ABSTRACT
Xenorhabdus nematophila bacteria are mutualistic symbionts of Steinernema carpocapsae nematodes and pathogens of insects. The X. nematophila global regulator Lrp controls the expression of many genes involved in both mutualism and pathogenic activities, suggesting a role in the transition between the two host organisms. We previously reported that natural populations of X. nematophila exhibit various levels of Lrp expression and that cells expressing relatively low levels of Lrp are optimized for virulence in the insect Manduca sexta. The adaptive advantage of the high-Lrp-expressing state was not established. Here we used strains engineered to express constitutively high or low levels of Lrp to test the model in which high-Lrp-expressing cells are adapted for mutualistic activities with the nematode host. We demonstrate that high-Lrp cells form more robust biofilms in laboratory media than do low-Lrp cells, which may reflect adherence to host tissues. Also, our data showed that nematodes cultivated with high-Lrp strains are more frequently colonized than are those associated with low-Lrp strains. Taken together, these data support the idea that high-Lrp cells have an advantage in tissue adherence and colonization initiation. Furthermore, our data show that high-Lrp-expressing strains better support nematode reproduction than do their low-Lrp counterparts under both in vitro and in vivo conditions. Our data indicate that heterogeneity of Lrp expression in X. nematophila populations provides diverse cell populations adapted to both pathogenic (low-Lrp) and mutualistic (high-Lrp) states.
IMPORTANCE Host-associated bacteria experience fluctuating conditions during both residence within an individual host and transmission between hosts. For bacteria that engage in evolutionarily stable, long-term relationships with particular hosts, these fluctuations provide selective pressure for the emergence of adaptive regulatory mechanisms. Here we present evidence that the bacterium Xenorhabdus nematophila uses various levels of the transcription factor Lrp to optimize its association with its two animal hosts, nematodes and insects, with which it behaves as a mutualist and a pathogen, respectively. Building on our previous finding that relatively low cellular levels of Lrp are optimal for pathogenesis, we demonstrate that, conversely, high levels of Lrp promote mutualistic activities with the Steinernema carpocapsae nematode host. These data suggest that X. nematophila has evolved to utilize phenotypic variation between high- and low-Lrp-expression states to optimize its alternating behaviors as a mutualist and a pathogen.
KEYWORDS: virulence modulation, phenotypic variation, symbiosis, colonization initiation
INTRODUCTION
Animals associate with microbes in relationships that can be mutualistic (beneficial), commensal (neutral), or pathogenic (harmful). Within all of these types of relationships, and during transitions between different hosts or natural environments, microbes can experience rapidly changing local conditions, such as fluctuating nutrient availability and dynamic host immune responses. Bacterial fitness is increased by adaptive responses to such changes. One mechanism by which bacteria can adapt to rapid environmental fluctuations is phenotypic variation, in which an isogenic population (derived from the same parental cell) comprises subpopulations exhibiting different phenotypes, each of which confers a selective advantage in a particular environment (1). Phenotypic variation can thereby eliminate the temporal lag associated with sense-and-response mechanisms during fluctuations in environment and/or host conditions.
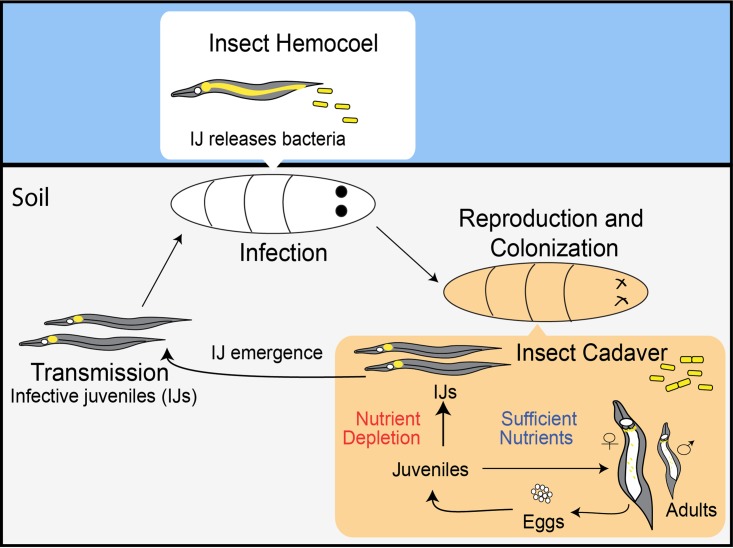
The life cycle of the bacterium Xenorhabdus nematophila alternates between a pathogenic phase in multiple types of insects and a mutualistic stage, specifically with the entomopathogenic nematode Steinernema carpocapsae, which transmits X. nematophila between insect hosts. During the transmission phase of the life cycle, X. nematophila bacteria are carried in the intestine, in a region termed the receptacle, of the nematode's soil-dwelling infective juvenile (IJ) stage (Fig. 1). The IJ invades the blood cavity of a host insect and releases X. nematophila, which helps suppress insect immunity and expresses virulence determinants that contribute to insect death. In the insect cadaver, bacteria secrete various enzymatic activities that degrade insect tissues, liberating nutrients that support nematode development and sexual reproduction. Progeny nematodes that experience nutrient depletion and high population densities will develop into IJs, colonized by their bacterial symbionts, and emerge from the insect cadaver into the soil to seek a new host to infect (2).
FIG 1.
Tripartite life cycle and mutualistic symbiosis of Xenorhabdus bacteria and Steinernema nematodes. During the transmission stage, IJ nematodes carry Xenorhabdus bacteria (shown in yellow) in an intestinal pocket known as the receptacle. In the infection stage, an IJ enters the insect larva and releases the bacterial symbiont; together, they kill the insect host. During the reproduction stage, bacteria replicate, colonize nematodes, and support nematode reproduction. Upon nutrient depletion, juvenile nematodes form IJs and emerge from the insect cadaver. See the text for additional details.
The adaptation of X. nematophila to the three general phases of the nematode's life cycle (infection, reproduction, and transmission) will directly impact its own fitness and the fitness of its symbiotic partner (Fig. 1) (3). Within each phase, X. nematophila experiences distinctive environmental conditions and various growth rates (4, 5). With regard to the nutrient composition and concentration, insect blood is rich in glucose and trehalose but limiting for iron (6, 7); the insect cadaver that supports the growth of X. nematophila to high densities appears to contain utilizable iron, lipids, and proteins (3, 7–9); and the IJ nematode receptacle, an environment in which X. nematophila experiences limited reproduction, contains growth-supporting levels of some amino acids and vitamins (e.g., serine, histidine, and pantothenate) but not others (threonine, paraminobenzoate, and pyridoxine) (10, 11).
The three different life cycle phases also offer distinctive surfaces and host molecules with which X. nematophila may interact. During infection of a living insect, X. nematophila encounters both constitutive and inducible immune factors such as melanin and antimicrobial peptides (12–14) and localizes to iron- and collagen-rich connective tissue in the midgut extracellular matrix (15). X. nematophila bacteria, but not other bacteria, adhere to the anterior intestinal region of developing and reproducing nematodes within the insect cadaver (16, 17), and in emerging IJs, X. nematophila cells are physically associated with a host-derived anucleate structure called the IVS (intravesicular structure), which is coated with wheat germ-reactive mucus-like material containing either N-acetylglucosamine or N-acetylneuraminic acid moieties (17). Although biofilm formation in X. nematophila has yet to be fully characterized, the species-specific association of X. nematophila with distinctive nematode tissues may involve biofilm-mediated attachment, similar to that which occurs in other mutualisms such as that between Vibrio fischeri bacteria and Euprymna scolopes squid during initiation of colonization (18).
To successfully navigate its life cycle and express the appropriate symbiotic activities, X. nematophila must sense and adapt to shifts in nutrient and host molecule identities. X. nematophila exhibits phenotypic variation that may contribute to this process. Two distinct but overlapping types of phenotypic switching have been observed in X. nematophila: variation between a “primary”- and “secondary”-form cell type and virulence modulation (VMO). Primary-form cells are typically observed in natural populations of Xenorhabdus bacteria, but prolonged growth under laboratory conditions leads to the appearance of secondary-form cells that no longer express multiple behaviors such as the ability to swim or swarm, bind the dye bromothymol blue, agglutinate red blood cells, and produce antibiotics and exoenzymes. Both primary and secondary forms of the bacteria colonize nematode IJs and kill insects, and the specific selective advantage of this phenomenon remains unclear (19–21). In VMO, a subpopulation of wild-type X. nematophila primary-form cells exhibits attenuated virulence, measured as the ability to kill insects after direct injection of the bacteria into the insect blood cavity. The VMO switch is spontaneous and reversible among bacterial colonies under laboratory conditions (22, 23). The adaptive benefit of a switch that results in a subpopulation of X. nematophila cells that are attenuated for virulence has not been investigated yet, but it may play a role in the reproductive and transmission stages of the X. nematophila life cycle.
In X. nematophila, both primary/secondary-form variation and VMO are controlled in part by the leucine-responsive regulatory protein (Lrp), a global regulatory transcription factor. Lrp homologs are small (∼19-kDa) regulatory proteins conserved across bacteria and archaea (24, 25). They are members of a larger conserved family of transcriptional regulators known as the feast/famine regulatory proteins, which are involved in nutrient adaptation (26). In X. nematophila, Lrp regulates more than 65% of the visible proteome and regulates genes that encode functions involved in pathogenesis, mutualism, and nutrient adaptation (27, 28). An lrp mutant has defects in suppressing insect immunity, shows reduced abilities to support nematode reproduction and colonize IJs, and has delayed growth relative to the wild type when transiting from nutrient-limiting to nutrient-rich conditions (27, 29). In addition, the lrp mutant shares many characteristics with secondary-form cells, including the inability to bind bromothymol blue dye and a reduced expression of exoenzymes such as lipase and protease. These lrp mutant phenotypes suggest that Lrp might be involved in the primary-to-secondary-form switch (27).
We recently reported that Lrp expression levels fluctuate among X. nematophila primary-form wild-type isolates and colonies and that this variation is responsible for the distinctive virulent and attenuated-virulence phenotypes observed in the VMO switch. When directly injected into M. sexta insect larvae, wild-type X. nematophila isolates naturally expressing either low or high relative levels of Lrp were virulent and immunosuppressive or virulence attenuated and nonimmunosuppressive, respectively. When Lrp expression levels were “fixed” by the plasmid-encoded constitutive expression of either low or high Lrp levels, we again observed virulent/suppressive and virulence-attenuated/nonsuppressive phenotypes, respectively, establishing a direct link between Lrp levels and the virulence phenotype. These findings suggest that in wild-type X. nematophila, variations in Lrp expression levels create heterogeneous subpopulations with respect to virulence phenotypes (23).
