ABSTRACT
Of the few predicted extracellular glycan-active enzymes, glycoside hydrolase family 13 subfamily 14 (GH13_14) pullulanases are the most common in human gut lactobacilli. These enzymes share a unique modular organization, not observed in other bacteria, featuring a catalytic module, two starch binding modules, a domain of unknown function, and a C-terminal surface layer association protein (SLAP) domain. Here, we explore the specificity of a representative of this group of pullulanases, Lactobacillus acidophilus Pul13_14 (LaPul13_14), and its role in branched α-glucan metabolism in the well-characterized Lactobacillus acidophilus NCFM, which is widely used as a probiotic. Growth experiments with L. acidophilus NCFM on starch-derived branched substrates revealed a preference for α-glucans with short branches of about two to three glucosyl moieties over amylopectin with longer branches. Cell-attached debranching activity was measurable in the presence of α-glucans but was repressed by glucose. The debranching activity is conferred exclusively by LaPul13_14 and is abolished in a mutant strain lacking a functional LaPul13_14 gene. Hydrolysis kinetics of recombinant LaPul13_14 confirmed the preference for short-branched α-glucan oligomers consistent with the growth data. Curiously, this enzyme displayed the highest catalytic efficiency and the lowest Km reported for a pullulanase. Inhibition kinetics revealed mixed inhibition by β-cyclodextrin, suggesting the presence of additional glucan binding sites besides the active site of the enzyme, which may contribute to the unprecedented substrate affinity. The enzyme also displays high thermostability and higher activity in the acidic pH range, reflecting adaptation to the physiologically challenging conditions in the human gut.
IMPORTANCE Starch is one of the most abundant glycans in the human diet. Branched α-1,6-glucans in dietary starch and glycogen are nondegradable by human enzymes and constitute a metabolic resource for the gut microbiota. The role of health-beneficial lactobacilli prevalent in the human small intestine in starch metabolism remains unexplored in contrast to colonic bacterial residents. This study highlights the pivotal role of debranching enzymes in the breakdown of starchy branched α-glucan oligomers (α-limit dextrins) by human gut lactobacilli exemplified by Lactobacillus acidophilus NCFM, which is one of the best-characterized strains used as probiotics. Our data bring novel insight into the metabolic preference of L. acidophilus for α-glucans with short α-1,6-branches. The unprecedented affinity of the debranching enzyme that confers growth on these substrates reflects its adaptation to the nutrient-competitive gut ecological niche and constitutes a potential advantage in cross-feeding from human and bacterial dietary starch metabolism.
KEYWORDS: glycoside hydrolase family 13, human gut microbiota, starch, starch binding modules
INTRODUCTION
The human gastrointestinal tract is inhabited by a vast, diverse, and dynamic microbial community (1), which is shaped by competition among the different taxa and selection by the host. Firmicutes and Bacteroidetes are the prevalent bacterial phyla of the human gut microbiota (HGM), followed by Actinobacteria, Proteobacteria, and Verrucomicrobia in healthy adults (2). This microbial community provides protection against enteric pathogens and endows the host with metabolic activities that are not encoded in the human genome. More importantly, the interplay between diet and the HGM is currently recognized as a major effector of the composition of this community (2, 3) and as a negotiator of human metabolism (4, 5). A key feature of the HGM is the ability to harvest energy from both host-derived and dietary glucans, particularly those resistant to digestion by human enzymes (6). Consequently, differential glycan metabolism is a key affecter of the microbiota composition (7).
Starch is the most abundant glycan in the human diet. This polysaccharide is a composite of two α-glucans: the linear α-1,4-glucan amylose and amylopectin, which possesses approximately 5% α-1,6-branch points with average branch lengths of 18 to 25 glucosyl moieties (8). Starch occurs naturally as supramolecular insoluble granules with semicrystalline regions (8). These granules differ in size and structural properties as well as digestibility by bacterial and human enzymes. Human digestive enzymes mainly target the α-1,4-glucosidic bonds but are less efficient in hydrolyzing α-1,6-branches in starch (9). The digestibility of starch varies considerably based on botanical origin, crystal packing, and processing (10). Significant amounts of dietary starch escape digestion in the upper gastrointestinal tract (resistant starch [RS]) (11) and are fermented in the colon by members of the gut microbiota (12–15). The small intestine, however, is dominated by bacteria from the Gram-positive Lactobacillaceae and the Gram-negative Enterobacteriaceae families (16). The former family contains human-gut-adapted lactobacilli from the Lactobacillus acidophilus group, many strains of which are used as probiotics (17).
The ability of health-beneficial bacteria from the Lactobacillus genus to grow on starch is limited to a few strains that grow on the soluble but not granular starch (18, 19). Growth on short starch-derived malto-oligosaccharides (α-gluco-oligosaccharides), however, is well established within this genus. Interestingly, an RS-rich diet appeared to boost the numbers of lactobacilli and levels of lactate production in the distal colon in rodent models (20, 21). A recent human study in rural Malawi children showed a similar increase in lactobacilli after RS intake (22). A possible explanation of these observations is cross-feeding on short α-glucans that are produced by primary starch degraders (14, 23). Cross-feeding requires efficient capture and transport systems and intracellular degrading enzymes reported to be conserved in this genus (24). The intracellular α-glucoside utilization machinery of acidophilus group lactobacilli (17) is relatively well understood (24–26). In contrast, the extracellular α-glucanolytic capabilities within this group of bacteria associated with a healthy gut microbiota are currently unexplored.
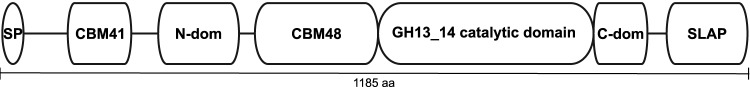
The commercial strain Lactobacillus acidophilus NCFM, which is used as a probiotic, is among the best studied of this taxonomic group (17). The genome of L. acidophilus NCFM (27) encodes nine enzymes of glycoside hydrolase family 13 (GH13), which harbors α-glucan-active enzymes according to the CAZy database (28). The only predicted α-glucan-active extracellular enzyme, however, is a pullulanase-type α-glucan debranching enzyme. This enzyme is multimodular, comprising an N-terminal starch binding domain of carbohydrate binding module family 41 (CBM41), followed by a domain of unknown function, a CBM48 domain, a GH13 subfamily 14 (GH13_14) catalytic module (28), and a C-terminal surface layer association protein (SLAP) (28, 29) (Fig. 1). Pullulanases catalyze hydrolysis of α-1,6-linked branches in glycogen, amylopectin, and other starch-derived glucans, as well as pullulan (Fig. 2) (30–32).
FIG 1.
Domain organization of LaPul13_14. Abbreviations: SP, signal peptide; CBM41, starch binding module of CBM41 in the CAZy database; N-dom, N-terminal domain of unknown function; CBM48, starch binding module of CBM48; GH13_14, catalytic module assigned to glycoside hydrolase family 13 subfamily 14; C-dom, C-terminal domain conserved in pullulanases of GH13; SLAP, surface layer association protein domain.
FIG 2.
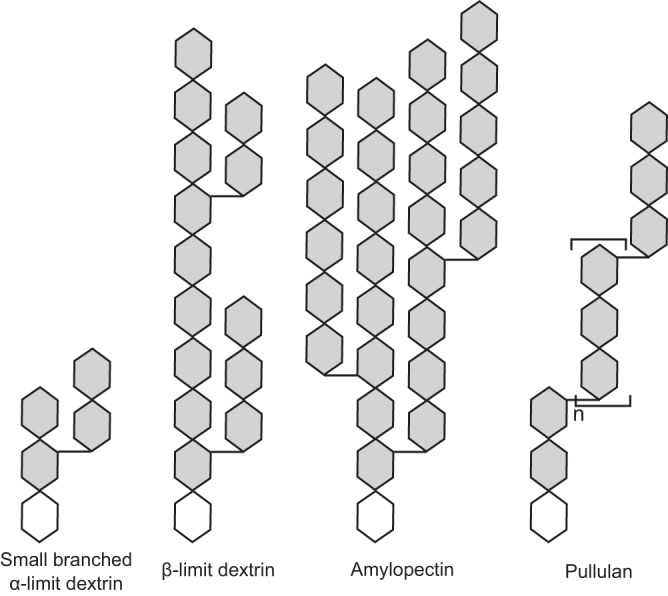
Schematic overview of α-glucans included in the present study. α-1,4-linked glucose units are shown as linear hexagons, and α-1,6-linkages are depicted as horizontal short segments between the glucosyl unit hexagons with the reducing end depicted as a white hexagon. The small branched α-limit dextrin resembles the one used in this study. β-Limit dextrin, produced from β-amylase hydrolysis of amylopectin, is used to provide experimental evidence for the preference of LaPul13_14 for short branches compared to amylopectin.
