ABSTRACT
Bacteriophage-based assays and biosensors rival traditional antibody-based immunoassays for detection of low-level Salmonella contaminations. In this study, we harnessed the binding specificity of the long tail fiber (LTF) from bacteriophage S16 as an affinity molecule for the immobilization, enrichment, and detection of Salmonella. We demonstrate that paramagnetic beads (MBs) coated with recombinant gp37-gp38 LTF complexes (LTF-MBs) are highly effective tools for rapid affinity magnetic separation and enrichment of Salmonella. Within 45 min, the LTF-MBs consistently captured over 95% of Salmonella enterica serovar Typhimurium cells from suspensions containing from 10 to 105 CFU · ml−1, and they yielded equivalent recovery rates (93% ± 5%, n = 10) for other Salmonella strains tested. LTF-MBs also captured Salmonella cells from various food sample preenrichments, allowing the detection of initial contaminations of 1 to 10 CFU per 25 g or ml. While plating of bead-captured cells allowed ultrasensitive but time-consuming detection, the integration of LTF-based enrichment into a sandwich assay with horseradish peroxidase-conjugated LTF (HRP-LTF) as a detection probe produced a rapid and easy-to-use Salmonella detection assay. The novel enzyme-linked LTF assay (ELLTA) uses HRP-LTF to label bead-captured Salmonella cells for subsequent identification by HRP-catalyzed conversion of chromogenic 3,3′,5,5′-tetramethylbenzidine substrate. The color development was proportional for Salmonella concentrations between 102 and 107 CFU · ml−1 as determined by spectrophotometric quantification. The ELLTA assay took 2 h to complete and detected as few as 102 CFU · ml−1 S. Typhimurium cells. It positively identified 21 different Salmonella strains, with no cross-reactivity for other bacteria. In conclusion, the phage-based ELLTA represents a rapid, sensitive, and specific diagnostic assay that appears to be superior to other currently available tests.
IMPORTANCE The incidence of foodborne diseases has increased over the years, resulting in major global public health issues. Conventional methods for pathogen detection can be laborious and expensive, and they require lengthy preenrichment steps. Rapid enrichment-based diagnostic assays, such as immunomagnetic separation, can reduce detection times while also remaining sensitive and specific. A critical component in these tests is implementing affinity molecules that retain the ability to specifically capture target pathogens over a wide range of in situ applications. The protein complex that forms the distal tip of the bacteriophage S16 long tail fiber is shown here to represent a highly sensitive affinity molecule for the specific enrichment and detection of Salmonella. Phage-encoded long tail fibers have huge potential for development as novel affinity molecules for robust and specific diagnostics of a vast spectrum of bacteria.
KEYWORDS: detection, Salmonella
INTRODUCTION
Salmonella strains are the second most frequent zoonotic pathogens recognized in Europe (1) and represent one of the most economically significant pathogens for food manufacturers around the world. Salmonellosis, the illness produced by Salmonella infection, causes an estimated 93.8 million cases of gastroenteritis and 155,000 deaths globally each year from contaminated food sources (2). Combined, Salmonella enterica subsp. enterica serovars Enteriditis (S. Enteriditis) and Typhimurium (S. Typhimurium) represent 60% and 30% of all reported cases of human salmonellosis in the European Union (3) and the United States (4), respectively. Therefore, the global food industry demands more stringent testing methods for Salmonella, in particular S. Enteriditis and S. Typhimurium, which in turn requires innovative research efforts. Traditional culture-based methods, such as the ISO horizontal method 6579-1:2017 (5), have been the gold standard for detecting Salmonella in food and animal feed (6). Although they demonstrate consistently high selectivity and sensitivity, culture-based methods are time consuming and therefore expensive, taking 3 to 5 days to enrich viable cells to detectable levels. This has led to the development and commercialization of a number of rapid screening methods and approaches aimed at decreasing enrichment time (7, 8). Immunomagnetic separation (IMS) appears very promising for streamlining enrichment steps. IMS involves capturing target organisms using antibody-coated magnetic beads (MBs) and separating the entire complex by applying a magnetic field (9). IMS simultaneously preenriches and concentrates the target organism, eliminating the need for lengthy, labor-intensive enrichment steps and, thus, reducing test times in comparison to those of conventional methods. IMS also separates bead-bound organisms from background contaminants for subsequent resuspension and downstream detection assays. The combination of IMS with PCR or enzyme-linked immunosorbent assay (ELISA) has previously been demonstrated to offer efficient enrichment and sensitive detection of pathogenic bacteria, including Escherichia coli O157, S. Typhimurium, S. Enteritidis, and Listeria monocytogenes (10–16).
One of the bottlenecks of IMS and other immunological-based methods is adopting a robust and easy-to-produce affinity molecule that can specifically recognize and capture the targeted bacterial species or genus. Clearly, antibodies are the most extensively used affinity molecules for biomolecular recognition and detection of pathogenic bacteria, spores, and bacterial toxins (17, 18). Although highly sensitive and widely available, antibodies are not always ideal for bacterial detection (17, 19). Immunological detection of Salmonella cells relies on antibodies raised against somatic O antigens of the cell wall lipopolysaccharide (LPS) or H antigens on the flagella of motile cells. Unfortunately, Escherichia coli, Citrobacter spp., Aeromonas spp., and others share some O-antigenic factors with Salmonella that can lead to cross-reactivity and false-positive identification (20). Additional limitations of antibodies include poor rates of recovery (21, 22), the inability to detect low-level contaminations (23), and sensitivity to pH, temperature, and proteases that can limit their in situ capacity to detect bacteria from clinical and highly complex food matrices (18, 24). Altogether, the limitations associated with using antibodies have emphasized the need for alternative affinity molecules with different physical characteristics and binding capabilities (24, 25).
Bacteriophages (phages) are ubiquitous viruses that specifically infect prokaryotic hosts. Phages and their encoded proteins have proven to be valuable tools in agriculture (26), biotechnology (27, 28), and clinical diagnostics (29, 30). Their inherent affinity for the individual target cells, which may be pathogens, has been exploited to develop detection assays using intact native or recombinant phages with binding tags as affinity markers (7, 31). For example, immobilized phages have been used in biosensors to detect Staphylococcus aureus (32) and Salmonella (33) down to 104 and 103 CFU · ml−1, respectively. In this respect, Salmonella phage P22-coated MBs have also been used for IMS-based enrichment of S. Typhimurium cells for successive immunoassay detection to a lower limit of 19 CFU · ml−1 (34), and modified T7 phage-coated MBs could enrich >86% of E. coli cells, resulting in a detection limit of 102 CFU · ml−1 (35). The limitations of phages as affinity molecules include their relatively large size in comparison to that of antibodies, the retention of enzymatic activity by their binding proteins, and their basal lytic activity for target cells, which releases cellular components that could hinder downstream detection (34). A simplified alternative to applying whole phages for bacterial detection is to utilize phage-encoded host interaction proteins as affinity molecules. Cell wall binding domains (CBDs) of phage endolysins (peptidoglycan hydrolases) have proven effective for detecting and enriching Gram-positive bacteria, such as Listeria (36, 37), Bacillus cereus, and Clostridium perfringens (38). Alternatively, phage receptor binding proteins (RBPs), which exist as long tail fibers (LTFs) or short tail spikes attached to the baseplate of tailed phages, have been applied for bacterial detection. The RBPs are of special interest since they represent the highly dedicated affinity molecules that initiate phage adsorption to the host cell. By recognizing specific polysaccharide and/or protein components on the bacterial surface, the RBPs confer the specificity that largely determines the infection range of a phage.
Previously, the tail spike RBP from phage P22 has been developed as a probe for detecting Salmonella in real-time biosensors (39) and, due to its inherent enzymatic activity, as a treatment against Salmonella colonization in chickens (40). Campylobacter phage RBPs have also been developed as probes for detecting the organism via surface plasmon resonance (SPR) biosensors (41), as well as for a cell agglutination assay (42).
We previously described the virulent, nontransducing phage S16 (43). This phage features a broad host range within the genus Salmonella, lysing 31/32 clinical isolates with no observed cross-reactivity (43) and surpassing other broad-range phages, such as Felix-O1, that are frequently used for Salmonella identification by plaque formation (44). Phage S16 has a “T-even” phage morphology and is closely related to phage T4, for which a robust model for phage adsorption has been established (45, 46, 67). Although genetically similar to T4, the phage S16 LTF features a phage T2-like architecture, composed of proteins gp34 to gp38 that extend in order from the phage baseplate to the distal fiber tip (45). During LTF maturation, gp37 trimerizes, assisted by the general chaperone gp57A. The C-terminal intramolecular chaperone domain of gp37 is then cleaved, prior to association of gp38 to its tip (43, 48). The adhesin gp38 itself offers a modular design that includes a C-terminal specificity domain that determines the binding range of phage S16 (47, 48).
In this study, we successfully produced a modified version of the phage S16 LTF distal tip, comprised of gp37 and gp38, as an affinity molecule for directional attachment to a solid surface for the magnetic separation and enrichment of Salmonella. In addition, we generated a detection probe consisting of the phage S16 LTF conjugated to horseradish peroxidase (HRP), which was combined with LTF-MB-based enrichment to produce a novel, rapid, and sensitive detection assay for Salmonella.
RESULTS
LTF-coated magnetic beads efficiently immobilize and enrich Salmonella cells.
