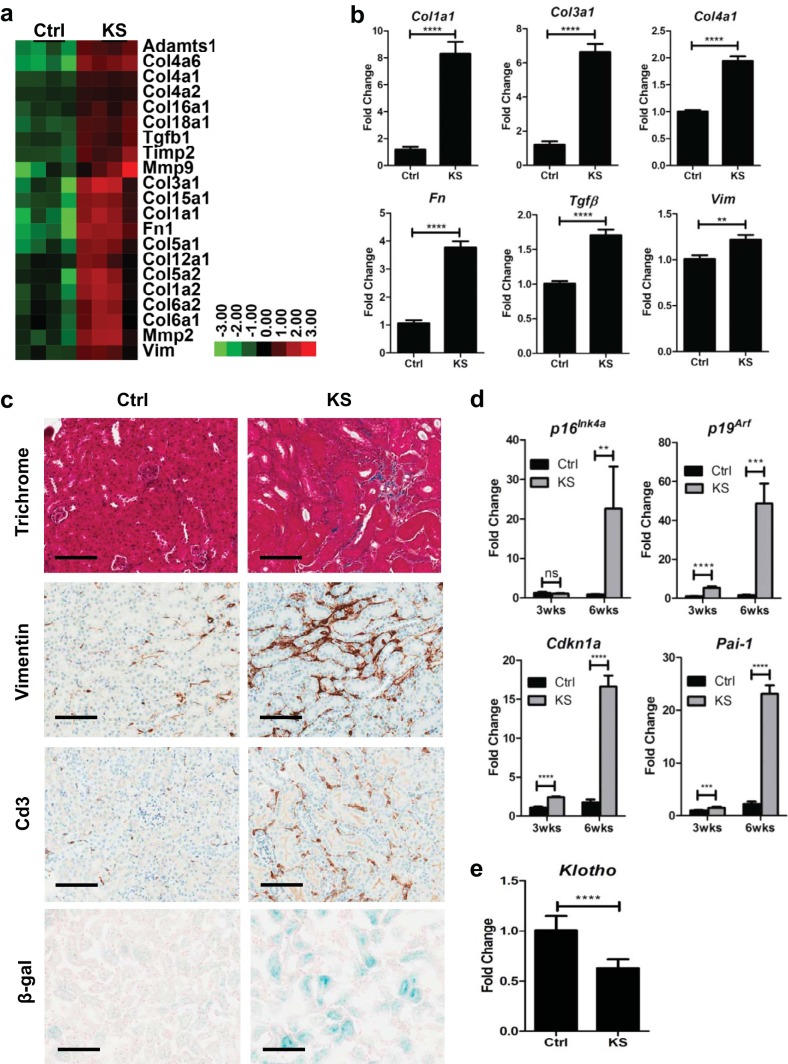
FIG 2.
Sav1 deletion in renal tubule cells activates profibrotic and cell senescence genes. (a) Heat map representation showing increased expression of genes associated with renal fibrosis in renal cortex tissue from KspCreER; Sav1f/f mice 6.7 weeks after treatment with vehicle (Ctrl) or tamoxifen (KS). (b) Real-time qRT-PCR analysis of total RNA isolated from renal cortex tissue of KspCreER; Sav1f/f mice 3 weeks after treatment with vehicle (Ctrl; n = 4) or tamoxifen (KS; n = 4). (c) Representative photomicrographs showing Ctrl (n = 6) and KS (n = 6) kidney sections 6 weeks after treatment, stained with Masson's trichrome and stained for vimentin, Cd3, and senescence-associated β-galactosidase (SA-β-Gal). (d) Relative expression of senescence genes assessed after treatment with vehicle (Ctrl; n = 4) or tamoxifen (KS; n = 4) for the indicated times. (e) Relative Klotho gene expression in Ctrl (n = 4) and KS (n = 4) mice as assessed by real-time qRT-PCR 3 weeks after treatment with vehicle or tamoxifen. All relative expression values from real-time qRT-PCR analyses were normalized to the 18S rRNA gene expression level. All vehicle-treated mice were given sunflower oil by oral gavage. Error bars in all graphs represent standard deviations. ns, not statistically significant; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001. Magnification, ×20 for all images; bars = 100 μm.
Aging has been linked to increased susceptibility to renal fibrosis (35–38). A multitude of studies have now clearly established that aging is not merely a phenotypic manifestation but consists of specific gene expression changes associated with cellular senescence (39–42). Prior studies have suggested a role for genes involved in cellular senescence in promoting CKD and fibrosis (38, 43–45). We examined KS mice for evidence of senescence and observed upregulated expression of key senescence markers (Fig. 2d). β-Galactosidase is another marker of cell senescence. Consistent with the activation of genes associated with senescence, senescence-associated β-galactosidase (SA-β-Gal) staining showed increased β-galactosidase levels in tissue sections from KS mice 6 weeks after treatment with tamoxifen compared to those for age-matched control littermates (Fig. 2c).
Klotho is a single-pass transmembrane protein previously termed the “anti-aging” gene. Defects in expression of the Klotho gene in mice were originally shown to result in a shortened life span and in aging-like phenotypes (46–48). Subsequent work later also identified a role for defective Klotho gene expression in renal fibrosis (49). Given that KS mice exhibit changes in gene expression previously documented to be associated with aging, we assessed Klotho expression by real-time qRT-PCR and found KS mice to have lower levels of Klotho gene expression than those of age-matched littermates (Fig. 2e). Collectively, our data support the hypothesis that loss of Sav1 in the renal epithelium induces changes in gene expression consistent with renal fibrosis, which is correlated with upregulated expression of genes associated with senescence.
Acute kidney injury induced by AA augments profibrotic gene expression and an accumulation of collagen in KS mice.
Mouse models have been instrumental in studying CKD brought forth by AKI. A number of methods are well established to provoke injury to mouse kidneys, including unilateral ureteral obstruction (UUO), ischemic reperfusion (IR), and aristolochic acid nephropathy (AAN) (50). Aristolochic acid (AA) belongs to a family of carcinogenic and nephrotoxic compounds (50). AAN, which involves intraperitoneal (i.p.) injection of AA, has been well documented to result in tubulointerstitial damage and is one of several widely accepted models for AKI studies (50). Given that gene expression changes observed in KS mouse kidneys suggest a predisposition to renal fibrosis (Fig. 2), we wanted to determine if loss of Sav1 affects the response to injury by AAN. We administered AA to Ctrl and KS mice at a dose of 5 mg/kg of body weight, which was previously shown to result in mild to negligible AKI in normal healthy mice (51). Intraperitoneal introduction of AA into KS mice 3 weeks after ablation of Sav1 expression resulted in dramatic increases in the expression levels of all profibrotic genes (Col1a1, Col3a1, Col4a1, Fn, Tgfβ1, and Vimentin) examined compared to those in AA-treated Ctrl mice with no Sav1 loss (Fig. 3a and b). Consistent with an increase in profibrotic gene expression, staining with Masson's trichrome revealed an accumulation of collagen at 3 weeks post-tamoxifen treatment that progressively increased at 6 and 13 weeks post-tamoxifen treatment compared to that in vehicle-treated control mice (Fig. 3c).
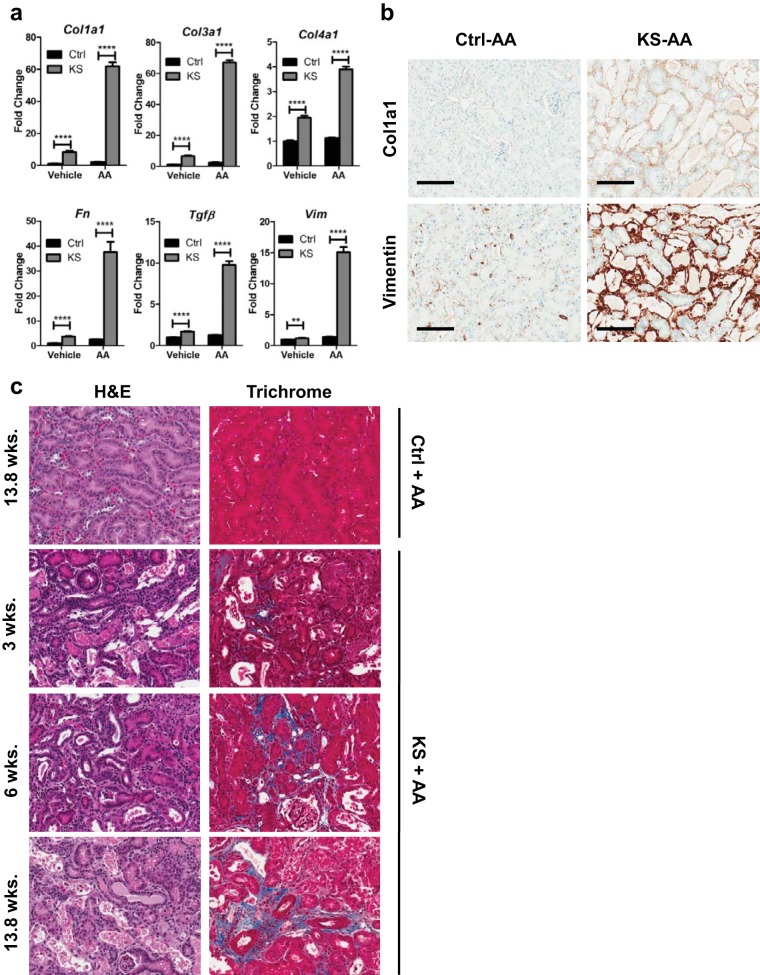
FIG 3.
