ABSTRACT
Acid-adapted strains of Escherichia coli K-12 W3110 were obtained by serial culture in medium buffered at pH 4.6 (M. M. Harden, A. He, K. Creamer, M. W. Clark, I. Hamdallah, K. A. Martinez, R. L. Kresslein, S. P. Bush, and J. L. Slonczewski, Appl Environ Microbiol 81:1932–1941, 2015, https://doi.org/10.1128/AEM.03494-14). Revised genomic analysis of these strains revealed insertion sequence (IS)-driven insertions and deletions that knocked out regulators CadC (acid induction of lysine decarboxylase), GadX (acid induction of glutamate decarboxylase), and FNR (anaerobic regulator). Each acid-evolved strain showed loss of one or more amino acid decarboxylase systems, which normally help neutralize external acid (pH 5 to 6) and increase survival in extreme acid (pH 2). Strains from populations B11, H9, and F11 had an IS5 insertion or IS-mediated deletion in cadC, while population B11 had a point mutation affecting the arginine activator adiY. The cadC and adiY mutants failed to neutralize acid in the presence of exogenous lysine or arginine. In strain B11-1, reversion of an rpoC (RNA polymerase) mutation partly restored arginine-dependent neutralization. All eight strains showed deletion or downregulation of the Gad acid fitness island. Strains with the Gad deletion lost the ability to produce GABA (gamma-aminobutyric acid) and failed to survive extreme acid. Transcriptome sequencing (RNA-seq) of strain B11-1 showed upregulated genes for catabolism of diverse substrates but downregulated acid stress genes (the biofilm regulator ariR, yhiM, and Gad). Other strains showed downregulation of H2 consumption mediated by hydrogenases (hya and hyb) which release acid. Strains F9-2 and F9-3 had a deletion of fnr and showed downregulation of FNR-dependent genes (dmsABC, frdABCD, hybABO, nikABCDE, and nrfAC). Overall, strains that had evolved in buffered acid showed loss or downregulation of systems that neutralize unbuffered acid and showed altered regulation of catabolism.
IMPORTANCE Experimental evolution of an enteric bacterium under a narrow buffered range of acid pH leads to loss of genes that enhance fitness above or below the buffered pH range, including loss of enzymes that may raise external pH in the absence of buffer. Prominent modes of evolutionary change involve IS-mediated insertions and deletions that knock out key regulators. Over generations of acid stress, catabolism undergoes reregulation in ways that differ for each evolving strain.
KEYWORDS: acid, Escherichia coli, experimental evolution, GABA, low pH, RNA polymerase, decarboxylase, fnr
INTRODUCTION
Enteric bacteria need to survive a wide range of pH values throughout the human intestinal tract. The pH of the intestinal tract ranges from 1.5 to 3.5 in the stomach, increases to 4 to 7 in the duodenum, reaches 7 to 9 in the jejunum, and decreases to pH 5 to 6 in the cecum (1). On a microscopic scale, extreme gradients of pH occur across the intestinal epithelium where enterobacteria adhere. To cope with such conditions, Escherichia coli maintains a cytoplasmic pH homeostasis of 7.3 to 7.8 during growth in an external pH range of 5 to 9 and survives extreme acid levels below pH 2.0 (2, 3). E. coli uses intricate regulatory networks such as acid resistance systems to survive this wide range of pH values (4). Several acid resistance systems of E. coli include decarboxylases of the amino acids lysine, arginine, glutamate/glutamine, and ornithine (5). Each amino acid-dependent decarboxylase catalyzes the decarboxylation of its respective amino acid by consuming a proton and releasing an amine or polyamine, i.e., cadaverine, agmatine, gamma-aminobutyric acid (GABA), and putrescine, respectively. The acid-dependent release of polyamines and of the neurotransmitters GABA and agmatine is of interest in studies of tumorigenesis (6) and the gut-brain axis (7). An important feature of acid-dependent decarboxylases is their different pH ranges for induction and acid consumption: lysine decarboxylase activity (CadA) peaks at approximately pH 6, arginine (Adi) has activity at pH 5, and glutamate (GadA and GadB) has activity below pH 5 (8, 9).
One way to investigate how E. coli adapts to contend with stress such as low pH is by experimental evolution (10–12). Experimental evolution of E. coli has been conducted under the stress of medium buffered at pH 4.6, the low end of the growth range for this microorganism (13). This pH range is relevant to the digestive tract as bacteria entering the stomach experience the in-between levels upon emerging from the inhibitory condition of stomach acid. Thus, via the fecal-oral route, enteric bacteria naturally undergo periods of selection at pH 4.6 to 5.0. The acid-evolved clones from our experiment (13) out-compete the ancestral strain growing at pH 4.6. Sequence analysis of acid-evolved clones reveals various categories of mutations relevant to pH response, such as single nucleotide polymorphisms (SNPs) in RNA polymerases rpoB and rpoC, as well as unexplained phenotypes such as loss of lysine decarboxylase activity.
While whole-genome sequencing provides a wealth of information about genetic differences between microorganisms, this type of analysis often fails to reveal complex mutations such as those mediated by insertion sequences (ISs). An updated genome resequencing pipeline, breseq, version 0.27.1, detects split-read alignments, where a single read maps to disjointed areas of the genome (14). In our analysis, breseq, version 0.27.1, revealed evidence of IS5-mediated interruptions and deletions, in particular, those that inactivate regulatory genes such as cadC, gadX, and fnr. Detecting these additional mutation types altered our view of the genomic changes in the acid-evolved clones.
Previously, we hypothesized that mutations in the acid-evolved clones could modulate the expression of acid-induced genes that are energetically wasteful under the conditions of the evolution experiment (13). Under our evolution conditions, the buffered environment would have prevented any rise in pH mediated by acid protection systems such as lysine decarboxylase; thus, the high expression level of CadA near pH 5 (15) would be energetically wasteful. We also hypothesized that the observed mutations in RNA polymerase (RNAP) subunit genes could have modulated the expression of decarboxylase systems. Lysine and arginine decarboxylases are among the most highly inducible pH-regulated genes (16, 17), and thus their overexpression could lead to a substantial energy cost.
To investigate the above hypotheses, we updated our genome sequence analysis, and we compared the evolved strains with respect to lysine, arginine, and glutamate decarboxylase activities. We tested constructs with reverted RNAP mutations for decarboxylase activity. We also performed transcriptome sequencing (RNA-seq) to analyze the overall expression patterns of our acid-evolved strains.
RESULTS
Genetic analysis of acid-evolved isolates.
Previously, we had conducted a 2,000-generation experimental evolution of 24 bacterial populations under moderately acidic conditions (pH 4.6 to 4.8). Two clones each from four independent bacterial populations (B11, F9, F11, and H9) were isolated for genomic analysis. The genomes of our acid-evolved strains (13) were assembled and compared with the E. coli K-12 W3110 reference genome using the breseq computational pipeline, version 0.23 (18). Our original results were largely consistent with those of an independent analysis using GATK tools at the Broad Institute that were available in 2014 (https://software.broadinstitute.org/gatk/). For the present work, we sought evidence for previously undetected mutations using an updated breseq, version 0.27.1 (19). Our revised analysis identified additional mutations including eight insertion knockouts as well as seven IS-mediated deletions (Table 1; see also Table S1 in the supplemental material). In addition to use of the updated pipeline, our revised analysis includes a less conservative view of mutation “calls” by the pipeline. In some cases, visual inspection of the sequence reads for a low-scoring call showed a mutation clearly present in all reads. We also compared the pairs of sister strains (strains from a shared population of the evolution experiment) for mutations called in one strain but not the other.
TABLE 1.