A further prediction from our previous study was that the VMO switch may contribute to X. nematophila adaptation during its transitions between hosts. The virulence-attenuated strains expressing fixed high levels of Lrp consistently exhibited increased antibiotic, protease, and lipase activities in vitro (23). Since these activities may promote degradation and protection of the insect cadaver (7, 30–32), we hypothesized that high-Lrp subpopulations may be adapted for the optimal support of the reproductive stage of symbiosis and that population heterogeneity with respect to Lrp levels contributes to adaptation between host environments. In this study, we sought to further explore the role of high- versus low-lrp-expressing populations in mutualism (reproduction and transmission) and mutualism-associated (biofilm formation) phenotypes.
RESULTS
Fixed high levels of Lrp confer optimal biofilm formation and mutualistic phenotypes in X. nematophila.
We hypothesized that, relative to X. nematophila low-lrp-expressing strains, those strains expressing high levels of Lrp (high-lrp strains) are optimized for mutualistic interactions with the S. carpocapsae nematode host. In various mutualistic microbial symbioses, biofilm formation is associated with tissue-specific colonization of the host animal (33, 34). To characterize the impact of various Lrp levels on X. nematophila biofilm formation, we measured adherence to the surfaces of borosilicate glass tubes, using previously described fixed-lrp strains of X. nematophila: an lrp-2::kan mutant carrying control, low-lrp, or high-lrp plasmids (23). After static incubation for 6 days in LB medium, all strains formed adherent populations at the liquid-air interface (Fig. 2A). Solubilization and quantification of stained biofilm material revealed that X. nematophila expressing high levels of Lrp had significantly more adherent cells than did either the no-lrp or low-lrp strain (Fig. 2B).
FIG 2.
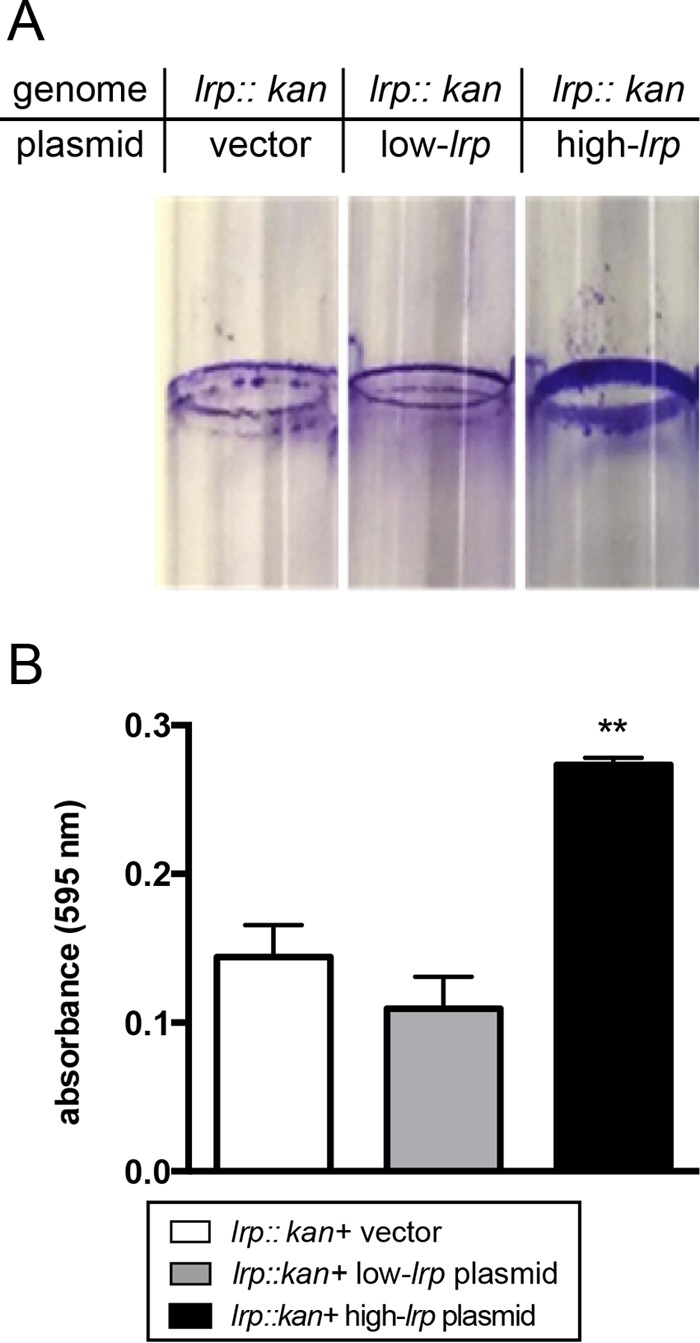
Biofilm formation of bacterial strains expressing fixed levels of Lrp. Bacterial strains expressing fixed levels of Lrp from plasmids were grown for 6 days in static culture to cultivate biofilms. (A) Biofilm materials were subsequently stained with crystal violet and visualized. (B) Stained materials were then solubilized and quantitated by using spectrophotometry. One-way ANOVA and Tukey's multiple-comparisons tests were used to establish statistical groups (**, P ≤ 0.01).
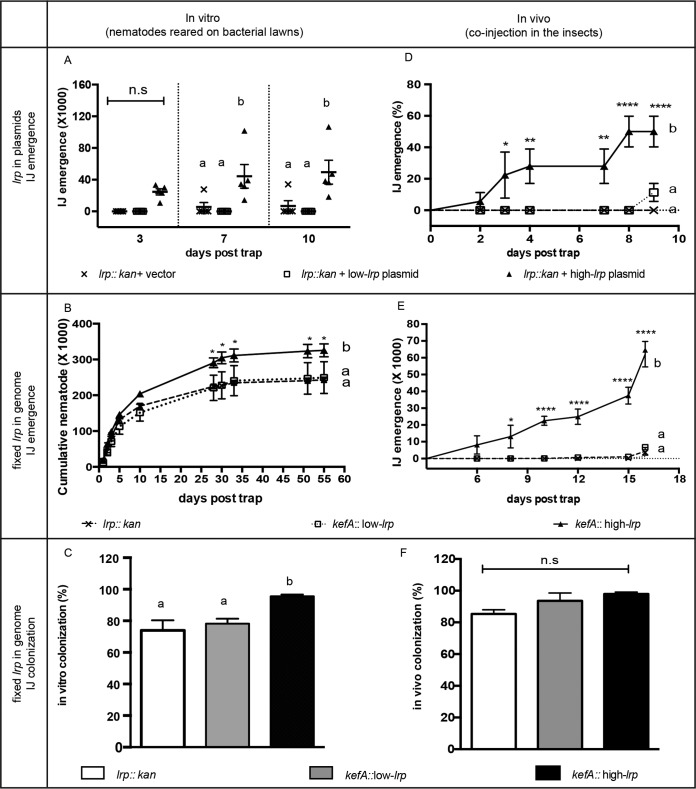
As a proxy for the mutualistic association of X. nematophila with nematodes, we next used the number of IJ nematodes produced after coculture on agar lawns of X. nematophila expressing plasmid-encoded fixed levels of Lrp (23). IJs that emerged from the bacterial lawns into water traps were counted over time. On days 7 and 10 post-water trap, the strain expressing plasmid-encoded high levels of Lrp (high-lrp plasmid strain) supported significantly higher IJ production than did either the no-lrp vector control or the low-lrp plasmid strain (Fig. 3A), supporting the idea that the high-lrp plasmid strain provided greater mutualistic benefits to the nematode host than did the other strains.
FIG 3.
Mutualistic symbiosis phenotypes in vitro (A to C) and in vivo (D to F) of IJ nematodes with bacterial strains expressing fixed levels of Lrp from either plasmids (A and D) or in-genome constructs (B, C, E, and F). Nematode fecundity is shown as cumulative IJs (A, B, and E) or percent productive infection (D). IJ colonization frequency was calculated as a percentage of colonized IJs among the emerged IJs (E and F). Data are shown as averaged measurements (n = 5 for panels A and E; n = 3 for panels B to D and F); error bars represent standard errors. Two-way ANOVA (A, B, D, and E) or one-way ANOVA (C and F) and Tukey's multiple-comparisons tests were used to establish statistical groups. Statistical significance and P value ranges are indicated by asterisks (n.s, not significant [P > 0.05]; *, P ≤ 0.05; **, P ≤ 0.01; ****, P ≤ 0.0001). Different letters are assigned to different statistical groups.
In our previous work, the high-lrp-, low-lrp-, and no-lrp-plasmid strains were used for short-term (<100-h) virulence assays in Manduca sexta insects (23). However, the use of a plasmid-based method to provide fixed levels of Lrp is not ideal for the long experimental times (10 to 64 days) required to monitor the full spectrum of mutualistic behaviors, since plasmid loss is likely to occur as antibiotic selection decays over time, ultimately masking phenotypes that may be associated with differing fixed levels of Lrp expression. To counter this issue, we constructed strains with genome-encoded fixed levels of lrp (in-genome fixed-lrp strains): the low-lrp and high-lrp constructs were integrated into the genome of the lrp-2::kan strain at the kefA gene, a locus that does not disrupt bacterium-nematode interactions (4), and the resulting strains were verified to express the expected relative levels of Lrp and virulence (see Materials and Methods for details; see also Table 1 and Fig. S1 in the supplemental material).
TABLE 1.
Bacterial strains used in this study
| Strain or plasmid | Relevant genetic characteristic(s) | Fluorescent phenotypea | Reference |
|---|---|---|---|
| Strains | |||
| HGB1059 | X. nematophila HGB800 lrp-2::kan | NA | 27 |
| HGB1685 | E. coli S17pir carrying plasmid pEH69 | GFP− RFP+ | This study |
| HGB1974 | E. coli S17pir carrying plasmid pMYC1 | GFP+ RFP+ | This study |
| HGB2261 | HGB1059 attTn7::PfliC-gfp | GFP− RFP+ | This study |
| HGB2262 | HGB2261 kefA::pMYC4 (low Lrp) | GFP− RFP+ | This study |
| HGB2263 | HGB2261 kefA::pMYC5 (high Lrp) | GFP+ RFP+ | This study |
| HGB2267 | E. coli S17λpir carrying plasmid pMYC4 (low Lrp) | NA | This study |
| HGB2268 | E. coli S17λpir carrying plasmid pMYC5 (high Lrp) | NA | This study |
| HGB1966 | X. nematophila lrp-2::kan plus pKV69 (vector) | NA | 23 |
| HGB1967 | X. nematophila lrp-2::kan plus pEH54 (low-Lrp plasmid) | NA | 23 |
| HGB1968 | X. nematophila lrp-2::kan plus pEH56 (high-Lrp plasmid) | NA | 23 |
| Plasmids | |||
| pEVS107 | ori6K mini-Tn7 delivery vector; Ermr Kanr | 64 | |
| pEH69 | pEVS107 Tn7-promoterless gfp/Plac-rfp; Ermr Kanr | This study | |
| pMYC1 | pEH69 with fliC promoter fused upstream of gfp | This study | |
| pKV69 | Multicopy mobilizable vector; Cmr Tetr | 65 | |
| pEH54 | pKV69 with lrp in the opposite orientation relative to Plac | 23 | |
| pEH56 | pKV69 with lrp under the control of Plac | 23 | |
| pMYC4 | ori6K-pEH54 with kefA fragment | This study | |
| pMYC5 | ori6K-pEH56 with kefA fragment | This study |
LB agar colony fluorescence was scored as + or − based on fluorescence microscopy. NA, not applicable.