In this study, we show that extracellular cell-attached pullulanase activity can be measured upon growth of L. acidophilus NCFM on a range of oligomeric and polymeric α-glucans, whereas this activity is repressed upon growth on glucose. We also establish the exclusive role of the debranching enzyme L. acidophilus Pul13_14 (LaPul13_14) in the degradation of branched α-glucans in this bacterium. The catalytic and binding properties of the recombinant LaPul13_14 were investigated, showing unprecedented substrate affinity and a clear preference toward short-branched α-glucan oligomers compared to amylopectin. These findings are consistent with the growth profile of L. acidophilus NCFM. Together, these findings highlight the role of LaPul13_14 and its homologues in mediating the utilization of branched oligomers derived from starch degradation in the human gut.
RESULTS
L. acidophilus growth on α-glucans and extracellular pullulanase activity.
To assess the α-glucan metabolic capabilities of L. acidophilus NCFM, growth was performed on glucose, α-1,4-linked maltooligosaccharides with a degree of polymerization (DP) of 2 to 4, amylopectin, β-limit dextrin possessing short branches (degradation product from hydrolysis of amylopectin by β-amylase), and pullulan (Fig. 2). L. acidophilus NCFM clearly preferred glucose followed by maltose, whereas the growth on the larger malto-oligosaccharides was much weaker (Fig. 3A). Among the branched polymeric α-glucans, only β-limit dextrin with the short branches (mainly maltosyl) seemed to sustain clear, albeit low, growth (Fig. 3A), whereas no significant growth was observed on pullulan and amylopectin (data not shown). Cells harvested in late log phase displayed the highest cell-associated pullulanase activity when maltose was used as a carbon source, but maltotriose, maltotetraose, and β-limit dextrin also resulted in a significant pullulanase activity (Fig. 3B). In contrast, no significant activity was detected when the cells were grown on either glucose or a 1:1 glucose-maltose mixture. No pullulanase activity was detected in the culture supernatant, i.e., all pullulanase activity measured was entirely associated with the cells.
FIG 3.
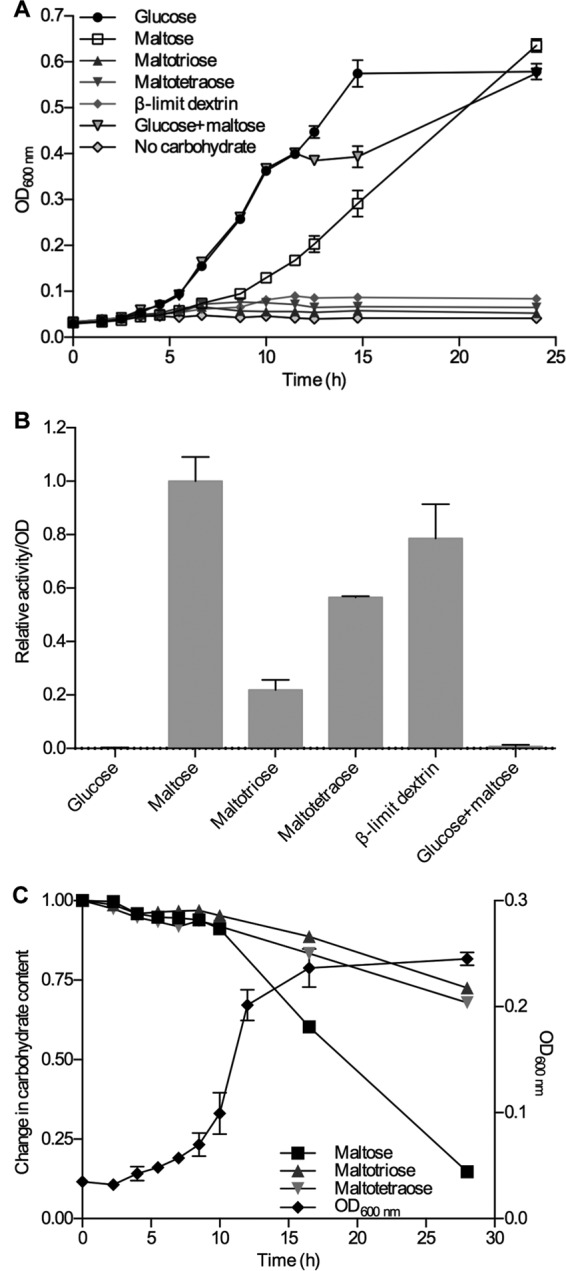
(A) Growth of L. acidophilus NCFM on different α-glucans. (B) Relative cell-associated pullulan-hydrolyzing activity of L. acidophilus NCFM cells harvested after 10 h of growth on the α-glucans shown in panel A. (C) Growth of L. acidophilus NCFM on a total 0.5% (wt/vol) mixture of maltose, maltotriose, and maltotetraose (1:1:1, based on weight). Its utilization of the substrates was analyzed by HPAEC-PAD. Only the debranched maltose (mainly) and maltotriose are used from β-limit dextrin during growth on this substrate, which explains the much lower degree of growth per mass of substrate compared to maltose.
The uptake preference of malto-oligosaccharides with a degree of polymerization of 2 to 4 by L. acidophilus NCFM was analyzed using a mixture of maltose, maltotriose, and maltotetraose as a carbon source, and the depletion of these saccharides in culture supernatants was monitored. Maltose was clearly the preferred substrate, but both maltotriose and maltotetraose were also taken up, albeit at a considerably lower rate (Fig. 3C).
To verify that the measured cell-associated pullulan-hydrolyzing activity stems from LaPul13_14, the gene encoding this enzyme was deleted and the L. acidophilus NCFM gene knockout (KO) strain was grown on glucose or maltose. No cell-associated pullulanase activity was measured from this strain, although the growth rate on maltose was much higher than that of the wild type (Fig. 4).
FIG 4.
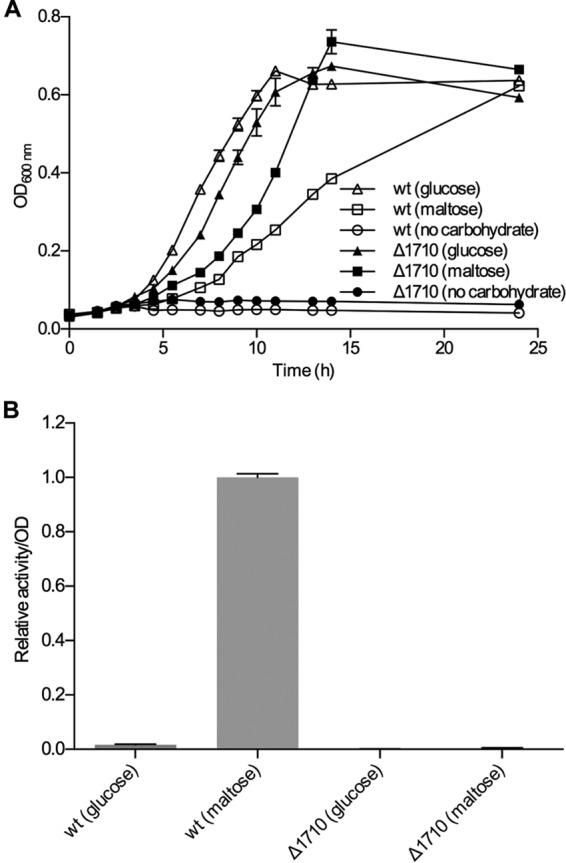
(A) Comparison of the growth of wild-type L. acidophilus NCFM and the LaPul13_14 gene deletion strain (ΔLBA1710) on glucose or maltose. (B) Cell-associated pullulan-hydrolyzing activity of cells harvested after 10 h of growth.
Enzymatic properties of LaPul13_14.
