ABSTRACT
Multidrug-resistant (MDR) Salmonella enterica can be spread from cattle to humans through direct contact with animals shedding Salmonella as well as through the food chain, making MDR Salmonella a serious threat to human health. The objective of this study was to use whole-genome sequencing to compare antimicrobial-resistant (AMR) Salmonella enterica serovars Typhimurium, Newport, and Dublin isolated from dairy cattle and humans in Washington State and New York State at the genotypic and phenotypic levels. A total of 90 isolates were selected for the study (37 S. Typhimurium, 32 S. Newport, and 21 S. Dublin isolates). All isolates were tested for phenotypic antibiotic resistance to 12 drugs using Kirby-Bauer disk diffusion. AMR genes were detected in the assembled genome of each isolate using nucleotide BLAST and ARG-ANNOT. Genotypic prediction of phenotypic resistance resulted in a mean sensitivity of 97.2 and specificity of 85.2. Sulfamethoxazole-trimethoprim resistance was observed only in human isolates (P < 0.05), while resistance to quinolones and fluoroquinolones was observed only in 6 S. Typhimurium isolates from humans in Washington State. S. Newport isolates showed a high degree of AMR profile similarity, regardless of source. S. Dublin isolates from New York State differed from those from Washington State based on the presence/absence of plasmid replicons, as well as phenotypic AMR susceptibility/nonsusceptibility (P < 0.05). The results of this study suggest that distinct factors may contribute to the emergence and dispersal of AMR S. enterica in humans and farm animals in different regions.
IMPORTANCE The use of antibiotics in food-producing animals has been hypothesized to select for AMR Salmonella enterica and associated AMR determinants, which can be transferred to humans through different routes. Previous studies have sought to assess the degree to which AMR livestock- and human-associated Salmonella strains overlap, as well as the spatial distribution of Salmonella's associated AMR determinants, but have often been limited by the degree of resolution at which isolates can be compared. Here, a comparative genomics study of livestock- and human-associated Salmonella strains from different regions of the United States shows that while many AMR genes and phenotypes were confined to human isolates, overlaps between the resistomes of bovine and human-associated Salmonella isolates were observed on numerous occasions, particularly for S. Newport. We have also shown that whole-genome sequencing can be used to reliably predict phenotypic resistance across Salmonella isolated from bovine sources.
KEYWORDS: whole-genome sequencing, zoonosis, dairy cattle, drug resistance evolution, genomics, nontyphoidal Salmonella
INTRODUCTION
Salmonella enterica is estimated to cause approximately 1.2 million illnesses and 450 deaths each year in the United States alone (1). While most individuals recover without medical intervention, severe infections require hospitalization and treatment with antimicrobials (1). An even greater challenge is posed when those infections are caused by antimicrobial-resistant (AMR) organisms. The Centers for Disease Control (CDC) estimates that 100,000 infections due to AMR nontyphoidal Salmonella occur in the United States annually and has designated AMR in nontyphoidal Salmonella as a serious threat to public health (2). More specifically, the World Health Organization (WHO) has listed fluoroquinolone-resistant nontyphoidal Salmonella as a global health concern (3).
Both the CDC and WHO have called for improved monitoring of AMR along the food chain, particularly in food-producing animals (2, 3). Due to concerns about the misuse of antimicrobials in farm animals, the farm is often viewed as a reservoir in which AMR can be acquired by bacteria that are then transmitted from animals to humans (4, 5). In this context, S. enterica becomes particularly relevant, as it can be transmitted between animal and human populations (6–8), as well as through food (9–11).
A number of studies have sought to assess the extent to which AMR is acquired by bacteria in livestock environments and subsequently transmitted to humans, and many have arrived at different conclusions (12–15). Often, the degree of resolution at which isolates can be compared is a limiting factor in determining the origin of a particular bacterial isolate and its AMR profile. Methods such as multilocus sequence typing (MLST), serotyping, and pulsed-field gel electrophoresis (PFGE) may not offer enough discriminatory power to detect differences between isolates from different sources or locations (16–18), while phenotypic testing of AMR may not distinguish between AMR mechanisms in different isolates (14).
The extent to which Salmonella and AMR genes associated with it are transmitted between animal and human sources remains unclear. The objective of this study was to use whole-genome sequencing (WGS) to compare AMR Salmonella enterica isolates previously serotyped as Typhimurium, Newport, or Dublin isolated from dairy cattle and humans in Washington State and New York State at the genotypic and phenotypic levels. In addition, correlations between AMR genotype and AMR phenotype were assessed. It was hypothesized that sources and geographic differences between Salmonella isolates could be elucidated at greater resolution through the implementation of WGS.
RESULTS
Overall distribution of SNPs, AMR genes, AMR phenotypes, and plasmid replicons.
Of the three serotypes studied, S. Typhimurium displayed the highest degree of phylogenetic diversity. Variant calling revealed a total number of 2,976 variants in the S. Typhimurium isolates, with 2,723 of those variants called as single nucleotide polymorphism (SNPs). In S. Newport, only 327 variants were called, 263 of which were SNPs. The fewest number of variants occurred in S. Dublin, with 183 variants, 131 of which were SNPs.
AMR genes belonging to 42 different groups were detected in the 90 genomes (see Table S2 in the supplemental material). The most common genes belonged to groups associated with resistance to penicillins (penicillin binding protein [PBP] gene), aminoglycosides [aac(6)-Iaa, strA, and strB], phenicols (floR), tetracyclines [tet(A) and tet(R)], cephalosporins (CMY), and sulfonamides (sul2) (Table 1). At the phenotypic level, all isolates displayed resistance or intermediate resistance to between 1 and 11 antimicrobials. The most common antimicrobial to which isolates were resistant was ampicillin (AMP), as 88 of 90 isolates were AMP resistant (Table 1). In addition, a total of 20 different plasmid replicons were detected in the genomes of the 90 isolates used in the study. The three most common replicons (ColRNAI, ColpVC, and IncA/C2) were each detected in over one-half of all isolates (Table 1). Several significant (P < 0.001) associations between plasmid replicons and AMR gene groups were observed, including the IncA/C2 replicon and gene groups CMY, floR, strA-strB, sul2, and tet(A)-tet(R) (see Table S3 in the supplemental material). These genes had previously been found on an IncA/C2 plasmid isolated from S. Newport (19).
TABLE 1.
Ranking of the five most common antimicrobial resistance (AMR) gene groups, phenotypic AMR profiles, and plasmid replicons for all serotypes, S. Typhimurium, S. Newport, and S. Dublina
| Rankb | All isolates (n = 90) | S. Typhimurium (n = 37) | S. Newport (n = 32) | S. Dublin (n = 21) |
|---|---|---|---|---|
| AMR gene groups | ||||
| 1 | aac(6)-Iaa, PBP gene (90) | aac(6)-Iaa, PBP gene (37) | aac(6)-Iaa, CMY, PBP gene, strA, strB, sul2, tet(A), tet(R) (32) | aac(6′)-Iaa, CMY, PBP gene, sul2 (21) |
| 2 | floR (72) | aadA (25) | floR (30) | strA, strB, tet(A), tet(R) (20) |
| 3 | CMY, tet(A), tet(R) (68) | floR (23) | aph(3″)-Ia (22) | floR (19) |
| 4 | sul2 (67) | sul1 (21) | aadA, dfrA, sul1 (3) | aph(3″)-Ia (18) |
| 5 | strA, strB (64) | aph(3″)-Ia (20) | blaTEM-1D (15) | |
| Phenotypic AMR profile | ||||
| 1 | AMP (88) | AMP (35) | AMC; AMP; CRO; FOX; STR; SX; TIO; TET (32) | AMP; CRO; TIO (21) |
| 2 | TET (82) | TET (31) | CHL (30) | AMC; FOX; SX (20) |
| 3 | AMC; SX (81) | STR (30) | SXT (3) | CHL; TET (19) |
| 4 | CHL; STR (72) | AMC; SX (29) | STR (10) | |
| 5 | CRO; TIO (71) | CHL (23) | SXT (1) | |
| Plasmid replicons | ||||
| 1 | ColRNAI (77) | ColRNAI (27) | ColRNAI; IncA/C2 (32) | IncX1 (21) |
| 2 | ColpVC (63) | IncFII(S) (25) | ColpVC (26) | IncA/C2 (20) |
| 3 | IncA/C2 (60) | ColpVC (20) | IncI1 (2) | ColRNAI (18) |
| 4 | IncFII(S) (36) | IncFIB(S) (17) | Col(BS512) (1) | ColpVC (17) |
| 5 | IncX1 (22) | IncI1 (10) | IncFII(S) (11) |
Numbers in parentheses indicate the number of isolates classified into a given AMR gene group or a given phenotypic AMR profile or the number of isolates carrying a given plasmid replicon.
Rank is based on the frequency of (i) AMR gene group presence, (ii) phenotypic AMR profile, and (iii) plasmid replicon presence.
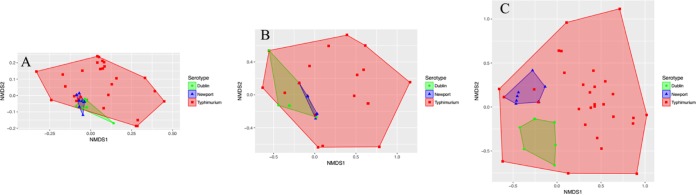
Serotypes were found to differ with regard to AMR gene sequences, phenotypic resistance/susceptibility, and the presence/absence of plasmid replicons when using analysis of similarity (ANOSIM) and/or permutational multivariate analysis of variance (PERMANOVA; P < 0.001 after a Holm-Bonferroni correction) (Table 2). Of the three serotypes studied, S. Typhimurium showed the widest range of AMR gene profiles, phenotypic AMR profiles, and plasmid replicon presence/absence profiles (Fig. 1).
TABLE 2.