We monitored the cumulative numbers of IJs emerging over a 55-day time course from agar lawns of in-genome fixed-lrp strains (Fig. 3B). Over this time period, high-lrp X. nematophila supported the production of significantly higher numbers of emerged IJs than the no-lrp and low-lrp strains, indicating similar mutualistic behaviors of the plasmid and in-genome lrp expression strains. Taken together, data from these experiments demonstrate that X. nematophila cells expressing high levels of Lrp are optimal for supporting nematode development into the transmission IJ stage.
A fundamental aspect of the S. carpocapsae-X. nematophila mutualism is the ability of the bacteria to colonize the IJ stage for transmission to the next insect host. To determine if Lrp expression levels impact this mutualistic trait, we monitored the frequency of X. nematophila colonization of the receptacles of emerged IJs (Fig. 3C). Colonization could be detected by using fluorescence microscopy based on the presence of the constitutively expressed red fluorescent protein (RFP) encoded in the kefA locus inserts (see Materials and Methods). Consistent with previously reported data (27), X. nematophila cells lacking lrp or expressing low levels of lrp had reduced colonization levels (∼75% and 78%, respectively) relative to those typically observed for the wild type (>90%) (4, 10, 35). In contrast, the high-lrp strain exhibited a wild-type colonization frequency of 96%, significantly higher than those of the no-lrp and low-lrp strains (Fig. 3C).
Bacteria expressing high levels of Lrp support high levels of mutualism in vivo.
The experiments presented thus far indicate that in vitro on agar plates, X. nematophila bacterial cultures expressing high levels of Lrp support both higher-level IJ production and a higher colonization frequency than strains expressing no or low levels of Lrp. In nature, nematode reproduction occurs within an insect cadaver. Given that Lrp is considered the “feast-or-famine” transcriptional regulator (26), the distinct nutritional environment of an insect cadaver relative to that of an agar plate may have an impact on the role of Lrp in supporting mutualistic phenotypes (11, 36). To test this possibility, we measured IJ reproduction and colonization after the injection of each X. nematophila fixed-Lrp strain and aposymbiotic IJ nematodes into individual G. mellonella insects (Fig. 3D and E). We chose G. mellonella rather than M. sexta (used for virulence assays) as an insect host (23), since the former has a less robust immune response and better supports nematode reproduction than the latter (our unpublished observations). Bacterial strains expressing no lrp, low lrp, and high lrp levels either on plasmids (Fig. 3D) or in the genome (Fig. 3E) were used to assess the role of Lrp in IJ production in vivo (i.e., in insect cadavers). As expected, 100% death was achieved in all insects at 36 h postinjection except for control insects injected with phosphate-buffered saline (PBS) (data not shown).
IJ reproduction was measured as the ability to produce IJ progeny from each insect (productive infection) (Fig. 3D) and the numbers of IJs produced from each infection (Fig. 3E). Consistent with data from the in vitro experiments described above, insects injected with high-lrp strains showed a significantly higher rate of productive infection (Fig. 3D) and higher numbers of cumulative progeny IJs (Fig. 3E) than did those injected with either the no-lrp or low-lrp strains. Similarly, a positively correlated trend between Lrp levels and colonization frequency was apparent (Fig. 3F). However, in contrast to the in vitro data, colonization frequencies of IJs that emerged from insects did not vary significantly depending on the strain, indicating that the nutritional environment of the insect cadaver may ameliorate the negative impact of low Lrp levels on colonization frequency, as we propose above. Overall, the colonization frequency of each strain was higher in vivo than in vitro, consistent with previously reported observations (11, 36).
The experiments described above relied upon IJ emergence as a proxy for nematode reproduction. To more fully explore the role of Lrp during nematode reproduction and development, G. mellonella insects were injected with nematodes and fixed-lrp-expression strains, and the resulting insect cadavers were dissected to enumerate nematodes from all developmental stages. When plasmid-based fixed-lrp-expression strains were used, total nematode counts were highly variable among individual insect cadavers, likely due to inconsistent maintenance of the lrp-carrying plasmids (see Fig. S4A in the supplemental material). In spite of this variability, overall, the trend showed that the high-lrp strain supported higher total nematode counts per insect than did the no-lrp and low-lrp strains.
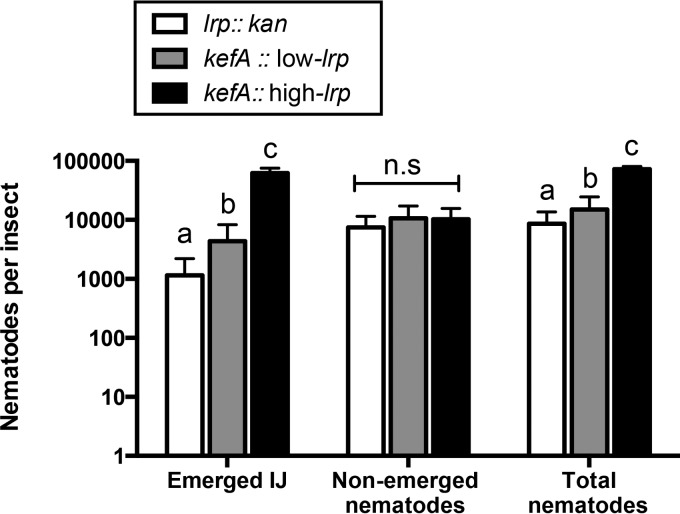
We next monitored numbers of emerged, nonemerged, and total (emerged and nonemerged) nematodes 23 days after coinjection with genome-encoded fixed-lrp-expression strains (Fig. 4 and Fig. S4B and S4C). The high-lrp strain supported significantly higher average nematode counts of both emerged IJs and total nematodes. The numbers of nonemerged nematodes were not statistically different among bacterial strains. Based on these data, we conclude that X. nematophila strains expressing high levels of Lrp are better able to support nematode progression into the IJ transmission stage than are strains expressing little or no Lrp.
FIG 4.
Nematode fecundity in Galleria insects shown as emerged IJs, nonemerged nematodes, and total nematodes. Data were obtained 23 days after coinjection of insects with aposymbiotic IJs and bacterial strains expressing fixed levels of Lrp. Numbers of nematodes per insect represent averages for each set of data (5 insects were used per bacterial strain in each biological replicate; n = 3), and error bars represent standard errors.
The number of emerging IJs reflects the success of multiple stages of the nematode life cycle: development through reproductive juvenile and adult stages, the fecundity of adults that mate and produce progeny, the development of progeny into the IJ transmission stage, and the dispersal of the IJs from the lawn or cadaver (Fig. 1). To predict if Lrp affects any of these stages, we created a simple mathematical model to simulate theoretical numbers of emerged, nonemerged, and total numbers of nematodes in comparison to the experimental data shown in Fig. 4. The model includes four adjustable parameters, each controlling one stage: nematode progeny per reproductive cycle (parameter a), IJ formation (parameter b), dispersal (parameter c), and life cycle progression (parameter d). The theoretical numbers of emerged IJs and nonemerged IJs and the total numbers of nematodes were simulated in Mathematica to test the following hypotheses: (i) a high Lrp level promotes only more progeny per reproductive cycle (parameter a), (ii) a high Lrp level promotes only a higher percentage of IJ formation per reproductive cycle (parameter b), (iii) a high Lrp level promotes only higher-level IJ dispersal or emergence (parameter c), and (iv) a high Lrp level promotes only faster reproduction and more reproductive cycles (parameter d).
First, we set each of the hypotheses to be true by increasing each parameter individually and continuously in Mathematica. The simulated outputs were compared to the output under the initial conditions, reflecting the theoretical prediction of each hypothesis (see Table 2). The predicted outcomes from the model were then compared to our experimental data to assess if these hypotheses could be true in our biological system (see Table 2). For instance, we set the first hypothesis to be true by increasing parameter a. The math model output was recorded as increased numbers of emerged IJs and nonemerged nematodes and total numbers of nematodes (see Table 2). Thus, the predicted outcome under either the first hypothesis or the opposite of the first hypothesis would not match our experimental data, suggesting that the first hypothesis cannot be the only explanation for the biological system.
TABLE 2.
Comparison of experimental and theoretical trends
| Type of data | Conditionc | Trend |
||
|---|---|---|---|---|
| Emerged IJs | Nonemerged nematodes | Total nematodes | ||
| Experimentala | High lrp vs low lrp levels | + | = | + |
| Theoreticalb | Increase of parameter a | + | + | + |
| Theoretical | Increase of parameter b | + then − | − | − |
| Theoretical | Increase of parameter c | + | − | = |
| Theoretical | Increase of parameter d | + | + | + |
Experimental data were extracted from data in Fig. 4 showing whether high-lrp strains support the production of higher (+) or equal (=) numbers of nematodes in each category in comparison to a low-lrp strain.
Theoretical trends were simulated based on the mathematical model (see Materials and Methods). An initial condition was set as theoretical low-lrp outcomes (using parameter values a = 3.3, b = 0.5, c = 0.2, and d = 5). Parameters a, b, c, and d were individually and continuously increased in Mathematica. By increasing each of the parameters, the trend of simulated outcomes in comparison to outcomes under initial conditions (emerged IJs, nonemerged nematodes, or total nematodes) was recorded as an increase (+), a decrease (−), or equal (=).
Parameter a, population expansion per reproductive cycle; parameter b, percentage of the population that forms IJs in the current reproductive cycle; parameter c, percentage of IJs that emerged in the current reproductive cycle; parameter d, number of reproductive cycles before insect dissection.
Our mathematical model indicates that high levels of Lrp expression are likely to promote both nematode reproduction and nematode dispersal (see Table 2). To show an example of theoretical prediction under a hypothesis combining the first three hypotheses, specific sets of parameter values (a = 3.3, b = 0.5, c = 0.2, and d = 5 for the low-lrp strain and a = 4.5, b = 0.7, c = 0.9, and d = 5 for the high-lrp strain) were chosen to simulate the math model and compared to the experimental data (see Fig. S3 in the supplemental material). The theoretical outcome matches our experimental data reasonably well, indicating the likelihood that a high Lrp expression level positively impacts multiple mutualistic behaviors that support the nematode life cycle.