Amino acid decarboxylase mutations and phenotypes of acid-evolved strains
| Straina | Lysine decarboxylase |
Arginine decarboxylase |
Glutamate decarboxylase |
|||
|---|---|---|---|---|---|---|
| Activityb | Mutation | Activity | Mutationd | Activity | Mutatione | |
| B11-1 | − | cadC::IS5 | − | adiY (C→G) | + | |
| B11-2 | − | Δ([cadA]–[cadC])c | − | adiY (C→G) | + | |
| F9-2 | + | + | − | Δ(gadX–slp) | ||
| F9-3 | + | + | − | Δ(gadX–slp) | ||
| F11-1 | − | cadC::IS5 | + | − | Δ(gadX–slp) | |
| F11-2 | − | Δ([ghoS]–cadC) | + | − | Δ(gadX–slp) | |
| H9-1 | − | cadC::IS5 | − | adiY ←IS5← adiA | + | |
| H9-2 | − | cadC::IS5 | − | adiY ←IS5← adiA | + | |
Strains were isolated from the experimentally evolved populations B11, F9, F11, and H9 (13).
Activity is shown as follows: +, the decarboxylase system is active; −, the decarboxylase system activity is decreased or absent.
Brackets indicate that the mutation takes place in the middle of the gene. Delta symbols indicate deletion. En dash indicates that deletion includes all genetic material between indicated genes.
IS5 with double arrows (←IS5←) denotes the insertion of an IS5 element into the intergenic region; the arrows indicate the direction of transcription of flanking genes.
Deletion of most or all of the Gad acid fitness island: gadA gadX gadW mdtF mdtE gadE hdeD hdeA hdeB yhiD dctR slp.
The newly identified mutations include IS-mediated knockouts of cadC (20), which encodes the pH-sensing transcriptional activator of cadA (lysine decarboxylase) and cadB (lysine-cadaverine antiporter) (21). The inserted regions consist of an IS5 element, 11 copies of which occur in the reference sequence of strain W3110 (22). The cadC::IS5 knockouts were found in sequenced strains from three of the four acid-evolved populations (B11, F11, and H9). For population B11, strain B11-1 showed the cadC::IS5 knockout, whereas B11-2 showed an IS5-mediated deletion that includes cadA and cadB {Δ([cadA]–[cadC]), where the brackets indicate that the mutation takes place in the middle of the gene}. Strain F11-2 showed an IS5-mediated deletion, Δ([ghoS]–cadC), at the same position as a cadA::IS5 insertion in the sister strain F11-1 (Tables 1 and S1). This more extensive deletion covers cadABC as well as the nutritional stress toxin-antitoxin system GhoT/GhoS (23). In all, we found four independent IS-mediated mutations in cadC among six strains. Populations B11 and H9 also show point mutations in regulators of arginine decarboxylase (13).
In addition, the strains from two populations (F9 and F11) showed deletions of all or part of the Gad acid fitness island, which includes gadA (glutamate decarboxylase) with its regulators gadX, gadY, and gadE, as well as extreme-acid periplasmic chaperones (hdeA and hdeB) and multidrug transporters (mdtE and mdtF) (24).
We thus found an overall pattern in which our strains that evolved at a constant narrow range of pH 4.6 to 4.8 experience strong selection pressure to lose or downregulate one or more of the acid-inducible degradative amino acid decarboxylases (Table 1). Population B11 lost decarboxylases for lysine and arginine, F9 lost glutamate, F11 lost lysine and glutamate, and H9 lost lysine and arginine.
Another major regulator, fnr, was lost only in population F9 (strains F9-2 and F9-3) within the IS-mediated deletion Δ([ynaI]–[ogt]) (Table S1). The most prominent anaerobic regulator of E. coli, Fnr normally activates genes under low-oxygen conditions (25–29).
Lysine and arginine decarboxylase activities correspond to the acid-evolved genotypes.
While the genetic evidence supports our hypothesis of decarboxylase loss, many mutations in our strains could contribute unknown effects to their phenotypes. We sought to confirm the phenotypes by conducting assays of lysine and arginine decarboxylase under conditions permitting pH rise in the medium, using a modified Møller broth test (13). This pH rise is believed to be the main mechanism by which the lysine, arginine, and glutamate decarboxylases contribute to growth in moderate acid (external pH 4.5 to 6.5) and to survival in extreme acid (external pH 1 to 3) (2, 4, 15).
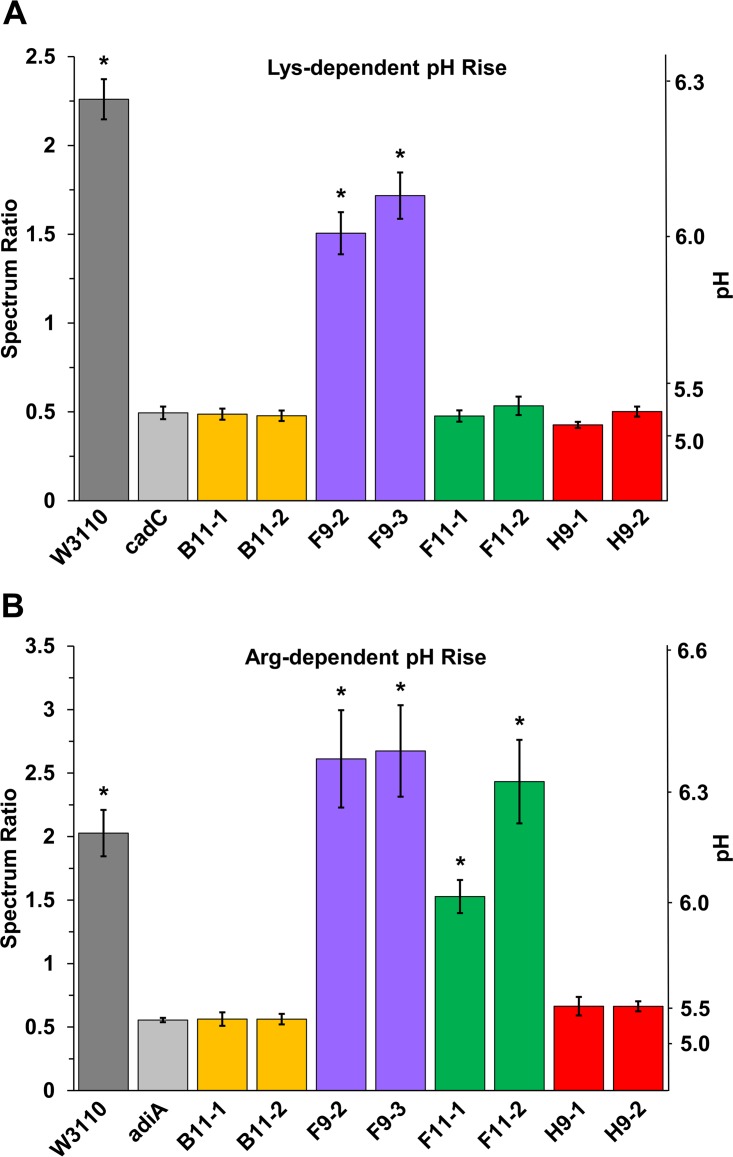
The lysine decarboxylase-associated pH rise was measured for our eight acid-evolved strains (Fig. 1A). Our modified Møller broth for the microplate test allowed precise measurement of the peak spectral intensities for the protonated and deprotonated forms of the dye bromocresol purple. The dye ratios were correlated with pH values by a standard curve (Fig. S1). In all trials, the strains showing cadC knockout or deletion showed 24-h endpoint pH values comparable to those of the control strain W3110 cadC::kan (Fig. 1A, light gray bar). Only strains F9-2 and F9-3 show decarboxylase activities significantly higher than those of the cadC knockout based on Tukey method comparison (P < 0.0001).
FIG 1.
pH rise of acid-evolved mutants cultured with lysine or arginine. Bacteria were inoculated in microplate wells containing 200 μl of modified Møller broth medium at pH 6.8 supplemented with 2.5 g/liter l-lysine (A) or at pH 5.5 supplemented with 2.5 g/liter l-arginine (B). The microplates were incubated for 24 h at 37°C. The terminal spectrum ratio for bromocresol purple absorbance (A570–590/A400–450) was converted to indicate pH (scale at right) (see Fig. S1). Error bars indicate standard errors of the means (n = 9). One-way ANOVA tests were run with the Tukey comparisons to identify significantly different groups. Asterisks indicate strains for which the results are significantly different from those of the knockout strain (P < 0.05).