ANOSIM and PERMANOVA statistics and their respective mean P valuesa
| Serotype(s) | Grouping factor/responseb | ANOSIM |
PERMANOVA |
||
|---|---|---|---|---|---|
| R statistic | Mean uncorrected P value | F statistic | Mean uncorrected P value | ||
| Antimicrobial resistance gene sequences | |||||
| All | Serotype | 0.234c | <0.001c | 15.598d | <0.001d |
| Typhimurium | Source | 0.079 | 0.040 | 2.937 | 0.020 |
| Typhimurium | Location | 0.045 | 0.105 | 2.093 | 0.074 |
| Newport | Source | 0.034 | 0.169 | 3.405 | 0.004 |
| Newport | Location | 0.241c | 0.002c | 3.185 | 0.008 |
| Dublin | Source | 0.041 | 0.188 | 1.578 | 0.231 |
| Dublin | Location | 0.145 | 0.064 | 5.366 | 0.004 |
| Phenotypic antimicrobial resistance/susceptibility profiles | |||||
| All | Serotype | 0.200c | <0.001c | 1.037 | 0.433 |
| Typhimurium | Source | 0.122 | 0.015 | 6.796 | 0.012 |
| Typhimurium | Location | −0.003 | 0.417 | 0.181 | 0.727 |
| Newport | Source | −0.030 | 1.000 | 1.739 | 0.053 |
| Newport | Location | 0.103 | 0.072 | 1.699 | 0.074 |
| Dublin | Source | 0.089 | 0.053 | 1.060 | 0.477 |
| Dublin | Location | 0.481c | <0.001c | 4.717d | <0.001d |
| Plasmid replicon presence/absence profiles | |||||
| All | Serotype | 0.350c | <0.001c | 21.800d | <0.001d |
| Typhimurium | Source | 0.025 | 0.201 | −0.299 | 0.853 |
| Typhimurium | Location | 0.107 | 0.009 | 6.077 | 0.011 |
| Newport | Source | −0.030 | 0.934 | 2.118 | 0.042 |
| Newport | Location | 0.098 | 0.074 | 1.572 | 0.105 |
| Dublin | Source | 0.040 | 0.146 | 1.521 | 0.116 |
| Dublin | Location | 0.408c | <0.001c | 4.466d | <0.001d |
Rows in boldface indicate that at least one test was significant (P < 0.05) after a Holm-Bonferroni correction was applied.
Grouping factors used were serotype (only for “All isolates”), source (bovine or human), and location (New York or Washington State).
Significant ANOSIM test (P < 0.05) after a Holm-Bonferroni correction was applied.
Significant PERMANOVA test (P < 0.05) after a Holm-Bonferroni correction was applied.
FIG 1.
Nonmetric multidimensional scaling (NMDS) plots for all isolates based on antimicrobial resistance (AMR) gene sequences (A), phenotypic antimicrobial resistance/susceptibility profiles (B), and presence/absence of plasmid replicons (C). Points represent isolates, while shaded regions and convex hulls correspond to isolate serotypes. For an interactive plot of these data, as well as interactive NMDS plots for individual serotypes, visit https://github.com/lmc297/2017_AEM_Figure_S2.
In silico AMR gene detection is correlated with phenotypic AMR patterns.
Genotypic and phenotypic AMR data were used to evaluate the ability of genotypic data to predict phenotypic resistance (Fig. 2). Ciprofloxacin (CIP) was not included in these analyses due to the rarity of resistant isolates in this data set (1 of the 90 isolates). Based on the 11 remaining antimicrobials, genotypic prediction of phenotypic resistance resulted in a mean sensitivity of 97.2% and specificity of 85.2% (Table 3). Genotypic prediction of phenotypic resistance to AMP, cefoxitin (FOX), chloramphenicol (CHL), streptomycin (STR), sulfisoxazole (SX), and tetracycline (TET) had a sensitivity of 100%, while the prediction of phenotypic resistance to AMP, ceftiofur (TIO), ceftriaxone (CRO), nalidixic acid (NAL), and trimethoprim-sulfamethoxazole (SXT) had a specificity of 100% (Table 3). With the exception of NAL, genotypic prediction of phenotypic resistance resulted in sensitivities greater than 90% for all drugs (Table 3). For all antimicrobials other than AMC, STR, SX, and TET, genotypic prediction of phenotypic resistance had specificity above 90% (Table 3). Consistent with these findings, significant differences in resistance (determined by the mean zone diameters from the Kirby-Bauer disk diffusion assays) were observed between isolates carrying at least one AMR gene conferring resistance to a given antimicrobial and those isolates that did not carry said AMR gene (P < 0.05 after a Holm-Bonferroni correction) (Table 4).
FIG 2.
Frequency of different phenotypic and genotypic resistance determinants for each serotype-source group (e.g., Salmonella Dublin isolates obtained from humans [S. Dublin Human]). Genotypic resistance was determined using nucleotide BLAST (blastn) and the ARG-ANNOT database; isolates were classified as having a resistant genotype if the AMR gene was detected by BLAST with a minimum coverage of 50% and a minimum sequence identity of 75%. Phenotypic resistance was tested using Kirby-Bauer disk diffusion. Percentages were calculated using the ratio of resistant isolates to total isolates in each serotype-source group (n = 17 for S. Typhimurium Bovine, n = 20 for S. Typhimurium Human, n = 14 for S. Newport Bovine, n = 18 for S. Newport Human, n = 10 for S. Dublin Bovine, and n = 11 for S. Dublin Human). Nalidixic acid (NAL)- and sulfamethoxazole-trimethoprim (SXT)-resistant isolates (6 and 12 of the 90 isolates, respectively) each had one isolate for which genotypic resistance did not correlate with phenotypic resistance.
TABLE 3.
Sensitivity and specificity of genotype predictions of resistant antimicrobial phenotype for all 90 Salmonella isolates in the study
| Antimicrobiala | Phenotype: resistant (n)b |
Phenotype: susceptible (n) |
Sensitivity (%) | Specificity (%) | ||
|---|---|---|---|---|---|---|
| Genotype: resistant | Genotype: susceptible | Genotype: resistant | Genotype: susceptible | |||
| AMC | 71 | 2 | 6 | 11 | 97.3 | 64.7 |
| AMP | 88 | 0 | 0 | 2 | 100.0 | 100.0 |
| FOX | 67 | 0 | 1 | 22 | 100.0 | 95.7 |
| TIO | 70 | 1 | 0 | 19 | 98.6 | 100.0 |
| CRO | 70 | 1 | 0 | 19 | 98.6 | 100.0 |
| CHL | 72 | 0 | 1 | 17 | 100.0 | 94.4 |
| NAL | 5 | 1 | 0 | 84 | 83.3 | 100.0 |
| STR | 72 | 0 | 17 | 1 | 100.0 | 5.6 |
| SX | 81 | 0 | 1 | 8 | 100.0 | 88.9 |
| SXT | 11 | 1 | 0 | 78 | 91.7 | 100.0 |
| TET | 82 | 0 | 1 | 7 | 100.0 | 87.5 |
| Overall | 97.2 | 85.2 | ||||
AMC, amoxicillin-clavulanic acid; AMP, ampicillin; FOX, cefoxitin; TIO, ceftiofur; CRO, ceftriaxone; CHL, chloramphenicol; NAL, nalidixic acid; STR, streptomycin; SX, sulfisoxazole; SXT, sulfamethoxazole/trimethoprim; TET, tetracycline.
Isolates that showed intermediate resistance to an antimicrobial are categorized as resistant.
TABLE 4.
Comparison of mean zone diameters between (i) Salmonella isolates with at least one AMR gene that has been known to confer resistance to a particular antimicrobial (ARG) and (ii) isolates with no genes known to confer resistance to that antimicrobiala
| Antimicrobial | 95% CI of MZDa (cm) |
|
|---|---|---|
| ARG absent | ARG present | |
| Aminopenicillins | ||
| Ampicillin | 25.4–25.6 | 0.0–0.02 |
| Amoxicillin-clavulanic acid | 13.9–18.7 | 9.2–11.0 |
| Chloramphenicol | 24.4–27.6 | 0.02–1.45 |
| Cephalosporins | ||
| Ceftiofur | 25.5–29.5 | 12.7–14.5 |
| Ceftriaxone | 29.7–34.5 | 13.4–15.5 |
| Cefoxitin | 23.2–27.5 | 8.4–10.2 |
| Streptomycin | 13.9–21.1 | 3.1–5.3 |
| Sulfonamides | ||
| Sulfisoxazole | 22.4–26.2 | 0.0–0.9 |
| Sulfamethoxazole-trimethoprim | 23.8–25.8 | 0–3.3 |
| Tetracycline | 19.0–26.5 | 2.0–4.2 |
MZD, mean zone diameter; CI, confidence interval. All P values were <0.0001.
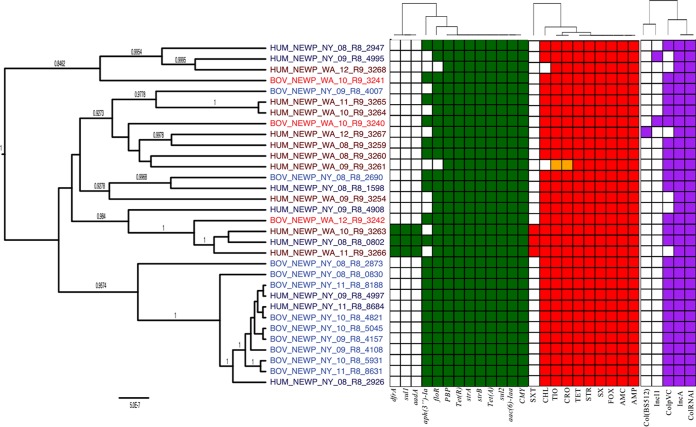
S. Typhimurium phylogeny, AMR genes, AMR phenotypes, and plasmid replicons.
A BEAST phylogeny of the 37 S. Typhimurium genomes separated these isolates into two major clades (Fig. 3; posterior probability, 1). One of these clades contained human isolates exclusively (n = 8), while the other major clade included 12 human and 17 bovine isolates (Fig. 3). Three isolates within this “mixed source” clade were particularly similar based on their AMR gene sequences: isolates BOV_TYPH_WA_09_R9_3247 (isolated from a dairy cow in Washington State in 2009), HUM_TYPH_WA_09_R9_3271 (isolated from a human in Washington State in 2009), and HUM_TYPH_NY_12_R9_0437 (isolated from a human in New York State in 2012) appeared to have highly similar AMR gene profiles (see Fig. S2 posted at https://github.com/lmc297/2017_AEM_Figure_S2). All AMR genes in these three isolates matched with 100% sequence identity except for tet(RG); HUM_TYPH_WA_09_R9_3271 tet(RG) differed from the other two isolates at nucleotide position 73.
FIG 3.
Phylogenetic tree of S. Typhimurium isolates constructed using BEAST. Gene groups for AMR genes detected in each genome sequence at more than 50% coverage and 75% identity using BLAST (blastn) and ARG-ANNOT are indicated in green. Antimicrobials to which each isolate is resistant are indicated in red, and intermediate resistance to an antimicrobial is indicated in orange. Plasmid replicons detected in each genome sequence using PlasmidFinder are indicated in purple. Branch lengths are reported in substitutions per site, while posterior probabilities are reported at tree nodes.
