ABSTRACT
The Bacillus thuringiensis strain HBF-18 (CGMCC 2070), containing two cry genes (cry8-like and cry8Ga), is toxic to Holotrichia oblita larvae. Both Cry8-like and Cry8Ga proteins are active against this insect pest, and Cry8-like is more toxic. To analyze the characteristics of the binding of Cry8-like and Cry8Ga proteins to brush border membrane vesicles (BBMVs) in H. oblita larvae, binding assays were conducted with a fluorescent DyLight488-labeled Cry8-like toxin. The results of saturation binding assays demonstrated that Cry8-like bound specifically to binding sites on BBMVs from H. oblita, and heterologous competition assays revealed that Cry8Ga shared binding sites with Cry8-like. Furthermore, Cry8-like-binding proteins in the midgut from H. oblita larvae were identified by pulldown assays and liquid chromatography-tandem mass spectrometry (LC-MS/MS). In addition, the H. oblita midgut transcriptome was assembled by high-throughput RNA sequencing and used for identification of Cry8-like-binding proteins. Eight Cry8-like-binding proteins were obtained from pulldown assays conducted with BBMVs. The LC-MS/MS data for these proteins were successfully matched with the H. oblita transcriptome, and BLASTX results identified five proteins as serine protease, transferrin-like, uncharacterized protein LOC658236 of Tribolium castaneum, ATPase catalytic subunit, and actin. These identified Cry8-like-binding proteins were different from those confirmed previously as receptors for Cry1A proteins in lepidopteran insect species, such as aminopeptidase, alkaline phosphatase, and cadherin.
IMPORTANCE Holotrichia oblita is one of the main soil-dwelling pests in China. The larvae damage the roots of crops, resulting in significant yield reductions and economic losses. H. oblita is difficult to control, principally due to its soil-dwelling habits. In recent years, some Cry8 toxins from Bacillus thuringiensis were shown to be active against this pest. Study of the mechanism of action of these Cry8 toxins is needed for their effective use in the control of H. oblita and for their future utilization in transgenic plants. Our work provides important basic data and promotes understanding of the insecticidal mechanism of Cry8 proteins against H. oblita larvae.
KEYWORDS: Bacillus thuringiensis, Cry8-like, Holotrichia oblita, binding proteins
INTRODUCTION
White grubs (larvae of the Scarabaeidae family) are important soil-dwelling insect pests throughout the world; the larvae feed on the roots of plants and the adults feed on the aerial parts of plants (1). In China, Holotrichia oblita is one of the main white grub pests and damages many crops, including sweet potatoes, soybeans, and peanuts, causing significant yield reductions and economic losses (2, 3). Control of white grubs is difficult, principally due to their soil-dwelling habits, which make it difficult to establish when and where white grubs are endangering plants before the crops show symptoms of damage, and the difficulty of delivering chemical pesticides against this pest (4, 5). Transgenic plants could be an alternative strategy to control soil-dwelling pests such as white grubs, since these plants produce insecticidal proteins continuously during the whole life cycle (6, 7). Insecticidal genes used in transgenic plants are mostly screened from Bacillus thuringiensis (Bt) bacteria (8). These Gram-positive, spore-forming bacteria produce different Cry proteins (insecticidal crystal proteins), which are highly specific against the target insects and are biodegradable, as the main virulence factors (9).
In previous work, Bt strain HBF-18 (CGMCC 2070) was shown to be toxic against H. oblita larvae and to contain two cry genes of the three-domain cry gene (3d-Cry) family, namely, cry8Ga (10) and cry8-like (11). The cry8Ga gene encodes a polypeptide of 1,157 amino acids, with a theoretical molecular size of 131.2 kDa (10), while the cry8-like gene encodes a polypeptide of 738 amino acids without a complete C-terminal crystal region, with a theoretical molecular size of 83.8 kDa (11). A Cry8-like hybrid protein constructed with the Cry8Ea C-terminal region fused to Cry8-like is expressed in Bt cells, forming crystals in the crystal-negative mutant Bt strain HD73−. The hybrid protein showed 3-fold higher activity against H. oblita larvae than did the Cry8Ga protein (11) and therefore is a candidate for the development of transgenic plants resistant to this insect pest.
To date, the mechanism of action of 3d-Cry proteins has been studied mainly with Cry1A toxins that are toxic to some lepidopteran insects. It is widely accepted that protease-activated Cry1A toxins must bind to specific receptors on the microvillus membranes of the midgut epithelium (12, 13). According to the pore formation model, the 3d-Cry proteins bind to specific receptors such as cadherin, aminopeptidase, and alkaline phosphatase, resulting in the formation of an oligomeric structure that is inserted into the midgut cell membrane, forming lytic pores and leading to cell death (14). In the signal transduction model, 3d-Cry proteins bind to the cadherin receptor, inducing a cell death signaling pathway (15). Based on their similar three-dimensional structures, similar toxic mechanisms for all 3d-Cry proteins were presumed (16). However, there is still a lack of data on the mode of action of Cry8-like and Cry8Ga proteins, which are similar to Cry8Ea, a typical 3d-Cry protein (17).
In a previous work, we identified the Cry8Ea-binding proteins from the related pest Holotrichia parallela by using pulldown assays and liquid chromatography-tandem mass spectrometry (LC-MS/MS) protein sequencing, identifying the protein sequences that matched the assembled midgut tissue transcriptome from H. parallela (18). Here, we analyzed the binding of Cry8-like and Cry8Ga to H. oblita midgut brush border membrane vesicles (BBMVs), and we identified the Cry8-like-binding proteins on the BBMVs of H. oblita larvae by a similar approach. We did not analyze the binding of Cry8Ea, since this toxin is nontoxic to H. oblita. In this work, we also report the new midgut tissue transcriptome sequence of H. oblita. Our work provides important basic data and promotes understanding of the insecticidal mechanism of Cry8 proteins against H. oblita larvae.
RESULTS
Binding assays with fluorescent DyLight488-labeled Cry8-like toxin.