For the same strains, we measured the pH rise associated with arginine decarboxylase (Fig. 1B). Strains B11-1 and B11-2 contain point mutations in the transcriptional activator adiY, which affects expression of arginine decarboxylase adiA (30). In addition, our new analysis showed that strain H9-1 contains an IS5 insertion found in the intergenic region between adiY and adiA (Table 1). Both H9 and B11 populations showed a significantly smaller arginine-dependent pH rise than that seen in W3110 (P < 0.001). The spectrum ratios (and inferred pH levels) were comparable to those of the negative-control strain, W3110 adiA::kan (P > 0.98). In contrast, the strains without adiY mutations showed a pH increase comparable to that of W3110. The strains F11-1, F11-2, F9-2, and F9-3 had a pH rise statistically equivalent to that of W3110 (P > 0.40).
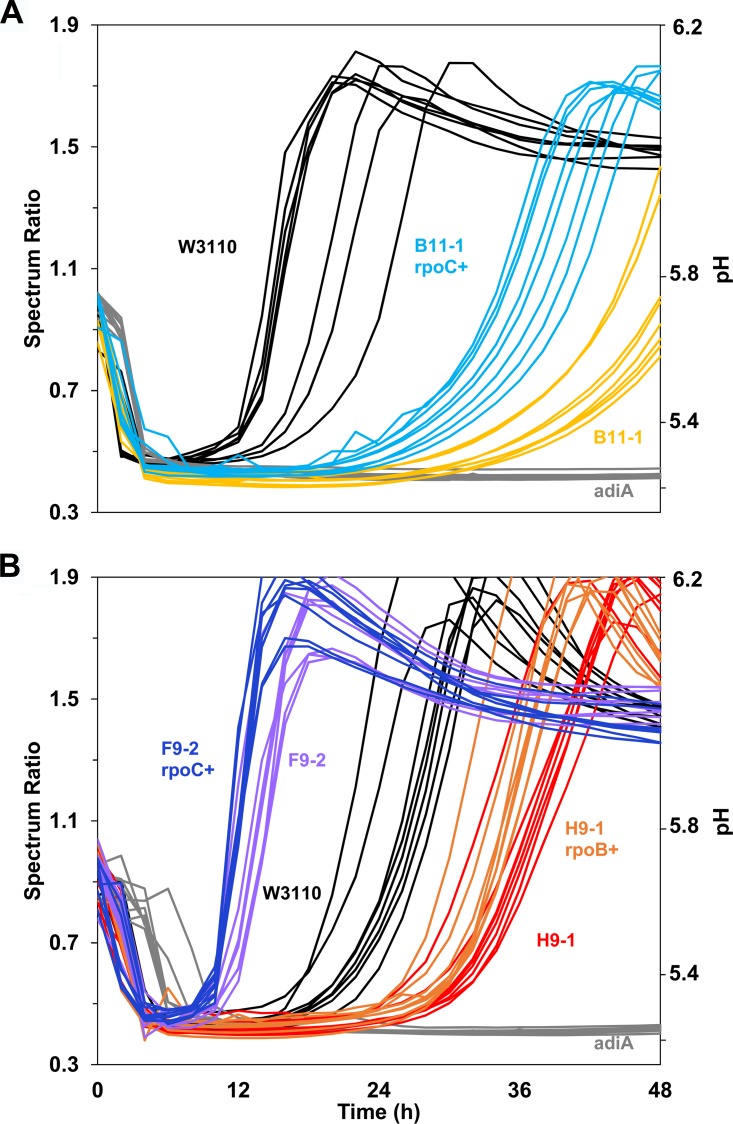
We also tested the hypothesis that the RNAP gene mutations found in our acid-evolved strains might modulate decarboxylase activities. For strains B11-1, F9-2, and H9-1, we replaced each mutant allele with an RNAP subunit with the reverted parental allele, using cotransduction of a linked Kanr marker. Each Kanr marker was converted to frt scar. The constructs thus obtained were B11-1 rpoC+ (JLSE0083thiG1A::frt) (13), F9-2 rpoC+ (JLSE0137thiH1C::frt), and H9-1 rpoB+ (JLSE0079thiH2C::frt). Of these constructs, B11-1 rpoC+ (JLSE0083thiG1A::frt) showed a modest recovery of arginine decarboxylase activity and pH rise compared to that of strain B11-1 (P < 0.0001) (Fig. 2A). Thus, the rpoC mutation V507L appears to have some modulatory effect on adi expression.
FIG 2.
Arginine decarboxylase assay. RpoC modulates arginine-dependent pH rise in strain B11-1 but not in strains F9-2 and H9-1. Unique rpo point mutations found in the acid-evolved strains were reverted to represent the original ancestral W3110 sequence for strain B11-1 (A) as well as strains H9-1 and F9-2 (B). The method described in the legend of Fig. 1B was modified for kinetic recording of absorbance ratios. Absorbance readings were taken every 2 h for 48 h. Replicates for each strain are shown as curves of the same color.
For comparison, we saw no such restoration of AdiA activity after rpo reversion in F9-2 or in H9-1 (Fig. 2B) (P > 0.5). The arginine-dependent pH rise actually increased for strain F9-2 compared to the level in the ancestor W3110; this is the only acid-evolved strain that showed a significant increase in any decarboxylase activity. Population H9 has an IS5 insertion in the intergenic region of adiA to adiY which may depress adiA expression; this insertion effect would be unlikely to be overcome by reversion of an rpo mutation. In contrast to H9-1, B11 has no IS5 insertion in the adi region, instead having a point mutation in the activator adiY. In B11, the adiY and rpoC point mutations may be exhibiting a combined effect to depress AdiA, which would explain the partial restoration we observed with rpoC reversion.
GABA production and extreme-acid survival correspond to the Gad genotypes.
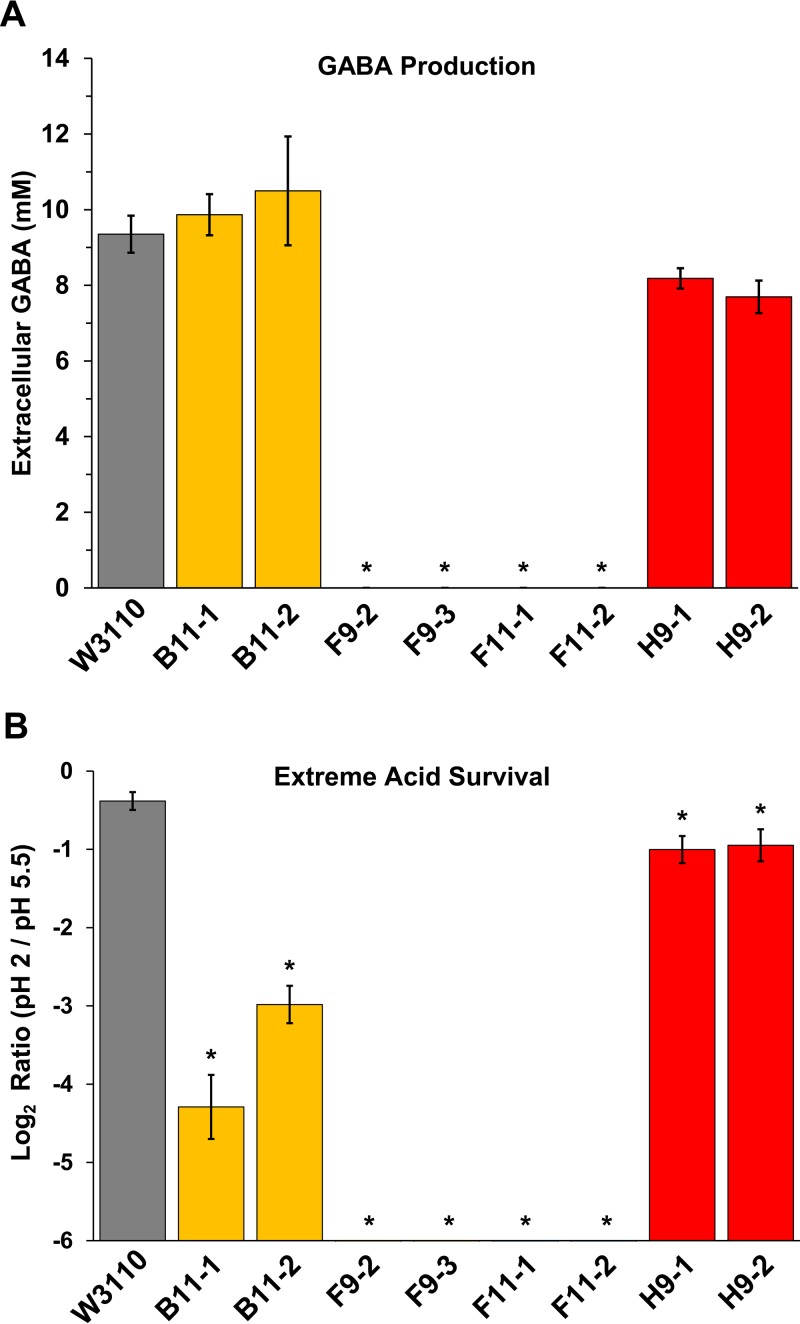
Given the large gad deletions we found in the F9 and F11 populations (Table 1), we sought evidence for glutamate decarboxylase activity. The GadA enzyme is generally expressed at around pH 5 but is most active under extreme-acid conditions such as pH 2 (4). Glutamate decarboxylase converts glutamate into GABA, which accepts a proton in the process, reducing pH stress (4). Glutamate decarboxylase activity can be inferred from GABA production measured by gas chromatography-mass spectrometry (GC-MS) (31). GABA was measured by GC-MS after anaerobic overnight cultures were exposed at pH 2.0. We found that strains F9-2, F9-3, F11-1, and F11-2 produced no detectable GABA (Fig. 3A). However, the other strains produced millimolar quantities of extracellular GABA, comparable to the level of strain W3110 (P > 0.05). Thus, we confirmed that the GABA-negative strains are the ones with deletions in the Gad acid fitness island.