LaPul13_14 was produced recombinantly in Escherichia coli and purified (yield, 0.26 mg g [wet weight] cells−1), and it migrated according to the predicted molecular mass of 129.4 kDa. The enzyme displayed a very high specific activity of 611 U mg−1 toward pullulan, corroborating the predicted specificity. The highest specific activity was measured at pH 5.0, and the enzyme retained more than 50% of its maximum activity in the pH range of 2.5 to 6.5. The highest activity was measured at 55°C (at pH 5.0), and the Arrhenius activation energy was determined to be 60.2 kJ mol−1.
Kinetic analysis.
The kinetic analysis performed on pullulan showed an exceptionally high catalytic efficiency (kcat/Km) of 10,368 ml s−1 mg−1 owing to a combination of a very low Km value (0.05 mg ml−1) and a high catalytic turnover number (kcat, 518 s−1) (Table 1). Among the branched substrates tested, the catalytic efficiency decreased with increasing substrate size and branch length. Thus, the catalytic efficiency on the branched-substrate β-limit dextrin, which has mainly maltosyl branches, was 14-fold higher than the corresponding value on amylopectin.
TABLE 1.
Hydrolysis kinetic parameters of LaPul13_14 toward oligomeric and polymeric α-1,6-branched glucans at 37°C and pH 5.0
| Substrate | Km (mg ml−1) | kcat (s−1) | kcat Km−1 (ml s−1 mg−1) | Normalized kcat Km−1 |
|---|---|---|---|---|
| Pullulan | 0.05 ± 0.004 | 518 ± 10.5 | 10,368 | 100 |
| Amylopectin | 0.37 ± 0.041 | 25 ± 0.7 | 67 | 0.6 |
| β-Limit dextrin | 0.20 ± 0.090 | 189 ± 15.8 | 945 | 9 |
| 62-α-d-Maltosyl maltotriose | 0.33 ± 0.040a | 378 ± 18.0 | 1,145b |
Millimolar.
Per millimolar per second.
Cyclodextrins are well-known starch mimic molecules that bind tightly in the active sites of pullulanases, resulting in apparent competitive inhibition (33). Interaction of α-, β-, and γ-cyclodextrins (α-, β-, and γ-CDs, respectively) with LaPul13_14 was determined by surface plasmon resonance (SPR) analysis. β- and γ-CDs had dissociation constants (KD) of 10.8 and 11.2 μM, respectively (Table 2). Interestingly, enzymatic inhibition kinetics analysis with pullulan revealed that the inhibition of LaPul13_14 with β-CD was not pure competitive inhibition but was mixed-type inhibition, giving a Ki of 35.9 μM. This is an average value of the binding to the active site and possibly other binding sites of lower affinity. The analysis of the binding of the enzyme to granular starch revealed only very low binding affinity, hampering a reliable quantitative measurement (see Fig. S1 in the supplemental material).
TABLE 2.
Binding of cyclodextrins to LaPul13_14 determined by surface plasmon resonance
| Cyclic ligand | pH | Temp (°C) | KD (μM) |
|---|---|---|---|
| α-CD | 5.0 | 25 | 89.0 |
| γ-CD | 5.0 | 25 | 11.2 |
| β-CD | 5.0 | 25 | 10.8 |
| β-CD | 5.0 | 15 | 7.2 |
| β-CD | 5.0 | 37 | 29.9 |
| β-CD | 7.0 | 25 | 21.8 |
DISCUSSION
SLAPs mediate attachment of glycan-active enzymes and other functionally important enzymes to the cell envelope in Lactobacillus.
L. acidophilus NCFM is one of the best-studied gut bacteria, which is ascribed health benefits and used as a commercial probiotic (34–37). The commercial and physiological relevance of L. acidophilus NCFM has spurred wide interest in the saccharide uptake and catabolism machinery of this organism to identify efficiently utilized glycans with potential as prebiotics (25, 38, 39). A few intracellular carbohydrate-active enzymes (CAZymes) from L. acidophilus NCFM have been characterized, including those active on maltose (26) and α-1,6-linked isomalto-oligosaccharides (24). However, insight is scarce into the extracellular CAZymes encoded by this bacterium and related human-gut-adapted lactobacilli (27). Recently, the first extracellular glycoside hydrolase from L. acidophilus NCFM was shown to be a β-N-acetylglucosaminidase autolysin essential for cell division (40). This enzyme harbors a surface layer association protein (SLAP) domain (Pfam family PF03217 [41]) that confers noncovalent attachment to the proteinaceous outermost surface layer common in several lactobacilli (42). SLAP domains also occur at the C termini of other functionally important proteins such as autolysins, fibronectin and mucin binding proteins, putative peptidases, nucleases, and glycoside hydrolases as well as polysaccharide lyases according to the Pfam database (41). This suggests that SLAP domains constitute a general cell attachment scaffold that is fused to a select set of activities destined for cell surface display. The prevalence of this domain in the S-layer exoproteome of distinct lactobacilli is also in agreement with this role (43).
In this study, we present the functional characterization of the SLAP domain-containing enzyme from L. acidophilus NCFM that is active on starch-derived dietary glucans.
LaPul13_14 is a cell-attached debranching enzyme that exclusively confers the utilization of branched α-glucans in L. acidophilus.
Growth experiments with L. acidophilus NCFM clearly showed a preference for glucose and maltose as the substrates. Low levels of growth were observed on the longer α-1,4-malto-oligosaccharides maltotriose and maltotetraose and on β-limit dextrin, which is a model substrate that contains only short α-1,6-branches (shown experimentally to be mainly 2- to 3-glucosyl units for the substrate used in the present study [Fig. 2]). In contrast, very little or no growth was observed on pullulan and amylopectin, which contain longer branches (data not shown). Since L. acidophilus NCFM is able to use only the debranched maltose from β-limit dextrin, the energy yield per mass of this substrate is much lower than that with growth on maltose. Therefore, the observed low level of growth on β-limit dextrin is in agreement with the debranching activity of the cells and the lack of α-amylase activity that is required for the full utilization of this substrate.
The poor growth on maltotriose and maltotetraose is, however, surprising as these substrates are predicted to be internalized through a maltodextrin-specific ATP-binding cassette (ABC) system (LBA1864 to LBA1867), which is conserved in acidophilus group lactobacilli (24). The corresponding transporter from the taxonomically related Gram-positive pathogen Streptococcus pneumoniae mediates the uptake of malto-oligosaccharides up to eight units with a preference for maltotetraose (44). The expression of the ABC uptake system may be affected by an inserted transposase (LBA1868) that separates the catabolic genes from the transporter in L. acidophilus NCFM but not in other lactobacilli (24). Nonetheless, the malto-oligosaccharide uptake profile reveals that maltotriose and maltotetraose are taken up, albeit at a significantly lower rate than maltose (Fig. 3C), in L. acidophilus NCFM cultures. Growth experiments on two additional L. acidophilus strains confirmed that the strain possessing the transposase grew similarly poorly as L. acidophilus NCFM, whereas the strain that lacks this insertion displayed better relative growth on maltotriose and maltotetraose (see Fig. S2 in the supplemental material).
Despite the weak growth, pullulanase activity was reliably measured during growth on maltotriose, maltotetraose, and β-limit dextrin but not on glucose (Fig. 3B). The pullulanase activity was detectable only in the cell fraction, providing evidence that the enzyme is cell attached. Thin-layer chromatography (TLC) analysis on samples from the pullulan assays with the cell fraction confirmed the pullulanase-type debranching activity, since the sole end product released was maltotriose. This precluded other enzymatic activities (e.g., neopullulanase or α-glucosidase) being responsible for the increase in reducing sugars from pullulan degradation (Fig. S3). The only predicted extracellular pullulanase in L. acidophilus NCFM is LaPul13_14 (locus tag LBA1710) (28). The inactivation of the gene encoding this enzyme abolished the cell-attached pullulanase activity, providing compelling evidence that LaPul13_14 is the sole extracellular α-glucan-debranching enzyme in L. acidophilus NCFM (Fig. 4).
The gene encoding LaPul13_14 resides on a separate locus than that of the maltodextrin utilization cluster (26). The repression of pullulanase activity in the presence of glucose is suggestive of regulation through global catabolite repression (45). Indeed, this enzyme was not identified in the exoproteome of L. acidophilus NCFM grown on glucose in a recent proteomic analysis of S-layer-associated proteins (43). A similar observation was made in a proteome analysis of L. acidophilus NCFM grown on raffinose, where LaPul13_14 was repressed in the presence of glucose (46) but was clearly detectable in the presence of raffinose, suggesting a level of constitutive expression of the enzyme in the absence of glucose.