S. carpocapsae nematodes associated with X. nematophila bacteria expressing high levels of Lrp are more fit than those expressing no or low levels of Lrp.
Symbiotic fitness between S. carpocapsae and X. nematophila depends on the production of colonized IJs from an infected cadaver and the abilities of these emerged IJs to reproduce in the next round of infection. The data described above demonstrate that when measuring individual parameters (percent productive infection [Fig. 3D], number of progeny IJs per infection [Fig. 3E and 4], and frequency of IJ colonization [Fig. 3D to F]), high levels of X. nematophila Lrp expression provide greater benefits than either the low-Lrp or no-Lrp strain. To obtain a representation of the overall fitness of the mutualistic pair, we first determined the total number of colonized IJs, versus the total number of uncolonized IJs (which are unlikely to be productive in future infections) (Fig. 5) (see Materials and Methods). In experiments conducted in vivo, while the frequencies of colonization among the total numbers of IJs observed were not significantly different among the three strains (Fig. 3F), our calculations revealed that the high-Lrp strain supported the production of a significantly higher overall number of colonized IJ progeny than do those injected with either the no-Lrp or low-Lrp strain (Fig. 5B). Thus, the reproduction of nematodes in the presence of X. nematophila expressing high levels of Lrp is more likely (relative to low- or no-lrp strains) to result in high numbers of IJ progeny capable of transmitting the symbiont to a new infection.
FIG 5.
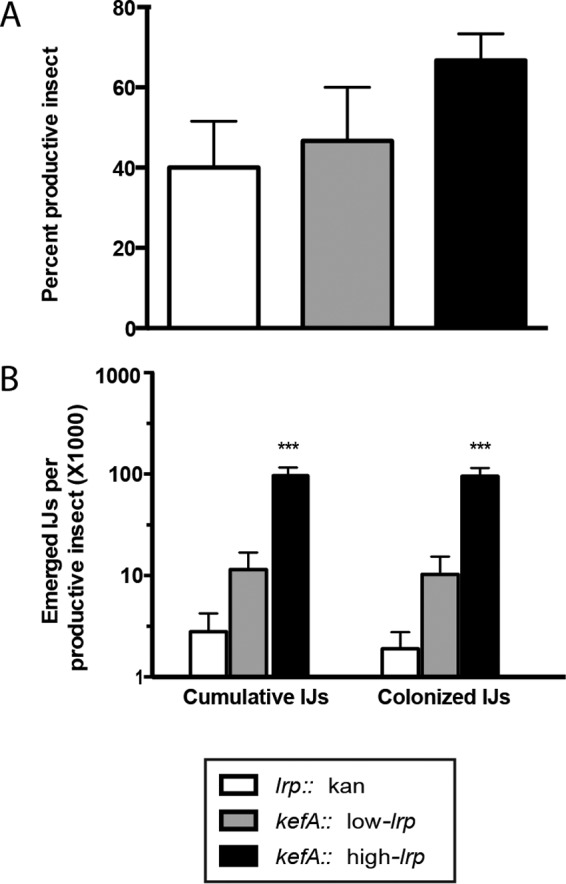
In vivo mutualism phenotypes. Bacterial strains with in-genome constructs expressing fixed levels of Lrp were coinjected into five Galleria insects per treatment. Insects that produced more than 50 emerged IJs were considered productive insects. (A) Percent productive insect is calculated as the percentage of productive insects out of the total number of insects infected per experiment. (B) Emerged and colonized IJs per productive insect were calculated as the colonization frequency multiplied by the number of emerged IJs per productive insect. Data are presented as means with standard errors (n = 3 biological replicates; each biological replicate is the average for five insects). One-way ANOVA was used to determine statistical significance (***, P ≤ 0.001).
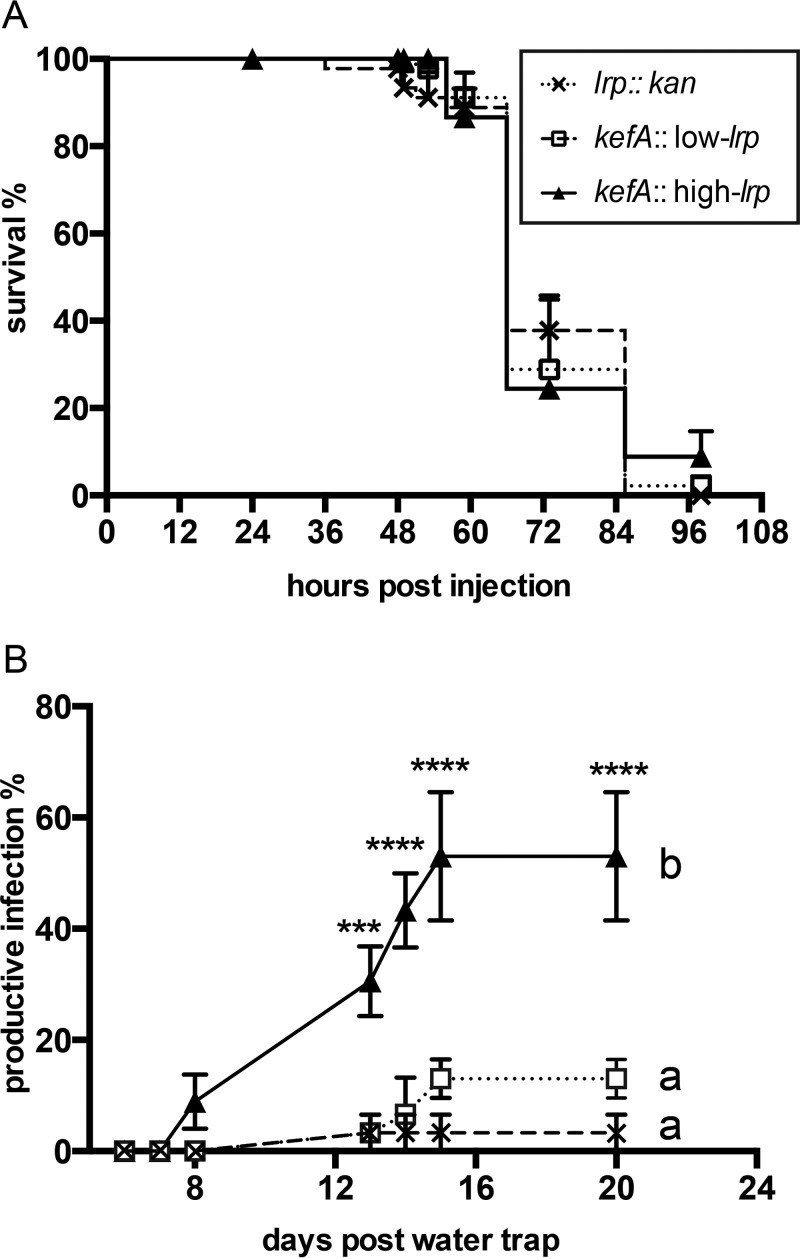
To assess the abilities of such emergent IJ populations to initiate a new round of infection, emerged IJs (from agar plates) were tested by using a sand trap assay, which measures successful infection (requiring host seeking, entry, recovery from the IJ stage, and release of bacteria), reproduction in the insect cadaver, and development into another generation of colonized IJs (37). We infected G. mellonella insects with IJs that had emerged from lawns of no-lrp, low-lrp, and high-lrp in-genome bacterial strains (Fig. 6). Infections were conducted with a modified sand trap assay (see Fig. S2C in the supplemental material) that incorporates the natural infection stages noted above. We did not observe significant differences among bacterial strains in insect survival postinfection, indicating that the ability to locate, infect, and kill G. mellonella was not influenced by the life history of the IJs with respect to the bacterial strain (Fig. 6A). As described above, we used percent productive infections as a measure of nematode reproduction. IJs derived from the high-lrp strain exhibited a significantly higher percentage of productive infections than did IJs derived from the no-lrp and low-lrp strains (Fig. 6B), indicating that the overall fitness of the S. carpocapsae-X. nematophila association is positively influenced by the expression of high levels of Lrp in the bacterial symbiont.
FIG 6.
Measurements of mutualistic interactions in natural infection of insects with colonized IJs. (A) Survival rate of Galleria insects after being naturally infected by IJs colonized with bacteria expressing fixed levels of Lrp in a sand trap. (B) Percent productive infection calculated as the percentage of insects that produce IJ nematodes in water traps. For each biological replicate, 15 insects were used for the sand trap assay, and 8 to 12 insect cadavers were used for water trapping of IJ nematodes. Data points represent averaged measurements (n = 3), and error bars represent standard errors. ***, P < 0.001; ****, P < 0.0001. Two-way ANOVA was used for statistical analysis, followed by Tukey’s multiple comparisons at each time point.
DISCUSSION
Our previous research provided a clear link between the transcription factor Lrp and the VMO phenomenon, in which a subpopulation of X. nematophila cells that express high levels of Lrp is attenuated for virulence and immunosuppression during initial infection of insect hosts (23). Here we provide evidence that the selective advantage of these high-Lrp-expressing strains is to support later stages of the X. nematophila life cycle, during which it provides mutualistic activities that support nematode reproduction and the development of the transmission IJ stage. Furthermore, high-Lrp cells are adapted for initiating the colonization of developing IJs and may have an advantage in colonizing host tissues, based on its robust biofilm-forming phenotype.
High-Lrp-expressing cells are adapted for mutualism.
The high-Lrp-expressing strain supports the production of higher numbers of IJs than strains lacking or expressing low levels of lrp, emerging from the cultivation assay (from bacterial lawns or from G. mellonella insects). In addition, our mathematical model indicates that high levels of Lrp positively influence both nematode reproduction and nematode dispersal from insect cadavers.
In both cases, the gene expression profile of the high-Lrp strain is likely responsible for the observed phenotypes. Indeed, our findings suggest that high and low levels of Lrp result in distinctive gene expression profiles that promote mutualism and virulence, respectively. As such, characterizing these profiles will facilitate the assignment of specific gene products and pathways to their functional roles in one symbiotic association or the other. The X. nematophila Lrp regulon has been characterized by using microarray analysis to compare gene expression in an lrp mutant to that in wild-type X. nematophila that was heterogeneous with respect to Lrp (23, 38, 39). Recent attempts in our laboratory to compare gene expression profiles of plasmid-encoded fixed-high- and low-Lrp strains were inconclusive, likely due to variability in plasmid maintenance (data not shown). Our creation of genome-encoded constitutive Lrp strains, as described here, will likely provide more consistent and interpretable data.