FIG 3.
GABA production and extreme-acid survival of acid-evolved strains. (A) GABA assays of anaerobic overnight cultures after exposure at pH 2.0 for 2 h, as measured by GC-MS (see Materials and Methods). ANOVA tests identified no significant difference in GABA production levels between strains W3110, B11, and H9. (B) Survival of overnight cultures after dilution and incubation at pH 2.0 for 2 h (see Materials and Methods). The y axis shows the log2 ratios of the colony counts at the indicated pH values. The axis cutoff (10−6) represents the lower limit of colony detection. One-way ANOVA tests were run with the Tukey method comparison to identify significantly different groups. Asterisks indicate strains that show significant difference in results from those of the ancestral strain W3110 (P < 0.05).
Strains that produce GABA should also test positive for the Gad acid survival system (4, 15). All the sequenced populations show a significant loss of survival at pH 2.0 (Fig. 3B). Strains from populations F9 (F9-2 and F9-3) and F11 (F11-1 and F11-2), which have deletions of the gad region, showed acid survival rates below our level of detection (log2 pH 2/pH 5.5 values below −6). Strains B11-1 and B11-2 survived at rates approximately 10% that of the ancestor (P < 0.0001). Strains H9-1 and H9-2 survived at approximately half the level of ancestral W3110 (P < 0.05). Thus, overall extreme-acid survival did correspond to the presence of the Gad system although other genetic factors may have affected the degree of acid survival. Because the strains were grown under moderate acid stress for 2,000 generations, mutations may have arisen that decreased extreme-acid survival. This loss of acid survival may represent a trade-off for another phenotype that improves growth.
Several mutations of interest that may have decreased acid survival in B11 were found, including ftsX (transporter subunit), hepA (RNA polymerase-associated helicase protein), and rpoC (RNA polymerase beta prime subunit) (Table S1). These genes function in the cell membrane or in transcription, so the mutations may affect the cell membrane permeability to protons, sensitivity to acid, or expression of acid response genes. H9 mutations that may have affected acid survival occurred in yej, inner membrane proteins, and in the intergenic region between rhaA and rhaD, the region within the l-rhamnose degradation pathway (Table S1).
Previously, we reported a similar loss of Gad during evolution in the presence of benzoic acid buffered at pH 6.5 (32). Some of the benzoate-evolved strains show loss of multidrug resistance. We therefore tested our acid-evolved strains for resistance to chloramphenicol but found no significant difference from the ancestral W3110 (data not shown).
RNA-seq of acid-evolved strains shows downregulated gad genes and upregulated anaerobic catabolism.
Many of the mutations in our acid-evolved strains impact regulatory proteins that could modulate transcription of a large number of products. We therefore used RNA-seq to compare the gene expression across the transcriptomes of four acid-evolved strains (B11-1, F9-2, F11-1, and H9-1), each from one of the four independently evolved populations (Tables S2 to S5). Each transcriptome was indexed to that of the ancestor, after culture at pH 4.8 (see details in Materials and Methods).
The four strains all showed unique patterns of differential transcription, with some features in common. Genes showing the greatest upregulation and downregulation are summarized in Table 2. Most of the upregulated genes involve anaerobic catabolism and transport. Strain B11-1 showed upregulation of citCDEFG (citrate fermentation), dcuAB (anaerobic C4-dicarboxylate transport), srlABDE (sorbitol permease and catabolism), tdcBCDEFG (anaerobic threonine catabolism), and ykgEFG (d-lactate catabolism). d-Lactate catabolism was upregulated in all four strains. Strain F11-1 also showed upregulation of glp operons (anaerobic glycerol catabolism), and strain H9-1 showed upregulation of glp as well as fdnGH (formate dehydrogenase).
TABLE 2.
Differential expression of acid-evolved strains compared to ancestral strain W3110
| Regulation of expression | Genes in the indicated straina |
|||
|---|---|---|---|---|
| B11-1 | F9-2 | F11-1 | H9-1 | |
| Upregulated ≥4-fold | citCDEFGX, dcuAB, srlABDE, tdcBCDEFG, ttdAB, ykgEFG | norVW, ykgEFG | {glpABC glpD glpFK glpTQ}, nirBD, ykgEG | fdnGH, {glpABC, glpTQ}, ykgEF |
| Downregulated ≥4-fold | {ariR, bhsA}, {argO, lysA, lysC}, cysACDHIJNPUW, {gadABCE, mdtEJ}, nrdHI, sufABC, yhiM, ymdF | dmsABC, fdnG, frdABCD, ΔgadBCEW, ΔhdeABD, hyaABCDEF, hybABO, hypABCD, napDF, narIJUYZ, nikABCDE, nrfAC, ybaST, yhiM, ynaIJ, ynfEFGKO | {argO, lysA, lysC, lysP}, ΔgadABCEWX, ΔhdeABD, ΔmdtEF, sufABCD, ybaST, ycaC, yciFG, yhiM, ymdF | appABC, {argO, lysA, lysC}, {gadABCEWX, hdeA, mdtEF, slp}, hyaABCDEF, osmEY, otsAB, sufABCDES, ybaST, yciFG, yhiM, ymdF |
Selected genes and regulons from Table S2 representing all genes upregulated or downregulated ≥4-fold relative to expression levels in W3110 and fitting the criterion of a P value of <0.001 as assessed by negative binomial tests in DESeq. RNA was isolated from bacteria cultured to log phase in LBKmal broth, pH 4.8. Delta symbols indicate gene deletions (Table S1). Genes within braces (boldface) are grouped together based on functional relevance.
A large number of genes were downregulated in the acid-evolved strains. Most strikingly, the Gad acid resistance genes, as well as multidrug transporters mdtEF, were downregulated in B11-1 and in H9-1, our two acid-evolved strains that retained the entire sequence of the Gad acid fitness island. All four strains showed downregulation of yhiM encoding a Gad-related acid stress membrane protein (33). Despite downregulation under the low-density culture condition for RNA-seq, stationary-phase cultures of B11-1 and H9-1 produced GABA at levels comparable to the level in the ancestor and showed only a slight decrease of extreme-acid resistance (Fig. 3). We infer that these strains retain Gad activity during stationary phase (the condition that normally shows highest Gad expression, for extreme-acid survival) while downregulating Gad expression during early log phase, where it incurs a fitness cost.