Initial attempts at binding assays were conducted with biotinylated Cry8-like, but biotinylation greatly affected the binding of Cry8-like to BBMVs (data not shown). As an alternative, DyLight488-labeled Cry8-like was used to perform binding assays. Chymotrypsin activation of Cry8-like protein resulted in a protease-resistant core fragment of approximately 66 kDa. The activated toxin was labeled, and the DyLight488-labeled Cry8-like protein (Fig. 1) was purified by anion-exchange chromatography and used for subsequent assays.
FIG 1.
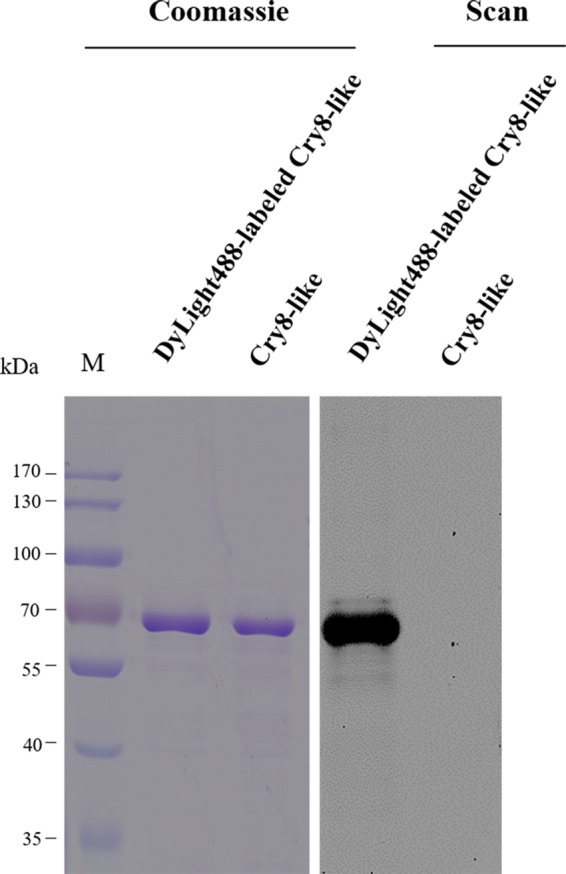
Analysis of Cry8-like protein purification and labeling with DyLight488. The purified protease-resistant core of Cry8-like protein was generated after digestion with chymotrypsin (Cry8-like). Then, this protein was labeled with the fluorescent DyLight488 dye (DyLight488-labeled Cry8-like). Scanning (Scan) for green fluorescence (excitation at 488 nm and emission at 518 nm) was performed to check the labeling of Cry8-like in a Typhoon Trio scanner. The numbers at the left indicate the molecular sizes (M) of protein markers, in kilodaltons.
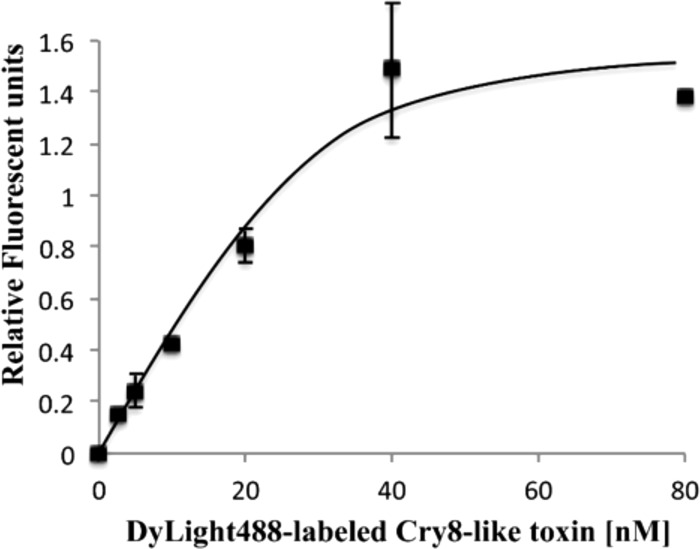
Specific binding was determined by incubating a fixed amount of BBMV proteins from H. oblita with increasing concentrations of DyLight488-labeled Cry8-like protein in the presence or absence of excess unlabeled Cry8-like. DyLight488-labeled Cry8-like exhibited specific and saturable binding to H. oblita BBMVs (Fig. 2). Nonspecific binding accounted for 20% of total binding.
FIG 2.
Specific binding of DyLight488-labeled Cry8-like to BBMVs from H. oblita. A fixed amount of BBMV proteins (2 μg) was incubated with increasing concentrations (from 0 to 80 nM) of DyLight488-labeled Cry8-like. Nonspecific binding was analyzed in the presence of a 100-fold molar excess of unlabeled homologous protein. Nonspecific binding was subtracted from total binding, resulting in specific binding, which is shown. Each data point represents the mean calculated from at least two independent experiments. Error bars represent standard errors.
Competition binding assays.
Competition binding experiments were conducted by incubating H. oblita BBMVs with a fixed amount of DyLight488-labeled Cry8-like in the presence of increasing concentrations of unlabeled homologous and heterologous competitors (Cry8-like and Cry8Ga, respectively). The binding of DyLight488-labeled Cry8-like protein to BBMVs of H. oblita was competitively inhibited by increasing concentrations of unlabeled Cry8-like competitor (Fig. 3), confirming that Cry8-like binds specifically to BBMVs from H. oblita. This binding was also competitively inhibited by unlabeled Cry8Ga competitor, suggesting that Cry8Ga shares binding sites on H. oblita BBMVs with Cry8-like protein (Fig. 3).
FIG 3.
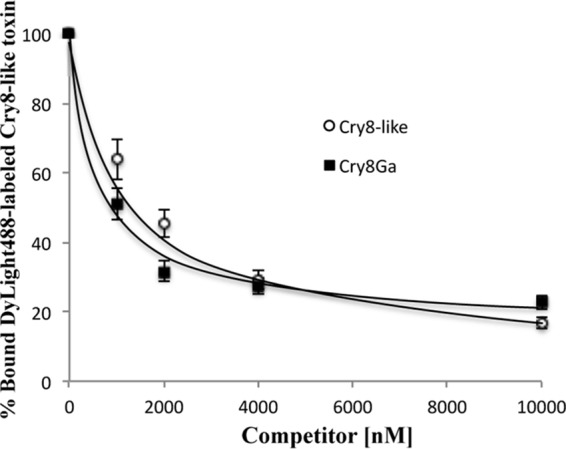
Homologous and heterologous competition binding experiments analyzing the binding of DyLight488-labeled Cry8-like to BBMVs from H. oblita. Curves represents the total binding of DyLight488-labeled Cry8-like (20 nM) to BBMV proteins (2 μg) with increasing amounts of unlabeled Cry8-like or Cry8Ga proteins as competitors, as indicated. After binding interactions, the proteins were analyzed by 10% SDS-PAGE and the gels were scanned for green fluorescence (excitation at 488 nm and emission at 518 nm) in a Typhoon Trio scanner. Densitometric analysis of the fluorescence signals collected from the gels was conducted with ImageJ software, to quantify binding. Each data point depicts the mean calculated from at least two independent experiments. Error bars depict standard errors.