Until such data are available, our knowledge of the Lrp regulon can still provide insights into the possible activities that contribute to the success of the high-Lrp strain in promoting mutualistic functions. Lrp regulates the expression of the flagellar regulon, which includes genes that encode lipase and protease (xlpA and prtA, respectively) (30). The xlpA-encoded lipase promotes nematode IJ production through an unknown mechanism, and lipase activity is expressed more highly in high-Lrp strains (23, 30, 31). Furthermore, flagellar motility and flagellar regulon-encoded activities appear to be induced during the transition between the virulence and reproduction stages of the X. nematophila life cycle (7, 30). Taken together, these findings suggest that high-Lrp strains may express elevated levels of the flagellar regulon, including the xlpA-encoded lipase that has a positive effect on emerging IJ population numbers. Nematode dispersal is a chemosensing-based behavior that can be a response to nematode-produced small molecules (40). Lrp may play a role in this behavior by regulating either the sensing or the production of small-molecule signals derived from insect tissues (such as ammonia), nematodes (such as pheromones), or bacteria (such as secondary metabolites) (40, 41). X. nematophila Lrp controls the production of numerous small molecules, many of which have an unknown biological function (23, 27, 39). An intriguing possibility that awaits further investigation is that one or more X. nematophila Lrp-dependent small molecules either trigger or enhance the dispersal behavior of the S. carpocapsae nematode host, thereby increasing its own fitness by ensuring transmission to the next insect host.
In addition to supporting the production of greater numbers of IJs, the high-Lrp-expressing strain is also optimal for colonizing these IJs, a process that is crucial to X. nematophila transmission to the next round of its life cycle (infection of a new insect host). The superior colonization levels of the high-Lrp-expressing strain were more apparent under in vitro cultivation conditions than under in vivo cultivation conditions. This might indicate that the G. mellonella cadaver environment contains or lacks specific molecules, such as particular amino acids, that influence the Lrp-dependent adaptation of X. nematophila from the reproduction stage to the transmission stage of its life cycle. For example, it is possible that within an insect cadaver but not on a spent-agar plate, there are metabolites that trigger the induction of colonization genes either independent of Lrp or in conjunction with it such that low levels of Lrp are sufficient to promote colonization.
The processes of X. nematophila colonization of both reproductive-stage and IJ-stage nematodes were described previously (4, 16), and each process involves visible bacterial attachment to specific internal nematode surfaces. To directly test the influence of Lrp levels on the general ability of X. nematophila to attach to a surface, we used a well-established laboratory biofilm formation assay. Consistent with our models in which biofilm formation is a component of the X. nematophila-S. carpocapsae association and in which high-Lrp strains are best adapted for mutualistic phenotypes, we found that high-Lrp strains form more robust biofilms than do low-Lrp or no-Lrp strains. Whether this biofilm phenotype corresponds specifically to an in vivo adherence phenotype remains to be addressed, a process that will be facilitated by the identification of conditions and Lrp-dependent genes that contribute to biofilm formation. Future detailed genetic and biochemical analyses of these pathways will yield insights into the symbiotic timeline, tissue tropism, and host environmental conditions experienced by X. nematophila as it interacts with its two invertebrate hosts.
Lrp-regulated virulence modulation is a switch between pathogenic and mutualistic phenotypes.
Data from the present study, combined with our previous data (23), establish that Lrp regulates bacterial mutualism in a reciprocal manner relative to virulence: a virulent strain expressing fixed low levels of Lrp has a defect in mutualistic interactions with nematodes, while a virulence-attenuated strain expressing fixed high levels of Lrp establishes optimal mutualism. Thus, the variation in Lrp levels in X. nematophila controls a phenotypic switch between pathogenic and mutualistic behaviors.
In many bacteria, phenotypic variation serves as a mechanism to confer adaptations under erratic or predictable environmental fluctuations. The fact that X. nematophila laboratory-grown populations are heterogeneous with respect to Lrp levels (and therefore symbiotic behavior) suggests that the switch is stochastic and that X. nematophila may be using a bet-hedging strategy to adapt to changing host environments (42, 43).
Whether X. nematophila has evolved stochastic bet-hedging or other strategies to optimize the fitness of itself and its nematode host remains to be tested. Regardless, our data indicate that Lrp is a key component of adaptation to host environments and that the overall cellular levels of Lrp dictate the symbiotic phenotype. Switching between a high (mutualistic)- and a low (virulence)-Lrp state by a cell could occur through random or stochastic noise, such as an uneven distribution of Lrp proteins in the two daughter cells during cell division. Alternatively, a bimodal or multimodal distribution of Lrp levels could be due to genetic, posttranscriptional, or posttranslational modifications that modulate the expression or stability of Lrp levels within the cell. Consistent with the latter possibility, our previously reported data demonstrated that lrp transcription is negatively autoregulated, as is true for Escherichia coli (23).
In uropathogenic E. coli (UPEC), Lrp controls a phenotypic on/off switch of Pap fimbriae, dictated by Lrp binding to the proximal or distal sites of the pap promoter (44). The Lrp regulatory output in E. coli may be modulated by Lrp multimerization, protein modification (e.g., acetylation), and interactions with metabolites and other regulators (45, 46). In turn, each of these control points may be influenced indirectly by the host environment and directly by the concentration of Lrp available within the cell. Based on the data that we present here, it seems likely that X. nematophila senses the nutritional environment of its hosts and responds, through various Lrp levels, to alter global gene expression. However, mechanistic details of this phenomenon remain to be examined. For example, do mutualistic and pathogenic subpopulations of X. nematophila coexist in a heterogeneous population that is subject to selective pressure from host environments for one phenotype or the other (i.e., the nematode environment selects for cells expressing mutualistic behaviors, while the insect environment selects for those with virulent traits), or does the host environment induce or prime one phenotype over the other, resulting in homogeneous populations expressing either high or low levels of Lrp that are adapted for a particular stage of the life cycle? The answers to these questions will require monitoring and quantification of Lrp and Lrp-dependent gene expression within individual bacterial cells in situ.
Conclusions.
Xenorhabdus bacteria have a complicated life history, in which they alternate between a pathogenic interaction with insects and a mutualistic interaction with Steinernema nematodes, and our work demonstrates that X. nematophila adapts to these changes by using an Lrp-dependent phenotypic switch. While the concept of host-associated microbes utilizing phenotypic variation to adapt to changing host/environmental conditions is well described, our work establishes that subtle variations in cellular concentrations of a global regulatory protein can dictate the outcome of an individual symbiotic interaction and the overall fitness of symbiosis partners. Furthermore, our work indicates that X. nematophila uses Lrp-dependent phenotypic variation to inversely coordinate mutualistic and pathogenic behaviors that are critical to its fitness but are potentially conflicting with each other (47). A similar inverse coordination may be a feature of other systems in which a symbiont, such as a vector-borne pathogen, encounters multiple distinct hosts during its life cycle. For example, Yersinia pestis alternates between flea and mammalian hosts, in which it expresses host-specific transcriptional profiles (48). The molecular mechanism controlling this global switch in Y. pestis has not been elucidated but may involve a phenotypic-variation phenomenon such as the one that we have described here.
The X. nematophila mutualism-to-pathogenesis phenotypic-variation phenomenon appears to be distinct from other well-established examples of phenotypic variation in parasitic symbioses that alternate between on and off states of virulence genes, such as the fim switch in pathogenic E. coli (49) and the ToxT switch in Vibrio cholerae (50). However, it is possible that the “off” state in these systems is coordinated with a reciprocal expression of genes that are adapted to other environments encountered by these pathogens. Experimental evolution selecting for commensalism in the nematode pathogen Pseudomonas aeruginosa resulted in attenuated virulence through a mutation in a global regulatory protein (51). It will be interesting to explore the function of this protein in the ancestral strain and to determine if it also controls a phenotypic switch between pathogenic and beneficial (or commensal) behaviors.
MATERIALS AND METHODS
Bacterial growth.
The bacterial strains and plasmids used in this study are listed in Table 1. Bacteria were grown in lysogeny broth (LB) culture medium (52) at either 30°C (for X. nematophila) or 37°C (for E. coli). The medium was either stored in the dark or supplemented with 0.1% sodium pyruvate (53). Xenorhabdus bacteria carrying plasmids were grown in culture medium supplemented with 7.5 μg/ml of tetracycline in order to maintain plasmids. Other antibiotics were used, as indicated, at the following concentrations: ampicillin (Amp) at 150 μg/ml, chloramphenicol (Cm) at 15 μg/ml (X. nematophila) and 30 μg/ml (E. coli), kanamycin (Kan) at 50 μg/ml, and erythromycin (Erm) at 200 μg/ml.
Plasmid and strain construction. (i) Construction of plasmid pEH69 (Tn7-promoterless gfp/Plac-rfp).
The Turbo RFP gene (rfp) was digested from pTurbo-RFP-B (Evrogen, Moscow, Russia) with NheI and XhoI and ligated into plasmid pEVS107 (54) (digested with SpeI, followed by the creation of blunt ends using T4 DNA polymerase [Promega]), generating plasmid pEH39. The green fluorescent protein (GFP) gene (gfp) was amplified from pVSV209 (55) by using PCR with the PrimeSTAR high-fidelity polymerase (Clontech Laboratories, Inc., Mountain View, CA) and primers CATGFP F (5′-AAAGGGCCCGCTTGCTCAATCAATCACCG-3′) and CATGFP R (5′-AAAGGGCCCGCCGCATAGTTAAGCCAGC-3′) and then introduced into pEH39 (digested with AvrII and filled in), generating plasmid pEH69.
(ii) Construction of plasmid pMYC1 (Tn7 PfliC-gfp/Plac-rfp).
The X. nematophila fliC promoter region was cloned upstream of the promoterless chloramphenicol resistance and gfp genes in pEH69 by PCR amplifying a 485-bp fragment containing the region upstream of and 12 bp within the fliC gene by using primers F2_PfliC AvrII (5′-CCTAGGCTTTTTCAGTTCTTCTGATGCTG-3′) and R2_PfliC AvrII (5′-CCTAGGGACTGATGCCATAGTAGAGTTCC-3′) (note that in all primers listed, any engineered restriction enzyme digestion sites are underlined) (Integrated DNA Technologies, Coralville, IA) and the PrimeSTAR high-fidelity polymerase (Clontech Laboratories, Inc., Mountain View, CA) according to the manufacturer's instructions. Both the PCR-amplified fragment and pEH69 were digested with AvrII and ligated (New England BioLabs, Inc., Ipswich, MA), generating plasmid pMYC1.
(iii) Construction of pMYC4 and pMYC5.