Acute kidney injury (AKI) with aristolochic acid (AA) drives activation of genes associated with renal fibrosis in KS mice. (a) KspCreER; Sav1f/f mice were given vehicle (sunflower oil) (Ctrl; n = 4) or tamoxifen (KS; n = 4) by oral gavage. Vehicle (DMSO and PBS) or aristolochic acid was introduced by intraperitoneal injection 3 weeks later to induce acute kidney injury. Real-time qRT-PCR analysis of total RNA isolated from renal cortex tissues was used to assess profibrotic gene expression. (b) Representative images of renal sections from the mice examined for panel a, showing localization of vimentin and Col1a1. (c) Representative H&E- and Masson's trichrome-stained images of renal sections from mice post-AAN. Mice were administered AA to induce AKI 3 weeks, 6 weeks, and 13.8 weeks after tamoxifen treatment or 13.8 weeks after vehicle (sunflower oil) treatment. **, P ≤ 0.01; ****, P ≤ 0.0001. Error bars in graphs represent standard deviations. Magnification, ×20 for all images; bars = 100 μm.
Senescence and SASP genes are further activated by AAN in KS mice.
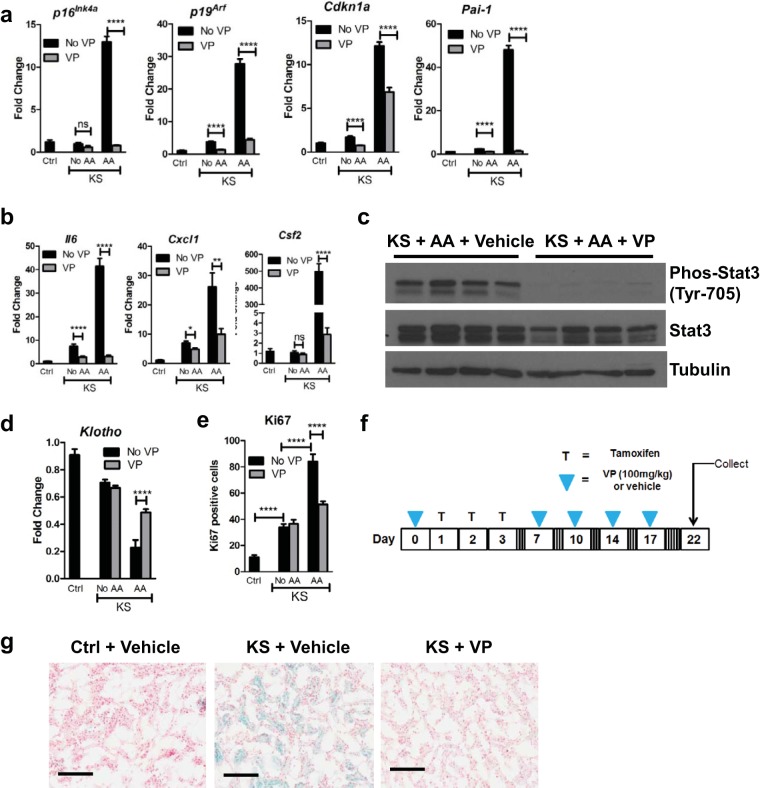
Senescent cells have been shown to have deleterious effects on organisms by contributing to age-related pathology through inducing expression of genes associated with senescence-associated secretory phenotypes (SASPs), including genes encoding interleukins, chemokines, and growth factors (52). Induction of profibrotic genes by AAN was also associated with dramatic increases in the expression levels of all senescence genes (p16ink4a, p19arf, Cdkn1a, and Pai-1) (Fig. 4a and b) examined. Moreover, activation of p53 was also observed by Western blotting (Fig. 4b). To determine whether senescence induced by Sav1 loss is associated with induction of SASP genes, we assessed the expression of select SASP genes after AAN in Ctrl and KS mice. We found all SASP genes assessed to be upregulated in KS mice compared to age-matched control mice, suggesting that Sav1 loss induces SASP genes upon AKI (Fig. 4c).
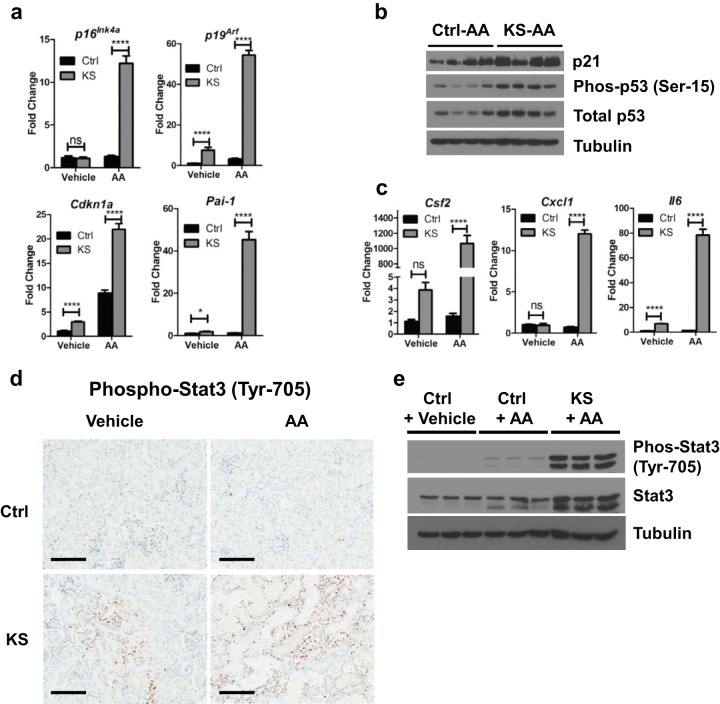
FIG 4.
Acute kidney injury (AKI) with aristolochic acid (AA) drives activation of genes associated with senescence and SASP in KS mice. KspCreER; Sav1f/f mice were given vehicle (sunflower oil) (Ctrl; n = 4) or tamoxifen (KS; n = 4) by oral gavage. Vehicle (DMSO and PBS) or aristolochic acid was introduced by intraperitoneal injection 3 weeks later to induce acute kidney injury. (a and b) Isolated renal cortex tissues were assessed for senescence genes by qRT-PCR (a) and Western blotting (b). (c) SASP gene expression levels were assessed by qRT-PCR. (d) Representative images of kidney sections from Ctrl (n = 6) and KS (n = 6) mice showing phospho-Stat3 localization. (e) Immunoblots showing Stat3 activation in Ctrl mice treated with vehicle (DMSO and PBS), Ctrl mice treated with AA, and KS mice treated with AA. ns, not statistically significant; *, P ≤ 0.05; ****, P ≤ 0.0001. Error bars in graphs represent standard deviations. Magnification, ×20 for all images; bars = 100 μm.
Cytokines such as interleukin-6 (Il6) are known to activate the signal transducers and activators of transcription (Stats) (53, 54). In particular, activation of Stat3 is known to be achieved through phosphorylation of its tyrosine 705 site by receptor tyrosine kinases (RTKs) and has been suggested to have a role in the progression of renal fibrosis (53–55). Although Stat3 activation from Sav1 loss alone was not obvious by Western blotting (data not shown), histological examination revealed an increase in phospho-Stat3 staining in the nuclei of renal tubule cells (Fig. 4d). We next asked whether loss of Sav1 in KS mice affects Stat3 activation upon AAN. Intraperitoneal injection of AA into Ctrl mice resulted in a minimal increase in phosphorylated (Tyr-705) Stat3, while AA injection into KS mice resulted in a dramatic increase in phosphorylated Stat3 expression (Fig. 4d and e). Our results suggest that Sav1 loss induces activation of Stat3 in association with a fibrosis phenotype that is further enhanced by AKI with AA.