Assembly of transcriptome.
To obtain the midgut tissue transcriptome of H. oblita, transcriptome sequencing (RNA-Seq) data were obtained from the third-instar larva midgut, as described in Materials and Methods. In total, 52,196,840 clean reads, with an accumulated length of 25,215,935,000 bp, were obtained after the removal of low-quality reads. All clean reads of high quality were assembled de novo, by the Trinity program, into 41,000 unigenes and 80,107,333 total bases, with an average length of 1,953.8 bp and a N50 of 3,039 bp. Among the unigenes, the longest and shortest unigenes were 31,852 bp and 201 bp, respectively (see Table S1 and Fig. S1 in the supplemental material).
All of the assembled unigenes were annotated by searching the NCBI nonredundant protein (Nr) database using BLASTX (E values of <10−5). The results showed that, among the 41,000 unigenes, 25,333 distinct sequences (61.79%) matched known proteins. For species distribution, the majority of matched unigenes (62.47%) had the greatest similarity to those of Tribolium castaneum, followed by Dendroctonus ponderosae (5.74%), Stegodyphus mimosarum (2.29%), and Zootermopsis nevadensis (2.15%). Unigenes from species of other organisms or classes had lower levels of similarity to those of H. oblita. A total of 6,930 unigenes, representing 27.36% of unique transcripts, had less than 2% similarity to those of other species (Fig. S2). We also compared the H. oblita transcriptome with the previously reported transcriptome of the related species H. parallela (18); of 41,000 unigenes identified in H. oblita, only 27.42% (11,243 unigenes) were shared with H. parallela (Fig. 4).
FIG 4.
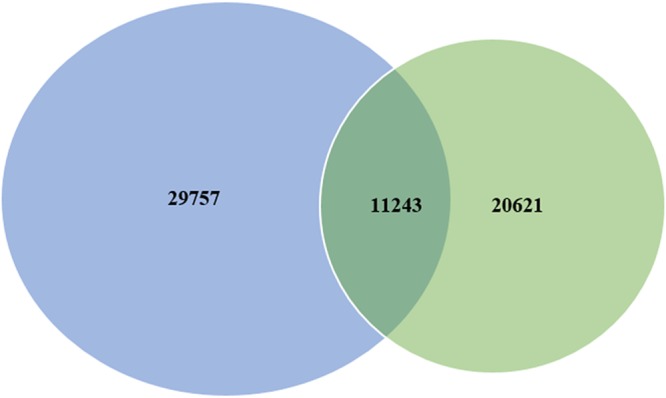
Comparison of the midgut tissue transcripts of H. oblita (blue) and H. parallela (green). The numbers given are unigenes.
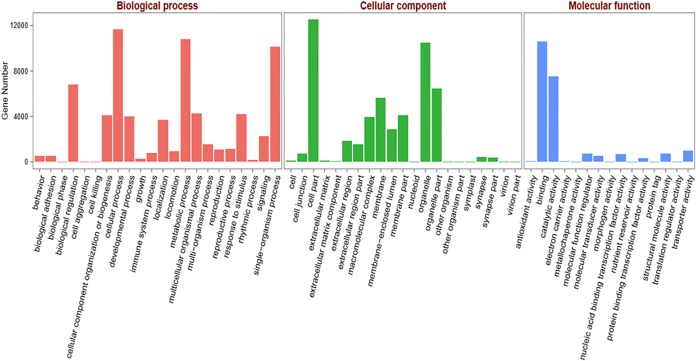
According to the BLASTX results from the Nr database search, functional classification of H. oblita unigenes was performed by Gene Ontology (GO) annotation. The GO terms were used to classify gene functions. In this study, among the 25,333 annotated unigenes in the Nr database, 16,318 were assigned GO terms; some were assigned multiple GO terms. These GO terms were divided into 58 subcategories, which were distributed in three categories (Fig. 5). Among the subcategories, 22 were distributed in the biological processes category, 21 in the cellular components category, and 15 in the molecular functions category. The majority of GO terms were distributed in the category of biological processes (16,820 terms [74.09%]), followed by cellular components (3,450 terms [15.20%]), and molecular functions (2,432 terms [10.71%]).
FIG 5.
GO assignments for the H. oblita transcriptome. The results were divided into three main categories, i.e., biological processes, cellular components, and molecular functions. The y axis represents the numbers of unigenes.
Detection of midgut Cry8-like-binding proteins in H. oblita.
To analyze the midgut Cry8-like-binding proteins in H. oblita, a pulldown assay was conducted as described in Materials and Methods. Solubilized BBMV proteins were incubated with Cry8-like-Sepharose beads to precipitate midgut Cry8-like-binding proteins. After incubation, the sample was washed by centrifugation to remove all unbound proteins. The precipitated proteins were dissociated by 5 min of boiling in loading buffer and were separated by 10% SDS-PAGE (Fig. 6). Eight bands of H. oblita midgut proteins that bound Cry8-like protein, ranging in molecular size from about 40 kDa to 200 kDa, were cut from the gel and subsequently analyzed by LC-MS/MS.
FIG 6.