Overall, 41 of the 42 AMR gene groups identified in the 90 isolates in this study were detected in S. Typhimurium (all except aadB; Fig. 3). The 37 S. Typhimurium isolates were distributed into 24 different genotypic MDR profiles, the most common of which was aac(6)-Iaa floR sul1 tet(RG) tet(G) blaCARB aadA PBP gene, which was found in 11% of S. Typhimurium genomes. In addition, between 2 and 7 unique plasmid replicons were detected per genome (Fig. 3). When ANOSIM and PERMANOVA were applied as metrics to assess clustering based on either AMR gene sequences or plasmid replicon presence/absence, there were no significant differences between bovine and human isolate clusters or between New York and Washington State clusters (P > 0.05 after a Holm-Bonferroni correction) (Table 2). While neither ANOSIM nor PERMANOVA found significant associations between AMR genes and either source or state after correcting for multiple testing (P > 0.05) (Table 2), Fisher's exact test indicated that the IncI1 replicon was more commonly detected in New York State isolates than in Washington State isolates (Table 5) (P < 0.05, after Holm-Bonferroni correction).
TABLE 5.
Odds ratios for association of AMR gene groups, antimicrobials, and plasmid replicons with source or location (only associations with P values of <0.05 are shown)a
| Characteristic | Serotype | Source/location favored by OR | OR | Uncorrected P value |
|---|---|---|---|---|
| Source | ||||
| Gene | ||||
| aac(3)-IIa | Typhimurium | Human | Infinity (only in humans) | 0.009 |
| floR | Typhimurium | Human | 5.42 | 0.021 |
| aph(3″)-Ia | Newport | Bovine | 0.0831 | 0.019 |
| Antimicrobial | ||||
| CHL | Typhimurium | Human | 5.42 | 0.021 |
| NAL | Typhimurium | Human | Infinity (only in humans) | 0.022 |
| SXT | Typhimurium | Human | Infinity (only in humans) | 0.004 |
| TET | Typhimurium | Human | Infinity (all human isolates) | 0.005 |
| STR | Dublin | Human | 9.28 | 0.030 |
| Plasmid | ||||
| IncA/C2 | Typhimurium | Human | 8.18 | 0.048 |
| ColpVC | Newport | Bovine | 0 (found in all bovine isolates) | 0.024 |
| Geographic location | ||||
| Gene | ||||
| blaTEM-1D | Typhimurium | WA | 4.60 | 0.045 |
| aph(3″)-Ia | Newport | NY | 0.172 | 0.049 |
| aadB | Dublin | WA | Infinity (found only in WA) | 0.005 |
| cmlA | Dublin | WA | Infinity (found only in WA) | 0.005 |
| Antimicrobial | ||||
| NAL | Typhimurium | WA | Infinity (found only in WA) | 0.020 |
| STR | Typhimurium | WA | 8.51 | 0.042 |
| SX | Typhimurium | WA | 10.8 | 0.019 |
| SXT | Typhimurium | WA | 9.36 | 0.042 |
| STR | Dublin | NY | 0.052 | 0.008 |
| Plasmid | ||||
| IncI1 | Typhimurium | NY | 0.0602 | 0.003 |
| IncP | Typhimurium | WA | Infinity (found only in WA) | 0.046 |
| IncFII(S) | Dublin | NY | 0 (present in all NY isolates) | 0.001 |
An odds ratio (OR) of infinity or 0 includes a short statement (in parentheses) that indicates which source or location was the driver for that OR (e.g., “only in humans” indicates that the given gene/phenotype/plasmid replicon was found in only human isolates and in none of the bovine isolates). WA, Washington State; NY, New York State. Values in boldface were significant (P < 0.05) after a Holm-Bonferroni correction was applied to the respective analysis.
At the phenotypic level, the number of antimicrobials to which S. Typhimurium isolates were resistant ranged from 1 to 11 (Fig. 3). The most common phenotypic resistance profiles for S. Typhimurium were AMC-AMP-CHL-SX-STR-TET and AMC-AMP-FOX-TIO-CRO, which were found in 27% and 11% of the isolates, respectively. When ANOSIM and PERMANOVA were used as metrics to assess clustering, no significant differences between bovine and human clusters or between New York and Washington State clusters formed by phenotypic resistance/susceptibility profiles were detected (P > 0.05 after a Holm-Bonferroni correction [Table 2]). However, when Fisher's exact test was used to test for differences at the individual antimicrobial level, resistance to SXT was seen only in human-associated S. Typhimurium isolates (P < 0.05 after a Holm-Bonferroni correction [Table 5]). In addition, all human-associated S. Typhimurium isolates were resistant to TET, while only 65% of bovine isolates were resistant to TET (P < 0.05 after a Holm-Bonferroni correction [Table 5]).
In addition to possessing the most diverse genotypic and phenotypic AMR profiles, S. Typhimurium was the only serotype in which resistance to NAL (a quinolone) and CIP (a fluoroquinolone) was observed. All isolates that were resistant to NAL and CIP originated from human clinical samples in Washington State (Fig. 3). qnr genes, which are plasmid-mediated quinolone resistance (PMQR) genes, were detected in the sequences of the two S. Typhimurium isolates that showed intermediate resistance to NAL (Table 6). For each of the four NAL-resistant isolates, point mutations were identified in the quinolone resistance-determining region (QRDR) of gyrA (Table 6). These nucleotide changes resulted in nonsynonymous amino acid changes (Asp87Asn, Asp87Tyr, and Ser83Tyr) that have been previously observed in quinolone-resistant Salmonella isolates (20). In addition, three of the four NAL-resistant isolates possessed oqxA and oqxB (Table 6). These genes encode the OqxAB multidrug efflux pump, which confers resistance to multiple agents, including low-level resistance to quinolones (21, 22).
TABLE 6.
S. Typhimurium isolates with qnr and/or oqx genes and/or point mutations in gyrA and/or gyrB and/or parCa
| Isolate | S/I/R status |
qnr and/or oqx gene(s) detected | Point mutationb detected in: |
|||
|---|---|---|---|---|---|---|
| NAL | CIP | gyrA | gyrB | parC | ||
| BOV_TYPH_NY_12_R8_9801 | S | S | None | 1641: T→G | WT | WT |
| BOV_TYPH_NY_12_R8_9815 | S | S | None | 1641: T→G | WT | WT |
| BOV_TYPH_NY_12_R8_9832 | S | S | None | 1641: T→G | WT | WT |
| HUM_TYPH_NY_11_R8_8073 | S | S | None | WT | 2202: G→A | WT |
| HUM_TYPH_NY_12_R9_0042 | S | S | None | WT | 2202: G→A | WT |
| HUM_TYPH_WA_08_R9_3269 | I | S | qnrS | WT | WT | 1713: C→T |
| HUM_TYPH_WA_08_R9_3270 | R | I | oqxA, oqxB | Asp87Tyr | WT | 1713: C→T |
| 259: G→T | ||||||
| HUM_TYPH_WA_09_R9_3271 | S | S | None | WT | 759: A→G | WT |
| HUM_TYPH_WA_10_R9_3273 | R | S | oqxA, oqxB | Ser83Tyr | WT | 1713: C→T |
| 248: C→A | ||||||
| HUM_TYPH_WA_10_R9_3274 | I | S | qnrB | WT | WT | WT |
| HUM_TYPH_WA_11_R9_3275 | R | S | oqxA, oqxB | Asp87Asn | WT | 1713: C→T |
| 259: G→A | ||||||
| HUM_TYPH_WA_11_R9_3276 | R | S | None | Asp87Asn | WT | 1713: C→T |
| 259: G→A | ||||||
| HUM_TYPH_WA_12_R9_3277 | S | S | None | WT | WT | 1713: C→T |
No point mutations were detected in parE.
Synonymous point mutations resulting in no amino acid change are shown as position nt→nt (e.g., 259: G→A); amino acid substitutions (for gyrA or gyrB and parC) are formatted as “position: reference base → alternate base”; WT, gene with no mutations.
S. Newport phylogeny, AMR genes, AMR phenotypes, and plasmid replicons.
Among the 19 S. Newport isolates from New York State, 11 clustered into a single, well-supported clade (posterior probability, 1) (Fig. 4). The inclusion of an additional isolate from New York State yielded a 12-isolate clade with a posterior probability of 0.9574.
FIG 4.
Phylogenetic tree of S. Newport isolates constructed using BEAST. Gene groups for AMR genes detected in each genome sequence at more than 50% coverage and 75% identity using BLAST (blastn) and ARG-ANNOT are indicated in green. Antimicrobials to which each isolate is resistant are indicated in red, and intermediate resistance to an antimicrobial is indicated in orange. Plasmid replicons detected in each genome sequence using PlasmidFinder are indicated in purple. Branch lengths are reported in substitutions per site, while posterior probabilities are reported at tree nodes.
The AMR gene profiles of the 32 S. Newport isolates showed a high degree of similarity, with only 5 different genotypic profiles (Fig. 4). The two most common genotypic profiles, i.e., aac(6)-Iaa floR CMY sul2 tet(A) aph(3″)-Ia strB strA tet(R) PBP gene and aac(6)-Iaa floR CMY sul2 tet(A) strB strA tet(R) PBP gene, were detected in 66% and 19% of S. Newport genomes, respectively. At the individual gene level, genes belonging to the aac(6)-Iaa, CMY, strA, strB, sul2, tet(A), tet(R), and PBP gene groups were detected in the sequences of all 32 isolates (Table 1). All S. Newport isolates had identical copies of each of these genes except for CMY, as a truncated version of the gene was detected in isolate BOV_NEWP_WA_10_R9_3240. In addition, the IncA/C2 and ColRNAI replicons were detected in all S. Newport genomes (Table 1). Neither ANOSIM nor PERMANOVA detected significant associations between AMR genes or plasmid replicon presence/absence and source after correcting for multiple testing (P > 0.05 after a Holm-Bonferroni correction [Table 2]). However, the AMR gene sequences of Washington State and New York State isolates were found to differ when ANOSIM was used as a metric (P < 0.05 after a Holm-Bonferroni correction [Table 2]). When Fisher's exact test was used to assess source and geographic associations at the individual gene level, genes belonging to the aph(3″)-Ia group were more commonly present in (i) S. Newport bovine isolates and (ii) isolates from New York State (P < 0.05 after a Holm-Bonferroni correction [Table 5]). Additionally, the ColpVC plasmid replicon was detected in all bovine S. Newport isolates and only 67% of the human isolates (P < 0.05 after a Holm-Bonferroni correction [Table 5]).