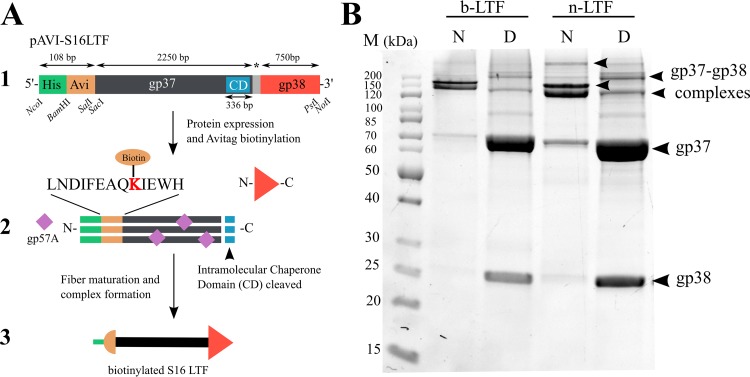
While thiol chemistry (39) and glutathione S-transferase tags (41) have been used previously for RBP attachment in biosensors, we employed biotin-streptavidin conjugation to provide stable and uniform attachment of the LTFs to the MB surface to maximize potential interactions with target cells. In order to streamline the purification and surface coupling of recombinant LTF molecules, adjacent polyhistidine and avidin tags were introduced to the N terminus of gp37 for immobilized-metal affinity chromatography (IMAC) purification and in vivo biotinylation, respectively (Fig. 1A). N-terminal biotinylation enables directional coupling of the LTF to the streptavidin-coated bead surface, preventing uncoordinated steric orientation of the gp38 adhesin and optimizing its display, i.e., pointing away from the bead surface. The biotinylated LTF (b-LTF) construct was coexpressed in E. coli strain BL21(DE3) in the presence of the biotin ligase BirA and the general tail fiber chaperone gp57A. The native gp38 adhesin, coexpressed from the same plasmid as the tagged gp37, attaches to the tip of gp37 during LTF formation (48). The mature gp37-gp38 complex was copurified by IMAC purification and demonstrated resistance to SDS treatment, requiring heat denaturation to separate the tightly associated gp37 and gp38 (Fig. 1B) (43).
FIG 1.
(A) Workflow for phage S16 LTF biotinylation and formation. (1) MCSI organization of pAVI-S16LTF encoding b-LTF; for n-LTF, the avidin tag is missing. CD, chaperone domain (112 C-terminal residues of gp37 form an intramolecular chaperone domain that is cleaved upon maturation); *, 27-bp intron separating gp37 and gp38; bp, base pairs. (2) Complex formation of gp37 and gp38, including biotinylation of Avitag for b-LTF. (3) Final b-LTF construct. (B) SDS-PAGE gel of purified samples of b-LTF and n-LTF. Samples were loaded directly (native [N]) or after denaturing for 10 min at 100°C (denatured [D]). In the native samples, gp37 and gp38 form higher-order complexes (final gp37-gp38 complex, ∼238 kDa). Denaturation separates the individual components (gp37, 71.0 kDa; gp38, 25.7 kDa). b-LTF and n-LTF fibers have the same SDS-PAGE band profiles, demonstrating uniformity between the constructs. M, PageRuler unstained protein ladder (PageRuler; ThermoFisher Scientific, USA).
The recombinant b-LTF was purified and conjugated to streptavidin-coated MBs to generate LTF-coated beads (LTF-MBs). The enrichment capacity of the LTF-MBs was tested using various densities of beads (1.7 × 107 to 3.5 × 108 beads · ml−1) incubated with 104 CFU · ml−1 S. Typhimurium strain DB7155 in different buffers. Following magnetic separation and washing, the resuspended LTF-MBs were surface plated on xylose lysine deoxycholate (XLD) agar in appropriate dilutions, and the plates incubated at 37°C overnight before colony enumeration. As previously found for bead-based Listeria separation (38), immobilization on beads did not affect the number of CFUs; i.e., the bead-bound bacteria remained viable and retained the same plating ability and growth pattern as freely suspended bacteria. This is explained by the fact that the number of beads always exceeded the number of cells to be recovered by more than 2 log.
In general, the phage S16 LTF-MBs displayed a wide functional range, recovering an average of 96% of Salmonella cells in the test volumes at pH 5 to 9 and 0 to 750 mM NaCl (see Table S1 in the supplemental material). There was a slight decrease in recovery in 1 M NaCl solution (90%); however, this remains a high recovery rate, viable for detection of Salmonella. As the difference in recovery rates from pH 7 to 8 was nominal, pH 7.4 was selected for consistency with the nonspecific enrichment buffer for detection of Salmonella in food according to ISO 6579-1:2017 (5). The wide functional range of the LTF-MBs is highly advantageous, considering the resilient and flexible nature of Salmonella, which allows it to survive under diverse conditions, requiring detection in very different food and clinical samples (49).
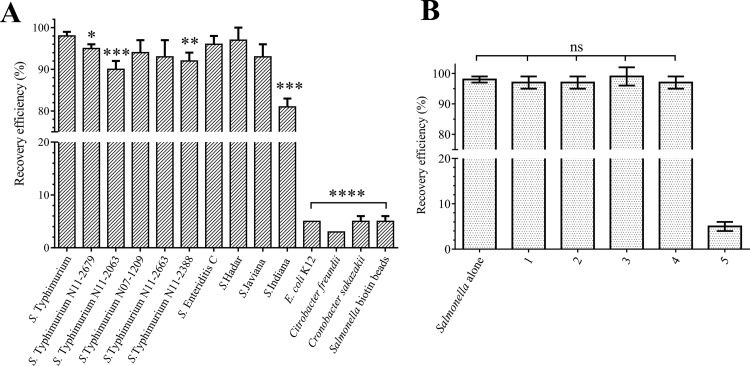
Using the optimized buffer (phosphate-buffered saline [PBS], pH 7.4, 125 mM NaCl, 0.1% Tween 20) and bead density (7.0 × 107 beads · ml−1), the LTF-MBs retained a superior recovery rate of over 98% when tested across a wide dilution range of 10 to 105 CFU · ml−1 S. Typhimurium DB7155 cells (Table 1). Both negative controls (E. coli K-12 recovery with LTF-MBs and S. Typhimurium DB7155 recovery with empty beads) featured less than 5% recovery, which was considered the unspecific background level. The remaining cells in the bead-bound fraction were assumed to result from nonspecific binding to the beads or incomplete washing. The high recovery rates observed with S. Typhimurium DB7155 were confirmed using nine additional Salmonella strains, as well as several monophasic S. Typhimurium isolates, where the LTF-MBs were capable of recovering between 81% and 97% of cells (Fig. 2A). The strict specificity of the LTF-MBs even among Gram-negative cells offering a similar surface structure was further demonstrated using E. coli, Cronobacter sakazakii, and Citrobacter freundii, which yielded background recovery rates of 5%, 5%, and 3%, respectively.
TABLE 1.
Recovery efficiencies of dilution series of S. Typhimurium using the LTF-MBs
| Type of beads and bacterial dilution (CFU · ml−1) | Mean recovery ± SD (%)a |
|---|---|
| LTF-MBs | |
| 101 | 98 ± 2 |
| 102 | 98 ± 0.1 |
| 103 | 98 ± 0.5 |
| 104 | 98 ± 1.5 |
| 105 | 98 ± 1 |
| Biotin-coated beadsb | |
| 105 | 5 ± 1 |
All experiments were performed in triplicate.
Biotin-coated MBs were used as a negative control to test for nonspecific recovery by the empty beads alone.
FIG 2.
(A) LTF-MB recovery efficiency was tested using the LTF-MB recovery protocol with 104 CFU · ml−1 of different bacteria. As a control, S. Typhimurium recovery was tested using biotin-coated MBs. The t test was used to calculate the statistical differences of individual recovery efficiencies compared to that of S. Typhimurium DB7155. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P value < 0.0001. (B) The recovery efficiency of S. Typhimurium is not affected by a mixed background flora containing common foodborne pathogens. S. Typhimurium recovery efficiency of LTF-MBs was also tested using S. Typhimurium alone. (1) Salmonella with E. coli K-12 at a ratio of 1:1. (2) Salmonella with E. coli K-12 at a ratio of 1:10. (3) Salmonella with E. coli K-12 at a ratio of 1:100. (4) Salmonella with Bacillus cereus ATCC 14579, Staphylococcus aureus NCTC 8325, Citrobacter freundii N0106, and Cronobacter sakazakii BAA894 at a ratio of 1:1:1:1:1. (5) S. Typhimurium recovery using biotin-passivated MBs as negative control. The differences in the recovery efficiencies of Salmonella alone or under conditions 1 to 4 were not significant (ns). Results are displayed as mean values ± SD.
Salmonella enrichment on the LTF-MBs is unaffected by background microflora.
The isolation of very low numbers of Salmonella in the presence of high levels of background flora can present a challenge during preenrichment. For IMS-based recovery from heterogeneous solutions, bead coatings must remain specific and sensitive for Salmonella. To test the recovery efficiency of the LTF-MBs in the presence of competing flora, a suspension of 104 CFU · ml−1 S. Typhimurium DB7155 was spiked with ratios of 1:1, 1:10, and 1:100 E. coli strain K-12 cells, as well as an equal-ratio mixture (1:1:1:1:1) of E. coli K-12 and other foodborne pathogens, i.e., Staphylococcus aureus, Citrobacter freundii, and Cronobacter sakazakii. The LTF-MBs were capable of recovering over 97% of S. Typhimurium DB7155 from all of the mixtures tested, with no significant difference between individual conditions as determined by one-way analysis of variance (ANOVA) (P = 0.707), indicating that being outnumbered by background flora does not affect LTF-MB-based enrichment (Fig. 2B). While cross-reactivity of Enterobacteriaceae is a common problem for antibody-based IMS of Salmonella strains (22, 50), this was not observed for the LTF-MBs.