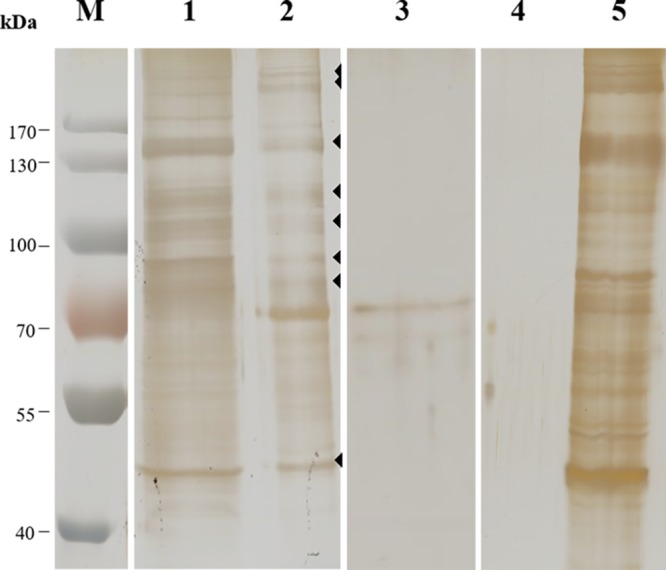
SDS-PAGE of midgut BBMV proteins from H. oblita. Lane 1, whole solubilized BBMV proteins from H. oblita; lane 2, H. oblita BBMV proteins bound to Cry8-like (the arrowheads indicate the protein bands bound to Cry8-like that were cut out and analyzed by LC-MS/MS); lane 3, Cry8-like-Sepharose beads without incubation with solubilized BBMV proteins; lane 4, BBMV proteins bound to beads without coupled Cry8-like; lane 5, unbound BBMV proteins after incubation with negative-control beads. The numbers at the left indicate the molecular sizes (M) of protein markers, in kilodaltons. The gel was silver stained.
Identification of midgut Cry8-like-binding proteins by LC-MS/MS.
The midgut Cry8-like-binding proteins in H. oblita were identified by LC-MS/MS, and the LC-MS/MS data were searched using Mascot MS/MS Ion Search, with the H. oblita midgut unigenes as the database. Table 1 shows the three proteins with better scores for each band. The identified unigenes with the better scores were searched for by BLASTX in the Nr database, and most of them (except the unigene comp46_seq0) demonstrated significant alignments. The BLASTX results showed that the Cry8-like-binding proteins in the H. oblita midgut were identified as serine protease, transferrin-like isoforms, uncharacterized protein LOC658236 of Tribolium castaneum, ATPase catalytic subunit, and actin (Table 1).
TABLE 1.
Identification of H. oblita Cry8-like-binding proteins
| Band no. | Mascot score | Unigene designation | BLAST score | Query cover (%) | E value | Identity (%) | GenBank accession no. | Description |
|---|---|---|---|---|---|---|---|---|
| 1 | 1,254 | comp46_seq0 | NAa | NA | NA | NA | NA | NA |
| 348 | comp410_seq1 | 4,590 | 88 | 0 | 93 | XP_018334652.1 | Predicted: spectrin α-chain isoform X7 (Agrilus planipennis) | |
| 338 | comp15_seq13 | 170 | 70 | 3e−40 | 25 | XP_013110288.1 | Predicted: serine protease 53-like (Stomoxys calcitrans) | |
| 2 | 1,208 | comp46_seq0 | NA | NA | NA | NA | NA | NA |
| 359 | comp15_seq13 | 170 | 70 | 3e−40 | 25 | XP_013110288.1 | Predicted: serine protease 53-like (Stomoxys calcitrans) | |
| 347 | comp8785_seq0 | 2,811 | 87 | 0 | 73 | XP_018318790.1 | Predicted: contactin-associated protein like 5-1 (Agrilus planipennis) | |
| 3 | 1,504 | comp15_seq13 | 170 | 70 | 3e−40 | 25 | XP_013110288.1 | Predicted: serine protease 53-like (Stomoxys calcitrans) |
| 359 | comp820_seq0 | 1,883 | 75 | 0 | 95 | XP_017784204.1 | Predicted: sodium/potassium-transporting ATPase subunit α isoform X9 (Nicrophorus vespilloides) | |
| 315 | comp2533_seq0 | 1,411 | 57 | 0 | 63 | XP_017773865.1 | Predicted: Golgi apparatus protein 1 (Nicrophorus vespilloides) | |
| 4 | 3,355 | comp2_seq10 | 310 | 25 | 2e−85 | 30 | XP_008199942.1 | Predicted: transferrin-like isoform X2 (Tribolium castaneum) |
| 1,105 | comp571_seq1 | 1,417 | 85 | 0 | 83 | XP_017773111.1 | Predicted: V-type proton ATPase 116-kDa subunit α isoform X3 (Nicrophorus vespilloides) | |
| 852 | comp15_seq13 | 170 | 70 | 3e−40 | 25 | XP_013110288.1 | Predicted: serine protease 53-like (Stomoxys calcitrans) | |
| 5 | 3,539 | comp1729_seq0 | 325 | 78 | 2e−94 | 31 | XP_008199942.1 | Predicted: transferrin-like isoform X2 (Tribolium castaneum) |
| 2,836 | comp162_seq0 | 515 | 88 | 5e−166 | 37 | XP_969735.1 | Predicted: uncharacterized protein LOC658236 (Tribolium castaneum) | |
| 1,868 | comp131_seq1 | 1,721 | 78 | 0 | 71 | XP_018331598.1 | Predicted: protein mesh isoform X3 (Agrilus planipennis) | |
| 6 | 5,477 | comp162_seq0 | 515 | 88 | 5e−166 | 37 | XP_969735.1 | Predicted: uncharacterized protein LOC658236 (Tribolium castaneum) |
| 2,164 | comp1259_seq0 | 1,066 | 90 | 0 | 73 | XP_973530.1 | Predicted: trifunctional enzyme subunit α, mitochondrial (Tribolium castaneum) | |
| 1,293 | comp2336_seq0 | 669 | 65 | 0 | 38 | XP_008198168.2 | Predicted: integrin alpha-PS3 (Tribolium castaneum) | |
| 7 | 3,580 | comp287_seq0 | 1,202 | 45 | 0 | 93 | XP_976188.1 | Predicted: V-type proton ATPase catalytic subunit A (Tribolium castaneum) |
| 2,612 | comp94_seq3 | 234 | 33 | 8e−63 | 34 | XP_017771966.1 | Predicted: vanin-like protein 2 (Nicrophorus vespilloides) | |
| 1,221 | comp177_seq0 | 1,162 | 46 | 0 | 95 | XP_018566266.1 | Predicted: 70-kDa heat shock protein cognate 3 (Anoplophora glabripennis) | |
| 8 | 4,567 | comp9_seq0 | 787 | 27 | 0 | 100 | NP_511052.1 | Actin 5C, isoform B (Drosophila melanogaster) |
| 1,422 | comp335_seq2 | 612 | 41 | 0 | 66 | XP_018570553.1 | Predicted: juvenile hormone epoxide hydrolase 1-like (Anoplophora glabripennis) | |
| 929 | comp924_seq0 | 511 | 76 | 6e−175 | 62 | XP_975769.1 | Predicted: cytochrome b-c1 complex subunit 2, mitochondrial (Tribolium castaneum) |
NA, not applicable (no match found).