Accumulation of Yap activity is observed in KS mice.
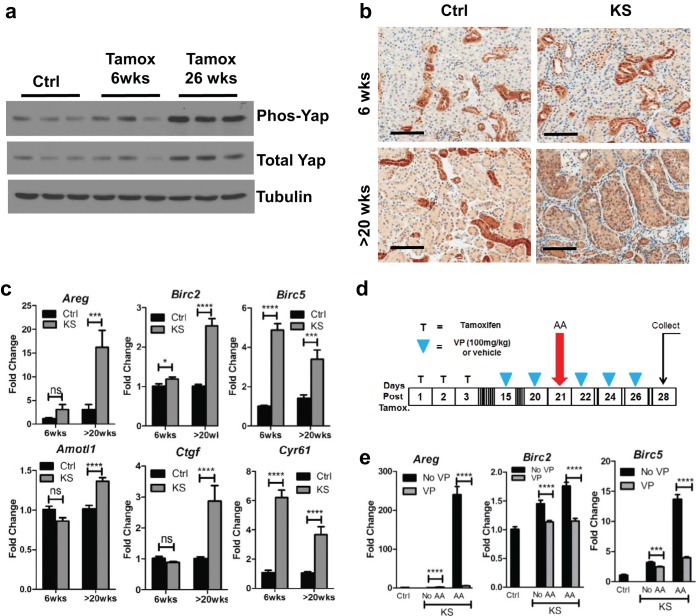
Sav1 has previously been documented to function as a scaffold protein that brings together kinases in the HIPPO signaling pathway to regulate phosphorylation and subsequent ubiquitin-mediated degradation of Yap (56, 57). Loss of Sav1 is thought to result in an accumulation of Yap in the nucleus, where it binds to other transcription factors, such as the TEAD family of transcription factors, to activate expression of target genes (56, 57). Previously published work suggested that Yap also plays a role in fibrosis (27, 28, 58, 59). We assessed Yap levels in KS mice compared to those in age-matched controls and saw progressive increases in Yap accumulation at 6 weeks and >20 weeks post-tamoxifen treatment (Fig. 5a and b). No changes in Yap levels were apparent between Ctrl mice of various ages (data not shown). Consistent with an accumulation of Yap protein resulting from Sav1 ablation, we also observed activation of Yap target genes after tamoxifen treatment of KS mice (Fig. 5c). As expected, we did not see any significant change in Yap mRNA levels upon inactivation of Sav1, since Yap is primarily regulated by ubiquitin-mediated degradation (data not shown).
FIG 5.
Conditional knockout of Sav1 in mouse renal tubule cells results in an accumulation of Yap activity that can be inhibited with verteporfin. (a) Immunoblots showing Yap expression in renal cortex tissue from KspCreER; Sav1f/f mice. Mice were given vehicle (Ctrl) or tamoxifen (KS) by oral gavage, and kidneys were collected at 6 weeks and 26 weeks posttreatment. Ctrl mice were age matched to KS mice, and kidneys were collected at 6 weeks post-tamoxifen treatment. (b) Representative images showing Yap accumulation in renal tissue from Ctrl or KS mice at the indicated time points post-tamoxifen or -vehicle (sunflower oil) treatment. (c) Real-time qRT-PCR analysis of total RNAs isolated from renal cortex tissues of Ctrl (n = 4) and KS (n = 4) mice. Mice were assessed for Yap target gene expression at the indicated time points. (d) Schematic showing treatment of Ctrl and KS mice with verteporfin, a Yap inhibitor, or with vehicle and induction of AKI with AA. (e) Real-time qRT-PCR analysis of total RNAs isolated from renal cortex tissues of Ctrl and KS mice after treatment with vehicle, AA, and/or VP as indicated (Ctrl, n = 3; KS plus no VP, n = 4; KS plus VP, n = 6; KS plus AA plus no VP, n = 5; KS plus AA plus VP, n = 6) for expression of selected Yap target genes. ***, P ≤ 0.001; ****, P ≤ 0.0001. Error bars represent standard deviations. Magnification, ×20 for all images; bars = 100 μm.
VP inhibition of AAN-induced TIF in KS mice coincides with inhibition of senescence and SASP gene activation and inhibition of Stat3 activation.
To determine if Yap plays a central role in phenotypes associated with tubulointerstitial fibrosis in the setting of Sav1 loss (KS mice), we tested the efficacy of the Yap inhibitor verteporfin (VP) in KS and Ctrl mice. VP is a small molecule belonging to the porphyrin family of compounds that was recently identified in a drug screen for inhibitors of YAP-TEAD activity (60). Further biochemical studies have shown that VP works by binding YAP and preventing its interaction with other transcription factors, such as TEAD (60). We tested the ability of VP to inhibit Yap target genes induced 3 weeks after Sav1 loss (Fig. 5d) and confirmed that VP treatment reduced the activation of Yap target genes (Fig. 5e).
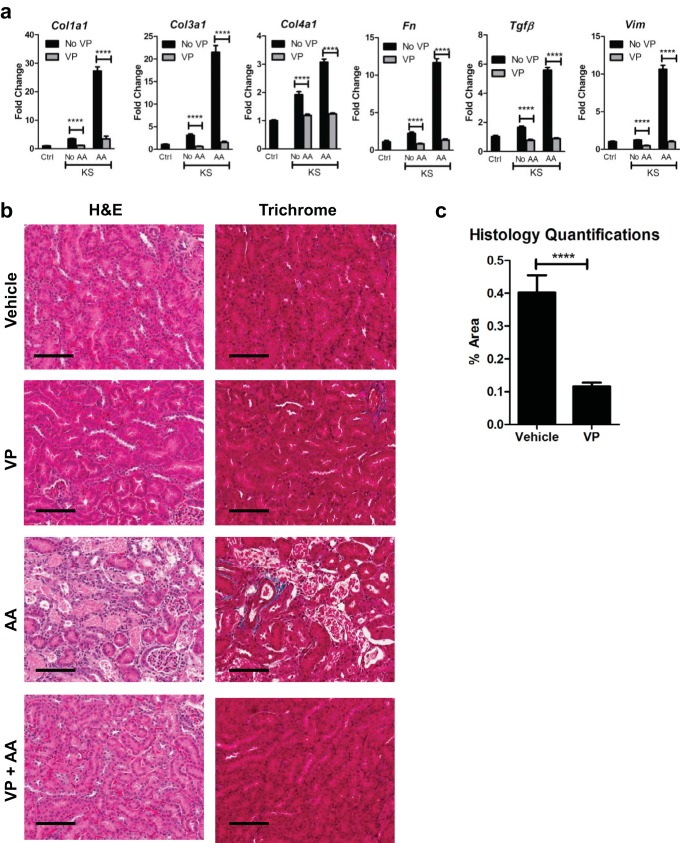
We tested the ability of VP to suppress the activation of profibrotic genes in KS and Ctrl mice after AAN. Consistent with our prior results, activation of profibrotic gene expression was observed in KS mice upon AAN compared to that in Ctrl mice (Fig. 6a). Ctrl and KS mice treated with AA but receiving VP treatment, however, showed levels of profibrotic gene expression comparable to basal levels (Fig. 6a). Masson's trichrome staining revealed less collagen deposition in mice that received VP, consistent with a reduction in profibrotic gene expression (Fig. 6b and c).
FIG 6.
Verteporfin inhibits profibrotic gene expression induced by Sav1 loss. AKI was induced in Ctrl and KS mice by AAN, and mice were treated with vehicle (DMSO and PBS) or VP. (a and b) Ctrl mice (n = 3) and KS mice treated with vehicle (DMSO and PBS; n = 3), VP (n = 6), AA (n = 5), or AA plus VP (n = 6) were assessed for expression of profibrotic genes (a) and stained with H&E and Masson's trichrome stain (b). (c) Bar graph representing the percent area of positive staining for collagen (blue staining) for kidneys from AA-treated KS mice that were treated with vehicle (n = 5) versus AA-treated KS mice that were treated with VP (n = 6). Three images were taken for each mouse section for quantitation using ImageJ, and the percentages of positive areas were averaged. Error bars represent standard deviations. ****, P ≤ 0.0001. Magnification, ×20 for all images; bars = 100 μm.