LTF-MBs can enrich Salmonella from contaminated food preenrichments.
Salmonella cells in food matrices are typically injured, low in numbers, and unevenly dispersed in the presence of much higher numbers of other Enterobacteriaceae (51). Preenrichment is used to resuscitate damaged cells and increase their availability for detection (52). Lengthy preenrichments can be shortened with IMS to specifically isolate and concentrate low levels of bacteria into small volumes suitable for detection assays (8). We tested the recovery efficiency of the LTF-MBs after preenrichment of six food samples artificially contaminated with 0, 1 to 10, 10, 100, or 1,000 CFU S. Typhimurium DB7155 per 25 g or ml. The foods were preenriched in a nonspecific medium, and Salmonella cells recovered using the LTF-MB recovery protocol. Contaminating Salmonella CFU were qualitatively detected in all samples, down to a limit of 10 CFU per 25 g or ml (Table 2). The lowest initial contamination rate tested, i.e., 1 to 10 CFU per 25 g or ml, could be detected in chicken, infant formula, milk, and chocolate milk, meeting the required detection limit for Salmonella according to the methods of the USDA Microbiology Laboratory Guidebook (53) and the FDA Bacteriological Analytical Manual (54) and significantly reducing detection times, from 72 h to 24 h.
TABLE 2.
LTF-MB enrichment of S. Typhimurium from preenriched food samples
| Food matrix | No. of CFU enriched from 25-g or -ml preenriched food samples artificially contaminated with indicated no. of coloniesa |
||||
|---|---|---|---|---|---|
| 0 | 1–10 | 10 | 100 | 1,000 | |
| Milk | × | + | ++ | +++ | +++ |
| Chocolate milk | × | + | ++ | +++ | +++ |
| RIF | × | + | ++ | +++ | +++ |
| Chicken | × | + | ++ | ++ | +++ |
| Celeryb | × | × | + | + | ++ |
| Alfalfa sproutsb | × | × | + | + | ++ |
After the LTF-MB recovery protocol was performed, the numbers of Salmonella cells captured were semiquantitatively determined by colony counting on XLD agar. +, 1 to 10 colonies; ++, 10 to 100 colonies; +++, over 100 colonies; ×, no colonies.
Celery and alfalfa sprouts required an additional overnight enrichment in RVS broth.
HRP-conjugated LTF offers a rapid enzyme-linked sandwich detection assay.
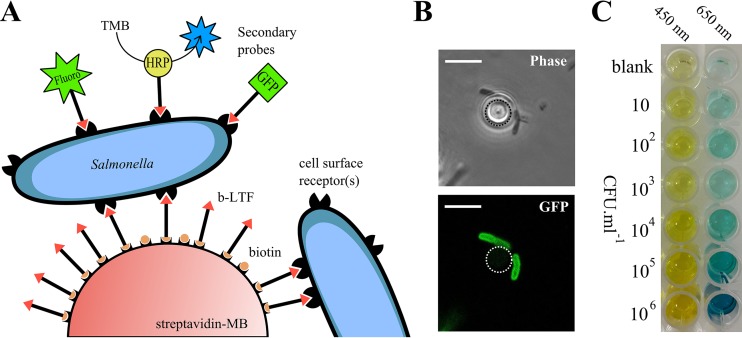
Colorimetric or fluorescent ELISA-based assays have been successfully combined with IMS for rapid identification and quantification of bacterial contaminations (12, 14, 15). We therefore assessed the functionality of the phage S16 LTF as a secondary probe to detect bead-bound Salmonella (Fig. 3A). As a proof of principle, b-LTF was conjugated with a fluorescent dye (fluoro-LTF) and used to label LTF-MB-captured Salmonella cells for direct observation by fluorescence microscopy, which provided quick, qualitative, and visual confirmation of Salmonella enrichment (Fig. 3B). Our attempts to quantify fluorescence by spectrophotometric measurement were hindered because the nonuniform distribution of LTF-MBs produced irregular spectrophotometric measurements (results not shown). We then designed a novel self-sandwich-like assay, the enzyme-linked long tail fiber assay (ELLTA), employing nonbiotinylated LTF (n-LTF) conjugated with horseradish peroxidase (HRP-LTF) as a detection probe. Following LTF-MB recovery, the beads were carefully washed and incubated for 30 min with the HRP-LTF probe, providing sufficient time to decorate bead-bound Salmonella cells. After removal of unbound HRP-LTF, the beads were incubated in a buffer containing 3,3′,5,5′-tetramethylbenzidine (TMB), where HRP-catalyzed conversion produced a blue color change, instantly visible by eye, indicating the presence of Salmonella. Finally, LTF-MBs were magnetically separated and the catalyzed TMB solution transferred to 96-well plates for photometric quantification (Fig. 3C).
FIG 3.
(A) Schematic representation of Salmonella enrichment and detection using LTF-MBs and LTF-based secondary probes with GFP (43), fluorescent dyes, or enzymes, as demonstrated by HRP-LTF. (B) Phase-contrast and fluorescence microscopy of two S. Typhimurium DB7155 cells bound to a single LTF-MB labeled with the Dylight 488-LTF secondary probe. Dotted line identifies the outline of the MB. Scale bar = 5 μm. (C) Results of ELLTA assay showing chromogenic conversion of TMB after using the LTF-based detection assay for amounts of 10 to 106 CFU · ml−1 S. Typhimurium DB7155. Absorbance and spectrophotometric quantification were determined at 650 nm and then at 450 nm following acidification with H2SO4.
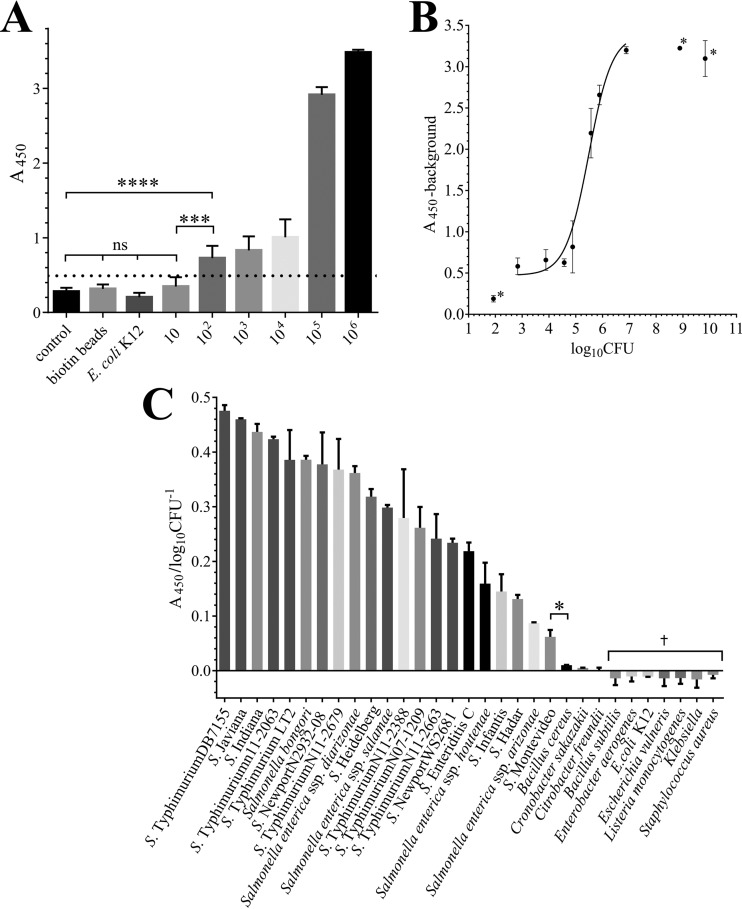
The sensitivity of ELLTA was determined using a dilution series of 10 to 108 CFU · ml−1 S. Typhimurium DB7155, as well as buffer-only (LTF-MBs incubated with no cells) and empty biotin-bead controls (Fig. 4A). A negligible amount of TMB conversion was observed for the buffer-only control, with an A450 value of 0.24 ± 0.05, suggesting that a residual amount of HRP-LTF remained after washing; this was set as the background value of the assay. ELLTA was sensitive and the results statistically significant down to 102 CFU · ml−1 (A450 of 0.77 ± 0.18; t test with buffer-only control, P < 0.001), similar to other phage- and phage-encoded protein-based assays with detection limits ranging between 10 and 104 CFU · ml−1 (17). While the value for the lowest Salmonella concentration tested, 10 CFU · ml−1 (A450 of 0.36 ± 0.10), was higher than the value for the blank-only control (A450 of 0.24), there was no statistical difference between the two values (P > 0.05). The limit of blank (LoB) and limit of detection (LoD) (Fig. 4A, dotted line) were calculated as an A450 of 0.32 and 0.49, respectively, using the approach outlined by Armbruster and Pry (55). The LoD is the lowest spectrophotometric measurement that can be distinguished from the LoB at which detection of Salmonella by ELLTA is reliable and practical. Therefore, A450 measurements above 0.49 should indicate a positive detection of Salmonella by this assay.