S. Newport isolates appeared even more similar at the phenotypic AMR level than at the genetic level. No significant source or geographic differences in MDR phenotype were observed when ANOSIM and PERMANOVA were used to assess clustering (P > 0.05 after a Holm-Bonferroni correction) (Table 2). All 32 S. Newport isolates were resistant to AMC, AMP, FOX, TIO, CRO, SX, STR, and TET, and only 3 different phenotypic profiles were detected (Fig. 4). The most common of these, AMC-AMP-FOX-TIO-CRO-CHL-SX-STR-TET, was carried by 27 of the 32 (84%) S. Newport isolates. Three isolates showed additional resistance to SXT; hence, the two most common profiles accounted for 30 of the 32 (94%) isolates. The three SXT-resistant isolates possessed aadA, dfrA, and sul1, which were not detected in any other S. Newport genomes (Fig. 4).
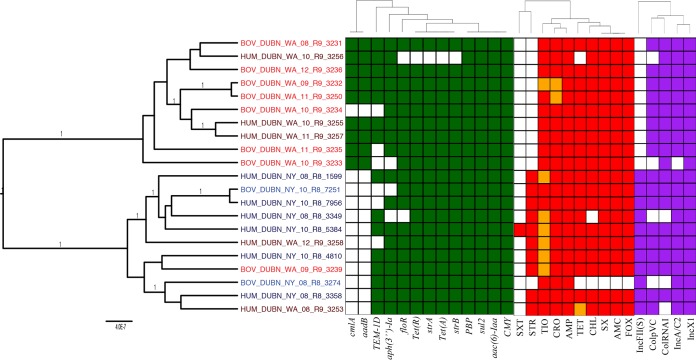
S. Dublin phylogeny, AMR genes, AMR phenotypes, and plasmid replicons.
S. Dublin isolates clustered into two separate clades with a posterior probability of 1, one of which consisted of 10 isolates exclusively from Washington State (referred to here as the “Washington State clade”) (Fig. 5). The other clade included all eight S. Dublin isolates from New York State and three isolates from Washington State (referred to here as the “mixed clade”) (Fig. 5). Both genotypic and phenotypic differences were observed between the two major clades. AMR genes aadB and cmlA, which were detected in all but 1 Washington State state clade isolate, were not detected in any of the mixed clade isolates (P < 0.05 after a Holm-Bonferroni correction) (Fig. 5). Not surprisingly, the frequencies at which these genes were detected in New York and Washington States were significantly different when Fisher's exact test was used (P < 0.05 after a Holm-Bonferroni correction) (Table 5). ANOSIM and PERMANOVA did not identify significant differences between S. Dublin geographic clusters formed by AMR gene sequences (Table 2). However, when ANOSIM and PERMANOVA were conducted using plasmid replicon presence/absence data, significant differences between New York and Washington State isolate clusters were observed for S. Dublin (P < 0.05 after a Holm-Bonferroni correction) (Table 2). In addition, when Fisher's exact test was used to test for possible geographic associations of individual plasmid replicons, the IncFII(S) replicon was detected only in mixed clade isolates, making it more commonly associated with isolates from New York State (P < 0.05 after a Holm-Bonferroni correction) (Fig. 5).
FIG 5.
Phylogenetic tree of S. Dublin isolates constructed using BEAST. Gene groups for AMR genes detected in each genome sequence at more than 50% coverage and 75% identity using BLAST (blastn) and ARG-ANNOT are indicated in green. Antimicrobials to which each isolate is resistant are indicated in red, and intermediate resistance to an antimicrobial is indicated in orange. Plasmid replicons detected in each genome sequence using PlasmidFinder are indicated in purple. Branch lengths are reported in substitutions per site, while posterior probabilities are reported at tree nodes.
Significant differences between New York and Washington State isolate clusters were observed for S. Dublin when ANOSIM and PERMANOVA were conducted using phenotypic resistance/susceptibility data (P < 0.05 after a Holm-Bonferroni correction) (Table 2). Despite the detection of both strA and strB in 20 of the 21 genomes (Table 1), STR resistance was observed only in isolates in the mixed clade (P < 0.05 after a Holm-Bonferroni correction) (Fig. 5). While the strB sequence was the same for the 20 isolates, the strA sequence showed a strong geographical association: all isolates in the Washington State clade possessed a truncated form of the gene, with the first 91 bp of the gene missing. Aside from this 91-bp deletion, the strA sequences were identical in all isolates. Overall, 11 isolates carried strB and a full-length strA; 10 of these isolates showed phenotypic STR resistance. However, 9 isolates carried strB and a truncated strA; all of these isolates were sensitive to STR. These data suggest that the presence of the truncated strA variant found here does not confer STR resistance and also suggest that the presence of only the strB variant found here, in the absence of a full-length strA, does not confer STR resistance.
The S. Dublin isolates were distributed into 8 different AMR genotypic profiles, with 33% of isolates genes belonging to the aac(6)-Iaa floR CMY sul2 tet(A) aph(3″)-Ia blaTEM-1D strB strA tet(R) PBP gene genotypic profile. The most common resistance genes in S. Dublin belonged to the aac(6)-Iaa, CMY, and sul2 groups, all of which were detected in all 21 S. Dublin isolates (Table 1). The sequences of these genes were identical for all S. Dublin isolates, regardless of source or geographic location. PBP gene was also detected in all 21 genomes (Table 1). PBP gene sequences for 20 isolates were identical; only the sequence of isolate BOV_DUBN_WA_09_R9_3239 differed by a single nucleotide from the 20 other sequences. In addition, the replicon for IncX1, which had been detected in only 1 S. Typhimurium isolate and no S. Newport isolates in this study, was detected in all 21 S. Dublin genomes (Fig. 5). At the phenotypic level, 6 different phenotypic profiles were observed. The two most common, AMC-AMP-FOX-TIO-CRO-CHL-SX-TET and AMC-AMP-FOX-TIO-CRO-CHL-SX-STR-TET, were observed in 43% and 38% of S. Dublin isolates, respectively.
DISCUSSION
Antimicrobial resistance in zoonotic and foodborne pathogens is considered to be one of the most serious threats to public health today (2, 3). The emergence and dispersal of AMR Salmonella are particularly problematic, due to (i) the fact that nontyphoidal Salmonella represents one of the most common causes of foodborne disease cases and associated deaths worldwide (23) and (ii) reports on the emergence and dispersal of different multidrug-resistant Salmonella strains (e.g., Salmonella Typhimurium DT104) (24–26). Studies of the relationships between AMR determinants and MDR strains found in humans and animals are often confounded by the selection of the isolates included in a given study, in which human and animal isolates may be of different serotypes, geographical locations, or temporal intervals. To further our understanding of AMR diversity and dispersal in Salmonella, we thus assembled and characterized a set of Salmonella isolates that (i) represented 3 serotypes associated with both human and bovine populations, (ii) were isolated over the same time frame (2008 to 2012), (iii) were matched by source (human or animal) so that approximately equal numbers of human and bovine isolates were selected from each serotype, and (iv) were matched by geographical location so that similar numbers of human and bovine isolates of the three different serotypes were obtained from each of the states of Washington and New York. Our data obtained from these isolates suggest that (i) WGS can be used to reliably predict phenotypic resistance across Salmonella isolates from both human and bovine sources, (ii) geographical differences can contribute to distinct, location-specific AMR patterns, and (iii) despite an overlap of AMR geno- and phenotypes, human and bovine isolates differ significantly based on a number of AMR-related geno- and phenotypic characteristics.
WGS can be used to predict phenotypic resistance in bovine and human-associated Salmonella Typhimurium, Newport, and Dublin with high sensitivity and specificity.
Our study reported here demonstrates that in silico AMR gene predictions are highly correlated with phenotypic resistance in Salmonella enterica Typhimurium, Newport, and Dublin, as AMR genotype correlated with AMR phenotype with an overall sensitivity and specificity of 97.2 and 85.2%, respectively. The ability to predict AMR phenotype from WGS data with high sensitivity and specificity has previously been observed in Salmonella enterica isolated from humans and retail meats (27) and S. Typhimurium from swine (28), as well as in other organisms, including Staphylococcus aureus (29, 30) Campylobacter spp. (31), and Mycobacterium tuberculosis (30). The results of our study further attest to the robustness of WGS in predicting resistance phenotypes in Salmonella enterica serotypes Typhimurium, Newport, and Dublin from both bovine and human sources. Verification of the ability of WGS to predict phenotypic AMR in bovine isolates is important, as isolates from different hosts can be facilitated by different mechanisms, as also shown here. Our data further support that as WGS becomes faster, cheaper, and more accessible, it may represent a valuable tool that could replace classical phenotypic AMR testing across human medical, public health, and veterinary fields.
In this study, the lowest sensitivity of predicting AMR phenotype from genotypic data occurred for NAL. This was not surprising, since the AMR phenotype prediction approach used here was based on the presence of genes that confer resistance to a given antibiotic. While AMR gene-based approaches generally work well, quinolone and fluoroquinolone resistance in particular can result from point mutations in housekeeping genes (e.g., gyrA) rather than from the presence of resistance genes, even though the presence of some resistance genes (e.g., PMQR genes) may also confer low-level resistance to quinolones and fluoroquinolones (20, 32). In our study, the two isolates that showed intermediate resistance to NAL possessed PMQR genes, but no mutations in housekeeping genes are known to confer resistance to quinolones. This is consistent with previous findings, in which isolates possessing PMQR genes have been shown to have reduced susceptibility to quinolones but were not clinically resistant (32). Of the four NAL-resistant isolates, three concurrently possessed PMQR genes and nonsynonymous mutations in the quinolone resistance-determining region (QRDR) of gyrA. One isolate that was NAL resistant due to the presence of only a nonsynonymous mutation in gyrA was falsely predicted to be NAL sensitive, due to an absence of quinolone resistance genes in its genome. This showcases that relying solely on gene presence/absence to predict AMR can result in reduced sensitivity. However, this drawback can be easily alleviated by incorporating SNP-based prediction of AMR (as now has been implemented in the ARG-ANNOT and CARD bioinformatic tools) (33, 34).
In this study, the lowest specificity of WGS-based AMR prediction was observed for STR, which accounted for more than one-half of all phenotype-susceptible/genotype-resistant (P−:G+) discrepancies. Here, more than 50% of these discrepancies were attributed to S. Dublin isolates from the Washington State clade, which carry an allele on a truncated strA that appeared to not confer STR resistance, while still being identified computationally as an STR resistance determinant. Similar discrepancies have been observed in a previous study (35) of Escherichia coli isolates from dairy calves; in this study, point mutations in strA were hypothesized to affect its ability to confer STR resistance. Additionally, a previous study that assessed phenotypic and genotypic resistance in nontyphoidal Salmonella isolated from retail meat and human clinical samples also found STR (P−:G+) discrepancies to be the most common (27). The authors of this previous study suggest that STR (P−:G+) discrepancies could be due to inaccurate clinical breakpoints for STR susceptibility in Salmonella, due in part to the fact that STR is not used to treat enteric infections (27). Overall, these findings suggest that refinement of WGS-based AMR prediction methods could benefit from the incorporation of tools that also classify specific allelic variants of resistance genes for their ability (or inability) to confer resistance. In the future, WGS-based AMR prediction tools that incorporate feedback from clinical use of antibiotics may even further improve the ability of WGS-based tools to predict the clinical outcome of treatment with a given antimicrobial.