DISCUSSION
Screening of Bt strains and cloning of novel insecticidal protein genes toxic to H. oblita larvae have resulted in the identification of Bt strains active against H. oblita larvae. One is the Bt strain HBF-18, which was identified in our laboratory (10), and the other is Bt strain B-JJX (19). Besides the cry8Ga and cry8-like genes, the cry8Ab gene cloned from Bt strain B-JJX was shown to be effective against H. oblita larvae (19). Study of the mechanism of action of these Cry8 proteins is needed for their effective use in the control of H. oblita and for their future utilization in transgenic plants.
The characteristics of Cry8-like and Cry8G binding to BBMV proteins of H. oblita larvae with fluorescent DyLight488-labeled Cry8-like showed that Cry8-like bound specifically to BBMVs from H. oblita, since homologous competition assays with unlabeled Cry8-like as the competitor confirmed the existence of specific binding sites for Cry8-like on H. oblita BBMVs. Heterologous competition assay with unlabeled Cry8Ga as the competitor revealed that Cry8Ga shared binding sites with Cry8-like on H. oblita BBMVs. However, the two proteins (Cry8-like and Cry8Ga) share only 29.8% sequence identity in domain II, which is the main determinant of specificity in the Cry protein family (20). Cry8-like shows 3-fold greater toxicity than Cry8Ga (11), confirming that Cry8-like would be the best choice for expression in transgenic plants for the control of H. oblita larvae.
Receptors play important roles in the activity of Cry proteins and in the determination of their insect specificity (13). Therefore, it is important to further understand the mechanism of action of these toxins, including the identification and characterization of Cry-toxin-binding proteins in the midgut of susceptible larvae. In this study, we identified Cry8-like-binding proteins in the midgut of H. oblita larvae with pulldown assays and protein sequencing by LC-MS/MS. In addition, a midgut transcriptome was constructed by high-throughput RNA sequencing, to enhance the accuracy of identification. Interestingly, the assembled transcriptome from H. oblita shared only 27% of its unigenes with the related insect species H. parallela (18). These data showed that, for accurate identification of Cry8-binding proteins in H. oblita, it was important to assemble the midgut transcriptome of this insect. Eight Cry8-like-binding proteins were successfully matched to H. oblita midgut unigenes (Table 1) by Mascot software, and five were identified by BLASTX. In a previous work, we identified 10 Cry8Ea-binding proteins from H. parallela using a similar strategy (18). In addition, Zhou et al. identified ABCC2 (ATP-binding cassette transporter subfamily C protein) as a Cry1Ac-binding protein in Helicoverpa armigera using the same methodology (21). These Cry8-like-binding proteins in H. oblita, as well as the Cry8Ea-binding H. parallela proteins (18), were different from those confirmed previously as receptors for Cry1A proteins in lepidopteran insect species, such as aminopeptidase (22), alkaline phosphatase (23), and cadherin (24).
H. oblita is not sensitive to Cry8E (the highest concentration tested was 100 μg/g soil); therefore, we expected that Cry8-like-binding proteins in H. oblita would differ from those identified previously as binding Cry8E in H. parallela (18). However, it is interesting to note that, in H. parallela and H. oblita, the same uncharacterized protein (LOC658236) was identified as a protein binding Cry8Ea (18) and Cry8-like (this work), suggesting that this protein could be important for Cry8 protein toxicity in these insects. Moreover, we analyzed potential transmembrane regions with the DAS-TMfilter server (http://mendel.imp.ac.at/sat/DAS/DAS.html) and identified a putative signal peptide and a transmembrane region located in residues 733 to 744 of the Tribolium homolog protein. This could suggest that this predicted protein may serve as a membrane-bound receptor for Cry8 proteins. In addition, we identified other Cry8-like-binding proteins in H. oblita that were not identified among the Cry8E-binding proteins from H. parallela, such as transferrin-like protein. We report here the identification of an insect transferrin as a Cry-binding protein (Table 1). Transferrins are glycoproteins that bind iron and control the levels of free iron in biological fluids; in insects, they have been shown to be involved in iron transport, as antioxidants, as vitellogenic proteins, and as antibiotic agents (25). Moreover, silencing of a transferrin gene in the lepidoptera Plutella xylostella resulted in lower tolerance to Bt infections (26). However, the roles of all Cry8-like-binding proteins identified in this study and the mode of action of Cry8-like still need to be analyzed by further functional analysis experiments, such as RNA interference silencing studies. The successful characterization of the binding proteins suggested that the transcriptome of the H. oblita midgut is important for understanding the insecticidal mechanism of Cry proteins, especially because there were insufficient genetic data in public databases (only 30 H. oblita sequences in the Nr database).
In conclusion, this study provides evidence to demonstrate that Cry8-like binds specifically to binding sites on H. oblita BBMVs and Cry8Ga shares binding sites with Cry8-like. In addition, we constructed a high-quality midgut transcriptome of H. oblita larvae, which will provide important basic sequence data for further studies, including studies of the mechanism of action of Cry toxins against H. oblita larvae. We showed here that the midgut transcriptome was useful for the identification of five Cry8-like-binding proteins that might be involved in the mode of action of this protein. Current experiments are focused on confirming the role of the Cry8-like-binding proteins of H. oblita larvae identified in this study in the mechanism of action of Cry8-like toxin.
MATERIALS AND METHODS
Insects.