FIG 4.
(A) ELLTA detection of 10 to 106 CFU · ml−1 S. Typhimurium DB7155, buffer only (control), and 105 CFU · ml−1 E. coli K-1 and an additional control of 105 CFU · ml−1 S. Typhimurium enrichment using biotin-coated beads with HRP-LTF detection. Dotted line indicates the LoD (A450 of 0.49). (B) A450 values are proportional to initial S. Typhimurium CFU per assay (log10 CFU · ml−1). Data were fitted using nonlinear regression (R2 = 0.961). *, data not included in the regression fit included (i) results for 101 CFU · ml−1, as not statistically different from the control, and (ii) A450 values for >7 log10 CFU · ml−1, as values were above the threshold of the spectrophotometer. (C) Results from ELLTA with Salmonella and non-Salmonella strains. †, A450 values for 7 non-Salmonella strains were below the A450 value for the buffer-only control used for background adjustment, giving negative A450/log10 CFU−1 values. (A to C) Experiments were performed in triplicate, and results are displayed as mean values ± SD. Statistical significance is shown as follows: *, P < 0.05; ***, P < 0.001; ****, P < 0.0001; ns, no significance.
Quantification and specific detection of Salmonella using ELLTA.
Taking into account that LTF-MB recovery captured 97% ± 4% of Salmonella cells over a 10 to 105 CFU · ml−1 range, the amount of cells bound to the LTF-MBs should correlate closely with the initially present Salmonella cell numbers per assay, and it may therefore be possible to quantify Salmonella counts based on TMB color development. The A450 values of a dilution series of 10 to 108 CFU · ml−1 S. Typhimurium DB7155 were background corrected (−0.24) and plotted against the initial cell numbers, log10 CFU · ml−1, which were calculated by CFU enumeration after plating the Salmonella dilution series (Fig. 4B). A nonlinear regression was fitted to the data, which revealed a clear correlation between absorbance and initial CFU · ml−1 per assay. Given the linear relationship of A450 values to log10 CFU · ml−1 between 105 and 107 CFU · ml−1, it appears feasible to utilize ELLTA for quick quantification of Salmonella cell counts.
We further tested the specificity of ELLTA with amounts of 105 CFU · ml−1 of cross-genus Salmonella strains from food and clinical isolates, including 5 monophasic S. Typhimurium strains, which are notoriously difficult to identify using routine biochemical and serological tests (56), and 10 non-Salmonella bacteria (Gram negative and Gram positive). All 21 Salmonella strains tested could be positively confirmed by visual inspection of the TMB color change, with all A450 measurements above the LoD for the assay (0.49), ranging between 0.62 and 3.02 (see Table S2 in the supplemental material). The 10 non-Salmonella strains tested had A450 measurements below the LoD for the assay, indicating no false-positive detection. After background correction, any inconsistencies in initial cell concentrations were corrected by dividing the A450 measurements by the initial cell numbers to provide a detection efficiency score (A450/log10 CFU · ml−1) by which individual strains could be ranked based on the ability of the HRP-LTF probe to detect bead-bound Salmonella (Fig. 4C; Table S2). While the 10 non-Salmonella bacteria gave detection efficiency scores that were close to zero or even negative, the Salmonella strains yielded efficiency scores of between 0.48 and 0.06 A450/log10 CFU · ml−1 for the most strongly detected (S. Typhimurium DB7155), and the most weakly detected (S. enterica subsp. enterica serovar Montevideo strain WS2678) strains tested. All the Salmonella strains were detected with statistically significant values (P < 0.05) compared with the values for the non-Salmonella strains. The strain-specific variability of the LTF-HRP probe affinity among different Salmonella strains is likely due to different binding capacities of the HRP-LTF to their cell surface receptors (43).
In conclusion, the combination of magnetic separation and self-sandwich detection in the ELLTA assay required only 2 h to complete and was capable of detecting S. Typhimurium at concentrations down to 102 CFU · ml−1, as well as 20 other Salmonella strains from across the genus, including other Salmonella enterica subspecies and Salmonella bongori, while producing no false positives.
DISCUSSION
The incidence of water- and foodborne illnesses remains a persistent global problem that greatly impedes socioeconomic development (57). Not only are consumers and products affected by pathogen contaminations, food production companies are also hit by huge loses in sales and equity due to product recalls or quarantined food supplies delayed due to time-ineffective bacterial identification tests. The demand for quicker and more reliable contaminant and quality controls of food has led to considerable progress in the development of rapid, high-throughput, and sensitive bacterial detection methods to replace the laborious and time-insensitive traditional culture-based detection methods (8). The use of affinity-based magnetic separation can simultaneously tackle two critical parameters to reduce detection assay time and increase sensitivity, by (i) enriching viable cell numbers to a detectable level and (ii) isolating cells from contaminants and debris found in food samples for more accurate downstream identification and characterization (51). Toward our aim to develop a sensitive and specific assay for bacterial enrichment that can outcompete the conventional application of antibodies for biomolecular recognition, the bacteriophage S16 LTF joins other approaches employing either whole phages (25, 32, 33, 58, 59) or phage-encoded proteins, such as endolysin cell wall-binding domains (CBDs) (36–38) and tail spike RBPs (39–41).
In this study, we demonstrate the extraordinary specificity of the phage S16 LTF-MBs for application as affinity molecules for Salmonella detection. The phage S16 LTF-MBs were capable of binding to and immobilizing all Salmonella strains tested, with a consistently high recovery rate (93% ± 5%, n = 10) and no cross-reactivity with other bacteria (<5%), including other related Enterobacteriaceae. Due to their high binding affinity, the binding and immobilization of cells by LTF-MBs was also highly effective, capturing essentially all (>98%) of 10 to 105 CFU · ml−1 S. Typhimurium cells from the tested suspensions. Besides not needing animals for their production and having less complicated production procedures than antibodies, phage-encoded host affinity proteins also demonstrate high degrees of resistance to pH, temperature, and protease damage (60). The phage S16 LTF-MBs were unaffected by excess background flora or variable buffer pH and salt concentrations (pH 4 to 9 and 0 to 1 M NaCl). This validates their suitability for in situ detection of Salmonella and seems superior to the more pH-sensitive and cross-reactivity-prone immunosorbent-based tests (17, 61).
Other bacteriophage recognition-based assays have been developed utilizing whole phage particles for SPR, bioluminescence, and magnetoelastic sensing; these can typically detect bacteria to lower limits of 102 to 103 CFU · ml−1 (17). Interestingly, by combining phage P22-based magnetic separation with anti-Salmonella antibodies, Laube et al. reported a detection limit of 19 CFU · ml−1 Salmonella, representing one of the most sensitive phage-based detection assays to date (34). However, it was also noted that the phage P22 particles used caused cell lysis (34). While phage-induced lysis can be combined with IMS to detect E. coli (62) and Salmonella (63) based on phage-encoded biomarker amplification, lysis may also be problematic if downstream identification requires the recovery of viable cells—an issue not associated with LTFs and other phage-encoded proteins with no inherent lytic activity. Moreover, single proteins or even protein complexes (such as gp37-gp38) are still much smaller than whole phage particles, less complicated to produce as a single entity, and easier to modify in order to alter or optimize their recognition properties (47, 64, 65). This was elegantly demonstrated by Singh et al., who observed a 6-fold increase in Salmonella immobilization in an SPR-based biosensor by removing the inherent endorhamnosidase activity of the phage P22 tail spike probe (39). We here demonstrate the ideal properties of phage S16 LTF-MBs for rapid detection of low-level Salmonella contaminations in foods. Following a short preenrichment, LTF-MBs were able to detect 1 to 10 CFU Salmonella in 25 g or ml of chicken, milk, chocolate milk, and infant formula samples (less than 1 CFU/g), demonstrating remarkable detection limits equivalent to or lower than those of other currently used tests (17).
To provide a complete, inexpensive, and easy to use Salmonella detection assay, we functionalized the phage S16 LTF as a secondary probe (HRP-LTF), used in combination with LTF-MB enrichment. This “self-sandwich”-based detection assay exploits the Salmonella selectivity of the phage S16 LTF to provide dual levels of Salmonella-specific interaction. The ELLTA assay only takes 2 h to complete, including all wash steps, and without enrichment or amplification is able to reliably detect S. Typhimurium DB7155 down to 102 CFU · ml−1, equivalent to the limits documented for other phage-based detection tests (17), including SPR biosensors based on other RBPs for detection of Salmonella (39) and Campylobacter jejuni (41). Similar to the phage S16 LTF, these RBP-based sensors also showed no cross-reactivity, a frequent feature of phage protein-based recognition.
The ELLTA assay provided rapid visual confirmation of Salmonella presence based on the colorimetric conversion of the chromogenic TMB substrate. The rate of conversion was highly proportional over a concentration of 105 to 107 CFU · ml−1 S. Typhimurium DB7155 cells, indicating that the test may deliver more than just a yes-or-no answer for Salmonella contamination by being applied in a semiquantitative way. Interestingly, there was a clear variation in the efficiency of detection (A450/log10 CFU · ml−1) across the range of Salmonella strains tested. Due to the consistently high recovery rates observed across different Salmonella strains (92% ± 2%, n = 10) (Fig. 2A), we believe this is not due to inconsistent enrichment by the LTF-MB recovery protocol. Instead, as each HRP-LTF interacts as an independent molecule, there is no additive effect of the multiple interactions that occur during bacterial capture by the LTF-MBs. Therefore, variations in labeling efficiency by the HRP-LTF likely originate from differences in receptor binding affinity from strain-specific epitope variation in the OmpC protein (43) and/or modifications to subsidiary recognition sites within the LPS core region.