Both phenotypic and genomic data show geographic differences in resistance-related characteristics for Salmonella, suggesting a need for location-specific AMR control strategies.
Our data show significant differences between New York and Washington State isolates with regard to AMR-relevant genotypic and phenotypic characteristics. Specifically, when ANOSIM and/or PERMANOVA were used as metrics, Washington and New York State isolates differed by (i) AMR gene sequences (in serotype Newport) and (ii) phenotypic resistance/susceptibility and plasmid replicon presence/absence (in serotype Dublin) (Table 2). In addition, a number of genes, antimicrobials, and plasmid replicons showed strong geographical associations, even after corrections for multiple testing (Table 5). For example, the presence of aadB and cmlA as well as STR resistance was associated with S. Dublin isolates from Washington State. In S. Typhimurium, the IncI1 plasmid replicon, which has been previously associated with extended-spectrum cephalosporin resistance in S. Typhimurium (36, 37), was more commonly detected in isolates from New York State. In S. Dublin, the IncFII(S) plasmid replicon was also more commonly detected in isolates from New York State; the IncFII(S) replicon, along with IncFIB(S), are characteristic of the Salmonella virulence plasmids (38) found in serotypes such as S. Typhimurium and S. Dublin, and it has been proposed that some virulence plasmids previously associated with S. Dublin have evolved from IncFII-like plasmids (39). The geographic differences observed for MDR-relevant genotypic and phenotypic characteristics suggest that different ecological factors and selective pressures may contribute to the development of AMR in different geographical locations (New York State and Washington State in our study here), suggesting a need for geographically specific interventions to effectively combat the spread of AMR. Our findings are consistent with previous studies that have shown that contemporary Salmonella antibiotic resistance patterns differ, even within a given country. For example, Davis et al. (40) showed that a specific MDR Salmonella Typhimurium strain emerged prior to 2000 in bovine populations in the Pacific Northwest (which includes Washington State) but was not found among contemporary isolates from the Northeast. Similarly, a large-scale WGS study of Salmonella Typhi isolates from across the world identified a specific MDR clone that emerged in Asia and Africa with subsequent inter- and intracontinental transmission events (41). Importantly, our findings are also consistent with a WGS-based study (42) of Escherichia coli O157 isolates from different sources (e.g., animals, humans, and the environment/food) and different countries and continents. This study reported significant genetic differences among isolates from different geographical regions and hypothesized that a combination of local emergence events and international transmission leads to a “patchwork” of geographically confined and widely distributed clades. This is similar to what we have observed, as we have identified certain geographic location-specific clones (e.g., a Washington State-specific Dublin clade that carries a truncated strA allele), as well as broadly distributed clonal groups with similar AMR profiles.
S. enterica isolates from humans contain a more diverse range of AMR genes and plasmid replicons than those isolated from bovine populations.
The development and spread of AMR have often been attributed to the misuse of antimicrobials in agricultural setting. However, the AMR profiles of Salmonella isolated from human infections cannot be fully explained by AMR in bovine isolates in this study alone. Here, resistance to CIP, NAL, and SXT were observed only in isolates from humans with salmonellosis. At the genotypic level, over one-half of the total of 42 AMR genes detected in this study were detected only in human isolates. Similar results were observed for plasmid replicons, as nearly one-half of the plasmid replicons detected were found only in human isolates. These results, along with the phylogenetic relationship of the isolates, suggest that some AMR genes are associated primarily with a particular host, with little overlap between species. Mather et al. (14) observed similar results for human- and animal-associated S. Typhimurium DT104: Salmonella isolates from humans and animals, as well as the AMR genes associated with them, were found to remain largely within their respective host populations, with little transmission from animals to humans and vice versa (14).
While many AMR genes and phenotypes were confined to the human isolates in this study, overlaps between the resistomes of bovine and human-associated Salmonella isolates were observed on numerous occasions, with the high degree of AMR sequence identity observed for S. Newport isolates serving as the most prominent example. This also is consistent with previous studies (43–45) that similarly described that certain clonal groups of AMR pathogens can be found in both humans and animals. However, further studies using WGS data from temporally sampled Salmonella enterica are needed to assess the spread of AMR Salmonella and the resistance genes associated with it in New York State and Washington State.
MATERIALS AND METHODS
Isolate selection.
A total of 93 Salmonella isolates were initially selected for the study. Bovine isolates originated from the Washington Animal Disease Diagnostic Laboratory (WADDL), the Washington State Zoonotic Research Unit, the Cornell Animal Health Diagnostic Center (Ithaca, NY), and Salmonella strains isolated from dairy cattle during previous research sampling at dairy farms. Isolates from human clinical specimens were obtained from the Washington State Department of Health Public Health Laboratory and from the New York State Department of Health Laboratory. Isolates were selected to (i) represent isolation dates between 2008 and 2012; (ii) represent one of the three serotypes of interest (Typhimurium, Newport, and Dublin, as determined using traditional serotyping; these serotypes were selected for their association with humans and cattle); and (iii) represent isolates that had previously been tested for phenotypic resistance to antimicrobials and were found to be resistant to at least one antimicrobial. Bovine isolates originated from fecal samples, independent of whether the host presented clinical signs of salmonellosis or not, while human isolates were from stool samples of patients presenting clinical signs of salmonellosis. Among the isolates that met these criteria, “redundant” isolates were filtered out (those known to come from the same animal/farm/farm visit), and selected isolates were chosen to represent approximately equal numbers of human and bovine isolates evenly distributed between New York State and Washington State. To ensure consistency between phenotypic testing methods, all of the isolates selected for this study were retested for phenotypic resistance using a single AMR testing method and a panel of antimicrobial drugs (see “Phenotypic AMR testing” below).
Following WGS (see “Whole-genome sequencing” below), seven isolates were found to belong to species/serotypes different from those to which they were initially assigned. One isolate that had been initially classified as S. enterica serotype Newport was found to belong to the genus Citrobacter. In addition, in silico multilocus sequence typing (MLST) and in silico serotyping using WGS data from the isolates (see “In silico serotyping and MLST” below) revealed that two of the isolates that had been classified as serotypes Typhimurium and Newport using traditional serotyping methods actually belonged to serotypes Give and Montevideo, respectively. These two isolates, as well as the Citrobacter isolate, were excluded from the study. Four isolates that were classified using traditional serotyping as Newport, Typhimurium, Typhimurium, and Dublin were reclassified as Dublin, Newport, Dublin, and Newport, respectively, and remained in the study under the new serotype classifications. A total of 90 isolates (37 S. Typhimurium, 32 S. Newport, and 21 S. Dublin isolates; see Table S1 in the supplemental material for details) were used in all subsequent analyses.
Phenotypic AMR testing.
The antimicrobial susceptibility of each Salmonella isolate was tested using a modified National Antimicrobial Resistance Monitoring System (NARMS) panel of 12 antimicrobial drugs. Susceptibility testing was performed using a Kirby-Bauer disk diffusion agar assay in accordance with the guidelines published by the Clinical and Laboratory Standards Institute (CLSI) and a methodology previously described (46, 47). Internal quality control was performed by the inclusion of E. coli ATCC 25922, which had previously been determined to be pansusceptible, as well as an E. coli isolate that had been previously characterized as positive for the blaCMY-2 gene and resistant to 9 of the antimicrobial agents tested. All isolates were tested using the following panel: ampicillin (AMP) at 10 μg, amoxicillin-clavulanic acid (AMC) at 20 and 10 μg, respectively, cefoxitin (FOX) at 30 μg, ceftiofur (TIO) at 30 μg, ceftriaxone (CRO) at 30 μg, chloramphenicol (CHL) at 30 μg, ciprofloxacin (CIP) at 5 μg, nalidixic acid (NAL) at 30 μg, streptomycin (STR) at 10 μg, tetracycline (TET) at 30 μg, sulfisoxazole (SX) at 250 μg, and trimethoprim-sulfamethoxazole (SXT) at 23.75 and 1.25 μg, respectively. Results of the disk diffusion test for the internal quality control strains were within the anticipated standards. Isolates were categorized as susceptible, intermediate, or resistant (SIR) by measuring the inhibition zone and using interpretive criteria and breakpoints established by the CLSI guidelines for each antimicrobial (46).
Whole-genome sequencing.
Isolates were plated on brain heart infusion (BHI) agar (Becton, Dickinson and Company, Franklin Lakes, NJ), grown for 24 h, and inoculated into 1.0 ml BHI broth in a Nunc U96 PP 2-ml DeepWell Natural plate (Fisher Scientific, Pittsburgh, PA). Following overnight incubation at 37°C, cells were pelleted by centrifugation at 3,320 relative centrifugal force (RCF) for 15 min. DNA extraction for the majority of isolates was performed with the DNeasy 96 blood and tissue kit (Qiagen, Valencia, CA) according to the manufacturer's specifications for high-throughput applications. DNA extraction for a smaller group of isolates was performed using the QIAamp DNA minikit (Qiagen, Valencia, CA) according to the manufacturer's protocol for bacteria. DNA was eluted in 50 μl Tris-HCl at pH 8.0 and stored at 4°C prior to sequencing. Following an initial spectrophotometry step to determine the optical density at 260 nm (OD260)/OD280 measurements, the genomic DNA from each isolate was quantified using a fluorescent nucleic acid dye (Picogreen; Invitrogen, Paisley, UK) and diluted to 200 pg/μl. Sequencing libraries were prepared using the Nextera XT DNA sample preparation kit and the associated Nextera XT Index kit with 96 indices (Illumina, Inc., San Diego, CA) according to the manufacturer's instructions. Pooled samples were sequenced with 2 lanes of an Illumina HiSeq 2500 rapid run with 2 × 100-bp paired-end sequencing.
Initial data processing and genome assembly.