H. oblita adults were obtained in Cangzhou, China. The females and males underwent mixed feeding with fresh leaves of Ulmus pumila (elm tree) in a rearing room maintained at 25°C and 18 to 20% relative humidity, with a 12-h light/12-h dark cycle. The eggs were hatched in soil, and the larvae were fed germinated wheat, in the same environment, until the third instar.
Protein preparation.
The Cry proteins (Cry8-like hybrid toxin and Cry8Ga) used in this study were produced in the recombinant HD8CLC8E and HD8G Bt strains, respectively. The recombinant host and the construction of the expression vector were described previously (10, 11). The HD8CLC8E and HD8G strains were grown in LB medium, in a shaker flask (220 rpm), at 30°C until cell lysis. Cry8 protoxin was digested for 2 h at 37°C with 10% chymotrypsin (wt/wt) in 50 mM Na2CO3-NaHCO3 (pH 10.0). The activated proteins were purified by anion-exchange chromatography on an ÄKTA avant system, at room temperature. The activated proteins were injected into a 5-ml HiTrap Q HP column (GE Healthcare) that had been preequilibrated with 5 column volumes (CVs) of equilibration buffer (50 mM Na2CO3-NaHCO3 [pH 10.0]). The column was then washed with 5 CVs of equilibration buffer. Elution was conducted with a linear gradient of 0 to 100% elution buffer (i.e., equilibration buffer with 1.0 M NaCl) within 20 min, at a flow rate of 2 ml/min, with 0.5 ml per fraction. The fractions containing the activated toxins were analyzed by 10% SDS-PAGE and pooled, and the gel was stained with Coomassie blue. The concentration of the target protein was measured with the Bradford method, with bovine serum albumin (BSA) as the standard.
Protein labeling.
Chymotrypsin-activated Cry8-like protein (1 mg) was labeled with the fluorescent DyLight488 antibody labeling kit (Thermo Scientific, Wilmington, DE, USA), as indicated by the manufacturer. Briefly, purified Cry8-like protein was incubated for 1 h at room temperature with the DyLight488 reagent in phosphate-buffered saline (PBS) (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4 [pH 7.4]), with protection from light. Free DyLight488 dye was removed using a 5-ml HiTrap desalting column (GE Healthcare). The labeled Cry8-like protein was quantified with the Bradford method, with BSA as the standard, and was stored in aliquots at −20°C until it was used for subsequent assays.
BBMV preparation.
Third-instar H. oblita larvae were used to prepare BBMVs. Midguts were dissected on ice, and gut contents were removed by washing with 0.9% NaCl, followed by washing with PBS. The dissected midgut tissue was frozen immediately in liquid nitrogen and was preserved at −80°C until required. BBMVs were prepared as reported (27). The total BBMV protein amount was quantified using the Bradford method, with BSA as the standard, and the BBMV preparations were stored at −80°C until they were used. To determine the quality of BBMV preparations, we estimated the enrichment of aminopeptidase N (APN) specific activity in the final purified BBMV preparations, compared with the level in the initial homogenate; the results showed 5-fold enrichment of APN activity.
Binding assays with DyLight488-labeled Cry8-like.
Saturation binding assays were conducted with the aim of analyzing specific protein binding and determining an appropriate amount of DyLight488-labeled Cry8-like for competition binding assays. Two micrograms of H. oblita BBMV proteins was incubated for 1 h at room temperature with increasing concentrations (0 to 80 nM) of DyLight488-labeled Cry8-like protein, in a final volume of 100 μl of binding buffer (PBS with 0.1% BSA and 0.1% Tween 20). After incubation, the protein bound to the BBMVs was separated from the free protein by centrifugation (14,000 × g for 10 min at 4°C), and the pellets were washed three times with 500 μl of cold binding buffer by using the same centrifugation conditions. The final pellets were suspended in 15 μl of loading buffer (PBS plus Laemmli lytic mixture) (28), boiled for 5 min, and analyzed by 10% SDS-PAGE. After electrophoretic separation, total binding was determined by scanning the gel for green fluorescence (excitation at 488 nm and emission at 518 nm) in a Typhoon Trio scanner (GE Healthcare). To determine nonspecific binding, an excess (100-fold) of unlabeled Cry8-like protein was added into the binding reaction. Total binding minus nonspecific binding is equal to specific binding. The experiments were repeated at least twice.
Competition binding assays.
Competition binding assays were conducted by incubating 2 μg of H. oblita BBMV proteins for 1 h at room temperature with 20 nM DyLight488-labeled Cry8-like protein in the presence of increasing concentrations (0 to 10,000 nM) of unlabeled homologous and heterologous competitors (Cry8-like protein and Cry8Ga protein, respectively), in a final volume of 100 μl of the same binding buffer. The samples were subsequently processed as described above.
Densitometric analysis of the fluorescent signals collected from the gel was conducted with ImageJ software to quantify binding. Competition binding experiments were replicated at least twice. Data were analyzed for significance and plotted with Microsoft Excel 2013.
Pulldown experiments.
To capture midgut BBMV Cry8-like-binding proteins, the protein buffer was changed to pulldown buffer (0.2 M NaHCO3, 0.5 M NaCl [pH 8.3]) with a 5-ml HiTrap desalting column (GE Healthcare), and Cry8-like was coupled to N-hydroxysuccinimide (NHS)-activated Sepharose (GE Healthcare), as described by the manufacturer. We chose NHS-activated Sepharose because it provides a spacer arm and is suitable for immobilizing protein and peptide ligands.
Pulldown experiments were conducted to capture Cry8-like-binding BBMV proteins as described previously (18). Two hundred micrograms of BBMV proteins was solubilized in solubilization buffer (20 mM Tris, 5 mM EGTA, 1% CHAPS [3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate], 150 mM NaCl [pH 7.5]), and 50 μl of Cry8-like-Sepharose beads was incubated with the 200 μg of solubilized BBMV proteins for 1 h at 4°C, in a final volume of 200 μl. The unbound BBMV proteins in the mixture were removed by centrifugation (1,000 × g for 1 min at 4°C). The beads were washed with washing buffer (PBS with 1.0 M NaCl) and subsequently washed with PBS to remove all free proteins. The remaining bound proteins were considered Cry8-like-binding proteins; they were separated from the beads by boiling in loading buffer for 5 min and subjected to 10% SDS-PAGE.