LaPul13_14 confers efficient targeting of small-branched α-glucans.
Given the dominance of starch in human nutrition, the metabolism of this glucan by the HGM has been subject to extensive studies. Ruminococcus bromii has been identified as the primary degrader of resistant starch in the human gut (23), although Bifidobacterium adolescentis strains were also reported to possess growth capabilities on this substrate (15). Major commensals from the Bacteroides genus (47) and the butyrate-producing Firmicutes member Eubacterium rectale (14) are other HGM members with considerable starch growth and degradation capabilities. Common to these bacteria is that they possess highly modular extracellular cell-attached enzymes with one or more catalytic modules and multiple carbohydrate binding modules (CBMs), which mediate tight binding to starch substrates.
Only a few Lactobacillus strains have been demonstrated to utilize only soluble starch (18, 19, 48). This is in agreement with the rare occurrence of genes encoding extracellular α-glucan enzymes in this genus (1.6%, or 11 out of 696 GH13 genes). Notably, only four Lactobacillus species encode these extracellular enzymes: L. acidophilus, Lactobacillus amylovorus, Lactobacillus plantarum, and Lactobacillus manihotivorans (Table S1). L. plantarum and L. amylophilus GV6 possess amylopullulanases (pullulanase type II) that degrade both α-1,4- and α-1,6-branches in starch (49–51). Notably, these starch-utilizing strains stem from other ecological niches than the human gut. In contrast, LaPul13_14 and its homologues that display an identical domain organization represent the main extracellular amylolytic activity in human gut lactobacilli (Table S1). This unique modular organization and the presence of the SLAP domain in gut lactobacilli raise a question on the importance of these enzymes in the gut niche.
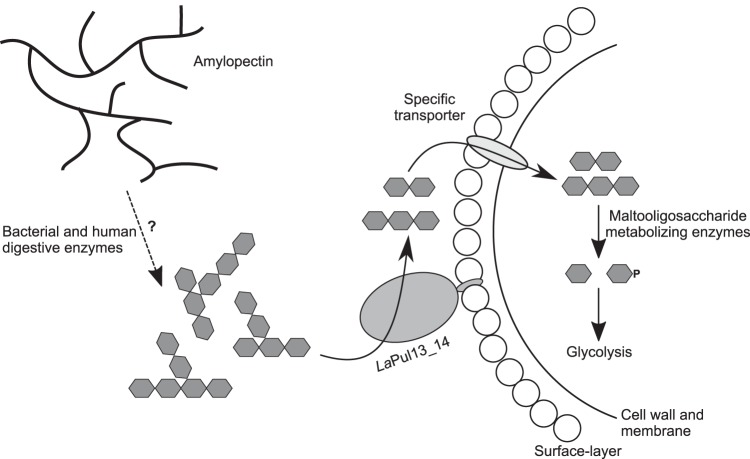
The lack of activity of LaPul13_14 on granular starch and the very weak binding to this substrate (Kd of >40 mg ml−1 [Fig. S1]) are in line with the lack of growth of L acidophilus NCFM on starch. In contrast, the enzyme binds the starch mimic β-CD with moderate affinity (Table 2). This molecule occupies the conserved +2 substrate binding subsite in pullulanases, thus acting as a competitive inhibitor (33). Our β-CD inhibition kinetics data on pullulan reveal an inhibition constant Ki of 36 μM, which is in the same range as the Kd obtained from surface plasmon resonance binding experiments (Table 2). More interestingly, the inhibition was not purely competitive but was of mixed nature (Fig. S4), indicative of the presence of additional α-glucan surface binding sites. The glucan binding residues in the CBM41 of the pullulanases from Streptococcus pneumoniae (52) are conserved in L. acidophilus NCFM. This makes the CBM41 a plausible candidate for the additional α-glucan binding in LaPul13_14. A possible rationale for this additional binding in LaPul13_14 is to increase substrate affinity by increasing the local substrate concentration in proximity to the active site. Strikingly, the Km of LaPul13_14 toward pullulan is the lowest reported for any debranching enzyme (Table S2), which attests to adaptation of the substrate affinity to the competitive human gut niche. The clear preference for substrates with short branches of about two to three glucose units (Table 1) may provide a metabolic advantage for L. acidophilus, which is able to take up and metabolize these substrates. Based on the kinetic parameters, the most likely substrates for LaPul13_14 are short-branched α-glucans that are generated from the action of α-amylases on starch or glycogen. Given the poor digestibility of branched α-glucans by human enzymes and the abundance of lactobacilli in the small intestine (16), this enzyme may act on branched oligomeric substrates from human degradation of dietary starch or glycogen (Fig. 5). Such an advantage may explain the enrichment of this debranching activity in human intestinal isolates. Taken all together, the study suggests that debranching enzymes of gut lactobacilli are evolved for efficient breakdown of short-branched α-glucans. The high substrate affinity may facilitate access to substrates that escape human or microbial metabolism of starch and glycogen. Further work is required to relate the in vivo functionality of these enzymes to gut adaptation of lactobacilli.
FIG 5.
Schematic overview of the extracellular α-glucan metabolism of L. acidophilus NCFM. The cell-attached pullulanase activity of LaPul13_14 mediates the debranching of preferentially smaller-branched oligomers that possibly are products of human or bacterial degradation of amylopectin, which is signified by a dashed line. The produced maltose and short maltooligosaccharides are taken up by one or more specific transporters and degraded intracellularly by the enzymes encoded by the malto-oligosaccharide utilization locus to produce β-glucose-1-phosphate and glucose, which enter glycolysis (26).
MATERIALS AND METHODS
Materials.
High-purity (>95%) chemicals and commercial enzymes were from Sigma-Aldrich, MO, USA, unless otherwise stated. Pullulan (>95%) and β-limit dextrin (>97%) were from Megazyme (Bray, Ireland). The maltotetraose used for the growth/induction experiment was an in-house preparation (purity of >92%; impurity was maltotriose). The branched α-limit dextrin used for the kinetic analysis (kind gift from the late Bent S. Enevoldsen) comprises a mixture of two isomers: 62-α-d-maltosyl-maltotriose and α-d-glucosyl-maltotetraose.
Bioinformatics analysis.
Sequences of LaPul13_14 homologues were extracted from the Carbohydrate-Active Enzymes database (CAZy; www.cazy.org) (28) using the CAZy tools provided by Alexander Holm Viborg (http://research.ahv.dk). Domain organization of the protein sequences was analyzed using a combination of batch searches in the Conserved Domain Database (29) and search in the Pfam protein family database (41). Identification of putative extracellular enzymes was done based on predictions from the SignalP 4.1 server (53).
Growth experiments.
L. acidophilus NCFM was grown on glucose, maltose, maltotriose, maltotetraose, a mixture of glucose and maltose (1:1, based on weight), β-limit dextrin, amylopectin, or pullulan. Growth was performed in 14 ml semidefined medium (SDM) (54) with 0.5% (wt/vol) carbohydrate in 15-ml conical culture tubes, which were inoculated with an overnight culture previously grown on SDM supplemented with 1% (wt/vol) glucose to an optical density at 600 nm (OD600) of 0.1 (0.04 as measured on 200 μl culture in a 96-well plate using a microplate reader). The cultures were incubated at 37°C, and 200-μl samples were withdrawn every 1 to 2 h for OD600 measurements. When the cultures reached late log phase (after approximately 10 h), two 1-ml samples per culture were collected and spun down (20,000 × g, 10 min, 4°C), and the cells and supernatant were assayed for enzymatic activity (see below). The growth experiments with the LaPul13_14 deletion mutant (ΔLBA1710; see below) were performed in a similar manner on glucose and maltose, and all growth experiments were performed in duplicates. The supernatants from the growth experiments were assayed for pullulanase activity using the standard reducing sugar assay described below, whereas the cells were washed twice with 0.9% NaCl before measurements of pullulanase activity (described below).