Illumina sequencing adapters and low-quality bases were trimmed using Trimmomatic version 0.32 for Nextera paired-end reads (48). FastQC version 0.11.2 was used to confirm that all adapter sequences had been removed and that the read quality was appropriate (49). Genomes were assembled de novo using SPAdes version 3.0.0, as SPAdes has been shown to produce few misassemblies and yield contigs with high N50 values when assembling bacterial genomes de novo from Illumina short reads (50). Genome coverage was determined using BBMap version 35.49 (51) and samtools version 0.1.19-96b5f2294a (52).
In silico serotyping and MLST.
To assess the results of traditional serotyping, in silico serotyping was performed using SeqSero and the assembled genome for each isolate (53). In addition, MLST was performed using the Short Read Sequence Typer 2 version 0.1.5 (SRST2) and the trimmed Illumina paired-end reads (54). Sequence types were associated with serotypes using the University of Warwick's MLST database for Salmonella (55).
In silico AMR gene detection.
AMR genes were detected in all 90 assembled genomes using nucleotide BLAST (blastn) version 2.4.0 (56) and the formatted ARG-ANNOT database included with SRST2 (33, 54). To prevent overlapping hits due to the presence of multiple alleles of the same gene in the database, one gene was selected from each SRST2-ARG-ANNOT gene group and used to build a reduced database (54). Genes that were detected using blastn and belonged to a particular gene group were categorized as being present in a genome if they were detected at ≥50% coverage and ≥75% nucleotide identity.
Initial phylogenetic tree construction and reference genome selection.
The closed chromosomal sequences of S. Typhimurium strain LT2 (RefSeq NC_003197.1), S. Newport strain SL254 (GenBank accession no. CP001113), and S. Dublin strain CT_02021853 (RefSeq NC_011205.1) were chosen as candidate reference sequences for reference-based SNP calling. To obtain an initial phylogeny of all isolates and determine if these candidate reference sequences clustered appropriately with the genomes of the isolates used in this study, a phylogenetic tree was constructed using the assembled genomes of all 90 isolates and the three candidate reference genomes using kSNP version 2.1.2 (57). Kchooser was used to determine an optimum k-mer size of 19 (57). This core SNP phylogeny based on the genomes of all 90 isolates used in the study, as well as three closed reference genomes from GenBank, clustered isolates into three distinct clades (see Fig. S1 in the supplemental material). As a result, all subsequent analyses were performed within each serotype clade to maximize resolution.
Reference-based variant calling.
Variant calling was performed within each of the three serotypes using the Cortex variant caller (cortex_var) (58). For S. Typhimurium isolates, S. Typhimurium strain LT2 was used as a reference genome. For S. Newport isolates, S. Newport strain SL254 was used as a reference, as all of the Newport isolates in this study were predicted to have the same sequence type (ST45) using SRST2 (59). For S. Dublin isolates, strain CT_02021853, which was used as a candidate reference in the initial phylogenetic tree, clustered relatively far from the closely related S. Dublin isolates used in this study. In order to obtain better resolution, variant calling was performed a second time using the contigs of isolate BOV_DUBN_WA_10_R9_3233 as a reference, as its assembly had the highest coverage of all of the S. Dublin isolates used in the study. An additional 11 SNPs were found using isolate BOV_DUBN_WA_10_R9_3233 as a reference; these SNPs were included in subsequent analyses.
SNPs were filtered from other variants using Plink/Seq version 0.10 (60), and recombination events were filtered out using Gubbins version 1.4.2 (61). Within each serotype, only SNPs at positions present in all genomes were used. MEGA6 was used to identify the best nucleotide substitution models for SNPs within each serotype (62). For S. Typhimurium, the general time-reversible (GTR) model was selected as the best model (63), while the Kimura 2-parameter model (64) was selected for both S. Newport and S. Dublin.
For each serotype, BEAST version 1.8.2 (65) was used to construct rooted phylogenetic trees. An ascertainment bias correction was applied to account for the use of solely variant sites (66). The best nucleotide substitution model, as determined by MEGA6, was used for each serotype, and base frequencies were estimated. Temporal signals, which were assessed using Path-O-Gen version 1.4 (now TempEst) (67), were not strong enough to estimate evolutionary rates using sampling dates (R < 0.10). As a result, the clock rate was set to 1.0 and tip dates were not used. For each serotype, combinations of either a strict or lognormal relaxed molecular clock (68) and either a coalescent constant size or Bayesian skyline population (69) were tested. Trees were constructed using chain lengths of 100 million generations, with sampling every 10,000 generations. Path sampling analyses (70, 71) were performed using 100 steps of 1 million generations, sampling every 1,000 generations. Bayes factors were calculated to determine which combination of molecular clock and population models best modeled each serotype. For S. Typhimurium and S. Newport, the best model used a relaxed molecular clock with a constant coalescent population model. For S. Dublin, the best model used a strict molecular clock with a constant coalescent population.
Plasmid replicon detection.
Plasmids replicons were detected in all whole-genome sequences using PlasmidFinder version 1.3 (38). An identity cutoff of 80% was used. PlasmidFinder was also used to confirm that plasmid replicons could not be detected in the chromosomal sequences of S. Typhimurium LT2, S. Newport SL254, and S. Dublin CT_02021853.
Statistical analyses.
Matrices were created using (i) the sequences of all AMR genes detected using blastn, (ii) phenotypic antimicrobial resistance/susceptibility, and (iii) the presence/absence of plasmid replicons detected using PlasmidFinder. For the phenotypic resistance matrix, isolates showing resistance or intermediate resistance to a particular antimicrobial, using NARMS breakpoints, were treated as resistant and given a value of 1, while susceptible isolates were given a value of 0. Fisher's exact tests were conducted to test whether a given AMR gene, AMR phenotype, or plasmid replicon was statistically associated with a particular source and/or geographic location using the fisher.test function in R version 3.3.0 (72). When performing Fisher's exact tests for each serotype category with n isolates, gene groups, AMR phenotypes, and plasmid replicons present in fewer than 3 and more than n − 3 isolates were not tested. A Holm-Bonferroni correction was applied to each test to correct for multiple comparisons (73). Additionally, Fisher's exact tests were used to test if any AMR gene groups were statistically associated with any plasmid replicons. Plasmid replicons present in fewer than 5 and more than n − 5 isolates were not tested, and a Bonferroni correction was applied to correct for multiple comparisons. Analysis of similarity (ANOSIM) (74) using the anosim function in the vegan package (75) in R was used to determine if the average ranks of within-serotype, within-source, and within-geographic-group distances were greater than or equal to the average ranks of between-group distances using AMR gene sequences, phenotypic resistance to a particular antimicrobial, and/or plasmid replicon presence/absence data (76). For ANOSIM simulations using AMR gene sequences, 5 runs of 10,000 permutations using unweighted unifrac distances (77) were conducted. For all ANOSIM simulations using phenotypic resistance/susceptibility and plasmid replicon presence/absence matrices, 5 runs of 10,000 permutations using Raup-Crick dissimilarities (78) were conducted. PERMANOVA (79) was performed to test whether the centroids of serotype, source, and geographic groups were equivalent for all groups (76) based on AMR gene sequences, phenotypic resistance to a particular antimicrobial, and/or plasmid replicon presence/absence using the adonis function in R's vegan package (75). Three runs of 10,000 permutations using unweighted unifrac distances were used to obtain mean PERMANOVA test statistics (F) and P values for AMR gene sequences, while three runs of 100,000 permutations and Raup-Crick distances were used for phenotypic resistance/susceptibility and plasmid replicon presence/absence data. The metaMDS function in the vegan package was used to perform nonmetric multidimensional scaling (NMDS) (80, 81) using monoMDS (75), a maximum of 10,000 random starts, and an appropriate distance metric (unweighted unifrac distances for AMR gene sequence data and Raup-Crick dissimilarities for phenotypic resistance/susceptibility and plasmid replicon presence/absence data). Interactive NMDS plots can be found at https://github.com/lmc297/2017_AEM_Figure_S2.
Descriptive analyses of the susceptible/intermediate/resistant (SIR) distribution of Salmonella isolates by antimicrobial drug and distribution of AMR phenotypes and genes were performed using PROC FREQ in SAS (SAS Institute Inc., USA). To evaluate the effect of presence or absence of resistance genes on the mean zone diameter (in centimeters) of the Kirby-Bauer disk diffusion test, multivariable mixed logistic regression models were fitted to the data using the Glimmix procedure of SAS. The independent variables (i) isolate source (bovine or human), (ii) isolation location (New York State or Washington State), and (iii) serotype were included in all models.
Accession number(s).
Paired-end reads for the 90 isolates used in this study have been deposited in the National Center for Biotechnology Information's (NCBI) Sequence Read Archive (SRA) under study accession number SRP068320.
Supplementary Material
ACKNOWLEDGMENTS
This material is based on work supported by the National Science Foundation Graduate Research Fellowship Program under grant no. DGE-1144153.
Research reported in this publication was supported by the Agriculture and Food Research Initiative Competitive Grant no. 2010-51110-21131 from the USDA National Institute of Food and Agriculture. The content is solely the responsibility of the authors and does not necessarily represent the official views of the USDA.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00140-17.