Identification of Cry8-like-binding proteins.
After silver staining, protein bands on the SDS-PAGE gel were cut out for subsequent identification by LC-MS/MS, as described elsewhere (21). The LC-MS/MS data were searched with Mascot MS/MS Ion Search, with the H. oblita midgut unigenes as the database.
Assembly of H. oblita midgut tissue transcriptome.
The H. oblita transcriptome was obtained by RNA-Seq using the Illumina HiSeq 2000 platform, as described previously for H. parallela (18). Third-instar H. oblita larvae were used for isolation of total RNA, using TRIzol reagent (Life Technologies, Carlsbad, CA, USA). Midguts were dissected on ice, and the gut contents were removed by washing with 0.9% NaCl. After transfer to a 1.5-ml Eppendorf tube, the midgut tissue was quickly homogenized in TRIzol reagent. The extraction and purification of total RNA were conducted according to the manufacturer's protocol. The quality of total RNA was analyzed with a NanoDrop ND-1000 spectrophotometer (Thermo Scientific), showing an A260/A280 ratio of 1.97.
RNA sequencing was conducted by RNA-Seq using the Illumina HiSeq 2000 platform, as described previously (18). After sequencing, the raw data were processed with Bcl2fastq conversion software (Illumina) and then referred to as “raw reads,” which were saved in the fastq format. Quality control of raw sequences was conducted to remove low-quality reads, as described previously (29). The resulting clean reads obtained from H. oblita midgut tissue were used for assembly of the transcriptome. De novo assembly was conducted with the Trinity program (30), as described previously (31).
Annotation of unigenes can provide functional information. For functional annotation, we first aligned unigene sequences with the NCBI Nr database with BLASTX, using an E value cutoff value of 10−5 to search for homologs. In addition, we performed GO annotation with the Blast2GO program (32), based on the results of Nr annotation. GO functional classification was performed with WEGO software (33) to determine the overall distribution of gene functions.
Accession number(s).
The clean reads produced from H. oblita midgut tissue, as reported here, were deposited in the NCBI Sequence Read Archive and assigned accession number SRR3211072.
Supplementary Material
ACKNOWLEDGMENTS
This work was partially funded by the National Natural Science Foundation of China (grants 31301731 and 31428020) and the National Science and Technology Major Project (grant 2014ZX08009-013B).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00541-17.
REFERENCES
- 1.Harrison JDG, Wingfield MJ. 2016. A taxonomic review of white grubs and leaf chafers (Coleoptera: Scarabaeidae: Melolonthinae) recorded from forestry and agricultural crops in sub-Saharan Africa. Bull Entomol Res 106:141–153. doi: 10.1017/S0007485315000565. [DOI] [PubMed] [Google Scholar]
- 2.Liang C, Guo W, Lu X, Li R, Yu H, Wang W, Zhao D, Xu D. 2015. The annual occurrence dynamics and its influencing factors of Holotrichia oblita (Faldermann) adults. Plant Protect 41:169–172. (In Chinese.) [Google Scholar]
- 3.Wei X, Xu X, Deloach CJ. 1995. Biological control of white grubs (Coleoptera: Scarabaeidae) by larvae of Promachus yesonicus (Diptera: Asilidae) in China. Biol Control 5:290–296. doi: 10.1006/bcon.1995.1036. [DOI] [Google Scholar]
- 4.Peters A. 2000. Susceptibility of Melolontha melolontha to Heterorhabditis bacteriophora, H. megidis and Steinernema glaseri. Integrated control of soil pest subgroup “Melolontha” conference proceeding IOBC/WPRS. Bull OILB/SROP 23:39–45. [Google Scholar]
- 5.Schnetter W, Mittermuller R, Froschle M. 1996. Control of the cockchafer Melolontha melolontha in the Kraichgau with NeemAzal-T/S. Bull OILB/SROP 19:95–99. [Google Scholar]
- 6.Betz FS, Hammond BG, Fuchs RL. 2000. Safety and advantages of Bacillus thuringiensis-protected plants to control insect pests. Regul Toxicol Pharmacol 32:156–173. doi: 10.1006/rtph.2000.1426. [DOI] [PubMed] [Google Scholar]
- 7.Shelton AM, Zhao JZ, Roush RT. 2002. Economic, ecological, food safety, and social consequences of the deployment of Bt transgenic plants. Annu Rev Entomol 47:845–881. doi: 10.1146/annurev.ento.47.091201.145309. [DOI] [PubMed] [Google Scholar]
- 8.Schnepf E, Crickmore N, Van Rie J, Lereclus D, Baum J, Feitelson J, Zeigler DR, Dean DH. 1998. Bacillus thuringiensis and its pesticidal crystal proteins. Microbiol Mol Biol Rev 62:775–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Raymond B, Johnston PR, Nielsen-LeRoux C, Lereclus D, Crickmore N. 2010. Bacillus thuringiensis: an impotent pathogen? Trends Microbiol 18:189–194. doi: 10.1016/j.tim.2010.02.006. [DOI] [PubMed] [Google Scholar]
- 10.Shu C, Yan G, Wang R, Zhang J, Feng S, Huang D, Song F. 2009. Characterization of a novel cry8 gene specific to Melolonthidae pests: Holotrichia oblita and Holotrichia parallela. Appl Microbiol Biotechnol 84:701–707. doi: 10.1007/s00253-009-1971-2. [DOI] [PubMed] [Google Scholar]
- 11.Bi Y, Zhang Y, Shu C, Crickmore N, Wang Q, Du L, Song F, Zhang J. 2015. Genomic sequencing identifies novel Bacillus thuringiensis Vip1/Vip2 binary and Cry8 toxins that have high toxicity to Scarabaeoidea larvae. Appl Microbiol Biotechnol 99:753–760. doi: 10.1007/s00253-014-5966-2. [DOI] [PubMed] [Google Scholar]
- 12.Bravo A, Jansens S, Peferoen M. 1992. Immunocytochemical localization of Bacillus thuringiensis insecticidal crystal proteins in intoxicated insects. J Invertebr Pathol 60:237–246. doi: 10.1016/0022-2011(92)90004-N. [DOI] [Google Scholar]