Interestingly, VP treatment also reduced the expression levels of the senescence (Fig. 7a) and SASP (Fig. 7b) genes examined. Pai-1 and p16Ink4a gene expression levels after AA-induced AKI were comparable to basal levels. Cdkn1a and p19Arf gene expression levels remained above basal levels but were still dramatically reduced even with AA treatment (Fig. 7a). Consistent with recently published work showing that VP inhibited Stat3 activation in colon cancer (61), VP inhibited Stat3 activation induced by AA in KS mice (Fig. 7c). Collectively, our results suggest that kidney fibrosis and associated changes in gene expression induced by Sav1 loss can be inhibited by verteporfin. Treatment with VP also reduced Ki67 staining and restored expression of Klotho expression in AA-treated KS mice compared to that in AA-treated Ctrl mice, although this was not observed in KS mice compared to Ctrl mice. These results suggest that VP may thwart fibrosis induced by Sav1 loss via mechanisms other than proliferation and Klotho gene expression (Fig. 7d and e).
FIG 7.
Inhibition of TIF by verteporfin (VP) coincides with inhibition of senescence and SASP gene expression and Stat3 activation in KS mice. (a and b) Senescence gene (a) and SASP gene (b) expression levels were measured for Ctrl and KS mice after AAN and/or VP treatment (Ctrl, n = 3; KS plus no VP, n = 4; KS plus VP, n = 6; KS plus AA plus no VP, n = 5; KS plus AA plus VP, n = 6). (c) Western blots comparing Stat3 and phosphorylated Stat3 in vehicle-treated KS mice, mice with AA-induced AKI, and VP-treated mice with AA-induced AKI. (d) Kidney sections from mice (Ctrl, n = 3; KS plus no VP, n = 4; KS plus VP, n = 6; KS plus AA plus no VP, n = 5; KS plus AA plus VP, n = 6) were assessed for Klotho gene expression by real-time qRT-PCR. (e) Kidney sections from mice (Ctrl, n = 3; KS plus no VP, n = 4; KS plus VP, n = 6; KS plus AA plus no VP, n = 5; KS plus AA plus VP, n = 6) were stained with Ki67. Three images were taken for each section, and Ki67-positive cells in tubules were counted and averaged. (f) Mice were administered one dosage of VP or vehicle (DMSO and PBS) followed by three consecutive daily treatments with tamoxifen or vehicle (sunflower oil). Mice were then administered VP or vehicle (DMSO and PBS) two times a week for 2 weeks before kidneys were harvested. (g) SA-β-Gal staining of kidney sections from Ctrl mice (n = 2), vehicle-treated KS mice (n = 3), and VP-treated KS mice (n = 3). Error bars in graphs represent standard deviations. ns, not statistically significant; *, P ≤ 0.05; **, P ≤ 0.01; ****, P ≤ 0.0001. Magnification, ×20 for all images; bars = 100 μm.
Given that VP treatment resulted in reduced expression of genes associated with senescence in KS mice, we determined whether VP treatment had an effect on increased β-galactosidase activity brought on by Sav1 loss. We previously observed that VP was not able to reverse SA-β-Gal activity that was already initiated by Sav1 loss (data not shown). We modified our protocol to initiate treatment with VP prior to inducing Sav1 loss with tamoxifen to determine if VP could instead prevent SA-β-Gal accumulation (Fig. 7f). As shown in Fig. 7g, treatment with VP before and after induction of Sav1 loss was able to prevent SA-β-Gal staining in KS mice (Fig. 7g).
DISCUSSION
Acute kidney injury resulting in chronic kidney disease (CDK) is a growing problem in the United States. Although it is well known that fibrosis is the final manifestation in the progression from CKD to end-stage renal disease (ESRD), the etiology underlying renal fibrosis is poorly understood. An understanding of the molecular mechanisms that contribute to fibrosis in the kidney is essential to the identification of potential candidates for targeted therapy. Seo et al. (30) recently published work showing that Sav1 loss plays a role in TIF induced by unilateral ureteral obstruction (UUO), consistent with our observations that Sav1 loss has a role in kidney fibrosis. Here we provide evidence that induction of fibrosis by Sav1 loss is linked to induction of senescence and Stat3 activation. Furthermore, we showed that verteporfin (VP), a Yap inhibitor, is effective at inhibiting fibrosis and that VP's efficacy in impeding fibrosis is correlated with its ability to inhibit the induction of senescence gene expression, SASPs, and Stat3 activation.
It was previously reported that knockdown of Sav1 in vitro in renal cell lines resulted in increased cell proliferation and anchorage-independent growth and that Sav1 knockout in the liver resulted in multifocal tumors (22), suggesting a role for Sav1 as a tumor suppressor gene (62–64). Although we did observe an increase in proliferation as indicated by Ki67 staining, consistent with a recent study (62), KS mice did not develop renal tumors within the time frame observed (∼32.3 weeks). While this may merely reflect a tissue-specific role of Sav1, with an oncogenic role in the liver but not in the kidney, we speculate that the induction of senescence and SASPs in vivo that we describe here might also serve as a significant barrier to neoplastic transformation of renal tubular cells.
Administration of VP to KS mice was able to inhibit the genetic and phenotypic manifestations of renal fibrosis induced by AA. Verteporfin (Visudyn; Novartis) is currently an FDA-approved drug administered intravenously to patients in photodynamic therapy to treat macular degeneration with minimal side effects. Our findings are consistent with recently published work showing the efficacy of VP for inhibiting Yap activity and renal fibrosis induced by UUO (65). Collectively, these results bring to light the potential for VP as a candidate pharmacological inhibitor to be administered to patients who may be at increased risk for AKI and subsequent renal fibrosis as a result of their existing medical condition or specific medical procedures and treatments which they are about to undergo (i.e., surgery, chemotherapy, etc.). VP also inhibited cell proliferation, as indicated by a reduction in Ki67-positive cells and restored Klotho expression induced by AA. Cell proliferation and reduced Klotho expression induced solely by Sav1 loss, however, were not affected, suggesting that VP may thwart fibrosis induced by Sav1 loss via other mechanisms.
Prior studies have suggested a correlation between the expression of senescence and aging genes and renal fibrosis. Reduced expression of the Klotho gene, for example, has been shown to play a role in TIF (49, 66), and repression of Klotho expression is tied to activated p16Ink4a expression (48). Moreover, it is now known that cell senescence also accommodates an induction of specific cytokines that comprise the SASP (52). SASP genes have been shown to play a role in inflammation and in facilitating epithelial-to-mesenchymal transition (EMT), two characteristics that are tightly linked to TIF progression (5, 6, 52). Cytokines such as Il6 are well known to induce Stat3 activation, which is also associated with the progression of TIF (55). In the KS mouse model, we observed that SA-β-Gal staining was not restricted to the cortex region of the kidney but was apparent throughout the kidney. While Ksp-cadherin is expressed to some degree in regions of the renal medulla, such as the loops of Henle and collecting ducts (31), we also hypothesize that induction of Sav1 loss in the proximal and distal tubular cells in the renal cortex results in paracrine signaling of SASPs that triggers senescence in other parts of the kidney. Inhibition of fibrosis by use of VP correlated with downregulated expression of senescence genes and inhibition of Stat3 activation. Collectively, our observations support a link between genes associated with senescence, Stat3 activation, and the manifestation of TIF.
Our observation that Sav1 loss in the renal epithelium does not result in decreased Yap phosphorylation as predicted by the canonical Hippo pathway is consistent with observations of Sav1 deletion in the liver (22) and suggests that there may be additional mechanisms of action from Sav1 loss that are involved in initiating the molecular events associated with fibrosis that have yet to be defined. The kinetics of induction of Yap activity following Sav1 loss also do not support a direct connection between Sav1 loss, Yap activation, and TIF. While Sav1 expression was undetectable by Western blotting with tissue lysates from the kidneys of KS mice immediately after tamoxifen treatment, we observed some restoration of Sav1 expression at later time points (∼15 to 20 weeks) post-tamoxifen treatment (data not shown). We hypothesize that this is a result of the transient nature of Cre expression driven by our tamoxifen-inducible Ksp-CreERT2 allele and the subsequent rejuvenation of kidney cells with intact Sav1. Yap levels seem to be highest at later time points post-tamoxifen treatment, when restored Sav1 expression is apparent. Although this may be a result of an expansion of Sav1-null cells, we are also open to the notion that at least in the context of kidney tissue, Sav1 expression may not function to promote degradation of Yap as the canonical HIPPO pathway predicts. We hypothesize instead that immediate Sav1 loss may irreversibly trigger a cascade of molecular events, independent of the HIPPO pathway, to promote activation of Yap and, in turn, to help to promote fibrosis.