Plasmids pMYC4 (low lrp) and pMYC5 (high lrp) were constructed by modifying plasmids pEH54 and pEH56 (23). The origins of replication in pEH54 and pEH56 were removed by using restriction digestion with ClaI and XmnI according to the manufacturer's directions (Promega, Madison, WI). The suicidal origin of replication oriR6K was PCR amplified from plasmid pEVS107 by using primers OriR6K_F1_XmnI (5′-GAATTTTTTCCCATGTCAGCCGTTAAGTGTTC-3′) and OriR6K_R1_ClaI (5′-ATCGATGAGGATCTGAAGATCAGCAGTTC-3′) (Integrated DNA Technologies, Coralville, IA) and standard PCR techniques with the PrimeSTAR high-fidelity polymerase (Clontech Laboratories, Inc., Mountain View, CA); it was then introduced into pEH54 and pEH56 to replace the origins of replication by using standard restriction digestion (ClaI and XmnI) and ligation (T4 DNA ligase; Promega, Madison, WI), generating pMYC2 (pEH54-oriR6K) and pMYC3 (pEH56-oriR6K). A fragment of the kefA gene from the X. nematophila (ATCC 19061) genome was PCR amplified by using primers sau_F1_ClaI (5′-ATCGATGATCTTGATGATTTGATTTGGGG-3′) and sau_R1_ClaI_long (5′-ATCGATGATCCAAGGCCATTGGAGG-3′) and introduced into pMYC2 and pMYC3 by standard restriction digestion (ClaI) and ligation, generating pMYC4 and pMYC5.
(iv) Creation of X. nematophila strains expressing low and high levels of Lrp from the genome.
To create low- and high-Lrp-expressing X. nematophila strains, pMYC4 (low-lrp donor plasmid) and pMYC5 (high-lrp donor plasmid) were inserted into the kefA gene of an lrp-2::kan mutant via biparental conjugation and homologous recombination, respectively. Plasmid integration at the kefA gene locus does not interfere with nematode colonization (4, 16, 56, 57). The Lrp-dependent fluorescence reporter pMYC1 (PfliC-gfp/Plac-rfp) was introduced into the attTn7 site of the genomes in both of the low-Lrp- and high-Lrp-expressing X. nematophila strains described above by triparental conjugation and Tn7 transposition (58, 59), creating low-lrp in-genome (low-lrp/Tn7-PfliC-gfp/Plac-rfp) and high-lrp in-genome (high-lrp/Tn7-PfliC-gfp/Plac-rfp) strains. Lrp expression levels in the low-lrp and high-lrp in-genome strains were initially screened by fluorescence microscopy using a Nikon Eclipse TE300 inverted microscope (Nikon, Melville, NY, USA) and confirmed to be positively correlated with GFP expression levels (Table 1) (23). The Lrp expression levels of the in-genome low- and high-lrp strains were further confirmed by semiquantitative Western blotting (see Fig. S1A in the supplemental material) performed by using previously reported methods (23).
The orientation and sequence accuracy of all introduced regions in the plasmid constructs in this research were verified by sequencing at the University of Wisconsin—Madison Biotechnology Center using BigDye version 3.1 (Applied Biosystems, Foster City, CA).
The virulence of the in-genome no-lrp, low-lrp, and high-lrp strains was tested by directly injecting bacterial cultures grown overnight into fourth-instar Manduca sexta larvae (23, 60). Consistent with previously reported data, the fixed-low-lrp in-genome strain is more virulent than either the no-lrp or high-lrp strain (Fig. S1B).
Biofilm assay.
For biofilm experiments, bacterial strains were grown overnight in LB from frozen stocks. Cultures grown overnight were diluted 1:100 in fresh LB medium and grown at room temperature under static incubation for 6 days. Biofilm material was stained for 15 min with 1% crystal violet and then drained and dried completely. Quantitative analyses of biofilm material followed, in which 70% ethanol was used to destain glass test tubes (with agitation). The resulting solution was quantitated by spectrophotometry at an optical density (OD) at 595 nm (61).
Nematode maintenance.
Conventional nematodes were propagated through Galleria mellonella larvae (Grubco, Hamilton, OH) and used for generating aposymbiotic nematodes. A new batch of conventional nematodes was used for each biological replicate. Aposymbiotic nematodes were generated from a standard axenic egg preparation from conventional nematodes (62). To generate aposymbiotic IJs, axenic eggs were then reared on X. nematophila colonization-deficient rpoS mutant lawns, and aposymbiotic nematodes were water trapped, surface sterilized, and confirmed by grinding and plating onto LB agar. Axenic nematodes were surface sterilized immediately before addition to bacterial lawns in colonization and fecundity assays (8).
In vitro nematode fecundity assays.
Cultures of X. nematophila were grown overnight at 30°C, spread onto lipid agar (15 ml per plate), and incubated at 25°C for 48 h to create bacterial lawns (see Fig. S2A in the supplemental material). In the experiments using engineered X. nematophila strains that carry multicopy plasmids, such as HGB 1966 (lrp-2::kan plus the no-lrp vector), HGB 1967 (lrp-2::kan plus the low-lrp plasmid), and HGB 1968 (lrp-2::kan plus the high-lrp plasmid) (23), lipid agar was supplemented with tetracycline (7.5 μg/ml) to maintain plasmids during bacterial growth. In the experiments using X. nematophila strains with in-genome insertions, no antibiotics were used when bacteria were growing. Approximately 5,000 aposymbiotic IJs were surface sterilized and seeded onto the bacterial lawns. Before 10 days post-nematode seeding, water traps were set up to collect emerging progeny nematodes. For plasmid-carrying strains expressing no lrp, low lrp, and high lrp levels (HGB 1966, HGB 1967, and HGB 1968, respectively), emerged IJs were counted on days 3, 7, and 10 post-water trap. At each time point, 0 to 60 nematodes were enumerated for each water trap and used for calculating cumulative IJs from each bacterial lawn. Cumulative IJs for each bacterial strain were represented by the averages of data from 3 technical replicates in each of the 5 to 6 biological replicates. For the no-lrp, low-lrp, and high-lrp in-genome construct strains (HGB 2261, HGB 2262, and HGB 2263, respectively), emerged IJs were counted on days 1, 2, 3, 5, 10, 28, 30, 33, 51, and 55 post-water trap. At each time point, 40 to 150 IJs were enumerated for each water trap and used for calculating cumulative IJs from each bacterial lawn. Cumulative IJs for each bacterial strain were represented by the averages of data from 5 technical replicates in each of the 3 biological replicates. IJs collected from in-genome construct strains expressing no lrp, low lrp, and high lrp levels (HGB 2261, HGB 2262, and HGB 2263, respectively) were stored for use in sand trap assays and colonization screens.
In vivo nematode fecundity assay. (i) Coinjection of bacteria and nematodes.
G. mellonella larvae (Grubco, Hamilton, OH) were used for bacterium-nematode coinjections (see Fig. S2B in the supplemental material). Cultures of bacteria grown overnight were serially diluted in PBS. Prior to injection, OD measurements and CFU counting by dilution plating were done to ensure that equal CFU counts were injected among bacterial strains. Aposymbiotic nematodes were surface sterilized, diluted in PBS, and combined with bacteria to approximately 50 IJs and 103 CFU in a 10-μl injection volume (37). HGB 1966, HGB 1967, and HGB 1968 were resuspended in PBS supplemented with 7.5 μg/ml tetracycline to maintain plasmids. Insect survival was monitored at 8- to 24-h intervals postinjection.
(ii) Assessment of progeny yield and percent productive infection.
A White trap was set up for each individual insect cadaver on day 7 postinjection (see Fig. S2B in the supplemental material) (63) and monitored for IJ emergence at 24-h intervals thereafter. The proportion of infected insects that yielded nematodes at each time point was calculated as percent productive infection. The number of IJs produced in each insect cadaver was also determined by removing water from the White trap and estimating the number of IJs per microliter. Samples of IJs were counted every other day for 14 days, and the total number of IJs per insect was estimated as the number of IJs per microliter multiplied by sample volume. Three biological replicates were performed (37). Insect cadavers that produced at least 50 IJs were scored as having productive infection, and those that did not were otherwise scored as having nonproductive infection.
(iii) Pepsin digestion of insect cadavers.
Pepsin digestion of infected insect cadavers was used to allow observation and counting of nonemerged nematodes (see Fig. S2B in the supplemental material). At various time points after coinjection of bacteria and nematodes, individual G. mellonella cadavers were rinsed in distilled water, dissected, and digested in a 10-ml pepsin solution (0.83% [wt/vol] pepsin [Carolina Biological, Burlington, NC], 24% [wt/vol] NaCl, and 2% [vol/vol] HCl in distilled water) (63) for 3 h at 37°C (32). Nematodes from all developmental stages were counted by using a dissecting scope.
Mathematical model of nematode reproduction and emergence.
We developed a simplified mathematical model to assess the relative contributions of development, reproduction, and emergence to our observed counts of nematodes from in vivo assays. The model was constructed under the assumptions that (i) all 50 aposymbiotic IJs injected into the insect survived and developed into adults; (ii) reproductive cycles were consecutive, with no overlaps; (iii) insect cadavers were not infected by competitive microbes such as fungi; (iv) carcass of dead nematodes did not decay; and (v) among different reproductive cycles, population expansion (parameter a), the percentage of the population that forms IJs (parameter b), and the percentage of IJs that emerged out of the insect cadaver (parameter c) were constant, except for the first reproduction cycle, in which no IJs are formed (b1 = 0; otherwise, b > 0) (16).
For the nth reproductive cycle, the total number of nematodes (including parental and progeny nematodes) from the current productive cycle is approximated as
The number of emerged IJs from the nth reproductive cycle is approximated by
The number of “trapped IJs” from only the nth reproductive cycle is approximated by
The number of nonemerged nematodes after the nth reproductive cycle is approximated by
After d rounds of reproductive cycles, the number of emerged IJs is approximated by
The number of nonemerged nematodes is approximated by
The total number of nematodes is approximated by
The theoretical numbers of emerged IJs (Em) and nonemerged IJs (Ne) and the total number of nematodes (T) were simulated in Mathematica (Wolfram Research, Champaign, IL). The initial condition was set by using the following parameter values: a = 3.3, b = 0.5, c = 0.2, and d = 5. To predict the outcomes of these hypotheses, we manipulated parameter values within ranges (2 < a < 20, 0 < b < 1, 0 < c < 1, and 2 < d < 10) and generated the simulated data (Table 2).
In vitro and in vivo colonization assays.