Additional growth experiments were performed to compare the growth of L. acidophilus NCFM to those of L. acidophilus DSM-20242, which possesses identical organization of the maltodextrin utilization cluster as L. acidophilus NCFM and is used as a control, and L. acidophilus DSM-9126, which possesses a typical maltodextrin utilization locus and lacks the inserted transposase present in both L. acidophilus NCFM and DSM-20242. Anaerobically grown cultures in MRS medium were transferred once in SDM with 1% glucose (anaerobic), and the resulting cultures were used to inoculate (1% or equivalent cell density for weaker-growing strains) the SDM-based growth medium (0.5% carbohydrate source, 200 μl per well, in duplicate wells and under duplicate conditions). Growth was monitored with a microtiter plate reader for 60 h.
Malto-oligosaccharide uptake.
L. acidophilus NCFM was grown on a total 0.5% (wt/vol) mixture of maltose, maltotriose, and maltotetraose (1:1:1, based on weight) as described above. Samples were collected during growth for OD600 measurements and oligosaccharide analysis, which was performed as follows: a 1-ml sample was spun down (20,000 × g, 10 min, 4°C), and 80 μl supernatant was diluted 100-fold in 0.1 M NaOH and sterile filtered through prerinsed (with MilliQ) 30-kDa filters (Amicon Ultraspin filters; Millipore, MA, USA) before the content of maltose, maltotriose, and maltotetraose was analyzed using high-performance anion-exchange chromatography with pulsed amperometric detection (HPAEC-PAD) using an ICS3000 system, equipped with a CarboPac PA200 anion-exchange column (Dionex Corporation, Sunnyvale, CA) using a 2-step sodium acetate gradient (0 to 7 min, 37.5 to 75 mM, and 7 to 30 min, 75 to 300 mM) and a constant concentration of 0.1 M NaOH at 25°C at 0.35 ml min−1. After each run, the column was regenerated with a constant concentration of 0.1 M NaOH and 400 mM sodium acetate for 5 min, followed by a gradient from 400 mM to 37.5 mM sodium acetate over 5 min, and finally an equilibration with 37.5 mM sodium acetate and 0.1 M NaOH for 5 min.
Construction of LBA1710 (LaPul13_14) deletion mutant.
The 3,555-bp lba1710 gene encoding LaPul13_14 in L. acidophilus NCFM was deleted using an upp-based counterselective gene replacement system (55). Briefly, a 3,489-bp in-frame deletion (98% of gene) within lba1710 was constructed by first PCR amplifying the 629-bp and 646-bp DNA segments flanking the upstream and downstream sequences of the lba1710 deletion target, respectively, using primer pairs 1710-1/1710-2 (5′-GTAATAGGATCCACAACAAGCTCAAGGGATTCA-3′ and 5′-AACACCTTTTGTTCCCCA-3′, respectively) and 1710-3/1710-4 (5′-TGGGGAACAAAAGGTGTTGCAGTGAATGTTGTTATTGAAG-3′ and 5′-TTAGTAGAATTCCTTGAGGGAGCTCAACTTTC-3′, respectively) (restriction sites are underlined), with Pfu UltraII HS DNA polymerase (Agilent Technologies, California, USA). Both purified PCR products were fused and amplified to generate copies of the Δlba1710 allele via splicing by overlap extension PCR (SOE-PCR) (56), using 10 ng of each PCR product as the amplification templates in a 50-μl PCR mixture with primer pair 1710-1/1710-4 (see above). Purified SOE-PCR products (1,275 bp) were digested with BamHI and EcoRI and ligated into compatible ends of the pTRK935 counterselectable integration vector. Construction of the resulting recombinant integration plasmid containing the Δlba1710 allele, designated pTRK1085, and the recovery of plasmid-free double recombinants with 5-fluorouracil were performed as described previously (55, 57). Double recombinants with the Δlba1710 allele were screened by colony PCR using primer pair 1710-1/1710-4. In-frame deletion and sequence integrity were confirmed by PCR and DNA sequencing using primer pair 1710-5/1710-6 (5′-TGAGCAAGTTAGCGCATCTG-3′ and 5′-GCTGGTGTTGCAGAAGTAG-3′, respectively), the primers in which specifically anneal to the flanking region of the lba1710 gene. One of the confirmed Δlba1710 deletion mutants, designated NCK2325, was selected for further studies.
Production and purification of recombinant LaPul13_14.
L. acidophilus NCFM genomic DNA, prepared as previously described (26), was used to amplify the LaPul13_14 gene (locus tag number LBA1710; GenBank accession number AAV43522.1), with the sense primer 5′-CTAGCTAGCGCAGAAACACCAGATGCTGG-3′ and the antisense primer 5′-CCGCTCGAGAGCTTTTACTTCAATAACAACATTC-3′ (restriction sites are underlined). The PCR amplicon encoding the mature peptide (3,468 bp) lacking the signal peptide (bp 1 to 105, corresponding to amino acid residues 1 to 35 [Fig. 1]) was cloned within the NheI and XhoI restriction sites in pET21a(+) (Novagen, Darmstadt, Germany) and transformed into Escherichia coli XL10-Gold Ultracompetent cells (Stratagene, California, USA) according to the manufacturer's protocols. Transformants harboring pET21a(+)-LaPul13_14 were selected on LB agar plates with 100 μg ml−1 ampicillin and verified by restriction analysis and full sequencing. E. coli Rosetta (DE3) cells (Invitrogen, USA) transformed with pET21a(+)-LaPul13_14 were used for production of the enzyme.
The enzyme was produced in a 5-liter bioreactor (Biostat B Plus; Sartorius Stedim, Germany) as described elsewhere (58) with the following modifications: 3.7 liters of defined medium was inoculated to an OD600 of 1.5 with an overnight culture grown in LB medium. The fermentation was carried out at 37°C until the OD600 reached 8, before decrease of the temperature to 15°C and induction of expression using 100 μM isopropyl-β-d-thiogalactopyranoside. Cells were harvested (6,000 × g, 20 min, 4°C) at an OD600 of 30.5 after 67 h of induction. The cell pellet was resuspended in buffer A (10 mM HEPES, pH 7.4, 25 mM imidazole, 40% glycerol, 0.5 M NaCl, 1 mM CaCl2, 0.005% [vol/vol] Triton X-100) and disrupted by high-pressure homogenization at 100 MPa. Disintegrated cells were treated with Benzonase nuclease (Invitrogen; 30 min at room temperature) and centrifuged twice (40,000 × g, 30 min, 4°C). The supernatant was filtered (0.45 μm) and loaded onto a 5-ml HisTrap HP column (GE Healthcare, Uppsala, Sweden). After washing with 10 column volumes of buffer A, including 26 mM imidazole, bound protein was eluted with a linear gradient from 3 to 80% buffer B (10 mM HEPES, pH 7.4, 400 mM imidazole, 40% glycerol, 0.5 M NaCl, 1 mM CaCl2). Fractions containing protein were pooled and concentrated (30-kDa Amicon Ultra spin filters; Millipore) to 5 ml, loaded onto a preequilibrated HiLoad 26/60 Superdex G200 column (GE Healthcare), and eluted with 50 mM morpholineethanesulfonic acid (MES), pH 6.0, 1 mM CaCl2, 20% glycerol, 150 mM NaCl at 0.75 ml min−1. Fractions containing LaPul13_14 were pooled and desalted on a HiPrep 26/10 desalting column (GE Healthcare) against 1 mM HEPES, pH 7.0. Desalted protein fractions were pooled and loaded onto a Resource Q column (6 ml; GE Healthcare) equilibrated with 10 mM HEPES, pH 7.0, at a flow rate of 2 ml min−1. Protein was eluted by a linear gradient (from 0 to 100% in 30 column volumes) of 10 mM HEPES, pH 7.0, 0.5 M NaCl. Fractions containing LaPul13_14 were pooled, concentrated, and buffer exchanged (30-kDa Amicon Ultra spin filters; Millipore) to 50 mM MES, pH 6.0, 20% glycerol, 0.5 mM CaCl2, 150 mM NaCl. The concentration of LaPul13_14 (SDS-PAGE) was determined spectrophotometrically using a molar extinction coefficient ε280 of 179,566 M−1 cm−1 as determined by amino acid analysis (59).
Enzyme activity assays and kinetics.