REFERENCES
- 1.Scallan E, Hoekstra RM, Angulo FJ, Tauxe RV, Widdowson MA, Roy SL, Jones JL, Griffin PM. 2011. Foodborne illness acquired in the United States—major pathogens. Emerg Infect Dis 17:7–15. doi: 10.3201/eid1701.P11101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.CDC. 2013. Antibiotic resistance threats in the United States, 2013. CDC, Atlanta, GA. [Google Scholar]
- 3.WHO. 2014. Antimicrobial resistance: global report on surveillance. WHO, Geneva, Switzerland. [Google Scholar]
- 4.Van Boeckel TP, Brower C, Gilbert M, Grenfell BT, Levin SA, Robinson TP, Teillant A, Laxminarayan R. 2015. Global trends in antimicrobial use in food animals. Proc Natl Acad Sci U S A 112:5649–5654. doi: 10.1073/pnas.1503141112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Silbergeld EK, Graham J, Price LB. 2008. Industrial food animal production, antimicrobial resistance, and human health. Annu Rev Public Health 29:151–169. doi: 10.1146/annurev.publhealth.29.020907.090904. [DOI] [PubMed] [Google Scholar]
- 6.Hendriksen SW, Orsel K, Wagenaar JA, Miko A, van Duijkeren E. 2004. Animal-to-human transmission of Salmonella Typhimurium DT104A variant. Emerg Infect Dis 10:2225–2227. doi: 10.3201/eid1012.040286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fey PD, Safranek TJ, Rupp ME, Dunne EF, Ribot E, Iwen PC, Bradford PA, Angulo FJ, Hinrichs SH. 2000. Ceftriaxone-resistant Salmonella infection acquired by a child from cattle. N Engl J Med 342:1242–1249. doi: 10.1056/NEJM200004273421703. [DOI] [PubMed] [Google Scholar]
- 8.Hoelzer K, Moreno Switt AI, Wiedmann M. 2011. Animal contact as a source of human non-typhoidal salmonellosis. Vet Res 42:34. doi: 10.1186/1297-9716-42-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.White DG, Zhao S, Sudler R, Ayers S, Friedman S, Chen S, McDermott PF, McDermott S, Wagner DD, Meng J. 2001. The isolation of antibiotic-resistant Salmonella from retail ground meats. N Engl J Med 345:1147–1154. doi: 10.1056/NEJMoa010315. [DOI] [PubMed] [Google Scholar]
- 10.Cody SH, Abbott SL, Marfin AA, Schulz B, Wagner P, Robbins K, Mohle-Boetani JC, Vugia DJ. 1999. Two outbreaks of multidrug-resistant Salmonella serotype Typhimurium DT104 infections linked to raw-milk cheese in Northern California. JAMA 281:1805–1810. doi: 10.1001/jama.281.19.1805. [DOI] [PubMed] [Google Scholar]
- 11.Hald T, Aspinall W, Devleesschauwer B, Cooke R, Corrigan T, Havelaar AH, Gibb HJ, Torgerson PR, Kirk MD, Angulo FJ, Lake RJ, Speybroeck N, Hoffmann S. 2016. World Health Organization estimates of the relative contributions of food to the burden of disease due to selected foodborne hazards: a structured expert elicitation. PLoS One 11:e0145839. doi: 10.1371/journal.pone.0145839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnson JR, Sannes MR, Croy C, Johnston B, Clabots C, Kuskowski MA, Bender J, Smith KE, Winokur PL, Belongia EA. 2007. Antimicrobial drug-resistant Escherichia coli from humans and poultry products, Minnesota and Wisconsin, 2002-2004. Emerg Infect Dis 13:838–846. doi: 10.3201/eid1306.061576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Price LB, Stegger M, Hasman H, Aziz M, Larsen J, Andersen PS, Pearson T, Waters AE, Foster JT, Schupp J, Gillece J, Driebe E, Liu CM, Springer B, Zdovc I, Battisti A, Franco A, Zmudzki J, Schwarz S, Butaye P, Jouy E, Pomba C, Porrero MC, Ruimy R, Smith TC, Robinson DA, Weese JS, Arriola CS, Yu F, Laurent F, Keim P, Skov R, Aarestrup FM. 2012. Staphylococcus aureus CC398: host adaptation and emergence of methicillin resistance in livestock. mBio 3(1):e00305-11. doi: 10.1128/mBio.00305-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mather AE, Reid SW, Maskell DJ, Parkhill J, Fookes MC, Harris SR, Brown DJ, Coia JE, Mulvey MR, Gilmour MW, Petrovska L, de Pinna E, Kuroda M, Akiba M, Izumiya H, Connor TR, Suchard MA, Lemey P, Mellor DJ, Haydon DT, Thomson NR. 2013. Distinguishable epidemics of multidrug-resistant Salmonella Typhimurium DT104 in different hosts. Science 341:1514–1517. doi: 10.1126/science.1240578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mather AE, Matthews L, Mellor DJ, Reeve R, Denwood MJ, Boerlin P, Reid-Smith RJ, Brown DJ, Coia JE, Browning LM, Haydon DT, Reid SW. 2012. An ecological approach to assessing the epidemiology of antimicrobial resistance in animal and human populations. Proc Biol Sci 279:1630–1639. doi: 10.1098/rspb.2011.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kwong JC, Mercoulia K, Tomita T, Easton M, Li HY, Bulach DM, Stinear TP, Seemann T, Howden BP. 2016. Prospective whole-genome sequencing enhances national surveillance of Listeria monocytogenes. J Clin Microbiol 54:333–342. doi: 10.1128/JCM.02344-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holmes A, Allison L, Ward M, Dallman TJ, Clark R, Fawkes A, Murphy L, Hanson M. 2015. Utility of whole-genome sequencing of Escherichia coli O157 for outbreak detection and epidemiological surveillance. J Clin Microbiol 53:3565–3573. doi: 10.1128/JCM.01066-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Taylor AJ, Lappi V, Wolfgang WJ, Lapierre P, Palumbo MJ, Medus C, Boxrud D. 2015. Characterization of foodborne outbreaks of Salmonella enterica serovar Enteritidis with whole-genome sequencing single nucleotide polymorphism-based analysis for surveillance and outbreak detection. J Clin Microbiol 53:3334–3340. doi: 10.1128/JCM.01280-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fricke WF, Welch TJ, McDermott PF, Mammel MK, LeClerc JE, White DG, Cebula TA, Ravel J. 2009. Comparative genomics of the IncA/C multidrug resistance plasmid family. J Bacteriol 191:4750–4757. doi: 10.1128/JB.00189-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cloeckaert A, Chaslus-Dancla E. 2001. Mechanisms of quinolone resistance in Salmonella. Vet Res 32:291–300. doi: 10.1051/vetres:2001105. [DOI] [PubMed] [Google Scholar]
- 21.Andres P, Lucero C, Soler-Bistue A, Guerriero L, Albornoz E, Tran T, Zorreguieta A, PMQR Group, Galas M, Corso A, Tolmasky ME, Petroni A. 2013. Differential distribution of plasmid-mediated quinolone resistance genes in clinical enterobacteria with unusual phenotypes of quinolone susceptibility from Argentina. Antimicrob Agents Chemother 57:2467–2475. doi: 10.1128/AAC.01615-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hansen LH, Jensen LB, Sorensen HI, Sorensen SJ. 2007. Substrate specificity of the OqxAB multidrug resistance pump in Escherichia coli and selected enteric bacteria. J Antimicrob Chemother 60:145–147. doi: 10.1093/jac/dkm167. [DOI] [PubMed] [Google Scholar]
- 23.WHO. 2015. WHO estimates of the global burden of foodborne diseases, 2007–2015 WHO, Geneva, Switzerland. [Google Scholar]
- 24.Helms M, Ethelberg S, Molbak K, DT104 Study Group. 2005. International Salmonella Typhimurium DT104 infections, 1992-2001. Emerg Infect Dis 11:859–867. doi: 10.3201/eid1106.041017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leekitcharoenphon P, Hendriksen RS, Le Hello S, Weill FX, Baggesen DL, Jun SR, Ussery DW, Lund O, Crook DW, Wilson DJ, Aarestrup FM. 2016. Global genomic epidemiology of Salmonella enterica serovar Typhimurium DT104. Appl Environ Microbiol 82:2516–2526. doi: 10.1128/AEM.03821-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ribot EM, Wierzba RK, Angulo FJ, Barrett TJ. 2002. Salmonella enterica serotype Typhimurium DT104 isolated from humans, United States, 1985, 1990, and 1995. Emerg Infect Dis 8:387–391. doi: 10.3201/eid0804.010202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McDermott PF, Tyson GH, Kabera C, Chen Y, Li C, Folster JP, Ayers SL, Lam C, Tate HP, Zhao S. 2016. Whole-genome sequencing for detecting antimicrobial resistance in nontyphoidal Salmonella. Antimicrob Agents Chemother 60:5515–5520. doi: 10.1128/AAC.01030-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zankari E, Hasman H, Kaas RS, Seyfarth AM, Agerso Y, Lund O, Larsen MV, Aarestrup FM. 2013. Genotyping using whole-genome sequencing is a realistic alternative to surveillance based on phenotypic antimicrobial susceptibility testing. J Antimicrob Chemother 68:771–777. doi: 10.1093/jac/dks496. [DOI] [PubMed] [Google Scholar]
- 29.Gordon NC, Price JR, Cole K, Everitt R, Morgan M, Finney J, Kearns AM, Pichon B, Young B, Wilson DJ, Llewelyn MJ, Paul J, Peto TE, Crook DW, Walker AS, Golubchik T. 2014. Prediction of Staphylococcus aureus antimicrobial resistance by whole-genome sequencing. J Clin Microbiol 52:1182–1191. doi: 10.1128/JCM.03117-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bradley P, Gordon NC, Walker TM, Dunn L, Heys S, Huang B, Earle S, Pankhurst LJ, Anson L, de Cesare M, Piazza P, Votintseva AA, Golubchik T, Wilson DJ, Wyllie DH, Diel R, Niemann S, Feuerriegel S, Kohl TA, Ismail N, Omar SV, Smith EG, Buck D, McVean G, Walker AS, Peto TE, Crook DW, Iqbal Z. 2015. Rapid antibiotic-resistance predictions from genome sequence data for Staphylococcus aureus and Mycobacterium tuberculosis. Nat Commun 6:10063. doi: 10.1038/ncomms10063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao S, Tyson GH, Chen Y, Li C, Mukherjee S, Young S, Lam C, Folster JP, Whichard JM, McDermott PF. 2016. Whole-genome sequencing analysis accurately predicts antimicrobial resistance phenotypes in Campylobacter spp. Appl Environ Microbiol 82:459–466. doi: 10.1128/AEM.02873-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hooper DC, Jacoby GA. 2015. Mechanisms of drug resistance: quinolone resistance. Ann N Y Acad Sci 1354:12–31. doi: 10.1111/nyas.12830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gupta SK, Padmanabhan BR, Diene SM, Lopez-Rojas R, Kempf M, Landraud L, Rolain JM. 2014. ARG-ANNOT, a new bioinformatic tool to discover antibiotic resistance genes in bacterial genomes. Antimicrob Agents Chemother 58:212–220. doi: 10.1128/AAC.01310-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jia B, Raphenya AR, Alcock B, Waglechner N, Guo P, Tsang KK, Lago BA, Dave BM, Pereira S, Sharma AN, Doshi S, Courtot M, Lo R, Williams LE, Frye JG, Elsayegh T, Sardar D, Westman EL, Pawlowski AC, Johnson TA, Brinkman FS, Wright GD, McArthur AG. 2017. CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res 45(D1):D566–D573. doi: 10.1093/nar/gkw1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Davis MA, Besser TE, Orfe LH, Baker KN, Lanier AS, Broschat SL, New D, Call DR. 2011. Genotypic-phenotypic discrepancies between antibiotic resistance characteristics of Escherichia coli isolates from calves in management settings with high and low antibiotic use. Appl Environ Microbiol 77:3293–3299. doi: 10.1128/AEM.02588-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Folster JP, Tolar B, Pecic G, Sheehan D, Rickert R, Hise K, Zhao S, Fedorka-Cray PJ, McDermott P, Whichard JM. 2014. Characterization of blaCMY plasmids and their possible role in source attribution of Salmonella enterica serotype Typhimurium infections. Foodborne Pathog Dis 11:301–306. doi: 10.1089/fpd.2013.1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Madec JY, Doublet B, Ponsin C, Cloeckaert A, Haenni M. 2011. Extended-spectrum beta-lactamase blaCTX-M-1 gene carried on an IncI1 plasmid in multidrug-resistant Salmonella enterica serovar Typhimurium DT104 in cattle in France. J Antimicrob Chemother 66:942–944. doi: 10.1093/jac/dkr014. [DOI] [PubMed] [Google Scholar]