Several operons implicated in the acid stress response were downregulated, including sufABC (Fe-S transporter) and hya and hyb (H2 consumption hydrogenases Hyd-1 and Hyd-2), all of which are acid-inducible under anaerobic conditions (17). Also downregulated were cysACD (sulfate transport) and ariR (regulator of acid resistance). Inactivation of these operons is associated with increased cell aggregation, which could contribute to the increased acid fitness in these strains (34, 35).
Strain F9-2, whose genome contains a deletion of the anaerobic regulator fnr, showed downregulation of a number of genes normally upregulated by Fnr under low-oxygen conditions (25, 28). The downregulated genes include dmsABC (dimethyl sulfoxide reductase), frdABCD (fumarate reductase), hybABO (Hyd-2), nikABCDE (nickel ABC transporter), and nrfAC (formate-dependent nitrite reductase). The downregulation of these genes implies that the genes show significant expression in the FNR+ ancestor W3110 when it is cultured at low pH.
DISCUSSION
Our analysis of acid-evolved genomes combines genomics, transcriptomics, and pH-modulating enzyme activities to identify the evolutionary selection forces impacting E. coli genomes under continual acid stress in semiaerobic microplate culture. The acid stress condition includes nutrient limitation as well as oxygen limitation caused by the physical geometry of the microplate. These conditions are relevant to the mammalian digestive tract, where enteric bacteria experience changes in pH as well as extreme gradients of oxygen across the intestinal epithelium (36).
Surprisingly, the overall picture is that genes highly upregulated by acid incur long-term fitness costs when the acid condition persists across many generations. In our acid-evolved genomes, different strains showed different combinations of loss or decrease of acid-inducible amino acid decarboxylase activities. Loss of decarboxylase activity occurred via three different mechanisms: IS knockouts of regulatory genes such as cadC; IS-mediated large deletions, particularly of the Gad regulon; and regulatory adjustment leading to downregulation of Gad activity at early log phase, such as that seen in the transcriptomes of the Gad-positive strains B11-1 and H9-1. The cause of most of the transcriptome modulation is unknown, but evidence for at least one possible mechanism comes from the modulation of arginine decarboxylase activity by the rpoC mutation in strain B11-1; the decarboxylase activity was partly restored in the B11-1 rpoC+ construct (Fig. 2). It is possible for RNAP mutations to offer a way for evolution to quickly fine-tune the expression of a large number of genes (37).
We imagine several possible causes of the loss of acid-induced genes. A comparable result was found in a study of heat-adapted E. coli clones (12) that show decreased heat shock gene expression. Genes that confer fitness under a transient extreme stress may actually be detrimental during prolonged exposure; thus, evolution selects for restoration of the nonstress physiology. Another consideration is that “acid stress” actually encompasses a wide range of concentrations of the hydronium ion and that systems conferring fitness at one pH may be less advantageous at a different acid level. Thus, the lysine decarboxylase might be advantageous for cells cultured at pH 6 but unhelpful at pH 4.6. Finally, the high concentration of buffer at low pH negates the advantage of decarboxylases that function to neutralize acid. Under nutrient limitation, without the advantage of neutralizing pH, the catabolic loss of amino acids would be strongly selected against.
The transcriptomes of our acid-evolved clones showed varied patterns of upregulation and downregulation of catabolic pathways. One common result for all four strains (Table 2) is the upregulation of ykgEFG, implying an important energy contribution from d-lactate catabolism. The fitness advantage of diverse catabolism implies that in our evolution experiment, carbon source utilization under limited oxygen may have had as great a selective force as pH. The cyclic transition through stationary phase and nutrient limitation may be an underappreciated contributor to fitness in classic evolution experiments where “daily dilution” always includes several hours of stationary phase.
The fnr deletion in strain F9-2 represents a divergent alternative path of evolution compared to the paths of our other three populations analyzed. It is unclear how the loss of anaerobic induction helps survival under our conditions of semiaerobic culture at low pH. One possibility is that turning off the consumption hydrogenases (hya and hyb) eliminates production of H+ ions that could increase acidification (17, 38, 39). Alternatively, some of the FNR-activated gene expression may have represented a wasteful energy expenditure, i.e., over 2,000 generations of evolution without extreme anaerobiosis. Thus, the loss of anaerobic response under a constant semiaerobic condition may be comparable to the loss of an extreme-acid response under a buffered acid condition.
It would be of interest to further establish the link between loss of decarboxylase activity and increased acid fitness. This could be accomplished with gene complementation although the construct would require testing to rule out construct-specific fitness effects. Alternatively, the mutated decarboxylase genes could be reverted to the wild-type sequence, as we have done with the rpo mutations in the acid-evolved strains. We would expect to see a decrease in moderate acid fitness when decarboxylase activity is restored.
Our findings on acid-influenced evolution of bacteria have implications for future studies of the human gut microbiome, in which ingested bacteria pass through the stomach where they experience an environment at pH 4 to 5 before entering the intestine. Interestingly, IS-inactivated cadC alleles are found in enteroinvasive strains of E. coli and Shigella (40). CadA inactivation is thought to occur in pathogens because cadaverine production lowers virulence. Another relevant observation is that acid-evolved bacteria show altered production of polyamines and GABA, which is of interest in the microbiome for carcinogenesis (6) and the gut-brain axis (7).
METHODS AND MATERIALS
Bacterial strains and medium.
Escherichia coli K-12 W3110 was the ancestral strain of all acid-evolved populations (13). All strains were cultured in Luria Bertani-potassium (LBK) broth, containing 10 g/liter tryptone, 5 g/liter yeast extract, and 7.45 g/liter KCl. The culture medium was buffered with either 100 mM homopiperazine-1,4-bis(2-ethanosulfonic acid) (HOMOPIPES) (pKa = 4.55, 8.12), 100 mM 2-(N-morpholino)ethanesulfonic acid (MES) (pKa = 5.96), or 100 mM 3-morpholinopropane-1-sulfonic acid (MOPS) (pKa = 7.20), depending on the desired pH of 4.4 to 4.8, 5.5 to 6.5, or 7.0, respectively. The pH of the medium was adjusted as necessary with either 5 M HCl or 5 M KOH. LBK medium buffered to the pH range of 4.5 to 4.8 was additionally supplemented with 10 g/liter dl-malic acid (pKa = 3.40, 5.11) (LBKmal) as described by Harden et al. (13). As both d- and l-malate may be catabolized by acid-consuming reactions (41, 42), supplementation with malic acid provided a potential resource to enhance acid tolerance.
Sequence analysis.
The sequence annotations of acid-evolved genes (13) were updated using breseq, version 0.27.1 (18, 19). This breseq version reveals IS-mediated insertions and deletions that are identified by inspection of unassigned new junctions and regions of missing coverage (43). Sequences of acid-evolved strains were compared to the E. coli K-12 W3110 reference sequence (NCBI GenBank accession number NC_007779.1) (22). Variant sequences present in our laboratory strain of W3110, referred to as W3110-D8, were filtered. Assembly and annotations of our evolved isolate sequences were visualized using the Integrative Genomics Viewer (IGV) from the Broad Institute. Sequence identity was confirmed by PCR amplification of selected mutations.
P1 phage transduction and strain construction.
Mutations were reverted to the ancestral K-12 W3110 sequence by cotransduction of linked markers, using P1 phage by standard procedures (44). Strains with desired deletions or functional insertions were ordered from the Keio collection (Coli Genetic Stock Center, Yale University) (45) and were introduced into the evolved strain of choice or the ancestral strain W3110. Constructs were confirmed by PCR amplification and Sanger sequencing of key alleles of donor and recipient.
FRT procedure.
Antibiotic resistance was eliminated from strains containing a Keio kan insertion allele by a flippase-FRT recombinase procedure as described in Datsenko and Wanner (46) using plasmid pCP20. The designation “frt” (flippase recognition target) is given to strains with the Keio insertion removed and indicates the remaining scar sequence (strains are listed in Table 3).
TABLE 3.