Determination of specific activity as well as kinetic parameters of LaPul13_14 was performed using a reducing sugar assay as previously described (33). In short; 1.1-ml reaction mixtures containing substrate (0.225 mg ml−1 pullulan for specific activity; 0.02 to 1 mg ml−1 pullulan and 0.1 to 10 mg ml−1 potato amylopectin dissolved in 8% [vol/vol] dimethyl sulfoxide [DMSO] or 0.225 to 9 mg ml−1 β-limit dextrin for kinetic analysis) and LaPul13_14 (0.05 to 1.5 nM) in assay buffer (20 mM sodium acetate, pH 5.0, 5 mM CaCl2, 0.005% Triton X-100) were incubated at 37°C, and aliquots (200 μl for pullulan and 100 μl for amylopectin and β-limit dextrin) were removed at five time points (3, 6, 9, 12, and 15 min) and added to 500 μl stop solutions (0.4 M sodium carbonate, pH 10.7, 2.5 mM CuSO2, 2.5 mM 4,4′-dicarboxy-1,2′-biquinoline, 6 mM l-serine). Milli-Q water was added to a final volume of 1 ml, and A540 was measured after a 30-min incubation at 80°C. The release of reducing sugars was quantified using a maltose standard. One activity unit (U) is defined as the amount of enzyme that releases 1 micromole of maltose reducing sugar equivalents per min from pullulan under assay conditions. The kinetic parameters Km and kcat were determined by fitting the Michaelis-Menten equation to the initial velocity data. The data obtained were analyzed using the Enzyme Kinetics module 1.0 of the program SigmaPlot 9.01 (Systat Software, Chicago, IL). Inhibition kinetics of LaPul13_14 by β-cyclodextrin (β-CD) were investigated using pullulan as the substrate. The kinetics assay was done as described above but with 50 μM β-CD included. The data obtained were analyzed by fitting competitive, noncompetitive, and mixed inhibition models to the data using the Enzyme Kinetics module 1.0 of the program SigmaPlot 9.01 (Systat Software, Chicago, IL), and the inhibition kinetics models were ranked based on χ2 of the fits.
The kinetic parameters of hydrolysis of the branched α-limit dextrin mixture described above (5 glucose units, maltosyl branch) by LaPul13_14 were determined using HPAEC-PAD. The starting reaction volume was 300 μl, including branched substrate (0.0625 to 1 mM) and 0.53 nM LaPul13_14 in 20 mM sodium acetate, pH 5.0, 5 mM CaCl2, 0.005% (vol/vol) Triton X-100. At four time points (3, 6, 9, and 12 min), 60-μl aliquots were drawn and mixed with 15 μl 0.5 M NaOH. The samples were spun (20,000 × g, 5 min, 4°C) before 65-μl samples were mixed with 65 μl of 0.1 M NaOH. The products were quantified based on peak areas from HPAEC-PAD analysis, which was performed as described above.
The described standard reducing sugar assay was also used to determine the pullulanase activity in the L. acidophilus culture supernatant and in the washed cell pellets, with the following exceptions: cells from 1 ml of culture in the late log phase (see above) were resuspended in 600 μl preheated (37°C) pullulan solution (0.4 mg ml−1 pullulan, 40 mM sodium acetate, pH 5.0, 0.5 mM CaCl2, 0.005% [vol/vol] Triton X-100), while 100 μl culture supernatant was mixed with 500 μl 0.48-mg ml−1 pullulan solution. Aliquots (100 μl) were drawn after 0, 1, 2, and 3 h. The samples were spun down at 4°C for 2 min, and 75 μl cell-free sample was mixed with 500 μl stop solution (0.4 M sodium carbonate, pH 10.7, 2.5 mM CuSO2, 2.5 mM 4,4′-dicarboxy-1,2′-biquinoline, 6 mM l-serine) and 425 μl MilliQ water. As a control, the degradation products from the assay with whole cells and pullulan were analyzed using thin-layer chromatography (TLC). Samples (6 μl) were drawn after 0 and 19 h of reaction and spotted directly onto a TLC Silica 60 F254 plate (Merck, Darmstadt, Germany), developed with isopropanol-ethyl acetate-water (60:20:20, vol/vol), and sprayed with 2% (wt/vol) orcinol in ethanol-H2SO4-water (80:10:10) followed by tarring at 300°C. The following standards were included on the gel (1 μl): 20 mM (either) glucose, maltose, maltotriose, or panose in water.
Temperature and pH activity profiles.
The reducing sugar assay described above was used for determining the dependence of initial reaction rates on temperature in the range 7 to 80°C using 0.46 nM LaPul13_14 and 0.225 mg ml−1 pullulan. The dependence of activity on pH in the range 2.0 to 8.5 was assayed using the reducing sugar assay described above in 20 mM (either) glycine (pH 2.0 to 3.5), sodium acetate (pH 3.5 to 5.5), MES (pH 5.5 to 7.0), or HEPES (7.0 to 8.5), all including 0.5 mM CaCl2 and 0.005% bovine serum albumin.
SPR binding analysis of cyclodextrins.
The affinity of LaPul13_14 toward α-, β-, and γ-CD was analyzed using surface plasmon resonance (SPR) on a Biacore T100 system (GE Healthcare). Random amine coupling was used to immobilize the enzyme on a CM5 sensor according to the manufacturer's protocol using 100 μg ml−1 protein in 10 mM sodium acetate, pH 4, 0.5 mM CaCl2, 1 mM β-CD to a final chip density of 5,477 response units (RU). The analysis comprised 100 s and 90 s for the association and dissociation phases, respectively, at a flow rate of 30 μl min−1 and 25°C at 19 β-CD concentrations (0.25 to 1,024 μM) as well as 18 α-CD and γ-CD concentrations (3 to 5,000 μM), all in 10 mM sodium acetate, pH 5.0, 150 mM NaCl, 0.005% (vol/vol) P20 surfactant. In the case of β-CD, the interaction was analyzed at 15°C and 37°C in addition to the 25°C standard analysis. Furthermore, the interaction with β-CD was also analyzed at pH 7.0 in 10 mM HEPES, pH 7.0, 150 mM NaCl, 0.005% P20 surfactant. A one-site binding model was fitted to the steady-state response blank and reference cell-corrected sensograms using the BIA evaluation software supplied with the instrument.
Starch binding assay.
Barley starch was washed three times in water followed by one time in assay buffer (40 mM sodium acetate, pH 5.0, 0.5 mM CaCl2, 0.005% [vol/vol] Triton X-100) overnight. Fifty microliters of LaPul13_14 diluted to 40 nM in assay buffer and 450 μl of washed starch suspension in reaction buffer (25, 50, 100, and 200 mg ml−1) were mixed and shaken vigorously at 4°C for 30 min. The mixture was subsequently centrifuged (20,000 × g, 5 min, 4°C), and 110 μl of supernatant was used for the standard reducing sugar assay described above using 0.72 mg ml−1 pullulan in assay buffer as the substrate. Samples were withdrawn at 0 and 10 min and directly transferred to stop solution, and the standard assay protocol was followed. The fraction of enzyme bound to starch was determined based on the activity in the supernatant relative to an enzyme sample without starch included.
Supplementary Material
ACKNOWLEDGMENTS
Alexander Holm Viborg is acknowledged for providing us with bioinformatics tools for CAZyme analyses (ahv.dk).
This work was supported by a grant from the Danish Strategic Research Council, Committee of Health and Nutrition, to the project Gene Discovery and Molecular Interactions in Prebiotics/Probiotics Systems: Focus on Carbohydrate Prebiotics. The Danish Council for Independent Research | Natural Sciences is thanked for an instrument grant for the SPR.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00402-17.