Additionally, the accumulation of Yap protein as determined by Western blotting is minimal at 3 weeks post-Sav1 ablation. As a result, the dramatic induction of profibrotic gene expression observed upon AAN at this time point cannot solely be attributed to Yap overexpression. Although the efficacy of VP treatment to inhibit fibrosis in our KS mouse model seems to suggest that Yap is a key mediator of kidney fibrosis, recent work revealed that VP inhibited Stat3 activation in colon cancer independently of VP's function as a Yap inhibitor (61). This observation brings to light the possibility that VP also inhibits fibrosis via other mechanisms in addition to its role as an inhibitor of Yap activity.
Collectively, our findings highlight the importance of Sav1 loss in inducing interstitial fibrosis and offer a model system for understanding the molecular mechanisms underlying renal fibrosis within which potential therapeutic opportunities can be explored.
MATERIALS AND METHODS
Genetically engineered mouse models.
KspCad-CreERT2 mice have been described previously (31). Sav1fl/fl mice were a kind gift from Randy Johnson (University of Texas) and have also been described previously (22). All experiments performed on mice have been approved by the University of North Carolina-Chapel Hill Institutional Animal Care and Use Committee (UNC-CH IACUC).
Immunoblotting conditions.
Cortex tissue was homogenized in EBC buffer (50 mM HEPES [pH 7.6], 250 mM NaCl, 0.1% NP-40, 5 mM EDTA [pH 8.0]) with set I and set II phosphatase inhibitors at 1× (Calbiochem) and protease inhibitors at 1× (Roche) by use of a TissueRuptor homogenizer (Qiagen). Protein concentrations were determined with Bio-Rad protein assay dye reagent concentrate (Bio-Rad). Proteins were resolved in SDS-PAGE gels and electrotransferred to polyvinylidene difluoride (PVDF) membranes. Western blotting was performed with the following antibodies from Cell Signaling: Yap (4912S), phospho-Yap (4911S), Sav1 (13301S), p21 (2946S), Stat3 (9139S), phospho-Stat3 (9145S), p53 (2524S), and phospho-p53 (9286S) antibodies.
Mouse RNA-seq.
A total of 200 to 1,000 ng of total RNA was used to prepare RNA libraries by use of a TruSeq Stranded mRNA sample prep kit (Illumina). Paired-end (75 bp) reads were sequenced on a NextSeq500 desktop sequencer by using a high-output flow cell kit (Illumina), yielding an average of over 28 million reads per sample. Quality control-passed reads were aligned to the mouse reference genome (mm9) by use of MapSplice (67). The alignment profile was determined by use of Picard Tools v1.64 (http://broadinstitute.github.io/picard/). Aligned reads were sorted and indexed by use of SAMtools, translated to transcriptome coordinates, and then filtered for indels, large inserts, and zero mapping quality by use of UBU v1.0 (https://github.com/mozack/ubu). Transcript abundance estimates for each sample were performed using RSEM, an expectation-maximization algorithm (68), using the UCSC known gene transcript and gene definitions. Raw RSEM read counts or all RNA-seq samples were normalized to the overall upper quartile (69).
RNA extraction and cDNA synthesis.
Cortex tissue was homogenized in RLT buffer (RNeasy kit; Qiagen) by use of a TissueRuptor homogenizer (Qiagen). RNA was extracted using an RNeasy kit (Qiagen) following the manufacturer's protocol. cDNA was synthesized with random primers, using an Im Pro-II reverse transcription system (Promega).
TaqMan real-time qRT-PCR.
TaqMan real-time qRT-PCR was performed following the standard manufacturer's protocol from Applied Biosystems. The following TaqMan probes were used: Col1a1 (Mm00801666_g1), Col3a1 (Mm01254476_m1), Col4a1 (Mm01210125_m1), Fn1 (Mm01256744_m1), Vim (Mm01333430_m1), Kl (Mm00502002_m1), Tgfb (Mm01178820_m1), Cdkn1a (Mm04205640_g1), Serpine1 (Mm00435860_m1), p16 (assay ID MP16-P163), p19 (assay ID MP19-P192), Ppara (Mm00440939_m1), Cpt1a (Mm01231183_m1), Birc2 (Mm00431811_m1), Birc5 (Mm00599749_m1), Areg (Mm01354339_m1), Ppargc1a (Mm01208835_m1), Csf2 (Mm1290062), Cxcl1 (Mm04207460), and Il16 (Mm00446190).
Statistical analyses.
The unpaired two-tailed t test was performed in GraphPad Prism to determine P values. Error bars on all graphs, representing standard deviations, were also calculated in GraphPad Prism.
Senescence-associated β-galactosidase staining.
Kidney tissue was flash frozen and embedded in OCT compound. The tissue was sectioned (5 μm) onto slides by the University of North Carolina (UNC) Tissue Procurement Facility and fixed in phosphate-buffered saline (PBS) (pH 6.0) containing 2% formaldehyde and 0.2% glutaraldehyde for 10 min. Slides were then rinsed in PBS (pH 6.0) and stained in a PBS (pH 6.0) solution with 0.005 M ferricyanide, 0.005 M ferrocyanide, 0.001 M MgSO4, and 1 mg/ml X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) at 37°C overnight. Slides were rinsed in water and then dipped in eosin for ∼5 min, rinsed in water, and dehydrated using 95% ethanol (3 times; 2 min each), 100% ethanol (3 times; 2 min each), and Histo-Clear (3 times; 2 min each). Slides were then covered with coverslips in Permount solution and dried. Tissue sectioning was performed by the UNC Tissue Procurement Facility. Images were scanned using an Aperio ScanScope by the UNC Translational Pathology Laboratory (TPL) Core Facility.
Histology.
Tissue (5 μm) was sectioned onto slides. Hematoxylin and eosin (H&E) and Masson's trichrome staining was performed by the UNC Center of Gastrointestinal Biology and Disease Histology Core Facility. Ki67 and Yap (4912S; Cell Signaling) staining was performed by the UNC TPL Core Facility. Vimentin (ab92547; Abcam), Cd3 (A0452; Dako), phospho-Stat3 (9145S; Cell Signaling), and Col1a1 (ab34710; Abcam) staining was performed by the UNC Animal Histology Core Facility. All images were scanned using an Aperio ScanScope by the UNC TPL. Quantifications were performed using ImageJ.
Reagents for animal studies. (i) Tamoxifen.
Conditional activation of Cre recombinase was induced by oral administration of tamoxifen (Sigma). Tamoxifen (100 mg) was resuspended in 100 μl of ethanol, followed by the addition of 1 ml of sunflower oil, to a final concentration of 100 mg/ml. The solution was sonicated at 4°C in a water bath sonicator until the tamoxifen was completely dissolved. Animals (8 to 9 weeks old) were administered 50 μl (5 mg) of tamoxifen solution per day for 3 consecutive days by oral gavage.
(ii) AA.
Aristolochic acid (AA) was suspended in 120 μl of dimethyl sulfoxide (DMSO) and then 880 μl of PBS (pH 7.4), to a final concentration of 2 mg/ml. The AA solution was administered to mice by intraperitoneal injection at a dosage of 5 mg/kg of body weight. Tissue was extracted for analysis 1 week after AA injection unless stated otherwise.
(iii) VP.
VP was resuspended in 100 μl DMSO and then 900 μl PBS (pH 7.4), to a final concentration of 10 mg/ml. The VP solution was vortexed vigorously for at least 20 min immediately prior to use. Mice were administered VP by intraperitoneal injection at a dosage of 100 mg/kg.
BUN and creatinine analyses.
Blood was collected into EDTA-free tubes. Plasma was isolated by centrifugation for 10 min at 2,000 × g. Blood urea nitrogen (BUN) and creatinine levels were assessed by the UNC Animal Clinical Chemistry and Gene Expression Laboratory Core Facility.
ACKNOWLEDGMENTS
We thank the UNC Center for Gastrointestinal Biology and Disease Histology Core Facility, the UNC Translational Pathology Laboratory, the UNC Tissue Procurement Facility, and the UNC Animal Histology Core Facility for their help with all the histology results presented in this article.
This work was funded in part by the North Carolina University Cancer Research Fund (UCRF).
We declare that we have no conflicts of interest.