Strains used in this study
| Strain name | Population isolate | Genotype | Source or reference |
|---|---|---|---|
| JLSE0079 | H9-1 | See Table S1 | 13 |
| JLSE0080 | H9-2 | See Table S1 | 13 |
| JLSE0083 | B11-1 | See Table S1 | 13 |
| JLSE0084 | B11-2 | See Table S1 | 13 |
| JLSE0091 | F11-1 | See Table S1 | 13 |
| JLSE0092 | F11-2 | See Table S1 | 13 |
| JLSE0137 | F9-2 | See Table S1 | 13 |
| JLSE0138 | F9-3 | See Table S1 | 13 |
| W3110 | Stock D8 | Fred Neidhardt | |
| JW5731-1 | ΔadiA743::kan | 45 | |
| JW4094-5 | ΔcadC760::kan | 45 | |
| JW3953-2 | ΔthiH760::kan | 45 | |
| JW5549-1 | ΔthiG761::kan | 45 | |
| JLSE0079thiH2C | JLSE0079(H9-1) ΔthiH760::kan rpoB+ | This study | |
| JLSE0079thiH2C::frt | JLSE0079(H9-1) ΔthiH760::frt rpoB+ | This study | |
| JLSE0083thiG1A | JLSE0083(B11-1) ΔthiG761::kan rpoC+ | 13 | |
| JLSE0083thiG1A::frt | JLSE0083(B11-1) ΔthiG761::frt rpoC+ | This study | |
| JLSE0137thiH1C | JLSE0137(F9-2) ΔthiH760::kan rpoC+ | This study | |
| JLSE0137thiH1C::frt | JLSE0137(F9-2) ΔthiH1C760::frt rpoC+ | This study |
Lysine and arginine decarboxylase assays and pH rise.
The pH rise associated with activity of lysine decarboxylase and arginine decarboxylase was measured using a procedure with Møller decarboxylase broth, with modification (47). Lysine decarboxylase broth contained 3 g/liter yeast extract, 1 g/liter d-glucose, 5 g/liter l-lysine, and 0.015 g/liter bromocresol purple, pH 6.8 ± 0.2. l-Lysine was replaced with l-arginine in the arginine decarboxylase broth at pH 5.5 ± 0.2. Strains were inoculated into a microplate containing aliquoted medium. The plate was sealed for anaerobiosis and incubated at 37°C for 24 h. The plate was inspected visually and verified by absorbance readings from 350 nm to 750 nm using a SpectraMax Plus 384 microplate reader (Molecular Devices). We established an absorbance ratio, R = Av/Ay, that reflects the pH shift as indicated by titration of the dye bromocresol purple. The ratio R was calculated from mean absorbance values, A570–590/A400–450. The pH conversion scale is shown in Fig. S1 in the supplemental material.
GABA assays.
The procedure for measuring GABA production via glutamate decarboxylase was modified from that of Creamer et al. and Ma et al. (32, 48). Strains were cultured overnight in LB medium (10 g/liter tryptone, 5 g/liter yeast extract, 100 mM NaCl, 10 mM glutamine) buffered with 100 mM MES, pH 5.5. Glutamine is converted to glutamate, the substrate of glutamate decarboxylase (49). Closed 9-ml screw-cap tubes nearly full of medium were incubated for 18 h at 37°C. The pH of each sample was lowered with HCl to pH 2.0 and incubated for 2 h with rotation. The supernatant from 1 ml of culture was filtered and prepared for GC-MS by EZfaast amino acid derivatization (50). GABA from the culture fluid was identified using National Institute of Standards and Technology (NIST) library analysis.
Extreme-acid survival assays.
The conditions for testing extreme-acid survival were based on the aerobic assay from Riggins et al. and Lin et al. (38, 51). Cultures were grown overnight (16 to 18 h) in LBK broth, buffered with 100 mM MES at pH 5.5 to upregulate acid response systems (4). Overnight cultures were diluted 1:200 in LBK broth at pH 2.0 and incubated at 37°C for 2 h. After exposure, cultures were serially diluted in M63 minimal medium [0.4 g/liter KH2PO4, 0.4 g/liter K2HPO4, 1 g/liter (NH4)2SO4, 7.45 g/liter KCl] buffered with 100 mM MOPS at pH 7.0 to a final dilution of 1:400,000. Control cultures at pH 5.5 were serially diluted to a final ratio of 1:400,000 in M63 buffered with 100 mM MOPS, pH 7.0. Fifty microliters of each final dilution was spread onto LBK agar plates and incubated at 37°C. Colony counts for each replicate were log transformed. The means of log2 values from each replicate are reported.
RNA purification and extraction.
Bacteria were cultured to stationary phase (16 to 18 h) in LBKmal broth (pH 4.8), diluted 1:50 into fresh medium at pH 4.8, and grown to early log phase as indicated by an optical density at 600 nm (OD600) of 0.2. Cultures were diluted 6:1 by volume into a 5% buffer-saturated phenol-ethanol solution and centrifuged (4 min at 5,000 rpm and 4°C). The supernatant was decanted, and centrifugation was repeated. The pellet was resuspended in Tris-EDTA (TE) buffer (100 μl) with 3 mg/ml lysozyme. To isolate RNA, a Qiagen RNeasy minikit was used according to the manufacturer's instructions, with the addition of one RNase-free DNase treatment for 15 min at room temperature. A second DNase treatment using MoBio DNase-Max was conducted according to the manufacturer's instructions.
Construction and sequencing of Illumina RNA-seq libraries.
The mRNA population from all RNA samples was enriched by depleting rRNA sequences, and strand-specific libraries were constructed as described earlier (52). Starting with 8 to 50 ng of rRNA-depleted RNA, random-primed cDNA synthesis was done using a ScriptSeq, version 2, RNA-seq library preparation kit (Epicenter, WI). cDNA was purified using an Agencourt AMPure XP system (Beckman Coulter, NJ). Libraries were amplified using a FailSafe PCR enzyme kit (Epicenter, WI). Typically 12 (50 to 100 ng of starting RNA) or 15 (8 ng of starting RNA) PCR cycles were used, and reverse primer from the kit was replaced with one of the ScriptSeq Index primers. After PCR amplification, libraries were purified and size selected (∼280 bp) using the Agencourt AMPure XP system (Beckman Coulter, NJ). Profiles of library insert sizes were verified on an Agilent Technology microfluidic platform using a DNA 1K chip (Agilent Technologies, Wilmington, DE). Libraries were sequenced on an Illumina NextSeq 500 (150-cycle paired-end mode).
Mapping and assembly of raw reads.
Sequences were initially analyzed using CLC Genomics software, version 6.0. Sequences with a quality score of less than 30 were discarded, the remaining sequences were trimmed, and sequences of less than 36 bp were discarded. Sequences were mapped to the E. coli W3110 genome, (NCBI accession number NC_007779.1) (22), and mapping was done using the following CLC genomics mapping parameters: mismatch, 1; insertion, 3; deletion, 3; length, 0.9; similarity, 0.95; auto-detect paired distances on and map randomly. The CLC RNA-seq was performed using the following parameters: mismatch, 2; length fraction, 0.9; similarity fraction, 0.95; strand specific selected; maximum 3 hits, 3; paired settings, 36 to 500; broken pairs counting selected. Only unique counts generated for individual genes were used as the starting data for all subsequent analyses.
RNA-seq analysis.
Differential expression analysis was performed using the R package DESeq (53). A multifactor design was used to control for batch effects. For each comparison, two models were fit to generalized linear models (GLM), one regressing the gene expression on both batch and strain and one regressing only on the batch. P values were determined by a chi-square test that compared the two GLM fits. Samples were normalized by size factor, and dispersions were estimated as described previously (53). Comparisons resulting in a P value of <0.001 with a log2 fold change of ≥2 or ≤ −2 were considered significant. Quality assessment was conducted using heat maps with variance-stabilized data and principal component analyses.
Statistical analysis.
Analysis of variance (ANOVA) with post hoc Tukey honestly significant differences (HSD) tests were used for between-strain statistical comparisons of Lys- and Arg-dependent pH rise, extreme-acid survival, and GABA production. The terminal spectrum ratios of bromocresol purple absorbance (A570–590/A400–450) for each strain were used for comparisons of Lys- and Arg-dependent pH rise. The time required to reach a spectrum ratio of 0.7 after 4 h was used for statistical comparisons in the kinetic decarboxylase assays. The log2 colony count ratios pre- and post-acid exposure were the values used to compare extreme-acid survival. Extracellular GABA levels (in millimolar amounts), as measured by GC-MS, were compared to assess differences in GABA production between strains.