The molecular details of the interaction of the phage S16 LTF with its target bacterial receptors are currently being investigated and are expected to support fine-tuning the protein-ligand interaction for future biotechnological applications. The distal-tip adhesin gp38, common to many T-even phages, represents the actual interface for host cell recognition and binding (48). The modular design of gp38 proteins is comprised of a highly variable C-terminal domain composed of glycine-rich motifs (GRMs) interlaced with a series of hypervariable segments (HVSs). The latter display high levels of plasticity (66) and are very susceptible to recombinational shuffling, which can modify the binding properties and, also, the phage host range (47, 48). Ultimately, structural characterization of gp38 and identification of the HVS receptor binding sites could permit directed modification of LTFs to generate affinity molecules with improved binding properties or altered host specificities. We envisage the application of phage S16 LTF and similar molecules as affinity reagents for any biorecognition events that require sensitive and selective attachment to bacterial pathogens. Despite their sheer abundance and genetic diversity in the environment, only a fraction of phages and their RBPs have yet been characterized, opening up a nearly limitless source of tail fibers for adaptation as affinity molecules.
MATERIALS AND METHODS
Bacterial strains.
All strains used in this study are listed in Table 3. The bacteria were grown in LB medium at 37°C under agitation. S. Typhimurium DB7155 and E. coli K-12 were used as the positive- and negative-control strains, respectively. E. coli strain XL1 Blue MRF′ cells (Stratagene, Basel, Switzerland) were used for all cloning steps and transformations. E. coli strains BL21(DE3) and Lemo21(DE3) (New England BioLabs [NEB], USA) were used for protein expression.
TABLE 3.
Bacterial strains used in this study
| Species, subspecies, or serovar | Phenotype | Strain | Sourcea |
|---|---|---|---|
| Salmonella serovars and species | |||
| S. Typhimurium | DB7155 | 1 | |
| S. Enteritidis C | 2 | ||
| S. enterica subsp. enterica serovar Hadar | WS 2691 | 1 | |
| S. enterica subsp. enterica serovar Heidelberg | N2743-08 | 1 | |
| S. enterica subsp. enterica serovar Indiana | 2 | ||
| S. enterica subsp. enterica serovar Infantis | 1 | ||
| S. enterica subsp. enterica serovar Javiana | N2427-08 | 3 | |
| S. Montevideo | WS2678 | 1 | |
| S. enterica subsp. enterica serovar Newport | WS2681 | 1 | |
| S. Newport | N2932-08 | 3 | |
| S. Typhimurium LT2 | ATCC 14028 | 4 | |
| S. Typhimurium | N11-2679 | 5 | |
| S. Typhimurium | N11-2063 | 5 | |
| S. Typhimurium | N07-1209 | 5 | |
| S. Typhimurium | N11-2663 | 5 | |
| S. Typhimurium | N11-2388 | 5 | |
| S. enterica subsp. arizonae | 56:z4,z23:− | N09-0860 | 3 |
| S. enterica subsp. diarizonae | 61:c:z35 | N09-2338 | 3 |
| S. enterica subsp. salamae | 30: l,z28:z6 | N09-2794 | 3 |
| S. enterica subsp. houtenae | 38:z4,z23:− | N09-2589 | 3 |
| S. bongori | 48:z35:− | N268-08 | 3 |
| Non-Salmonella species | |||
| Bacillus cereus | ATCC 14579 | 1 | |
| Bacillus cereus | HER 1399 | 1 | |
| Bacillus atrophaeus (subtilis) | ATCC 9372 | 1 | |
| Citrobacter freundii | N 0106 | 1 | |
| Cronobacter sakazakii | ATCC BAA-894 | 1 | |
| Cronobacter sakazakii | ATCC 29544 | 1 | |
| Enterobacter aerogenes | DSM 30053 | 1 | |
| Escherichia coli | LPS chemotype K-12 | CGSC4401 | 6 |
| Escherichia vulneris | DSM 4564 | 1 | |
| Klebsiella pneumoniae | 1319 | 1 | |
| Listeria monocytogenes | Serovar 1/2c | WSLC 1001 | 1 |
| Staphylococcus aureus | NCTC 8325 | 1 | |
| Staphylococcus aureus | ATCC 19685 | 1 |
Sources of strains: 1, laboratory stock; 2, University of Würzburg, Würzburg, Germany; 3, National Reference Centre for Enteropathogenic Bacteria and Listeria (NENT) (Zurich, Switzerland); 4, Thilo Fuchs (Technical University of Munich, Munich, Germany); 5, Roger Stephan and Herbert Hächler (University of Zurich, Zurich, Switzerland) (56); 6, Coli Genetic Stock Center (CGSC) (Yale University, New Haven, CT, USA).
Generation of the LTF constructs.
The biotinylated LTF (b-LTF) was created from a pETDUET-1 plasmid backbone containing birA, encoding biotin ligase, between the NdeI and XhoI sites of multiple cloning site II (MCSII). An Avitag polypeptide (N′-AGGGLNDIFEAQKIEWHEVDEL-C′)-encoding sequence was generated as a double-stranded DNA (dsDNA) segment using Avi1 and Avi2 oligonucleotides (Table 4), designed with a 3′ overhang suitable for ligation to SalI-digested dsDNA. Avi1 and Avi2 at 5 μM were combined in a final volume of 10 μl, heated to 100°C for 5 min, and annealed at room temperature to form a dsDNA AviTag. Bicistronic gp37-gp38 was amplified from a whole S16 phage template using primers LTF1 and LTF2, which contained a SalI site at the 5′ end of gp37. This PCR product was digested with SalI and ligated to the double-stranded AviTag oligonucleotide, using T4 ligase at room temperature for 10 min. The fused product was used as a template with primers LTF2 and LTF3 to amplify across the Avitag LTF fusion product, generating an Avitag-g37-g38 PCR product. This was subsequently digested and inserted between the BamHI and PstI sites of MCSI in pETDUET-1_birA to generate pAVI-S16LTF (Fig. 1A; Table 4). The pETDUET-1 backbone was also used for generating the nonbiotinylated LTF (n-LTF). Phage S16 gp57A was cloned using primers LTF4 and LTF5, digested, and ligated between NdeI and XhoI of MCSII. Bicistronic gp37-gp38 was amplified from a whole S16 phage template using primers LTF6 and LTF7, digested, and inserted between BamHI and NotI of MCSI to generate pHIS-S16LTF (Table 4). All cloned plasmids were transformed into E. coli XL1 Blue MRF′ cells, purified, and sequenced to ensure correct insertions.
TABLE 4.
Plasmids and primers used in this study
| Plasmid or primer | Description or sequence (5′ to 3′) | Reference or source |
|---|---|---|
| Plasmids | ||
| pRM4 | pBAD18 Cmr; gp57A (SacI/SalI) | 43 |
| pAVI-S16LTF | pETDUET Ampr; MCSI:AVI-LTF (BamHI/PstI); MCSII:BirA (NdeI/XhoI) | This study |
| pHIS-S16LTF | pETDUET Ampr backbone; MCSI:LTF (BamHI/NotI); MCSII:S16 gp57A chaperone (NdeI/XhoI) | This study |
| Primers | ||
| Avi1 | GATCCGGCGGGTGGCGGTCTGAACGACATCTTCGAGGCTCAGAAAATCGAATGGCACGAA | |
| Avi2 | TCGACTTCGTGCCATTCGATTTTCTGAGCCTCGAAGATGTCGTTCAGACCGCCACCCGCCG | |
| LTF1_SalI | ATTCGTCGACGAGCTCATGGCTACTATAAA | |
| LTF2_PstI | TTTTTTCTGCAGTTATAACCAAGAACCAGCAATATTACC | |
| LTF3_BamHI | GCCAGGATCCGGCGGGTGGC | |
| LTF4_NdeI | CGCCATATGACTGATAAAATTAAACAGCT | |
| LTF5_XhoI | CCGCTCGAGTCATTCATCATCCGGCGTT | |
| LTF6_BamHI | CGCGGATCCGGAGAATCTGTATTTCCAGGGAATGGCTACTATAAAACAAATACAA | |
| LTF7_NotI | ATAGTTTAGCGGCCGCTTATAACCAAGAACCAGCAATA | |
Protein production and purification.