REFERENCES
- 1.LeBleu VS, Taduri G, O'Connell J, Teng Y, Cooke VG, Woda C, Sugimoto H, Kalluri R. 2013. Origin and function of myofibroblasts in kidney fibrosis. Nat Med 19:1047–1053. doi: 10.1038/nm.3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zeisberg M, Neilson EG. 2010. Mechanisms of tubulointerstitial fibrosis. J Am Soc Nephrol 21:1819–1834. doi: 10.1681/ASN.2010080793. [DOI] [PubMed] [Google Scholar]
- 3.Edeling M, Ragi G, Huang S, Pavenstadt H, Susztak K. 2016. Developmental signalling pathways in renal fibrosis: the roles of Notch, Wnt and Hedgehog. Nat Rev Nephrol 12:426–439. doi: 10.1038/nrneph.2016.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boutet A, De Frutos CA, Maxwell PH, Mayol MJ, Romero J, Nieto MA. 2006. Snail activation disrupts tissue homeostasis and induces fibrosis in the adult kidney. EMBO J 25:5603–5613. doi: 10.1038/sj.emboj.7601421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grande MT, Sanchez-Laorden B, Lopez-Blau C, De Frutos CA, Boutet A, Arevalo M, Rowe RG, Weiss SJ, Lopez-Novoa JM, Nieto MA. 2015. Snail1-induced partial epithelial-to-mesenchymal transition drives renal fibrosis in mice and can be targeted to reverse established disease. Nat Med 21:989–997. doi: 10.1038/nm.3901. [DOI] [PubMed] [Google Scholar]
- 6.Lovisa S, LeBleu VS, Tampe B, Sugimoto H, Vadnagara K, Carstens JL, Wu CC, Hagos Y, Burckhardt BC, Pentcheva-Hoang T, Nischal H, Allison JP, Zeisberg M, Kalluri R. 2015. Epithelial-to-mesenchymal transition induces cell cycle arrest and parenchymal damage in renal fibrosis. Nat Med 21:998–1009. doi: 10.1038/nm.3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boffa JJ, Lu Y, Placier S, Stefanski A, Dussaule JC, Chatziantoniou C. 2003. Regression of renal vascular and glomerular fibrosis: role of angiotensin II receptor antagonism and matrix metalloproteinases. J Am Soc Nephrol 14:1132–1144. doi: 10.1097/01.ASN.0000060574.38107.3B. [DOI] [PubMed] [Google Scholar]
- 8.Huby AC, Rastaldi MP, Caron K, Smithies O, Dussaule JC, Chatziantoniou C. 2009. Restoration of podocyte structure and improvement of chronic renal disease in transgenic mice overexpressing renin. PLoS One 4:e6721. doi: 10.1371/journal.pone.0006721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kavvadas P, Weis L, Abed AB, Feldman DL, Dussaule JC, Chatziantoniou C. 2013. Renin inhibition reverses renal disease in transgenic mice by shifting the balance between profibrotic and antifibrotic agents. Hypertension 61:901–907. doi: 10.1161/HYPERTENSIONAHA.111.00639. [DOI] [PubMed] [Google Scholar]
- 10.Mauer M, Zinman B, Gardiner R, Suissa S, Sinaiko A, Strand T, Drummond K, Donnelly S, Goodyer P, Gubler MC, Klein R. 2009. Renal and retinal effects of enalapril and losartan in type 1 diabetes. N Engl J Med 361:40–51. doi: 10.1056/NEJMoa0808400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Musso G, Cassader M, Cohney S, De Michieli F, Pinach S, Saba F, Gambino R. 2016. Fatty liver and chronic kidney disease: novel mechanistic insights and therapeutic opportunities. Diabetes Care 39:1830–1845. doi: 10.2337/dc15-1182. [DOI] [PubMed] [Google Scholar]
- 12.Kottgen A, Pattaro C, Boger CA, Fuchsberger C, Olden M, Glazer NL, Parsa A, Gao X, Yang Q, Smith AV, O'Connell JR, Li M, Schmidt H, Tanaka T, Isaacs A, Ketkar S, Hwang SJ, Johnson AD, Dehghan A, Teumer A, Pare G, Atkinson EJ, Zeller T, Lohman K, Cornelis MC, Probst-Hensch NM, Kronenberg F, Tonjes A, Hayward C, Aspelund T, Eiriksdottir G, Launer LJ, Harris TB, Rampersaud E, Mitchell BD, Arking DE, Boerwinkle E, Struchalin M, Cavalieri M, Singleton A, Giallauria F, Metter J, de Boer IH, Haritunians T, Lumley T, Siscovick D, Psaty BM, Zillikens MC, Oostra BA, Feitosa M, Province M, de Andrade M, Turner ST, Schillert A, Ziegler A, Wild PS, Schnabel RB, Wilde S, Munzel TF, Leak TS, Illig T, Klopp N, Meisinger C, Wichmann HE, Koenig W, Zgaga L, Zemunik T, Kolcic I, Minelli C, Hu FB, Johansson A, Igl W, Zaboli G, Wild SH, Wright AF, Campbell H, Ellinghaus D, Schreiber S, Aulchenko YS, Felix JF, Rivadeneira F, Uitterlinden AG, Hofman A, Imboden M, Nitsch D, Brandstätter A, Kollerits B, Kedenko L, Mägi R, Stumvoll M, Kovacs P, Boban M, Campbell S, Endlich K, Völzke H, Kroemer HK, Nauck M, Völker U, Polasek O, Vitart V, Badola S, Parker AN, Ridker PM, Kardia SL, Blankenberg S, Liu Y, Curhan GC, Franke A, Rochat T, Paulweber B, Prokopenko I, Wang W, Gudnason V, Shuldiner AR, Coresh J, Schmidt R, Ferrucci L, Shlipak MG, van Duijn CM, Borecki I, Krämer BK, Rudan I, Gyllensten U, Wilson JF, Witteman JC, Pramstaller PP, Rettig R, Hastie N, Chasman DI, Kao WH, Heid IM, Fox CS. 2010. New loci associated with kidney function and chronic kidney disease. Nat Genet 42:376–384. doi: 10.1038/ng.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Okada Y, Terao C, Ikari K, Kochi Y, Ohmura K, Suzuki A, Kawaguchi T, Stahl EA, Kurreeman FA, Nishida N, Ohmiya H, Myouzen K, Takahashi M, Sawada T, Nishioka Y, Yukioka M, Matsubara T, Wakitani S, Teshima R, Tohma S, Takasugi K, Shimada K, Murasawa A, Honjo S, Matsuo K, Tanaka H, Tajima K, Suzuki T, Iwamoto T, Kawamura Y, Tanii H, Okazaki Y, Sasaki T, Gregersen PK, Padyukov L, Worthington J, Siminovitch KA, Lathrop M, Taniguchi A, Takahashi A, Tokunaga K, Kubo M, Nakamura Y, Kamatani N, Mimori T, Plenge RM, Yamanaka H, Momohara S, Yamada R, Matsuda F, Yamamoto K. 2012. Meta-analysis identifies nine new loci associated with rheumatoid arthritis in the Japanese population. Nat Genet 44:511–516. doi: 10.1038/ng.2231. [DOI] [PubMed] [Google Scholar]
- 14.Chan SW, Lim CJ, Chen L, Chong YF, Huang C, Song H, Hong W. 2011. The hippo pathway in biological control and cancer development. J Cell Physiol 226:928–939. doi: 10.1002/jcp.22435. [DOI] [PubMed] [Google Scholar]
- 15.Pan D. 2010. The Hippo signaling pathway in development and cancer. Dev Cell 19:491–505. doi: 10.1016/j.devcel.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao B, Li L, Lei Q, Guan KL. 2010. The Hippo-YAP pathway in organ size control and tumorigenesis: an updated version. Genes Dev 24:862–874. doi: 10.1101/gad.1909210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ziosi M, Baena-López LA, Grifoni D, Froldi F, Pession A, Garoia F, Trotta V, Bellosta P, Cavicchi S, Pession A. 2010. dMyc functions downstream of Yorkie to promote the supercompetitive behavior of Hippo pathway mutant cells. PLoS Genet 6:e1001140. doi: 10.1371/journal.pgen.1001140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cordenonsi M, Zanconato F, Azzolin L, Forcato M, Rosato A, Frasson C, Inui M, Montagner M, Parenti Anna R, Poletti A, Daidone Maria G, Dupont S, Basso G, Bicciato S, Piccolo S. 2011. The Hippo transducer TAZ confers cancer stem cell-related traits on breast cancer cells. Cell 147:759–772. doi: 10.1016/j.cell.2011.09.048. [DOI] [PubMed] [Google Scholar]