Colonization assays were performed by using bacterial strains with the in-genome constructs HGB 2261 (no lrp), HGB 2262 (low lrp levels), and HGB 2263 (high lrp levels) both using lipid agar (in vitro) and in G. mellonella (in vivo). IJs emerged from either bacterial lawns (see Fig. S2A in the supplemental material) or insect cadavers (Fig. S2B) after coinjection with bacteria, and nematodes were collected at 7 days post-water trap. For each cadaver, ∼100 to 150 IJs (a minimum of 50 IJs was necessary) were paralyzed by using 2 nM levamisole and screened for bacterial colonization, indicated by red fluorescence from constitutive RFP expression, by using a Nikon Eclipse TE300 inverted microscope (Nikon, Melville, NY, USA). Percent colonization was calculated as (number of colonized IJs/total number of IJs counted) × 100 (27). Percent colonization values from three water traps were used as technical replicates and averaged for each bacterial strain. Five biological replicates were performed. A minimum of 50 IJs was deemed necessary to screen for colonization.
Sand trap assay.
To conduct sand trap assays, 6 g of silica sand (Meeco, Seattle, WA) was distributed into individual 60- by 15-mm-diameter petri dishes (see Fig. S2C in the supplemental material). Infective juvenile nematodes colonized with the various in-genome test strains were inoculated at approximately 100 IJs per plate, and 6 G. mellonella larvae were added to each plate. Insects were monitored for survival and placed into individual White traps at about 7 days postdeath. Emergent nematodes were monitored every 24 h, and the percentage of insects that yielded nematodes was recorded as percent productive infection.
Statistical analysis.
Data were processed and analyzed in PRISM6 using one-way or two-way analysis of variance (ANOVA) followed by Tukey's multiple-comparison test among bacterial strains, with each time point as an independent comparison. Samples that showed no significant differences among them were grouped by the same letter. Statistical significance and P value ranges are indicated by asterisks.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants to H.G.-B. from the National Science Foundation (IOS-1353674 and IOS-0950873) and the UW-Madison U.S. Department of Agriculture (USDA) Hatch Multi-State Research Formula Fund (WIS01582) and by grants to E.A.H. from the Mississippi INBRE program (P20 GM103476) via the National Institutes of General Medical Sciences. M.C. was supported by a UW-Madison Louis and Elsa Thomsen Wisconsin Distinguished Graduate Fellowship and a Department of Bacteriology Michael Foster Predoctoral Fellowship.
We thank David H. Brown (Colorado College) for his lectures, encouragement, and guidance in mathematical modeling of biological systems.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00276-17.
REFERENCES
- 1.van der Woude MW, Bäumler AJ. 2004. Phase and antigenic variation in bacteria. Clin Microbiol Rev 17:581–611. doi: 10.1128/CMR.17.3.581-611.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Herbert EE, Goodrich-Blair H. 2007. Friend and foe: the two faces of Xenorhabdus nematophila. Nat Rev Microbiol 5:634–646. doi: 10.1038/nrmicro1706. [DOI] [PubMed] [Google Scholar]
- 3.Richards GR, Goodrich-Blair H. 2009. Masters of conquest and pillage: Xenorhabdus nematophila global regulators control transitions from virulence to nutrient acquisition. Cell Microbiol 11:1025–1033. doi: 10.1111/j.1462-5822.2009.01322.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Martens EC, Heungens K, Goodrich-Blair H. 2003. Early colonization events in the mutualistic association between Steinernema carpocapsae nematodes and Xenorhabdus nematophila bacteria. J Bacteriol 185:3147–3154. doi: 10.1128/JB.185.10.3147-3154.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Orchard SS, Goodrich-Blair H. 2004. Identification and functional characterization of a Xenorhabdus nematophila oligopeptide permease. Appl Environ Microbiol 70:5621–5627. doi: 10.1128/AEM.70.9.5621-5627.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Phalaraksh C, Lenz EM, Lindon JC, Nicholson JK, Farrant RD, Reynolds SE, Wilson ID, Osborn D, Weeks JM. 1999. NMR spectroscopic studies on the haemolymph of the tobacco hornworm, Manduca sexta: assignment of 1H and 13C NMR spectra. Insect Biochem Mol Biol 29:795–805. doi: 10.1016/S0965-1748(99)00053-3. [DOI] [Google Scholar]
- 7.Jubelin G, Pagès S, Lanois A, Boyer M-H, Gaudriault S, Ferdy J-B, Givaudan A. 2011. Studies of the dynamic expression of the Xenorhabdus FliAZ regulon reveal atypical iron-dependent regulation of the flagellin and haemolysin genes during insect infection. Environ Microbiol 13:1271–1284. doi: 10.1111/j.1462-2920.2011.02427.x. [DOI] [PubMed] [Google Scholar]
- 8.Vivas EI, Goodrich-Blair H. 2001. Xenorhabdus nematophilus as a model for host-bacterium interactions: rpoS is necessary for mutualism with nematodes. J Bacteriol 183:4687–4693. doi: 10.1128/JB.183.16.4687-4693.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Forst S, Dowds B, Boemare N, Stackebrandt E. 1997. Xenorhabdus and Photorhabdus spp.: bugs that kill bugs. Annu Rev Microbiol 51:47–72. doi: 10.1146/annurev.micro.51.1.47. [DOI] [PubMed] [Google Scholar]
- 10.Martens EC, Russell FM, Goodrich-Blair H. 2005. Analysis of Xenorhabdus nematophila metabolic mutants yields insight into stages of Steinernema carpocapsae nematode intestinal colonization. Mol Microbiol 58:28–45. doi: 10.1111/j.1365-2958.2005.04742.x. [DOI] [PubMed] [Google Scholar]
- 11.Flores-Lara Y, Renneckar D, Forst S, Goodrich-Blair H, Stock P. 2007. Influence of nematode age and culture conditions on morphological and physiological parameters in the bacterial vesicle of Steinernema carpocapsae (Nematoda: Steinernematidae). J Invertebr Pathol 95:110–118. doi: 10.1016/j.jip.2007.01.006. [DOI] [PubMed] [Google Scholar]
- 12.Crawford JM, Portmann C, Zhang X, Roeffaers MBJ, Clardy J. 2012. Small molecule perimeter defense in entomopathogenic bacteria. Proc Natl Acad Sci U S A 109:10821–10826. doi: 10.1073/pnas.1201160109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hwang J, Park Y, Kim Y, Hwang J, Lee D. 2013. An entomopathogenic bacterium, Xenorhabdus nematophila, suppresses expression of antimicrobial peptides controlled by Toll and Imd pathways by blocking eicosanoid biosynthesis. Arch Insect Biochem Physiol 83:151–169. doi: 10.1002/arch.21103. [DOI] [PubMed] [Google Scholar]
- 14.Casanova-Torres ÁM, Goodrich-Blair H. 2013. Immune signaling and antimicrobial peptide expression in Lepidoptera. Insects 4:320–338. doi: 10.3390/insects4030320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sicard M, Brugirard-Ricaud K, Page S, Lanois A, Boemare NE, Brehe M, Givaudan A, Gpia L. 2004. Stages of infection during the tripartite interaction between Xenorhabdus nematophila, its nematode vector, and insect hosts. Appl Environ Microbiol 70:6473–6480. doi: 10.1128/AEM.70.11.6473-6480.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chaston JM, Murfin KE, Heath-Heckman EA, Goodrich-Blair H. 2013. Previously unrecognized stages of species-specific colonization in the mutualism between Xenorhabdus bacteria and Steinernema nematodes. Cell Microbiol 15:1545–1559. doi: 10.1111/cmi.12134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martens EC, Goodrich-Blair H. 2005. The Steinernema carpocapsae intestinal vesicle contains a subcellular structure with which Xenorhabdus nematophila associates during colonization initiation. Cell Microbiol 7:1723–1735. doi: 10.1111/j.1462-5822.2005.00585.x. [DOI] [PubMed] [Google Scholar]
- 18.Yip ES, Grublesky BT, Hussa EA, Visick KL. 2005. A novel, conserved cluster of genes promotes symbiotic colonization and σ54-dependent biofilm formation by Vibrio fischeri. Mol Microbiol 57:1485–1498. doi: 10.1111/j.1365-2958.2005.04784.x. [DOI] [PubMed] [Google Scholar]
- 19.Forst S, Clarke D. 2002. Bacteria-nematode symbiosis, p 57–77. In Gaugler R. (ed), Entomopathogenic nematology. CABI, Wallingford, United Kingdom. [Google Scholar]
- 20.Volgyi A, Fodor A, Szentirmai A, Forst S. 1998. Phase variation in Xenorhabdus nematophilus. Appl Environ Microbiol 64:1188–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Givaudan A, Baghdiguian S, Lanois A, Boemare N. 1995. Swarming and swimming changes concomitant with phase variation in Xenorhabdus nematophilus. Appl Environ Microbiol 61:1408–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Park Y, Herbert EE, Cowles CE, Cowles KN, Menard ML, Orchard SS, Goodrich-Blair H. 2007. Clonal variation in Xenorhabdus nematophila virulence and suppression of Manduca sexta immunity. Cell Microbiol 9:645–656. doi: 10.1111/j.1462-5822.2006.00815.x. [DOI] [PubMed] [Google Scholar]
- 23.Hussa EA, Casanova-Torres ÁM, Goodrich-Blair H. 2015. The global transcription factor Lrp controls virulence modulation in Xenorhabdus nematophila. J Bacteriol 197:3015–3025. doi: 10.1128/JB.00272-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Platko JV, Calvo JM. 1993. Mutations affecting the ability of Escherichia coli Lrp to bind DNA, activate transcription, or respond to leucine. J Bacteriol 175:1110–1117. doi: 10.1128/jb.175.4.1110-1117.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tani TH, Khodursky A, Blumenthal RM, Brown PO, Matthews RG. 2002. Adaptation to famine: a family of stationary-phase genes revealed by microarray analysis. Proc Natl Acad Sci U S A 99:13471–13476. doi: 10.1073/pnas.212510999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yokoyama K, Ishijima SA, Clowney L, Koike H, Aramaki H, Tanaka C, Makino K, Suzuki M. 2006. Feast/famine regulatory proteins (FFRPs): Escherichia coli Lrp, AsnC and related archaeal transcription factors. FEMS Microbiol Rev 30:89–108. doi: 10.1111/j.1574-6976.2005.00005.x. [DOI] [PubMed] [Google Scholar]
- 27.Cowles KN, Cowles CE, Richards GR, Martens EC, Goodrich-Blair H. 2007. The global regulator Lrp contributes to mutualism, pathogenesis and phenotypic variation in the bacterium Xenorhabdus nematophila. Cell Microbiol 9:1311–1323. doi: 10.1111/j.1462-5822.2006.00873.x. [DOI] [PubMed] [Google Scholar]