E. coli Lemo21(DE3) cells were cotransformed with pRM4 (encoding gp57A) and pAVI-S16LTF or transformed with pHIS-S16LTF alone for b-LTF or n-LTF expression, respectively. LB medium supplemented with 1.5 mM l-rhamnose was inoculated with overnight cultures, and the cultures grown with agitation at 37°C until reaching an optical density at 600 nm (OD600) of 0.6. The cultures were cooled to 20°C, and for b-LTF expression, 50 μM l-biotin was added. The cultures were induced with 400 μM isopropyl-β-d-thiogalactopyranoside (IPTG) and 1 mM l-arabinose and incubated for 16 h with agitation at 20°C. The cells were harvested by centrifugation (5,500 × g for 15 min), resuspended in PBS-T buffer (50 mM dihydrogen phosphate, 130 mM sodium chloride, 0.1% Tween 20, pH 7.4) at 4°C, and lysed using a Stansted pressure cell homogenizer (Stansted Fluid Power, UK). The cell extract was centrifuged (16,000 × g for 75 min) prior to immobilized-metal affinity chromatography (IMAC) using low-density Ni-nitrilotriacetic acid (NTA) resin (Chemie Brunschwig, Basel, Switzerland) and finally dialyzed into 25 mM Tris buffer, pH 7.4. The final protein yields were between 6 and 8 mg/liter LB medium. Purified proteins were analyzed for purity by SDS-PAGE. Here, 10-μl amounts of 1-mg · ml−1 concentrated protein samples were boiled for 10 min and electrophoresed on TGX stain-free precast gels (Bio-Rad, USA). Protein bands were visualized via UV absorbance (280 nm) using a Gel Doc XR+ imaging system (Bio-Rad) and additionally stained with InstantBlue Coomassie stain (Expedeon, USA) for contaminant determination.
b-LTF coating of streptavidin-coated MBs.
Batches of 200 μl of streptavidin-coated MB (10 mg · ml−1) (M-270 Dynabeads; Thermo Fisher Scientific, USA) were transferred into 2-ml Eppendorf tubes and washed three times with 1 ml PBS-T for 5 min. Beads were magnetically separated using a MagnaSphere magnetic stand (Promega AG, Dübendorf, Switzerland), and the wash solution removed. Following the manufacturer's specifications to ensure maximum conjugation, 1 μg of b-LTF was added to every 1 μl of MBs and the mixture incubated in PBS-T on an overhead rotator for 1 h at room temperature. The beads were separated, and the supernatant was measured by UV absorbance at 280 nm to confirm conjugation had occurred. The beads were washed twice with PBS-T, and the remaining streptavidin sites were blocked by incubation with 5 mM biotin and 2% bovine serum albumin (BSA) in PBS-T buffer. As a negative control, empty biotin-blocked beads were used. Finally, the functionalized beads were resuspended in PBS-T to the initial volume used for conjugation and stored at 4°C in PBS-T containing 0.01% sodium azide for up to 6 months, without loss in recovery efficiency.
LTF-MB recovery protocol.
Log-phase bacterial cultures were serially diluted to the desired concentration in PBS-T. Aliquots of 200 μl of a diluted bacterial sample were combined with 20 μl of LTF-MBs in a 2-ml Eppendorf tube and incubated on an overhead rotator for 45 min at room temperature. The beads were magnetically separated, and the supernatant removed. The whole supernatant fraction was then plated on LB agar plates for direct enumeration of the unbound bacterial fraction (UBF). The LTF-MBs were resuspended in 200 μl PBS-T, washed for 5 min on an overhead rotator, and magnetically separated, and the whole wash fraction (WF) was plated for enumeration. The washed LTF-MBs were resuspended in 200 μl of PBS-T and plated after dilution onto LB agar to give the bound bacterial fraction (BBF). Recovery efficiency is reported as the percentage determined by dividing the BBF by the total cell number (BBF plus UBF plus WF). To calculate Salmonella recovery efficiency from a mixed background flora, samples at all steps were plated onto xylose lysine deoxycholate (XLD) agar plates (Oxoid, Basingstoke, UK). This allowed direct enumeration of Salmonella cells (colonies with black centers) from background flora. All experiments were performed in triplicate.
Fluorescence microscopy.
Conjugation of b-LTF to the fluorescent dye streptavidin-Dylight 488 (Thermo Fisher Scientific, USA) to produce fluoro-LTF was performed according to the manufacturer's instructions. Following the above-described LTF-MB recovery protocol, beads were resuspended in 200 μl PBS-T containing 5 μg fluoro-LTF, incubated for 45 min, washed in PBS-T, and resuspended in 200 μl PBS-T. A small aliquot (4 to 5 μl) of the bead suspension was then used for fluorescence confocal laser microscopy at ×100 magnification using an oil immersion lens (Leica TCS SPE with Leica CTR 4000).
ELLTA.
For the enzyme-linked LTF assay (ELLTA), conjugation of n-LTF to an activated horseradish peroxidase (EZ-link plus; Thermo Fisher Scientific, USA) was performed following the manufacturer's specifications to produce HRP-linked LTF molecules (HRP-LTF). HRP-LTF was then adjusted to 1 mg · ml−1 in PBS-T, pH 7.4 (2% BSA) and stored at 4°C. Labeling of cells with the HRP-LTF probe was performed directly after the LTF-MB recovery protocol. Beads were resuspended in 1 ml PBS-T (2% BSA), 1 μg HRP-LTF was added, and the mixture was incubated for 30 min on an overhead rotator. The beads were magnetically separated, and the supernatant removed. The beads were then washed three times for 5 min with 1 ml PBS-T. A total of 100 μl soluble TMB (Merck Millipore, Schaffhausen, Switzerland) was gently mixed with the beads by pipetting, and the mixture left at room temperature to react in the dark for 15 min. The beads were magnetically captured, and the TMB solution (100 μl) was pipetted into a well of a 96-well, clear, flat-bottom Nunc plate (Thermo Fisher Scientific, USA). An equal volume (100 μl) of 0.3 M sulfuric acid H2SO4 was added to stop the TMB reaction, acidify the medium, and convert the blue chromogen to the higher-absorbance (450 nm) yellow chromogen. The converted TMB was then quantified at 450 nm absorbance using a POLARStar omega spectrophotometer (BMG Labtech, Germany). The limit of blank (LoB) and limit of detection (LoD) were calculated as follows: LoB = mean of background + (1.645 × standard deviation [SD] of background), and LoD = LoB + (1.645 × SD of 101 CFU · ml−1 sample) (55).
Statistical analysis.
Results are shown as means ± standard deviations. The statistical analyses were performed using Prism version 7.02 (GraphPad, San Diego, CA). The LTF-MB recovery efficiencies shown in Fig. 2B were compared using one-way analysis of variance (ANOVA). Student's t test was used to compare the LTF-MB recovery efficiencies shown in Fig. 2A and the ELLTA absorbance measurements shown in Fig. 4. For all tests, a confidence level of 95% was used.
Artificial contamination of food.
All six food samples listed in Table 2 were purchased locally, stored according to recommendations for food type, and tested for natural contamination according to ISO 6579-1:2017 (5) to exclude Salmonella background contamination. Infant formula was reconstituted (RIF) according to the manufacturer's recommendations using sterile deionized water. An overnight culture of S. Typhimurium DB7155 was serially diluted in buffered peptone water, pH 7.4 (BPW), and added to food samples to allow bacteria to integrate into the food matrix at 1 to 10, 100, or 1,000 CFU per 25 g or ml. For solid foods, 25-g portions were soaked for 1 h in diluted bacterial suspensions in a sterile petri dish under a sterile hood at room temperature with regular turning. Excess liquid was removed, and the samples dried for 30 min at room temperature under a sterile fume hood before immediate testing.
Food preenrichment and recovery with the LTF-MB recovery protocol.
For preenrichment, 25 g or ml of each food sample was mixed 1:10 with BPW, pH 7.4, to a total volume of 250 ml and incubated for 6 h at 37°C with vigorous shaking. Celery and alfalfa sprouts were additionally preenriched as follows: 100-μl samples of the BPW enrichment were added to 10 ml RVS broth (Oxoid, Basingstoke, UK) and incubated overnight at 37°C with vigorous shaking. One hundred microliters each of the BPW- or RVS-enriched samples was combined with 20 μl LTF-MBs and 100 μl PBS-T, and the LTF-MB recovery protocol was used. Here, quantification and calculation of recovery rates were omitted, as the initial preenrichment step prevented accurate correlation with initial contamination levels. Instead, the beads were plated on XLD agar and incubated overnight at 37°C for qualitative analysis of Salmonella colonies.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Stefan Miller (Regensburg, Germany) for advice regarding the production and functional characterization of phage proteins and to Roger Stephan and Herbert Hächler (University of Zurich, Zurich, Switzerland) for valuable discussions concerning the detection of Salmonella and for providing several Salmonella strains used in this study.
This project was funded by grant number 16756 N from the AiF/FEI, Bundesministerium für Wirtschaft und Technologie, Berlin, Germany.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00277-17.