REFERENCES
- 1.Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R. 2012. Diversity, stability and resilience of the human gut microbiota. Nature 489:220–230. doi: 10.1038/nature11550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA, Biddinger SB, Dutton RJ, Turnbaugh PJ. 2014. Diet rapidly and reproducibly alters the human gut microbiome. Nature 505:559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martens EC. 2016. Fibre for the future. Nature 529:158–159. doi: 10.1038/529158a. [DOI] [PubMed] [Google Scholar]
- 4.Sonnenburg JL, Bäckhed F. 2016. Diet-microbiota interactions as moderators of human metabolism. Nature 535:56–64. doi: 10.1038/nature18846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hur KY, Lee M-S. 2015. Gut microbiota and metabolic disorders. Diabetes Metab J 39:198–203. doi: 10.4093/dmj.2015.39.3.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Flint HJ, Scott KP, Duncan SH, Louis P, Forano E. 2012. Microbial degradation of complex carbohydrates in the gut. Gut Microbes 3:289–306. doi: 10.4161/gmic.19897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koropatkin NM, Cameron EA, Martens EC. 2012. How glycan metabolism shapes the human gut microbiota. Nat Rev Microbiol 10:323–335. doi: 10.1038/nrmicro2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tester RF, Karkalas J, Qi X. 2004. Starch—composition, fine structure and architecture. J Cereal Sci 39:151–165. doi: 10.1016/j.jcs.2003.12.001. [DOI] [Google Scholar]
- 9.Møller MS, Goh YJ, Viborg AH, Andersen JM, Klaenhammer TR, Svensson B, Abou Hachem M. 2014. Recent insight in α-glucan metabolism in probiotic bacteria. Biologia (Bratisl) 69:713–721. [Google Scholar]
- 10.Blazek J, Gilbert EP. 2010. Effect of enzymatic hydrolysis on native starch granule structure. Biomacromolecules 11:3275–3289. doi: 10.1021/bm101124t. [DOI] [PubMed] [Google Scholar]
- 11.Zhang G, Hamaker BR. 2009. Slowly digestible starch: concept, mechanism, and proposed extended glycemic index. Crit Rev Food Sci Nutr 49:852–867. doi: 10.1080/10408390903372466. [DOI] [PubMed] [Google Scholar]
- 12.Fuentes-Zaragoza E, Sánchez-Zapata E, Sendra E, Sayas E, Navarro C, Fernández-López J, Pérez-Alvarez JA. 2011. Resistant starch as prebiotic: a review. Starch 63:406–415. doi: 10.1002/star.201000099. [DOI] [Google Scholar]
- 13.Cockburn DW, Koropatkin NM. 2016. Polysaccharide degradation by the intestinal microbiota and its influence on human health and disease. J Mol Biol 428:3230–3252. doi: 10.1016/j.jmb.2016.06.021. [DOI] [PubMed] [Google Scholar]
- 14.Cockburn DW, Orlovsky NI, Foley MH, Kwiatkowski KJ, Bahr CM, Maynard M, Demeler B, Koropatkin NM. 2015. Molecular details of a starch utilization pathway in the human gut symbiont Eubacterium rectale. Mol Microbiol 95:209–230. doi: 10.1111/mmi.12859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leitch ECM, Walker AW, Duncan SH, Holtrop G, Flint HJ. 2007. Selective colonization of insoluble substrates by human faecal bacteria. Environ Microbiol 9:667–679. doi: 10.1111/j.1462-2920.2006.01186.x. [DOI] [PubMed] [Google Scholar]
- 16.Donaldson GP, Lee SM, Mazmanian SK. 2016. Gut biogeography of the bacterial microbiota. Nat Rev Microbiol 14:20–32. doi: 10.1038/nrmicro3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bull M, Plummer S, Marchesi J, Mahenthiralingam E. 2013. The life history of Lactobacillus acidophilus as a probiotic: a tale of revisionary taxonomy, misidentification and commercial success. FEMS Microbiol Lett 349:77–87. doi: 10.1111/1574-6968.12293. [DOI] [PubMed] [Google Scholar]
- 18.Petrova P, Petrov K, Stoyancheva G. 2013. Starch-modifying enzymes of lactic acid bacteria—structures, properties, and applications. Starch 65:34–47. doi: 10.1002/star.201200192. [DOI] [Google Scholar]
- 19.Velikova P, Stoyanov A, Blagoeva G, Popova L, Petrov K, Gotcheva V, Angelov A, Petrova P. 2016. Starch utilization routes in lactic acid bacteria: new insight by gene expression assay. Starch 68:1–8. doi: 10.1002/star.201500151. [DOI] [Google Scholar]
- 20.Wang X, Brown IL, Khaled D, Mahoney MC, Evans AJ, Conway PL. 2002. Manipulation of colonic bacteria and volatile fatty acid production by dietary high amylose maize (amylomaize) starch granules. J Appl Microbiol 93:390–397. doi: 10.1046/j.1365-2672.2002.01704.x. [DOI] [PubMed] [Google Scholar]
- 21.Le Blay GM, Michel CD, Blottière HM, Cherbut CJ. 2003. Raw potato starch and short-chain fructo-oligosaccharides affect the composition and metabolic activity of rat intestinal microbiota differently depending on the caecocolonic segment involved. J Appl Microbiol 94:312–320. doi: 10.1046/j.1365-2672.2003.01836.x. [DOI] [PubMed] [Google Scholar]
- 22.Ordiz MI, May TD, Mihindukulasuriya K, Martin J, Crowley J, Tarr PI, Ryan K, Mortimer E, Gopalsamy G, Maleta K, Mitreva M, Young G, Manary MJ. 2015. The effect of dietary resistant starch type 2 on the microbiota and markers of gut inflammation in rural Malawi children. Microbiome 3:37. doi: 10.1186/s40168-015-0102-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ze X, Duncan SH, Louis P, Flint HJ. 2012. Ruminococcus bromii is a keystone species for the degradation of resistant starch in the human colon. ISME J 6:1535–1543. doi: 10.1038/ismej.2012.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Møller MS, Fredslund F, Majumder A, Nakai H, Poulsen J-CN, Lo Leggio L, Svensson B, Abou Hachem M. 2012. Enzymology and structure of the GH13_31 glucan 1,6-α-glucosidase that confers isomaltooligosaccharide utilization in the probiotic Lactobacillus acidophilus NCFM. J Bacteriol 194:4249–4259. doi: 10.1128/JB.00622-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Andersen JM, Barrangou R, Abou Hachem M, Lahtinen SJ, Goh Y-J, Svensson B, Klaenhammer TR. 2012. Transcriptional analysis of prebiotic uptake and catabolism by Lactobacillus acidophilus NCFM. PLoS One 7:e44409. doi: 10.1371/journal.pone.0044409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nakai H, Baumann MJ, Petersen BO, Westphal Y, Schols H, Dilokpimol A, Abou Hachem M, Lahtinen SJ, Duus JØ, Svensson B. 2009. The maltodextrin transport system and metabolism in Lactobacillus acidophilus NCFM and production of novel α-glucosides through reverse phosphorolysis by maltose phosphorylase. FEBS J 276:7353–7365. doi: 10.1111/j.1742-4658.2009.07445.x. [DOI] [PubMed] [Google Scholar]
- 27.Altermann E, Russell WM, Azcarate-Peril MA, Barrangou R, Buck BL, McAuliffe O, Souther N, Dobson A, Duong T, Callanan M, Lick S, Hamrick A, Cano R, Klaenhammer TR. 2005. Complete genome sequence of the probiotic lactic acid bacterium Lactobacillus acidophilus NCFM. Proc Natl Acad Sci U S A 102:3906–3912. doi: 10.1073/pnas.0409188102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B. 2014. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res 42:490–495. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marchler-Bauer A, Derbyshire MK, Gonzales NR, Lu S, Chitsaz F, Geer LY, Geer RC, He J, Gwadz M, Hurwitz DI, Lanczycki CJ, Lu F, Marchler GH, Song JS, Thanki N, Wang Z, Yamashita RA, Zhang D, Zheng C, Bryant SH. 2015. CDD: NCBI's conserved domain database. Nucleic Acids Res 43:D222–D226. doi: 10.1093/nar/gku1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bertoldo C, Antranikian A. 2002. Starch-hydrolyzing enzymes from thermophilic archaea and bacteria. Curr Opin Chem Biol 6:151–160. doi: 10.1016/S1367-5931(02)00311-3. [DOI] [PubMed] [Google Scholar]
- 31.Domań-Pytka M, Bardowski J. 2004. Pullulan degrading enzymes of bacterial origin. Crit Rev Microbiol 30:107–121. doi: 10.1080/10408410490435115. [DOI] [PubMed] [Google Scholar]