- 28.Cowles KN, Goodrich-Blair H. 2005. Expression and activity of a Xenorhabdus nematophila haemolysin required for full virulence towards Manduca sexta insects. Cell Microbiol 7:209–219. doi: 10.1111/j.1462-5822.2005.00536.x. [DOI] [PubMed] [Google Scholar]
- 29.Martens EC. 2005. Initiation and maintenance of Steinernema carpocapsae nematode colonization by Xenorhabdus nematophila bacteria. PhD thesis University of Wisconsin—Madison, Madison, WI. [Google Scholar]
- 30.Park D, Forst S. 2006. Co-regulation of motility, exoenzyme and antibiotic production by the EnvZ-OmpR-FlhDC-FliA pathway in Xenorhabdus nematophila. Mol Microbiol 61:1397–1412. doi: 10.1111/j.1365-2958.2006.05320.x. [DOI] [PubMed] [Google Scholar]
- 31.Richards GR, Goodrich-Blair H. 2010. Examination of Xenorhabdus nematophila lipases in pathogenic and mutualistic host interactions reveals a role for xlpA in nematode progeny production. Appl Environ Microbiol 76:221–229. doi: 10.1128/AEM.01715-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mitani DK, Kaya HK, Goodrich-Blair H. 2004. Comparative study of the entomopathogenic nematode, Steinernema carpocapsae, reared on mutant and wild-type Xenorhabdus nematophila. Biol Control 29:382–391. doi: 10.1016/j.biocontrol.2003.07.005. [DOI] [Google Scholar]
- 33.Bogino PC, Oliva MDLM, Sorroche FG, Giordano W. 2013. The role of bacterial biofilms and surface components in plant-bacterial associations. Int J Mol Sci 14:15838–15859. doi: 10.3390/ijms140815838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morris AR, Visick KL. 2010. Control of biofilm formation and colonization in Vibrio fischeri: a role for partner switching? Environ Microbiol 12:2051–2059. doi: 10.1111/j.1462-2920.2010.02269.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cowles CE, Goodrich-Blair H. 2008. The Xenorhabdus nematophila nilABC genes confer the ability of Xenorhabdus spp. to colonize Steinernema carpocapsae nematodes. J Bacteriol 190:4121–4128. doi: 10.1128/JB.00123-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goetsch M, Owen H, Goldman B, Forst S. 2006. Analysis of the PixA inclusion body protein of Xenorhabdus nematophila. J Bacteriol 188:2706–2710. doi: 10.1128/JB.188.7.2706-2710.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Murfin KE, Lee MM, Klassen JL, McDonald BR, Larget B, Forst S, Stock SP, Currie CR, Goodrich-Blair H. 2015. Xenorhabdus bovienii strain diversity impacts coevolution and symbiotic maintenance with Steinernema spp. nematode hosts. mBio 6:e00076-15. doi: 10.1128/mBio.00076-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cowles CE, Goodrich-Blair H. 2006. nilR is necessary for co-ordinate repression of Xenorhabdus nematophila mutualism genes. Mol Microbiol 62:760–771. doi: 10.1111/j.1365-2958.2006.05400.x. [DOI] [PubMed] [Google Scholar]
- 39.Engel Y, Windhorst C, Lu X, Goodrich-Blair H, Bode HB. 2017. The global regulators Lrp, LeuO, and HexA control secondary metabolism in entomopathogenic bacteria. Front Microbiol 8:209. doi: 10.3389/fmicb.2017.00209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kaplan F, Alborn HT, von Reuss SH, Ajredini R, Ali JG, Akyazi F, Stelinski LL, Edison AS, Schroeder FC, Teal PE. 2012. Interspecific nematode signals regulate dispersal behavior. PLoS One 7:e38735. doi: 10.1371/journal.pone.0038735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.San-Blas E, Pirela D, García D, Portillo E. 2014. Ammonia concentration at emergence and its effects on the recovery of different species of entomopathogenic nematodes. Exp Parasitol 144:1–5. doi: 10.1016/j.exppara.2014.05.014. [DOI] [PubMed] [Google Scholar]
- 42.Veening J-W, Smits WK, Kuipers OP. 2008. Bistability, epigenetics, and bet-hedging in bacteria. Annu Rev Microbiol 62:193–210. doi: 10.1146/annurev.micro.62.081307.163002. [DOI] [PubMed] [Google Scholar]
- 43.Veening J-W, Stewart EJ, Berngruber TW, Taddei F, Kuipers OP, Hamoen LW. 2008. Bet-hedging and epigenetic inheritance in bacterial cell development. Proc Natl Acad Sci U S A 105:4393–4398. doi: 10.1073/pnas.0700463105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Van Der Woude M, Braaten B, Low D. 1996. Epigenetic phase variation of the pap operon in Escherichia coli. Trends Microbiol 4:5–9. doi: 10.1016/0966-842X(96)81498-3. [DOI] [PubMed] [Google Scholar]
- 45.Koike H, Ishijima SA, Clowney L, Suzuki M. 2004. The archaeal feast/famine regulatory protein: potential roles of its assembly forms for regulating transcription. Proc Natl Acad Sci U S A 101:2840–2845. doi: 10.1073/pnas.0400109101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Unoarumhi Y, Blumenthal RM, Matson JS. 2016. Evolution of a global regulator: Lrp in four orders of γ-Proteobacteria. BMC Evol Biol 16:111. doi: 10.1186/s12862-016-0685-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Morran LT, Penley MJ, Byrd VS, Meyer AJ, O'Sullivan TS, Bashey F, Goodrich-Blair H, Lively CM. 2016. Nematode-bacteria mutualism: selection within the mutualism supersedes selection outside of the mutualism. Evolution 70:687–695. doi: 10.1111/evo.12878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vadyvaloo V, Jarrett C, Sturdevant DE, Sebbane F, Hinnebusch BJ. 2010. Transit through the flea vector induces a pretransmission innate immunity resistance phenotype in Yersinia pestis. PLoS Pathog 6:e1000783. doi: 10.1371/journal.ppat.1000783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Corcoran CP, Dorman CJ. 2009. DNA relaxation-dependent phase biasing of the fim genetic switch in Escherichia coli depends on the interplay of H-NS, IHF and LRP. Mol Microbiol 74:1071–1082. doi: 10.1111/j.1365-2958.2009.06919.x. [DOI] [PubMed] [Google Scholar]
- 50.Nielsen AT, Dolganov NA, Rasmussen T, Otto G, Miller MC, Felt SA, Torreilles S, Schoolnik GK. 2010. A bistable switch and anatomical site control Vibrio cholerae virulence gene expression in the intestine. PLoS Pathog 6:e1001102. doi: 10.1371/journal.ppat.1001102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jansen G, Crummenerl LL, Gilbert F, Mohr T, Pfefferkorn R, Thänert R, Rosenstiel P, Schulenburg H. 2015. Evolutionary transition from pathogenicity to commensalism: global regulator mutations mediate fitness gains through virulence attenuation. Mol Biol Evol 32:2883–2896. doi: 10.1093/molbev/msv160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Miller JH. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 53.Xu J, Hurlbert RE. 1990. Toxicity of irradiated media for Xenorhabdus spp. Appl Environ Microbiol 56:815–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McCann J, Stabb EV, Millikan DS, Ruby EG. 2003. Population dynamics of Vibrio fischeri during infection of Euprymna scolopes. Appl Environ Microbiol 69:5928–5934. doi: 10.1128/AEM.69.10.5928-5934.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dunn AK, Millikan DS, Adin DM, Bose JL, Stabb EV. 2006. New rfp- and pES213-derived tools for analyzing symbiotic Vibrio fischeri reveal patterns of infection and lux expression in situ. Appl Environ Microbiol 72:802–810. doi: 10.1128/AEM.72.1.802-810.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bhasin A, Chaston JM, Goodrich-Blair H. 2012. Mutational analyses reveal overall topology and functional regions of NilB, a bacterial outer membrane protein required for host association in a model of animal-microbe mutualism. J Bacteriol 194:1763–1776. doi: 10.1128/JB.06711-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chaston JM, Suen G, Tucker SL, Andersen AW, Bhasin A, Bode E, Bode HB, Brachmann AO, Cowles CE, Cowles KN, Darby C, de Léon L, Drace K, Du Z, Givaudan A, Herbert Tran EE, Jewell KA, Knack JJ, Krasomil-Osterfeld KC, Kukor R, Lanois A, Latreille P, Leimgruber NK, Lipke CM, Liu R, Lu X, Martens EC, Marri PR, Médigue C, Menard ML, Miller NM, Morales-Soto N, Norton S, Ogier J-C, Orchard SS, Park D, Park Y, Qurollo BA, Sugar DR, Richards GR, Rouy Z, Slominski B, Slominski K, Snyder H, Tjaden BC, van der Hoeven R, Welch RD, Wheeler C, Xiang B, Barbazuk B, et al. 2011. The entomopathogenic bacterial endosymbionts Xenorhabdus and Photorhabdus: convergent lifestyles from divergent genomes. PLoS One 6:e27909. doi: 10.1371/journal.pone.0027909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bao Y, Lies DP, Fu H, Roberts GP. 1991. An improved Tn7-based system for the single-copy insertion of cloned genes into chromosomes of Gram-negative bacteria. Gene 109:167–168. doi: 10.1016/0378-1119(91)90604-A. [DOI] [PubMed] [Google Scholar]
- 59.Forst SA, Tabatabai N. 1997. Role of the histidine kinase, EnvZ, in the production of outer membrane proteins in the symbiotic-pathogenic bacterium Xenorhabdus nematophilus. Appl Environ Microbiol 63:962–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hussa E, Goodrich-Blair H. 2012. Rearing and injection of Manduca sexta larvae to assess bacterial virulence. J Vis Exp 70:e4295. doi: 10.3791/4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Merritt JH, Kadouri DE, O'Toole GA. 2005. Growing and analyzing static biofilms. Curr Protoc Microbiol Chapter 1:Unit 1B.1. doi: 10.1002/9780471729259.mc01b01s00. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Murfin KE, Chaston J, Goodrich-Blair H. 2012. Visualizing bacteria in nematodes using fluorescent microscopy. J Vis Exp 68:4298. doi: 10.3791/4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kaya HK, Stock SP. 1997. Techniques in insect nematology, p 281–324. In Lacey L. (ed), Manual of techniques in insect pathology. Academic Press, San Diego, CA. [Google Scholar]
- 64.Stabb EV, Ruby EG. 2002. RP4-based plasmids for conjugation between Escherichia coli and members of the Vibrionaceae. Methods Enzymol 358:413–426. doi: 10.1016/S0076-6879(02)58106-4. [DOI] [PubMed] [Google Scholar]
- 65.Visick KL, Skoufos LM. 2001. Two-component sensor required for normal symbiotic colonization of Euprymna scolopes by Vibrio fischeri. J Bacteriol 183:835–842. doi: 10.1128/JB.183.3.835-842.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.