REFERENCES
- 1.Di Cesare A, Losasso C, Barco L, Eckert EM, Conficoni D, Sarasini G, Corno G, Ricci A. 2016. Diverse distribution of toxin-antitoxin II systems in Salmonella enterica serovars. Sci Rep 6:28759. doi: 10.1038/srep28759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Majowicz SE, Musto J, Scallan E, Angulo FJ, Kirk M, O'Brien SJ, Jones TF, Fazil A, Hoekstra RM. 2010. The Global burden of nontyphoidal Salmonella gastroenteritis. Clin Infect Dis 50:882–889. doi: 10.1086/650733. [DOI] [PubMed] [Google Scholar]
- 3.European Food Safety Authority and European Centre for Disease Prevention and Control. 2015. The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2013: EU summary report on zoonoses, zoonotic agents and food-borne outbreaks 2013. EFSA J 13:3991. doi: 10.2903/j.efsa.2015.3991. [DOI] [Google Scholar]
- 4.Centers for Disease Control and Prevention (CDC). 2013. National Salmonella surveillance annual report, 2011. US Department of Health and Human Services, CDC, Atlanta, GA. [Google Scholar]
- 5.International Organization for Standardization. 2017. Microbiology of the food chain—horizontal method for the detection, enumeration and serotyping of Salmonella—part 1: detection of Salmonella spp. ISO 6579-1:2017 International Organization for Standardization, Geneva, Switzerland. [Google Scholar]
- 6.Schönenbrücher V, Mallinson ET, Bülte M. 2008. A comparison of standard cultural methods for the detection of foodborne Salmonella species including three new chromogenic plating media. Int J Food Microbiol 123:61–66. doi: 10.1016/j.ijfoodmicro.2007.11.064. [DOI] [PubMed] [Google Scholar]
- 7.Singh A, Arutyunov D, Szymanski CM, Evoy S. 2012. Bacteriophage based probes for pathogen detection. The Analyst 137:3405–3421. doi: 10.1039/c2an35371g. [DOI] [PubMed] [Google Scholar]
- 8.Lee K-M, Runyon M, Herrman TJ, Phillips R, Hsieh J. 2015. Review of Salmonella detection and identification methods: aspects of rapid emergency response and food safety. Food Control 47:264–276. doi: 10.1016/j.foodcont.2014.07.011. [DOI] [Google Scholar]
- 9.Cudjoe KS, Krona R, Olsen E. 1994. IMS: a new selective enrichment technique for detection of Salmonella in foods. Int J Food Microbiol 23:159–165. doi: 10.1016/0168-1605(94)90049-3. [DOI] [PubMed] [Google Scholar]
- 10.Yu H, Bruno JG. 1996. Immunomagnetic-electrochemiluminescent detection of Escherichia coli O157 and Salmonella Typhimurium in foods and environmental water samples. Appl Environ Microbiol 62:587–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ivnitski D, Abdel-Hamid I, Atanasov P, Wilkins E. 1999. Biosensors for detection of pathogenic bacteria. Biosens Bioelectron 14:599–624. doi: 10.1016/S0956-5663(99)00039-1. [DOI] [PubMed] [Google Scholar]
- 12.Mansfield LP, Forsythe SJ. 2000. The detection of Salmonella using a combined immunomagnetic separation and ELISA end-detection procedure. Lett Appl Microbiol 31:279–283. doi: 10.1046/j.1472-765x.2000.00811.x. [DOI] [PubMed] [Google Scholar]
- 13.Madonna AJ, Basile F, Furlong E, Voorhees KJ. 2001. Detection of bacteria from biological mixtures using immunomagnetic separation combined with matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Rapid Commun Mass Spectrom 15:1068–1074. doi: 10.1002/rcm.344. [DOI] [PubMed] [Google Scholar]
- 14.Hibi K, Abe A, Ohashi E, Mitsubayashi K, Ushio H, Hayashi T, Ren H, Endo H. 2006. Combination of immunomagnetic separation with flow cytometry for detection of Listeria monocytogenes. Anal Chim Acta 573:158–163. doi: 10.1016/j.aca.2006.03.022. [DOI] [PubMed] [Google Scholar]
- 15.Qiu J, Zhou Y, Chen H, Lin J-M. 2009. Immunomagnetic separation and rapid detection of bacteria using bioluminescence and microfluidics. Talanta 79:787–795. doi: 10.1016/j.talanta.2009.05.003. [DOI] [PubMed] [Google Scholar]
- 16.Jayamohan H, Gale BK, Minson B, Lambert CJ, Gordon N, Sant HJ. 2015. Highly sensitive bacteria quantification using immunomagnetic separation and electrochemical detection of guanine-labeled secondary beads. Sensors 15:12034–12052. doi: 10.3390/s150512034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Singh A, Poshtiban S, Evoy S. 2013. Recent advances in bacteriophage based biosensors for food-borne pathogen detection. Sensors 13:1763–1786. doi: 10.3390/s130201763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Velusamy V, Arshak K, Korostynska O, Oliwa K, Adley C. 2010. An overview of foodborne pathogen detection: in the perspective of biosensors. Biotechnol Adv 28:232–254. doi: 10.1016/j.biotechadv.2009.12.004. [DOI] [PubMed] [Google Scholar]
- 19.Dwivedi HP, Jaykus L-A. 2011. Detection of pathogens in foods: the current state-of-the-art and future directions. Crit Rev Microbiol 37:40–63. doi: 10.3109/1040841X.2010.506430. [DOI] [PubMed] [Google Scholar]
- 20.Muldoon MT, Teaney G, Jingkun LI, Onisk DV, Stave JW. 2007. Bacteriophage-based enrichment coupled to immunochromatographic strip-based detection for the determination of Salmonella in meat and poultry. J Food Prot 70:2235–2242. doi: 10.4315/0362-028X-70.10.2235. [DOI] [PubMed] [Google Scholar]
- 21.Eriksson E, Aspan A. 2007. Comparison of culture, ELISA and PCR techniques for Salmonella detection in faecal samples for cattle, pig and poultry. BMC Vet Res 3:21. doi: 10.1186/1746-6148-3-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.de Cássia dos Santos da Conceição R, Moreira ÂN, Ramos RJ, Goularte FL, Carvalhal JB, Aleixo JAG. 2008. Detection of Salmonella sp in chicken cuts using immunomagnetic separation. Braz J Microbiol 39:173–177. doi: 10.1590/S1517-83822008000100034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kumar S, Balakrishna K, Singh GP, Batra HV. 2005. Rapid detection of Salmonella typhi in foods by combination of immunomagnetic separation and polymerase chain reaction. World J Microbiol Biotechnol 21:625–628. doi: 10.1007/s11274-004-3553-x. [DOI] [Google Scholar]
- 24.Tombelli S, Minunni M, Mascini M. 2007. Aptamers-based assays for diagnostics, environmental and food analysis. Biomol Eng 24:191–200. doi: 10.1016/j.bioeng.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 25.Schmelcher M, Loessner MJ. 2014. Application of bacteriophages for detection of foodborne pathogens. Bacteriophage 4:e28137. doi: 10.4161/bact.28137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gill JJ. 2016. 118 phage applications in animal agriculture and food safety. J Anim Sci 94(Suppl 1):57. doi: 10.2527/ssasas2015-118. [DOI] [Google Scholar]
- 27.Olsen EV, Sorokulova IB, Petrenko VA, Chen I-H, Barbaree JM, Vodyanoy VJ. 2006. Affinity-selected filamentous bacteriophage as a probe for acoustic wave biodetectors of Salmonella Typhimurium. Biosens Bioelectron 21:1434–1442. doi: 10.1016/j.bios.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 28.Nanduri V, Balasubramanian S, Sista S, Vodyanoy VJ, Simonian AL. 2007. Highly sensitive phage-based biosensor for the detection of β-galactosidase. Anal Chim Acta 589:166–172. doi: 10.1016/j.aca.2007.02.071. [DOI] [PubMed] [Google Scholar]
- 29.Schofield DA, Sharp NJ, Westwater C. 2012. Phage-based platforms for the clinical detection of human bacterial pathogens. Bacteriophage 2:105–283. doi: 10.4161/bact.19274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haq IU, Chaudhry WN, Akhtar MN, Andleeb S, Qadri I. 2012. Bacteriophages and their implications on future biotechnology: a review. Virol J 9:9. doi: 10.1186/1743-422X-9-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ripp S. 2010. Bacteriophage-based pathogen detection. Adv Biochem Eng Biotechnol 118:65–83. doi: 10.1007/10_2009_7. [DOI] [PubMed] [Google Scholar]
- 32.Balasubramanian S, Sorokulova IB, Vodyanoy VJ, Simonian AL. 2007. Lytic phage as a specific and selective probe for detection of Staphylococcus aureus—a surface plasmon resonance spectroscopic study. Biosens Bioelectron 22:948–955. doi: 10.1016/j.bios.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 33.Lakshmanan RS, Guntupalli R, Hu J, Petrenko VA, Barbaree JM, Chin BA. 2007. Detection of Salmonella Typhimurium in fat free milk using a phage immobilized magnetoelastic sensor. Sens Actuators B Chem 126:544–550. doi: 10.1016/j.snb.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 34.Laube T, Cortés P, Llagostera M, Alegret S, Pividori MI. 2014. Phagomagnetic immunoassay for the rapid detection of Salmonella. Appl Microbiol Biotechnol 98:1795–1805. doi: 10.1007/s00253-013-5434-4. [DOI] [PubMed] [Google Scholar]
- 35.Wang Z, Wang D, Chen J, Sela DA, Nugen SR. 2016. Development of a novel bacteriophage based biomagnetic separation method as an aid for sensitive detection of viable Escherichia coli. Analyst 141:1009–1016. doi: 10.1039/C5AN01769F. [DOI] [PubMed] [Google Scholar]