- 38.Carattoli A, Zankari E, Garcia-Fernandez A, Voldby Larsen M, Lund O, Villa L, Moller Aarestrup F, Hasman H. 2014. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother 58:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chu C, Feng Y, Chien AC, Hu S, Chu CH, Chiu CH. 2008. Evolution of genes on the Salmonella Virulence plasmid phylogeny revealed from sequencing of the virulence plasmids of S. enterica serotype Dublin and comparative analysis. Genomics 92:339–343. doi: 10.1016/j.ygeno.2008.07.010. [DOI] [PubMed] [Google Scholar]
- 40.Davis MA, Besser TE, Eckmann K, MacDonald K, Green D, Hancock DD, Baker KN, Warnick LD, Soyer Y, Wiedmann M, Call DR. 2007. Multidrug-resistant Salmonella Typhimurium, Pacific Northwest, United States. Emerg Infect Dis 13:1583–1586. doi: 10.3201/eid1310.070536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wong VK, Baker S, Pickard DJ, Parkhill J, Page AJ, Feasey NA, Kingsley RA, Thomson NR, Keane JA, Weill FX, Edwards DJ, Hawkey J, Harris SR, Mather AE, Cain AK, Hadfield J, Hart PJ, Thieu NT, Klemm EJ, Glinos DA, Breiman RF, Watson CH, Kariuki S, Gordon MA, Heyderman RS, Okoro C, Jacobs J, Lunguya O, Edmunds WJ, Msefula C, Chabalgoity JA, Kama M, Jenkins K, Dutta S, Marks F, Campos J, Thompson C, Obaro S, MacLennan CA, Dolecek C, Keddy KH, Smith AM, Parry CM, Karkey A, Mulholland EK, Campbell JI, Dongol S, Basnyat B, Dufour M, Bandaranayake D, Naseri TT, Singh SP, Hatta M, Newton P, Onsare RS, Isaia L, Dance D, Davong V, Thwaites G, Wijedoru L, Crump JA, De Pinna E, Nair S, Nilles EJ, Thanh DP, Turner P, Soeng S, Valcanis M, Powling J, Dimovski K, Hogg G, Farrar J, Holt KE, Dougan G. 2015. Phylogeographical analysis of the dominant multidrug-resistant H58 clade of Salmonella Typhi identifies inter- and intracontinental transmission events. Nat Genet 47:632–639. doi: 10.1038/ng.3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Strachan NJ, Rotariu O, Lopes B, MacRae M, Fairley S, Laing C, Gannon V, Allison LJ, Hanson MF, Dallman T, Ashton P, Franz E, van Hoek AH, French NP, George T, Biggs PJ, Forbes KJ. 2015. Whole genome sequencing demonstrates that geographic variation of Escherichia coli O157 genotypes dominates host association. Sci Rep 5:14145. doi: 10.1038/srep14145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Spoor LE, McAdam PR, Weinert LA, Rambaut A, Hasman H, Aarestrup FM, Kearns AM, Larsen AR, Skov RL, Fitzgerald JR. 2013. Livestock origin for a human pandemic clone of community-associated methicillin-resistant Staphylococcus aureus. mBio 4(4):e00356-13. doi: 10.1128/mBio.00356-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ward MJ, Gibbons CL, McAdam PR, van Bunnik BA, Girvan EK, Edwards GF, Fitzgerald JR, Woolhouse ME. 2014. Time-scaled evolutionary analysis of the transmission and antibiotic resistance dynamics of Staphylococcus aureus clonal complex 398. Appl Environ Microbiol 80:7275–7282. doi: 10.1128/AEM.01777-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Madec JY, Haenni M, Nordmann P, Poirel L. 2017. ESBL/AmpC- and carbapenemase-producing Enterobacteriaceae in animals: a threat for humans? Clin Microbiol Infect doi: 10.1016/j.cmi.2017.01.013. [DOI] [PubMed] [Google Scholar]
- 46.CLSI. 2012. Performance standards for antimicrobial susceptibility testing; twenty-second informational supplement. M100-D22, 22nd ed Clinical Laboratory Standard Institute, Wayne, PA. [Google Scholar]
- 47.CLSI. 2013. Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals approved standard—fourth edition. VET01-A4, 3rd ed Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 48.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Babraham Bioinformatics. 2014. FastQC v. 0.11.2. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
- 50.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bushnell B. 2015. BBMap v. 35.49, https://sourceforge.net/projects/bbmap/.
- 52.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup. 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang S, Yin Y, Jones MB, Zhang Z, Deatherage Kaiser BL, Dinsmore BA, Fitzgerald C, Fields PI, Deng X. 2015. Salmonella serotype determination utilizing high-throughput genome sequencing data. J Clin Microbiol 53:1685–1692. doi: 10.1128/JCM.00323-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Inouye M, Dashnow H, Raven LA, Schultz MB, Pope BJ, Tomita T, Zobel J, Holt KE. 2014. SRST2: rapid genomic surveillance for public health and hospital microbiology labs. Genome Med 6:90. doi: 10.1186/s13073-014-0090-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.University of Warwick. 2015. Salmonella enterica MLST database. University of Warwick, Coventry, England. [Google Scholar]
- 56.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gardner SN, Hall BG. 2013. When whole-genome alignments just won't work: kSNP v2 software for alignment-free SNP discovery and phylogenetics of hundreds of microbial genomes. PLoS One 8:e81760. doi: 10.1371/journal.pone.0081760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Iqbal Z, Caccamo M, Turner I, Flicek P, McVean G. 2012. De novo assembly and genotyping of variants using colored de Bruijn graphs. Nat Genet 44:226–232. doi: 10.1038/ng.1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sangal V, Harbottle H, Mazzoni CJ, Helmuth R, Guerra B, Didelot X, Paglietti B, Rabsch W, Brisse S, Weill FX, Roumagnac P, Achtman M. 2010. Evolution and population structure of Salmonella enterica serovar Newport. J Bacteriol 192:6465–6476. doi: 10.1128/JB.00969-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.PLINK/Seq.2014. PLINK/Seq v. 0.10. https://atgu.mgh.harvard.edu/plinkseq/.
- 61.Croucher NJ, Page AJ, Connor TR, Delaney AJ, Keane JA, Bentley SD, Parkhill J, Harris SR. 2015. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res 43:e15. doi: 10.1093/nar/gku1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tavare S. 1986. Some probabilistic and statistical problems in the analysis of DNA sequences, p 57–86. In Miura RM. (ed), Lectures on mathematics in the life sciences, vol 17 American Mathematical Society, Providence, RI. [Google Scholar]
- 64.Kimura M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 65.Drummond AJ, Suchard MA, Xie D, Rambaut A. 2012. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol 29:1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rambaut A. 2013. Analysis of variable sites only in BEAST or MrBayes. https://groups.google.com/forum/#!topic/beast-users/V5vRghILMfw.
- 67.Rambaut A, Lam TT, Max Carvalho L, Pybus OG. 2016. Exploring the temporal structure of heterochronous sequences using TempEst (formerly Path-O-Gen). Virus Evol 2(1):vew007. doi: 10.1093/ve/vew007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Drummond AJ, Ho SY, Phillips MJ, Rambaut A. 2006. Relaxed phylogenetics and dating with confidence. PLoS Biol 4:e88. doi: 10.1371/journal.pbio.0040088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Drummond AJ, Rambaut A, Shapiro B, Pybus OG. 2005. Bayesian coalescent inference of past population dynamics from molecular sequences. Mol Biol Evol 22:1185–1192. doi: 10.1093/molbev/msi103. [DOI] [PubMed] [Google Scholar]
- 70.Baele G, Li WL, Drummond AJ, Suchard MA, Lemey P. 2013. Accurate model selection of relaxed molecular clocks in Bayesian phylogenetics. Mol Biol Evol 30:239–243. doi: 10.1093/molbev/mss243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Baele G, Lemey P, Bedford T, Rambaut A, Suchard MA, Alekseyenko AV. 2012. Improving the accuracy of demographic and molecular clock model comparison while accommodating phylogenetic uncertainty. Mol Biol Evol 29:2157–2167. doi: 10.1093/molbev/mss084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.R Core Team. 2016. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: https://www.R-project.org/. [Google Scholar]
- 73.Holm S. 1979. A simple sequentially rejective multiple test procedure. Scand J Statistics 6:65–70. [Google Scholar]
- 74.Clarke KR. 1993. Non-parametric multivariate analyses of changes in community structure. Austral J Ecol 18:117–143. doi: 10.1111/j.1442-9993.1993.tb00438.x. [DOI] [Google Scholar]
- 75.Oksanen J, Blanchet FG, Friendly M, Kindt R, Legendre P, McGlinn D, Minchin PR, O'Hara RB, Simpson GL, Solymos P, Stevens MHH, Szoecs E, Helene Wagner H. 2016. vegan: community ecology package, v2.4-0. https://CRAN.R-project.org/package=vegan.
- 76.Anderson MJ, Walsh DCI. 2013. PERMANOVA, ANOSIM, and the Mantel test in the face of heterogeneous dispersions: what null hypothesis are you testing? Ecol Monographs 83:557–574. doi: 10.1890/12-2010.1. [DOI] [Google Scholar]
- 77.Lozupone C, Knight R. 2005. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol 71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chase JM, Kraft NJB, Smith KG, Vellend M, Inouye BD. 2011. Using null models to disentangle variation in community dissimilarity from variation in α-diversity. Ecosphere 2:1–11. doi: 10.1890/ES10-00117.1. [DOI] [Google Scholar]
- 79.Anderson MJ. 2001. A new method for non-parametric multivariate analysis of variance. Austral Ecol 26:32–46. [Google Scholar]
- 80.Kruskal JB. 1964. Multidimensional scaling by optimizing goodness of fit to a nonmetric hypothesis. Psychometrika 29:1–27. doi: 10.1007/BF02289565. [DOI] [Google Scholar]
- 81.Kruskal JB. 1964. Nonmetric multidimensional scaling: a numerical method. Psychometrika 29:115–129. doi: 10.1007/BF02289694. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.