- 32.Møller MS, Henriksen A, Svensson B. 2016. Structure and function of α-glucan debranching enzymes. Cell Mol Life Sci 73:2619–2641. doi: 10.1007/s00018-016-2241-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vester-Christensen MB, Abou Hachem M, Naested H, Svensson B. 2010. Secretory expression of functional barley limit dextrinase by Pichia pastoris using high cell-density fermentation. Protein Expr Purif 69:112–119. doi: 10.1016/j.pep.2009.08.016. [DOI] [PubMed] [Google Scholar]
- 34.Sanders ME, Klaenhammer TR. 2001. Invited review: the scientific basis of Lactobacillus acidophilus NCFM functionality as a probiotic. J Dairy Sci 84:319–331. doi: 10.3168/jds.S0022-0302(01)74481-5. [DOI] [PubMed] [Google Scholar]
- 35.O'Flaherty S, Klaenhammer TR. 2010. The role and potential of probiotic bacteria in the gut, and the communication between gut microflora and gut/host. Int Dairy J 20:262–268. doi: 10.1016/j.idairyj.2009.11.011. [DOI] [Google Scholar]
- 36.Leyer GJ, Li S, Mubasher ME, Reifer C, Ouwehand AC. 2009. Probiotic effects on cold and influenza-like symptom incidence and duration in children. Pediatrics 124:e172–. doi: 10.1542/peds.2008-2666. [DOI] [PubMed] [Google Scholar]
- 37.Ringel-Kulka T, Palsson OS, Maier D, Carroll I, Galanko JA, Leyer G, Ringel Y. 2011. Probiotic bacteria Lactobacillus acidophilus NCFM and Bifidobacterium lactis Bi-07 versus placebo for the symptoms of bloating in patients with functional bowel disorders: a double-blind study. J Clin Gastroenterol 45:518–525. doi: 10.1097/MCG.0b013e31820ca4d6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Andersen JM, Barrangou R, Abou Hachem M, Lahtinen S, Goh YJ, Svensson B, Klaenhammer TR. 2011. Transcriptional and functional analysis of galactooligosaccharide uptake by lacS in Lactobacillus acidophilus. Proc Natl Acad Sci U S A 108:17785–17790. doi: 10.1073/pnas.1114152108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barrangou R, Azcarate-Peril MA, Duong T, Conners SB, Kelly RM, Klaenhammer TR. 2006. Global analysis of carbohydrate utilization by Lactobacillus acidophilus using cDNA microarrays. Proc Natl Acad Sci U S A 103:3816–3821. doi: 10.1073/pnas.0511287103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Johnson BR, Klaenhammer TR. 2016. AcmB is an S-layer-associated β-N-acetylglucosaminidase and functional autolysin in Lactobacillus acidophilus NCFM. Appl Environ Microbiol 82:5687–5697. doi: 10.1128/AEM.02025-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Finn RD, Coggill P, Eberhardt RY, Eddy SR, Mistry J, Mitchell AL, Potter SC, Punta M, Qureshi M, Sangrador-Vegas A, Salazar GA, Tate J, Bateman A. 2016. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res 44:D279–D285. doi: 10.1093/nar/gkv1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hynönen U, Palva A. 2013. Lactobacillus surface layer proteins: structure, function and applications. Appl Microbiol Biotechnol 97:5225–5243. doi: 10.1007/s00253-013-4962-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Johnson BR, Hymes J, Sanozky-Dawes R, DeCrescenzo Henriksen E, Barrangou R, Klaenhammer TR. 2015. Conserved S-layer-associated proteins revealed by exoproteomic survey of S-layer-forming lactobacilli. Appl Environ Microbiol 82:134–145. doi: 10.1128/AEM.01968-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Abbott DW, Higgins MA, Hyrnuik S, Pluvinage B, Lammerts van Bueren A, Boraston AB. 2010. The molecular basis of glycogen breakdown and transport in Streptococcus pneumoniae. Mol Microbiol 77:183–199. doi: 10.1111/j.1365-2958.2010.07199.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mahr K, Hillen W, Titgemeyer F. 2000. Carbon catabolite repression in Lactobacillus pentosus: analysis of the ccpA region. Appl Environ Microbiol 66:277–283. doi: 10.1128/AEM.66.1.277-283.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Celebioglu HU, Ejby M, Majumder A, Købler C, Goh YJ, Schmidt B, O'Flaherty S, Abou Hachem M, Lahtinen SJ, Jacobsen S, Klaenhammer TR, Brix S, Mølhave K, Svensson B. 2016. Differential proteome and cellular adhesion analyses of the probiotic bacterium Lactobacillus acidophilus NCFM grown on raffinose—an emerging prebiotic. Proteomics 16:1361–1375. doi: 10.1002/pmic.201500212. [DOI] [PubMed] [Google Scholar]
- 47.Cameron EA, Maynard MA, Smith CJ, Smith TJ, Koropatkin NM, Martens EC. 2012. Multidomain carbohydrate-binding proteins involved in Bacteroides thetaiotaomicron starch metabolism. J Biol Chem 287:34614–34625. doi: 10.1074/jbc.M112.397380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee HS, Gilliland SE, Carter S. 2001. Amylolytic cultures of Lactobacillus acidophilus: potential probiotics to improve dietary starch utilization. J Food Sci 66:338–344. doi: 10.1111/j.1365-2621.2001.tb11343.x. [DOI] [Google Scholar]
- 49.Kim J-H, Sunako M, Ono H, Murooka Y, Fukusaki E, Yamashita M. 2008. Characterization of gene encoding amylopullulanase from plant-originated lactic acid bacterium, Lactobacillus plantarum L137. J Biosci Bioeng 106:449–459. doi: 10.1263/jbb.106.449. [DOI] [PubMed] [Google Scholar]
- 50.Kim J-H, Sunako M, Ono H, Murooka Y, Fukusaki E, Yamashita M. 2009. Characterization of the C-terminal truncated form of amylopullulanase from Lactobacillus plantarum L137. J Biosci Bioeng 107:124–129. doi: 10.1016/j.jbiosc.2008.10.019. [DOI] [PubMed] [Google Scholar]
- 51.Vishnu C, Naveena BJ, Altaf M, Venkateshwar M, Reddy G. 2006. Amylopullulanase—a novel enzyme of L. amylophilus GV6 in direct fermentation of starch to L(+) lactic acid. Enzyme Microb Technol 38:545–550. doi: 10.1016/j.enzmictec.2005.07.010. [DOI] [Google Scholar]
- 52.Lammerts van Bueren A, Ficko-Blean E, Pluvinage B, Hehemann J-H, Higgins MA, Deng L, Ogunniyi AD, Stroeher UH, El Warry N, Burke RD, Czjzek M, Paton JC, Vocadlo DJ, Boraston AB. 2011. The conformation and function of a multimodular glycogen-degrading pneumococcal virulence factor. Structure 19:640–651. doi: 10.1016/j.str.2011.03.001. [DOI] [PubMed] [Google Scholar]
- 53.Petersen TN, Brunak S, von Heijne G, Nielsen H. 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 54.Kimmel SA, Roberts RF. 1998. Development of a growth medium suitable for exopolysaccharide production by Lactobacillus delbrueckii ssp. bulgaricus RR. Int J Food Microbiol 40:87–92. doi: 10.1016/S0168-1605(98)00023-3. [DOI] [PubMed] [Google Scholar]
- 55.Goh YJ, Azcárate-Peril MA, O'Flaherty S, Durmaz E, Valence F, Jardin J, Lortal S, Klaenhammer TR. 2009. Development and application of a upp-based counterselective gene replacement system for the study of the S-layer protein SlpX of Lactobacillus acidophilus NCFM. Appl Environ Microbiol 75:3093–3105. doi: 10.1128/AEM.02502-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Horton RM, Hunt HD, Ho SN, Pullen JK, Pease LR. 1989. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene 77:61–68. doi: 10.1016/0378-1119(89)90359-4. [DOI] [PubMed] [Google Scholar]
- 57.Goh YJ, Klaenhammer TR. 2010. Functional roles of aggregation-promoting-like factor in stress tolerance and adherence of Lactobacillus acidophilus NCFM. Appl Environ Microbiol 76:5005–5012. doi: 10.1128/AEM.00030-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fredslund F, Abou Hachem M, Larsen RJ, Sørensen PG, Coutinho PM, Lo Leggio L, Svensson B. 2011. Crystal structure of α-galactosidase from Lactobacillus acidophilus NCFM: insight into tetramer formation and substrate binding. J Mol Biol 412:466–480. doi: 10.1016/j.jmb.2011.07.057. [DOI] [PubMed] [Google Scholar]
- 59.Barkholt V, Jensen AL. 1989. Amino acid analysis: determination of cysteine plus half-cystine in proteins after hydrochloric acid hydrolysis with a disulfide compound as additive. Anal Biochem 177:318–322. doi: 10.1016/0003-2697(89)90059-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.