- 19.Nicolay BN, Bayarmagnai B, Islam ABMMK, Lopez-Bigas N, Frolov MV. 2011. Cooperation between dE2F1 and Yki/Sd defines a distinct transcriptional program necessary to bypass cell cycle exit. Genes Dev 25:323–335. doi: 10.1101/gad.1999211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao B, Li L, Wang L, Wang CY, Yu J, Guan KL. 2012. Cell detachment activates the Hippo pathway via cytoskeleton reorganization to induce anoikis. Genes Dev 26:54–68. doi: 10.1101/gad.173435.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao B, Wei X, Li W, Udan RS, Yang Q, Kim J, Xie J, Ikenoue T, Yu J, Li L, Zheng P, Ye K, Chinnaiyan A, Halder G, Lai ZC, Guan KL. 2007. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev 21:2747–2761. doi: 10.1101/gad.1602907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu L, Li Y, Kim SM, Bossuyt W, Liu P, Qiu Q, Wang Y, Halder G, Finegold MJ, Lee JS, Johnson RL. 2010. Hippo signaling is a potent in vivo growth and tumor suppressor pathway in the mammalian liver. Proc Natl Acad Sci U S A 107:1437–1442. doi: 10.1073/pnas.0911427107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baldwin C, Garnis C, Zhang L, Rosin MP, Lam WL. 2005. Multiple microalterations detected at high frequency in oral cancer. Cancer Res 65:7561–7567. [DOI] [PubMed] [Google Scholar]
- 24.Modena P, Lualdi E, Facchinetti F, Veltman J, Reid JF, Minardi S, Janssen I, Giangaspero F, Forni M, Finocchiaro G, Genitori L, Giordano F, Riccardi R, Schoenmakers EF, Massimino M, Sozzi G. 2006. Identification of tumor-specific molecular signatures in intracranial ependymoma and association with clinical characteristics. J Clin Oncol 24:5223–5233. doi: 10.1200/JCO.2006.06.3701. [DOI] [PubMed] [Google Scholar]
- 25.Snijders AM, Schmidt BL, Fridlyand J, Dekker N, Pinkel D, Jordan RC, Albertson DG. 2005. Rare amplicons implicate frequent deregulation of cell fate specification pathways in oral squamous cell carcinoma. Oncogene 24:4232–4242. doi: 10.1038/sj.onc.1208601. [DOI] [PubMed] [Google Scholar]
- 26.Yokoyama T, Osada H, Murakami H, Tatematsu Y, Taniguchi T, Kondo Y, Yatabe Y, Hasegawa Y, Shimokata K, Horio Y, Hida T, Sekido Y. 2008. YAP1 is involved in mesothelioma development and negatively regulated by Merlin through phosphorylation. Carcinogenesis 29:2139–2146. doi: 10.1093/carcin/bgn200. [DOI] [PubMed] [Google Scholar]
- 27.Liu F, Lagares D, Choi KM, Stopfer L, Marinkovic A, Vrbanac V, Probst CK, Hiemer SE, Sisson TH, Horowitz JC, Rosas IO, Fredenburgh LE, Feghali-Bostwick C, Varelas X, Tager AM, Tschumperlin DJ. 2015. Mechanosignaling through YAP and TAZ drives fibroblast activation and fibrosis. Am J Physiol Lung Cell Mol Physiol 308:L344–L357. doi: 10.1152/ajplung.00300.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mannaerts I, Leite SB, Verhulst S, Claerhout S, Eysackers N, Thoen LF, Hoorens A, Reynaert H, Halder G, van Grunsven LA. 2015. The Hippo pathway effector YAP controls mouse hepatic stellate cell activation. J Hepatol 63:679–688. doi: 10.1016/j.jhep.2015.04.011. [DOI] [PubMed] [Google Scholar]
- 29.McNeill H, Reginensi A. 2017. Lats1/2 regulate Yap/Taz to control nephron progenitor epithelialization and inhibit myofibroblast formation. J Am Soc Nephrol 28:852–861. doi: 10.1681/ASN.2016060611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seo E, Kim WY, Hur J, Kim H, Nam SA, Choi A, Kim YM, Park SH, Chung C, Kim J, Min S, Myung SJ, Lim DS, Kim YK. 2016. The Hippo-Salvador signaling pathway regulates renal tubulointerstitial fibrosis. Sci Rep 6:31931. doi: 10.1038/srep31931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lantinga-van Leeuwen IS, Leonhard WN, van de Wal A, Breuning MH, Verbeek S, de Heer E, Peters DJ. 2006. Transgenic mice expressing tamoxifen-inducible Cre for somatic gene modification in renal epithelial cells. Genesis 44:225–232. doi: 10.1002/dvg.20207. [DOI] [PubMed] [Google Scholar]
- 32.Bottinger EP, Bitzer M. 2002. TGF-beta signaling in renal disease. J Am Soc Nephrol 13:2600–2610. doi: 10.1097/01.ASN.0000033611.79556.AE. [DOI] [PubMed] [Google Scholar]
- 33.Meng XM, Nikolic-Paterson DJ, Lan HY. 2016. TGF-beta: the master regulator of fibrosis. Nat Rev Nephrol 12:325–338. doi: 10.1038/nrneph.2016.48. [DOI] [PubMed] [Google Scholar]
- 34.Kang HM, Ahn SH, Choi P, Ko YA, Han SH, Chinga F, Park AS, Tao J, Sharma K, Pullman J, Bottinger EP, Goldberg IJ, Susztak K. 2015. Defective fatty acid oxidation in renal tubular epithelial cells has a key role in kidney fibrosis development. Nat Med 21:37–46. doi: 10.1038/nm.3762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Anderson S, Eldadah B, Halter JB, Hazzard WR, Himmelfarb J, Horne FM, Kimmel PL, Molitoris BA, Murthy M, O'Hare AM, Schmader KE, High KP. 2011. Acute kidney injury in older adults. J Am Soc Nephrol 22:28–38. doi: 10.1681/ASN.2010090934. [DOI] [PubMed] [Google Scholar]
- 36.Grams ME, Chow EK, Segev DL, Coresh J. 2013. Lifetime incidence of CKD stages 3–5 in the United States. Am J Kidney Dis 62:245–252. doi: 10.1053/j.ajkd.2013.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rosner MH. 2013. Acute kidney injury in the elderly. Clin Geriatr Med 29:565–578. doi: 10.1016/j.cger.2013.05.001. [DOI] [PubMed] [Google Scholar]
- 38.Yang HC, Fogo AB. 2014. Fibrosis and renal aging. Kidney Int Suppl (2011) 4:75–78. doi: 10.1038/kisup.2014.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Childs BG, Durik M, Baker DJ, van Deursen JM. 2015. Cellular senescence in aging and age-related disease: from mechanisms to therapy. Nat Med 21:1424–1435. doi: 10.1038/nm.4000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim WY, Sharpless NE. 2006. The regulation of INK4/ARF in cancer and aging. Cell 127:265–275. doi: 10.1016/j.cell.2006.10.003. [DOI] [PubMed] [Google Scholar]
- 41.Krishnamurthy J, Torrice C, Ramsey MR, Kovalev GI, Al-Regaiey K, Su L, Sharpless NE. 2004. Ink4a/Arf expression is a biomarker of aging. J Clin Invest 114:1299–1307. doi: 10.1172/JCI22475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lopez-Otin C, Blasco MA, Partridge L, Serrano M, Kroemer G. 2013. The hallmarks of aging. Cell 153:1194–1217. doi: 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Clements ME, Chaber CJ, Ledbetter SR, Zuk A. 2013. Increased cellular senescence and vascular rarefaction exacerbate the progression of kidney fibrosis in aged mice following transient ischemic injury. PLoS One 8:e70464. doi: 10.1371/journal.pone.0070464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.DiRocco DP, Bisi J, Roberts P, Strum J, Wong KK, Sharpless N, Humphreys BD. 2014. CDK4/6 inhibition induces epithelial cell cycle arrest and ameliorates acute kidney injury. Am J Physiol Renal Physiol 306:F379–F388. doi: 10.1152/ajprenal.00475.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ferenbach DA, Bonventre JV. 2015. Mechanisms of maladaptive repair after AKI leading to accelerated kidney ageing and CKD. Nat Rev Nephrol 11:264–276. doi: 10.1038/nrneph.2015.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kuro-o M, Matsumura Y, Aizawa H, Kawaguchi H, Suga T, Utsugi T, Ohyama Y, Kurabayashi M, Kaname T, Kume E, Iwasaki H, Iida A, Shiraki-Iida T, Nishikawa S, Nagai R, Nabeshima YI. 1997. Mutation of the mouse klotho gene leads to a syndrome resembling ageing. Nature 390:45–51. doi: 10.1038/36285. [DOI] [PubMed] [Google Scholar]