Accession number(s).
Genomic sequence and alignment data have been deposited in the NCBI Sequence Read Archive (SRA) under accession number SRP041420. RNA-seq data have been deposited in the NCBI SRA under accession number SRP095404 (see Table S6 for sample descriptions).
Supplementary Material
ACKNOWLEDGMENTS
We thank Jeff Barrick, Michael Harden, Kerry Rouhier, Peter Lund, and John Foster for valuable discussions. Greg Gillen conducted excellent enzyme assays.
This work was funded by grant MCB-1613278 from the National Science Foundation and by the Kenyon College Summer Science Scholars.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00442-17.
REFERENCES
- 1.Fallingborg J. 1999. Intraluminal pH of the human gastrointestinal tract. Dan Med Bull 46:183–196. [PubMed] [Google Scholar]
- 2.Slonczewski JL, Fujisawa M, Dopson M, Krulwich TA. 2009. Cytoplasmic pH Measurement and Homeostasis in Bacteria and Archaea. Adv Microb Physiol 55:1–79. doi: 10.1016/S0065-2911(09)05501-5. [DOI] [PubMed] [Google Scholar]
- 3.Krulwich TA, Sachs G, Padan E. 2011. Molecular aspects of bacterial pH sensing and homeostasis. Nat Rev Microbiol 9:330–343. doi: 10.1038/nrmicro2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Foster JW. 2004. Escherichia coli acid resistance: tales of an amateur acidophile. Nat Rev Microbiol 2:898–907. doi: 10.1038/nrmicro1021. [DOI] [PubMed] [Google Scholar]
- 5.Lund P, Tramonti A, De Biase D. 2014. Coping with low pH: molecular strategies in neutralophilic bacteria. FEMS Microbiol Rev 38:1091–1125. doi: 10.1111/1574-6976.12076. [DOI] [PubMed] [Google Scholar]
- 6.Holmes E, Li JV, Athanasiou T, Ashrafian H, Nicholson JK. 2011. Understanding the role of gut microbiome-host metabolic signal disruption in health and disease. Trends Microbiol 19:349–359. doi: 10.1016/j.tim.2011.05.006. [DOI] [PubMed] [Google Scholar]
- 7.Collins SM, Surette M, Bercik P. 2012. The interplay between the intestinal microbiota and the brain. Nat Rev Microbiol 10:735–742. doi: 10.1038/nrmicro2876. [DOI] [PubMed] [Google Scholar]
- 8.Kanjee U, Houry W. 2013. Mechanisms of acid resistance in Escherichia coli. Annu Rev Microbiol 67:65–81. doi: 10.1146/annurev-micro-092412-155708. [DOI] [PubMed] [Google Scholar]
- 9.Capitani G, De Biase D, Aurizi C, Gut H, Bossa F, Grutter MG. 2003. Crystal structure and functional analysis of Escherichia coli glutamate decarboxylase. EMBO J 22:4027–4037. doi: 10.1093/emboj/cdg403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.LaCroix RA, Sandberg TE, O'Brien EJ, Utrilla J, Ebrahim A, Guzman GI, Szubin R, Palsson BO, Feist AM. 2015. Use of adaptive laboratory evolution to discover key mutations enabling rapid growth of Escherichia coli K-12 MG1655 on glucose minimal medium. Appl Environ Microbiol 81:17–30. doi: 10.1128/AEM.02246-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lenski RE, Travisano M. 1994. Dynamics of adaptation and diversification: a 10,000-generation experiment with bacterial populations. Proc Natl Acad Sci U S A 91:6808–6814. doi: 10.1073/pnas.91.15.6808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sandberg TE, Pedersen M, LaCroix RA, Ebrahim A, Bonde M, Herrgard MJ, Palsson BO, Sommer M, Feist AM. 2014. Evolution of Escherichia coli to 42°C and subsequent genetic engineering reveals adaptive mechanisms and novel mutations. Mol Biol Evol 31:2647–2662. doi: 10.1093/molbev/msu209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harden MM, He A, Creamer K, Clark MW, Hamdallah I, Martinez KA, Kresslein RL, Bush SP, Slonczewski JL. 2015. Acid-adapted strains of Escherichia coli K-12 obtained by experimental evolution. Appl Environ Microbiol 81:1932–1941. doi: 10.1128/AEM.03494-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deatherage DE, Barrick JE. 2014. Identification of mutations in laboratory-evolved microbes from next-generation sequencing data using breseq. Methods Mol Biol 1151:165–188. doi: 10.1007/978-1-4939-0554-6_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kanjee U, Gutsche I, Ramachandran S, Houry WA. 2011. The enzymatic activities of the Escherichia coli basic aliphatic amino acid decarboxylases exhibit a pH zone of inhibition. Biochemistry 50:9388–9398. doi: 10.1021/bi201161k. [DOI] [PubMed] [Google Scholar]
- 16.Maurer LM, Yohannes E, Bondurant SS, Radmacher M, Slonczewski JL. 2005. pH regulates genes for flagellar motility, catabolism, and oxidative stress in Escherichia coli K-12. J Bacteriol 187:304–319. doi: 10.1128/JB.187.1.304-319.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hayes ET, Wilks JC, Sanfilippo P, Yohannes E, Tate DP, Jones BD, Radmacher MD, BonDurant SS, Slonczewski JL. 2006. Oxygen limitation modulates pH regulation of catabolism and hydrogenases, multidrug transporters, and envelope composition in Escherichia coli K-12. BMC Microbiol 6:89. doi: 10.1186/1471-2180-6-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barrick JE, Colburn G, Deatherage DE, Traverse CC, Strand MD, Borges JJ, Knoester DB, Reba A, Meyer AG. 2014. Identifying structural variation in haploid microbial genomes from short-read resequencing data using breseq. BMC Genomics 15:1039. doi: 10.1186/1471-2164-15-1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deatherage DE, Traverse CC, Wolf LN, Barrick JE. 2014. Detecting rare structural variation in evolving microbial populations from new sequence junctions using breseq. Front Genet 5:468. doi: 10.3389/fgene.2014.00468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eichinger A, Haneburger I, Koller C, Jung K, Skerra A. 2011. Crystal structure of the sensory domain of Escherichia coli CadC, a member of the ToxR-like protein family. Protein Sci 20:656–669. doi: 10.1002/pro.594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Neely MN, Olson ER. 1996. Kinetics of expression of the Escherichia coli cad operon as a function of pH and lysine. J Bacteriol 178:5522–5528. doi: 10.1128/jb.178.18.5522-5528.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hayashi K, Morooka N, Yamamoto Y, Fujita K, Isono K, Choi S, Ohtsubo E, Baba T, Wanner BL, Mori H, Horiuchi T. 2006. Highly accurate genome sequences of Escherichia coli K-12 strains MG1655 and W3110. Mol Syst Biol 2:2006.0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cheng HY, Soo VWC, Islam S, McAnulty MJ, Benedik MJ, Wood TK. 2014. Toxin GhoT of the GhoT/GhoS toxin/antitoxin system damages the cell membrane to reduce adenosine triphosphate and to reduce growth under stress. Environ Microbiol 16:1741–1754. doi: 10.1111/1462-2920.12373. [DOI] [PubMed] [Google Scholar]
- 24.Mates AK, Sayed AK, Foster JW. 2007. Products of the Escherichia coli acid fitness island attenuate metabolite stress at extremely low pH and mediate a cell density-dependent acid resistance. J Bacteriol 189:2759–2768. doi: 10.1128/JB.01490-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Constantinidou C, Hobman JL, Griffiths L, Patel MD, Penn CW, Cole JA, Overton TW. 2006. A reassessment of the FNR regulon and transcriptomic analysis of the effects of nitrate, nitrite, NarXL, and NarQP as Escherichia coli K12 adapts from aerobic to anaerobic growth. J Biol Chem 281:4802–4815. doi: 10.1074/jbc.M512312200. [DOI] [PubMed] [Google Scholar]