- 36.Schmelcher M, Shabarova T, Eugster MR, Eichenseher F, Tchang VS, Banz M, Loessner MJ. 2010. Rapid multiplex detection and differentiation of Listeria cells by use of fluorescent phage endolysin cell wall binding domains. Appl Environ Microbiol 76:5745–5756. doi: 10.1128/AEM.00801-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Walcher G, Stessl B, Wagner M, Eichenseher F, Loessner MJ, Hein I. 2010. Evaluation of paramagnetic beads coated with recombinant Listeria phage endolysin-derived cell-wall-binding domain proteins for separation of Listeria monocytogenes from raw milk in combination with culture-based and real-time polymerase chain reaction-based quantification. Foodborne Pathog Dis 7:1019–1024. doi: 10.1089/fpd.2009.0475. [DOI] [PubMed] [Google Scholar]
- 38.Kretzer JW, Lehmann R, Schmelcher M, Banz M, Kim K-P, Korn C, Loessner MJ. 2007. Use of high-affinity cell wall-binding domains of bacteriophage endolysins for immobilization and separation of bacterial cells. Appl Environ Microbiol 73:1992–2000. doi: 10.1128/AEM.02402-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Singh A, Arya SK, Glass N, Hanifi-Moghaddam P, Naidoo R, Szymanski CM, Tanha J, Evoy S. 2010. Bacteriophage tailspike proteins as molecular probes for sensitive and selective bacterial detection. Biosens Bioelectron 26:131–138. doi: 10.1016/j.bios.2010.05.024. [DOI] [PubMed] [Google Scholar]
- 40.Waseh S, Hanifi-Moghaddam P, Coleman R, Masotti M, Ryan S, Foss M, MacKenzie R, Henry M, Szymanski CM, Tanha J. 2010. Orally administered P22 phage tailspike protein reduces Salmonella colonization in chickens: prospects of a novel therapy against bacterial infections. PLoS One 5:e13904. doi: 10.1371/journal.pone.0013904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Singh A, Arutyunov D, McDermott MT, Szymanski CM, Evoy S. 2011. Specific detection of Campylobacter jejuni using the bacteriophage NCTC 12673 receptor binding protein as a probe. Analyst 136:4780–4786. doi: 10.1039/c1an15547d. [DOI] [PubMed] [Google Scholar]
- 42.Javed MA, Poshtiban S, Arutyunov D, Evoy S, Szymanski CM. 2013. Bacteriophage receptor binding protein based assays for the simultaneous detection of Campylobacter jejuni and Campylobacter coli. PLoS One 8:e69770. doi: 10.1371/journal.pone.0069770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marti R, Zurfluh K, Hagens S, Pianezzi J, Klumpp J, Loessner MJ. 2013. Long tail fibres of the novel broad-host-range T-even bacteriophage S16 specifically recognize Salmonella OmpC. Mol Microbiol 87:818–834. doi: 10.1111/mmi.12134. [DOI] [PubMed] [Google Scholar]
- 44.Whichard JM, Weigt LA, Borris DJ, Li LL, Zhang Q, Kapur V, Pierson FW, Lingohr EJ, She Y-M, Kropinski AM, Sriranganathan N. 2010. Complete Genomic Sequence of Bacteriophage Felix O1. Viruses 2:710–730. doi: 10.3390/v2030710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bartual SG, Otero JM, Garcia-Doval C, Llamas-Saiz AL, Kahn R, Fox GC, van Raaij MJ. 2010. Structure of the bacteriophage T4 long tail fiber receptor-binding tip. Proc Natl Acad Sci U S A 107:20287–20292. doi: 10.1073/pnas.1011218107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Taylor NMI, Prokhorov NS, Guerrero-Ferreira RC, Shneider MM, Browning C, Goldie KN, Stahlberg H, Leiman PG. 2016. Structure of the T4 baseplate and its function in triggering sheath contraction. Nature 533:346–352. doi: 10.1038/nature17971. [DOI] [PubMed] [Google Scholar]
- 47.Tétart F, Desplats C, Krisch HM. 1998. Genome plasticity in the distal tail fiber locus of the T-even bacteriophage: recombination between conserved motifs swaps adhesin specificity. J Mol Biol 282:543–556. doi: 10.1006/jmbi.1998.2047. [DOI] [PubMed] [Google Scholar]
- 48.Trojet SN, Caumont-Sarcos A, Perrody E, Comeau AM, Krisch HM. 2011. The gp38 adhesins of the T4 superfamily: a complex modular determinant of the phage's host specificity. Genome Biol Evol 3:674–686. doi: 10.1093/gbe/evr059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lin J, Lee IS, Frey J, Slonczewski JL, Foster JW. 1995. Comparative analysis of extreme acid survival in Salmonella Typhimurium, Shigella flexneri, and Escherichia coli. J Bacteriol 177:4097–4104. doi: 10.1128/jb.177.14.4097-4104.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mansfield LP, Forsythe SJ. 2001. The detection of Salmonella serovars from animal feed and raw chicken using a combined immunomagnetic separation and ELISA method. Food Microbiol 18:361–366. doi: 10.1006/fmic.2001.0416. [DOI] [Google Scholar]
- 51.Jasson V, Baert L, Uyttendaele M. 2011. Detection of low numbers of healthy and sub-lethally injured Salmonella enterica in chocolate. Int J Food Microbiol 145:488–491. doi: 10.1016/j.ijfoodmicro.2011.01.031. [DOI] [PubMed] [Google Scholar]
- 52.Sandel MK, Wu Y-FG, McKillip JL. 2003. Detection and recovery of sublethally-injured enterotoxigenic Staphylococcus aureus. J Appl Microbiol 94:90–94. doi: 10.1046/j.1365-2672.2003.01807.x. [DOI] [PubMed] [Google Scholar]
- 53.FSIS, USDA. 2014. Isolation and identification of Salmonella from meat, poultry, pasteurized egg, and catfish products and carcass and environmental sponges. Microbiology laboratory guidebook, 4.08. USDA, Athens, GA. [Google Scholar]
- 54.Andrews WH, Jacobson A, Hammack T. 2011. Bacteriological analytical manual, chapter 5: Salmonella FDA, Silver Spring, MD. [Google Scholar]
- 55.Armbruster DA, Pry T. 2008. Limit of blank, limit of detection and limit of quantitation. Clin Biochem Rev 29:S49–S52. [PMC free article] [PubMed] [Google Scholar]
- 56.Gallati C, Stephan R, Hächler H, Malorny B, Schroeter A, Nüesch-Inderbinen M. 2013. Characterization of Salmonella enterica subsp. enterica serovar 4,[5],12:i:- clones isolated from human and other sources in Switzerland between 2007 and 2011. Foodborne Pathog Dis 10:549–554. doi: 10.1089/fpd.2012.1407. [DOI] [PubMed] [Google Scholar]
- 57.Nyachuba DG. 2010. Foodborne illness: is it on the rise? Nutr Rev 68:257–269. doi: 10.1111/j.1753-4887.2010.00286.x. [DOI] [PubMed] [Google Scholar]
- 58.Sun W, Brovko L, Griffiths M. 2001. Use of bioluminescent Salmonella for assessing the efficiency of constructed phage-based biosorbent. J Ind Microbiol Biotechnol 27:126–128. doi: 10.1038/sj.jim.7000198. [DOI] [PubMed] [Google Scholar]
- 59.Tolba M, Minikh O, Brovko LY, Evoy S, Griffiths MW. 2010. Oriented immobilization of bacteriophages for biosensor applications. Appl Environ Microbiol 76:528–535. doi: 10.1128/AEM.02294-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Van Dorst B, Mehta J, Bekaert K, Rouah-Martin E, De Coen W, Dubruel P, Blust R, Robbens J. 2010. Recent advances in recognition elements of food and environmental biosensors: a review. Biosens Bioelectron 26:1178–1194. doi: 10.1016/j.bios.2010.07.033. [DOI] [PubMed] [Google Scholar]
- 61.Jeníková G, Pazlarová J, Demnerová K. 2000. Detection of Salmonella in food samples by the combination of immunomagnetic separation and PCR assay. Int Microbiol 3:225–229. [PubMed] [Google Scholar]
- 62.Martelet A, L'Hostis G, Nevers M-C, Volland H, Junot C, Becher F, Muller BH. 2015. Phage amplification and immunomagnetic separation combined with targeted mass spectrometry for sensitive detection of viable bacteria in complex food matrices. Anal Chem 87:5553–5560. doi: 10.1021/ac504508a. [DOI] [PubMed] [Google Scholar]
- 63.Favrin SJ, Jassim SA, Griffiths MW. 2001. Development and optimization of a novel immunomagnetic separation-bacteriophage assay for detection of Salmonella enterica serovar Enteritidis in broth. Appl Environ Microbiol 67:217–224. doi: 10.1128/AEM.67.1.217-224.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yoichi M, Abe M, Miyanaga K, Unno H, Tanji Y. 2005. Alteration of tail fiber protein gp38 enables T2 phage to infect Escherichia coli O157:H7. J Biotechnol 115:101–107. doi: 10.1016/j.jbiotec.2004.08.003. [DOI] [PubMed] [Google Scholar]
- 65.Mahichi F, Synnott AJ, Yamamichi K, Osada T, Tanji Y. 2009. Site-specific recombination of T2 phage using IP008 long tail fiber genes provides a targeted method for expanding host range while retaining lytic activity. FEMS Microbiol Lett 295:211–217. doi: 10.1111/j.1574-6968.2009.01588.x. [DOI] [PubMed] [Google Scholar]
- 66.Drexler K, Riede I, Montag D, Eschbach M-L, Henning U. 1989. Receptor specificity of the Escherichia coli T-even type phage Ox2: mutational alterations in host range mutants. J Mol Biol 207:797–803. doi: 10.1016/0022-2836(89)90245-3. [DOI] [PubMed] [Google Scholar]
- 67.Hu B, Margolin W, Molineux IJ, Liu J. 2015. Structural remodeling of bacteriophage T4 and host membranes during infection initiation. Proc Natl Acad Sci USA 112:E4919–E4928. doi: 10.1073/pnas.1501064112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.