- 47.Kurosu H, Yamamoto M, Clark JD, Pastor JV, Nandi A, Gurnani P, McGuinness OP, Chikuda H, Yamaguchi M, Kawaguchi H, Shimomura I, Takayama Y, Herz J, Kahn CR, Rosenblatt KP, Kuro-o M. 2005. Suppression of aging in mice by the hormone Klotho. Science 309:1829–1833. doi: 10.1126/science.1112766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sato S, Kawamata Y, Takahashi A, Imai Y, Hanyu A, Okuma A, Takasugi M, Yamakoshi K, Sorimachi H, Kanda H, Ishikawa Y, Sone S, Nishioka Y, Ohtani N, Hara E. 2015. Ablation of the p16(INK4a) tumour suppressor reverses ageing phenotypes of klotho mice. Nat Commun 6:7035. doi: 10.1038/ncomms8035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Satoh M, Nagasu H, Morita Y, Yamaguchi TP, Kanwar YS, Kashihara N. 2012. Klotho protects against mouse renal fibrosis by inhibiting Wnt signaling. Am J Physiol Renal Physiol 303:F1641–F1651. doi: 10.1152/ajprenal.00460.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Debelle FD, Vanherweghem JL, Nortier JL. 2008. Aristolochic acid nephropathy: a worldwide problem. Kidney Int 74:158–169. doi: 10.1038/ki.2008.129. [DOI] [PubMed] [Google Scholar]
- 51.Susnik N, Sorensen-Zender I, Rong S, von Vietinghoff S, Lu X, Rubera I, Tauc M, Falk CS, Alexander WS, Melk A, Haller H, Schmitt R. 2014. Ablation of proximal tubular suppressor of cytokine signaling 3 enhances tubular cell cycling and modifies macrophage phenotype during acute kidney injury. Kidney Int 85:1357–1368. doi: 10.1038/ki.2013.525. [DOI] [PubMed] [Google Scholar]
- 52.Coppe JP, Patil CK, Rodier F, Sun Y, Munoz DP, Goldstein J, Nelson PS, Desprez PY, Campisi J. 2008. Senescence-associated secretory phenotypes reveal cell-nonautonomous functions of oncogenic RAS and the p53 tumor suppressor. PLoS Biol 6:2853–2868. doi: 10.1371/journal.pbio.0060301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chai EZ, Shanmugam MK, Arfuso F, Dharmarajan A, Wang C, Kumar AP, Samy RP, Lim LH, Wang L, Goh BC, Ahn KS, Hui KM, Sethi G. 2016. Targeting transcription factor STAT3 for cancer prevention and therapy. Pharmacol Ther 162:86–97. doi: 10.1016/j.pharmthera.2015.10.004. [DOI] [PubMed] [Google Scholar]
- 54.Chuang PY, He JC. 2010. JAK/STAT signaling in renal diseases. Kidney Int 78:231–234. doi: 10.1038/ki.2010.158. [DOI] [PubMed] [Google Scholar]
- 55.Bienaime F, Muorah M, Yammine L, Burtin M, Nguyen C, Baron W, Garbay S, Viau A, Broueilh M, Blanc T, Peters D, Poli V, Anglicheau D, Friedlander G, Pontoglio M, Gallazzini M, Terzi F. 2016. Stat3 controls tubulointerstitial communication during CKD. J Am Soc Nephrol 27:3690–3705. doi: 10.1681/ASN.2015091014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tapon N, Harvey KF, Bell DW, Wahrer DC, Schiripo TA, Haber DA, Hariharan IK. 2002. salvador promotes both cell cycle exit and apoptosis in Drosophila and is mutated in human cancer cell lines. Cell 110:467–478. doi: 10.1016/S0092-8674(02)00824-3. [DOI] [PubMed] [Google Scholar]
- 57.Wu S, Huang J, Dong J, Pan D. 2003. hippo encodes a Ste-20 family protein kinase that restricts cell proliferation and promotes apoptosis in conjunction with salvador and warts. Cell 114:445–456. doi: 10.1016/S0092-8674(03)00549-X. [DOI] [PubMed] [Google Scholar]
- 58.Piersma B, Bank RA, Boersema M. 2015. Signaling in fibrosis: TGF-beta, WNT, and YAP/TAZ converge. Front Med (Lausanne) 2:59. doi: 10.3389/fmed.2015.00059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xu J, Li PX, Wu J, Gao YJ, Yin MX, Lin Y, Yang M, Chen DP, Sun HP, Liu ZB, Gu XC, Huang HL, Fu LL, Hu HM, He LL, Wu WQ, Fei ZL, Ji HB, Zhang L, Mei CL. 2016. Involvement of the Hippo pathway in regeneration and fibrogenesis after ischaemic acute kidney injury: YAP is the key effector. Clin Sci (Lond) 130:349–363. doi: 10.1042/CS20150385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu-Chittenden Y, Huang B, Shim JS, Chen Q, Lee SJ, Anders RA, Liu JO, Pan D. 2012. Genetic and pharmacological disruption of the TEAD-YAP complex suppresses the oncogenic activity of YAP. Genes Dev 26:1300–1305. doi: 10.1101/gad.192856.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang H, Ramakrishnan SK, Triner D, Centofanti B, Maitra D, Gyorffy B, Sebolt-Leopold JS, Dame MK, Varani J, Brenner DE, Fearon ER, Omary MB, Shah YM. 2015. Tumor-selective proteotoxicity of verteporfin inhibits colon cancer progression independently of YAP1. Sci Signal 8:ra98. doi: 10.1126/scisignal.aac5418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kai T, Tsukamoto Y, Hijiya N, Tokunaga A, Nakada C, Uchida T, Daa T, Iha H, Takahashi M, Nomura T, Sato F, Mimata H, Ikawa M, Seto M, Matsuura K, Moriyama M. 2016. Kidney-specific knockout of Sav1 in the mouse promotes hyperproliferation of renal tubular epithelium through suppression of the Hippo pathway. J Pathol 239:97–108. doi: 10.1002/path.4706. [DOI] [PubMed] [Google Scholar]
- 63.Matsuura K, Nakada C, Mashio M, Narimatsu T, Yoshimoto T, Tanigawa M, Tsukamoto Y, Hijiya N, Takeuchi I, Nomura T, Sato F, Mimata H, Seto M, Moriyama M. 2011. Downregulation of SAV1 plays a role in pathogenesis of high-grade clear cell renal cell carcinoma. BMC Cancer 11:523. doi: 10.1186/1471-2407-11-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Donninger H, Allen N, Henson A, Pogue J, Williams A, Gordon L, Kassler S, Dunwell T, Latif F, Clark GJ. 2011. Salvador protein is a tumor suppressor effector of RASSF1A with Hippo pathway-independent functions. J Biol Chem 286:18483–18491. doi: 10.1074/jbc.M110.214874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Szeto SG, Narimatsu M, Lu M, He X, Sidiqi AM, Tolosa MF, Chan L, De Freitas K, Bialik JF, Majumder S, Boo S, Hinz B, Dan Q, Advani A, John R, Wrana JL, Kapus A, Yuen DA. 2016. YAP/TAZ are mechanoregulators of TGF-β-Smad signaling and renal fibrogenesis. J Am Soc Nephrol 27:3117–3128. doi: 10.1681/ASN.2015050499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sugiura H, Yoshida T, Shiohira S, Kohei J, Mitobe M, Kurosu H, Kuro-o M, Nitta K, Tsuchiya K. 2012. Reduced Klotho expression level in kidney aggravates renal interstitial fibrosis. Am J Physiol Renal Physiol 302:F1252–F1264. doi: 10.1152/ajprenal.00294.2011. [DOI] [PubMed] [Google Scholar]
- 67.Wang K, Singh D, Zeng Z, Coleman SJ, Huang Y, Savich GL, He X, Mieczkowski P, Grimm SA, Perou CM, MacLeod JN, Chiang DY, Prins JF, Liu J. 2010. MapSplice: accurate mapping of RNA-seq reads for splice junction discovery. Nucleic Acids Res 38:e178. doi: 10.1093/nar/gkq622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li B, Dewey CN. 2011. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bullard JH, Purdom E, Hansen KD, Dudoit S. 2010. Evaluation of statistical methods for normalization and differential expression in mRNA-Seq experiments. BMC Bioinformatics 11:94. doi: 10.1186/1471-2105-11-94. [DOI] [PMC free article] [PubMed] [Google Scholar]