- 26.Salmon K, Hung S, Mekjian K, Baldi P, Hatfield GW, Gunsalus RP. 2003. Global gene expression profiling in Escherichia coli K12: the effects of oxygen availability and FNR. J Biol Chem 278:29837–29855. doi: 10.1074/jbc.M213060200. [DOI] [PubMed] [Google Scholar]
- 27.Kang Y, Weber KD, Qiu Y, Kiley PJ, Blattner FR. 2005. Genome-wide expression analysis indicates that FNR of Escherichia coli K-12 regulates a large number of genes of unknown function. J Bacteriol 187:1135–1160. doi: 10.1128/JB.187.3.1135-1160.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kumar R, Shimizu K. 2011. Transcriptional regulation of main metabolic pathways of cyoA, cydB, fnr, and fur gene knockout Escherichia coli in C-limited and N-limited aerobic continuous cultures. Microb Cell Fact 10:3. doi: 10.1186/1475-2859-10-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Myers KS, Yan H, Ong IM, Chung D, Liang K, Tran F, Kele° S, Landick R, Kiley PJ. 2013. Genome-scale analysis of Escherichia coli FNR reveals complex features of transcription factor binding. PLoS Genet 9:e1003565. doi: 10.1371/journal.pgen.1003565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stim-Herndon KP, Flores TM, Bennett GN. 1996. Molecular characterization of adiY, a regulatory gene which affects expression of the biodegradative acid-induced arginine decarboxylase gene (adiA) of Escherichia coli. Microbiology 142:1311–1320. doi: 10.1099/13500872-142-5-1311. [DOI] [PubMed] [Google Scholar]
- 31.Moroni F, Bianchi C, Tanganelli S, Moneti GBL. 1981. The release of gamma-aminobutyric acid, glutamate, and acetylcholine from striatal slices: a mass fragmentographic study. J Neurochem 36:1691–1697. doi: 10.1111/j.1471-4159.1981.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 32.Creamer K, Ditmars F, Basting P, Kunka K, Hamdallah I, Bush S, Scott Z, He A, Penix S, Gonzales A, Eder E, Camperchioli D, Berndt A, Clark M, Rouhier K, Slonczewski J. 2017. Benzoate and salicylate tolerant strains lose antibiotic resistance during laboratory evolution of Escherichia coli K-12. Appl Environ Microbiol 83:e02736-16. doi: 10.1128/AEM.02736-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nguyen T, Sparks-Thissen R. 2012. The inner membrane protein, YhiM, is necessary for Escherichia coli (E. coli) survival in acidic conditions. Arch Microbiol 194:637–641. doi: 10.1007/s00203-012-0798-x. [DOI] [PubMed] [Google Scholar]
- 34.Rossi E, Motta S, Mauri P, Landini P. 2014. Sulfate assimilation pathway intermediate phosphoadenosine 5′-phosphosulfate acts as a signal molecule affecting production of curli fibres in Escherichia coli. Microbiology 160:1832–1844. doi: 10.1099/mic.0.079699-0. [DOI] [PubMed] [Google Scholar]
- 35.Lee J, Page R, García-Contreras R, Palermino JM, Zhang XS, Doshi O, Wood TK, Peti W. 2007. Structure and function of the Escherichia coli protein YmgB: a protein critical for biofilm formation and acid-resistance. J Mol Biol 373:11–26. doi: 10.1016/j.jmb.2007.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zheng L, Kelly CJ, Colgan SP. 2015. Physiologic hypoxia and oxygen homeostasis in the healthy intestine. A review in the theme: cellular responses to hypoxia. Am J Physiol Cell Physiol 309:C350–C360. doi: 10.1152/ajpcell.00191.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Herring CD, Raghunathan A, Honisch C, Patel T, Applebee MK, Joyce AR, Albert TJ, Blattner FR, van den Boom D, Cantor CR, Palsson BØ. 2006. Comparative genome sequencing of Escherichia coli allows observation of bacterial evolution on a laboratory timescale. Nat Genet 38:1406–1412. doi: 10.1038/ng1906. [DOI] [PubMed] [Google Scholar]
- 38.Riggins DP, Narvaez MJ, Martinez KA, Harden MM, Slonczewski JL. 2013. Escherichia coli K-12 survives anaerobic exposure at pH 2 without RpoS, Gad, or hydrogenases, but shows sensitivity to autoclaved broth products. PLoS One 8:e56796. doi: 10.1371/journal.pone.0056796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Noguchi K, Riggins DP, Eldahan KC, Kitko RD, Slonczewski JL. 2010. Hydrogenase-3 contributes to anaerobic acid resistance of escherichia coli. PLoS One 5:e10132. doi: 10.1371/journal.pone.0010132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Casalino M, Latella MC, Prosseda G, Colonna B. 2003. CadC is the preferential target of a convergent evolution driving enteroinvasive Escherichia coli toward a lysine decarboxylase-defective phenotype. Infect Immun 71:5472–5479. doi: 10.1128/IAI.71.10.5472-5479.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lukas H, Reimann J, Kim Bin O, Grimpo J, Unden G. 2010. Regulation of aerobic and anaerobic d-malate metabolism of Escherichia coli by the LysR-type regulator DmlR (YeaT). J Bacteriol 192:2503–2511. doi: 10.1128/JB.01665-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim Bin O, Reimann J, Lukas H, Schumacher U, Grimpo J, Dünnwald P, Unden G. 2009. Regulation of tartrate metabolism by TtdR and relation to the DcuS-DcuR-regulated C4-dicarboxylate metabolism of Escherichia coli. Microbiology 155:3632–3640. doi: 10.1099/mic.0.031401-0. [DOI] [PubMed] [Google Scholar]
- 43.Tenaillon O, Rodríguez-Verdugo A, Gaut RL, McDonald P, Bennett AF, Long AD, Gaut BS. 2012. The molecular diversity of adaptive convergence. Science 335:457–461. doi: 10.1126/science.1212986. [DOI] [PubMed] [Google Scholar]
- 44.Deininger KNW, Horikawa A, Kitko RD, Tatsumi R, Rosner JL, Wachi M, Slonczewski JL. 2011. A requirement of TolC and MDR efflux pumps for acid adaptation and GadAB induction in Escherichia coli. PLoS One 6:e18960. doi: 10.1371/journal.pone.0018960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2:2006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Møller V. 1955. Simplified tests for some amino acid decarboxylases and for the arginine dihydrolase system. Acta Pathol Microbiol Scand 36:158–172. [DOI] [PubMed] [Google Scholar]
- 48.Ma Z, Gong S, Richard H, Tucker DL, Conway T, Foster JW. 2003. GadE (YhiE) activates glutamate decarboxylase-dependent acid resistance in Escherichia coli K-12. Mol Microbiol 49:1309–1320. doi: 10.1046/j.1365-2958.2003.03633.x. [DOI] [PubMed] [Google Scholar]
- 49.Lu P, Ma D, Chen Y, Guo Y, Chen G-Q, Deng H, Shi Y. 2013. l-Glutamine provides acid resistance for Escherichia coli through enzymatic release of ammonia. Cell Res 23:635–644. doi: 10.1038/cr.2013.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Badawy A. 2012. The EZ:Faast family of amino acid analysis kits: application of the GC-FID kit for rapid determination of plasma tryptophan and other amino acids. Methods Mol Biol 828:153–164. doi: 10.1007/978-1-61779-445-2_14. [DOI] [PubMed] [Google Scholar]
- 51.Lin J, In Soo Lee Frey J, Slonczewski JL, Foster JW. 1995. Comparative analysis of extreme acid survival in Salmonella typhimurium, Shigella flexneri, and Escherichia coli. J Bacteriol 177:4097–4104. doi: 10.1128/jb.177.14.4097-4104.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Smith AD, Yan X, Chen C, Dawson HD, Bhagwat AA. 2016. Understanding the host-adapted state of Citrobacter rodentium by transcriptomic analysis. Arch Microbiol 198:353–362. doi: 10.1007/s00203-016-1191-y. [DOI] [PubMed] [Google Scholar]
- 53.Anders S, Huber W. 2010. Differential expression analysis for sequence count data. Genome Biol 11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.