- 13.Pardo-Lopez L, Soberon M, Bravo A. 2013. Bacillus thuringiensis insecticidal three-domain Cry toxins: mode of action, insect resistance and consequences for crop protection. FEMS Microbiol Rev 37:3–22. doi: 10.1111/j.1574-6976.2012.00341.x. [DOI] [PubMed] [Google Scholar]
- 14.Soberon M, Gill SS, Bravo A. 2009. Signaling versus punching hole: how do Bacillus thuringiensis toxins kill insect midgut cells? Cell Mol Life Sci 66:1337–1349. doi: 10.1007/s00018-008-8330-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang X, Candas M, Griko NB, Taussig R, Bulla LA Jr. 2006. A mechanism of cell death involving an adenylyl cyclase/PKA signaling pathway is induced by the Cry1Ab toxin of Bacillus thuringiensis. Proc Natl Acad Sci U S A 103:9897–9902. doi: 10.1073/pnas.0604017103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bravo A, Gomez I, Porta H, Garcia-Gomez BI, Rodriguez-Almazan C, Pardo L, Soberon M. 2013. Evolution of Bacillus thuringiensis Cry toxins insecticidal activity. Microb Biotechnol 6:17–26. doi: 10.1111/j.1751-7915.2012.00342.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guo S, Ye S, Liu Y, Wei L, Xue J, Wu H, Song F, Zhang J, Wu X, Huang D, Rao Z. 2009. Crystal structure of Bacillus thuringiensis Cry8Ea1: an insecticidal toxin toxic to underground pests, the larvae of Holotrichia parallela. J Struct Biol 168:259–266. doi: 10.1016/j.jsb.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 18.Shu C, Tan S, Yin J, Soberon M, Bravo A, Liu C, Geng L, Song F, Li K, Zhang J. 2015. Assembling of Holotrichia parallela (dark black chafer) midgut tissue transcriptome and identification of midgut proteins that bind to Cry8Ea toxin from Bacillus thuringiensis. Appl Microbiol Biotechnol 99:7209–7218. doi: 10.1007/s00253-015-6755-2. [DOI] [PubMed] [Google Scholar]
- 19.Zhang Y, Zheng G, Tan J, Li C, Cheng L. 2013. Cloning and characterization of a novel cry8Ab1 gene from Bacillus thuringiensis strain B-JJX with specific toxicity to scarabaeid (Coleoptera: Scarabaeidae) larvae. Microbiol Res 168:512–517. doi: 10.1016/j.micres.2013.03.003. [DOI] [PubMed] [Google Scholar]
- 20.Adang MJ, Crickmore N, Jurat-Fuentes JL. 2014. Diversity of Bacillus thuringiensis crystal toxins and mechanism of action. Adv Insect Physiol 47:39–87. doi: 10.1016/B978-0-12-800197-4.00002-6. [DOI] [Google Scholar]
- 21.Zhou Z, Wang Z, Liu Y, Liang G, Shu C, Song F, Zhou X, Bravo A, Soberon M, Zhang J. 2016. Identification of ABCC2 as a binding protein of Cry1Ac on brush border membrane vesicles from Helicoverpa armigera by an improved pull-down assay. Microbiologyopen 5:659–669. doi: 10.1002/mbo3.360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Knight PJ, Carroll J, Ellar DJ. 2004. Analysis of glycan structures on the 120 kDa aminopeptidase N of Manduca sexta and their interactions with Bacillus thuringiensis Cry1Ac toxin. Insect Biochem Mol Biol 34:101–112. doi: 10.1016/j.ibmb.2003.09.007. [DOI] [PubMed] [Google Scholar]
- 23.Jurat-Fuentes JL, Adang MJ. 2004. Characterization of a Cry1Ac-receptor alkaline phosphatase in susceptible and resistant Heliothis virescens larvae. Eur J Biochem 271:3127–3135. doi: 10.1111/j.1432-1033.2004.04238.x. [DOI] [PubMed] [Google Scholar]
- 24.Nagamatsu Y, Koike T, Sasaki K, Yoshimoto A, Furukawa Y. 1999. The cadherin-like protein is essential to specificity determination and cytotoxic action of the Bacillus thuringiensis insecticidal CryIAa toxin. FEBS Lett 460:385–390. doi: 10.1016/S0014-5793(99)01327-7. [DOI] [PubMed] [Google Scholar]
- 25.Geiser DL, Winzerling JJ. 2012. Insect transferrins: multifunctional proteins. Biochim Biophys Acta 1820:437–451. doi: 10.1016/j.bbagen.2011.07.011. [DOI] [PubMed] [Google Scholar]
- 26.Kim J, Kim Y. 2010. A viral histone H4 suppresses expression of a transferrin that plays a role in the immune response of the diamondback moth, Plutella xylostella. Insect Mol Biol 19:567–574. doi: 10.1111/j.1365-2583.2010.01014.x. [DOI] [PubMed] [Google Scholar]
- 27.Wolfersberger M, Luethy P, Maurer A, Parenti P, Sacchi F, Giordana B, Hanozet G. 1987. Preparation and partial characterization of amino acid transporting brush border membrane vesicles from the larval midgut of the cabbage butterfly (Pieris brassicae). Comp Biochem Physiol A Physiol 86:301–308. doi: 10.1016/0300-9629(87)90334-3. [DOI] [Google Scholar]
- 28.Laemmli UK. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 29.Zhang W, Song W, Zhang Z, Wang H, Yang M, Guo R, Li M. 2014. Transcriptome analysis of Dastarcus helophoroides (Coleoptera: Bothrideridae) using Illumina HiSeq sequencing. PLoS One 9:e100673. doi: 10.1371/journal.pone.0100673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q. 2011. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu Y, Shen D, Zhou F, Wang G, An C. 2014. Identification of immunity-related genes in Ostrinia furnacalis against entomopathogenic fungi by RNA-seq analysis. PLoS One 9:e86436. doi: 10.1371/journal.pone.0086436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M. 2005. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21:3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- 33.Ye J, Fang L, Zheng H, Zhang Y, Chen J, Zhang Z, Wang J, Li S, Li R, Bolund L. 2006. WEGO: a web tool for plotting GO annotations. Nucleic Acids Res 34:W293–W297. doi: 10.1093/nar/gkl031. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.