Abstract
The present study aimed to investigate the expression and prognostic significance of insulin-like growth factor binding protein 6 (IGFBP-6) in gastric adenocarcinoma. The expression of IGFBP-6 was examined in 263 specimens from gastric adenocarcinoma patients using reverse transcription-quantitative polymerase chain reaction (RT-qPCR), western blotting and immunohistochemical (IHC) staining. The association between IGFBP-6 expression, clinicopathological factors and clinical outcomes was investigated. Akaike information criterion (AIC) and Harrell's concordance index (c-index) were used to evaluate the accuracy of the predictive prognosis. RT-qPCR and western blotting results showed that IGFBP-6 mRNA expression was lower in the tumors compared with that in adjacent non-tumor tissues. IGFBP-6 showed significantly decreased expression in 170 out of 263 patients based on IHC data and this was associated with a larger tumor size (P<0.001) and poorly-differentiated adenocarcinoma (P=0.001), as well as with palliative gastrectomy (P=0.015). Additionally, decreased expression of IGFBP-6 was associated with stage T3/4a/4b disease and lymph node-positive metastasis (P<0.001). The association between decreased expression and a poor prognosis was revealed by Kaplan-Meier curves. Cox regression model identified IGFBP-6 as an independent prognostic factor. The prognostic value of the model with IGFBP-6 expression (AIC, 924.881; c-index, 0.878) was superior to that without IGFBP-6 expression (AIC, 947.164; c-index, 0.825). In conclusion, IGFBP-6 involves the development and progression of gastric adenocarcinoma, and its decreased expression predicts poor clinical outcomes.
Keywords: insulin-like growth factor binding protein 6, expression, gastric carcinoma, prognosis
Introduction
Gastric carcinoma, a gastrointestinal malignancy, is the second leading cause of cancer-associated mortality worldwide (1). There are more new gastric cancer cases every year in China than in any other country (2). Despite advances in the diagnosis and treatment strategies, the 5-year overall survival rate of gastric cancer is <25%, which makes the prognosis for gastric cancer patients one of the poorest among all malignant cancer types (3).
Gastric cancer is a type of highly heterogeneous malignancy in terms of histology and genetics; therefore, it is difficult to forecast the clinical outcomes using traditional histological classification (4). Current clinical treatments for gastric cancer, such as surgery, radiotherapy and chemotherapy, have limited efficacy, thus highlighting the importance of advanced treatment strategies to prolong patient survival. The occurrence and development of gastric cancer are identified to be a multistage process with multigene regulation involving oncogenes and tumor suppressor genes. It has been reported recently that certain novel oncogenes and suppressor genes contribute to the early diagnosis and targeted therapy of gastric cancer (5,6). An in-depth study on the molecular mechanisms of gastric carcinogenesis and progression is important to develop novel therapeutic targets and to improve clinical outcomes (5,7–10).
A number of cellular processes are involved in the regression or progression of gastric cancer. Among them, the insulin-like growth factor (IGF) signaling pathway plays a significant role. The IGF system, as an important modifier of tumor cell proliferation, growth and survival, is composed of IGF-I and -II and their receptors, the IGF-I receptor and IGF-II/mannose-6-phosphate receptor, and a family of six high-affinity IGF binding proteins (11). IGF-I and IGF-II are known to stimulate growth, differentiation and the proliferative abilities of various tumor cells. IGF dysregulation is involved in tumor development, and increased IGF activity has been detected in certain tumors (5). IGF-II acts as an autocrine growth factor in certain tumors and tumor cell lines, and contributes to tumorigenesis. The higher expression level and rate of IGF-II compared with IGF-I in local tumor tissues has been reported in several studies (12–17), revealing the more significant role of IGF-II in gastric carcinogenesis and development in comparison to IGF-I.
IGF binding protein (IGFBP)-6 is a member of a family that includes six high-affinity IGFBPs, which participate in the regulation of the activities of IGF and IGF-independent effects (18). IGFBP-6 is unique among the IGFBPs due to its 20- to 100-fold higher binding affinity for IGF-II than IGF-I (19–25), making it a relatively distinctive IGF-II inhibitor. IGFBP-6 can inhibit IGF-II actions in vitro and in vivo by preventing IGF-II from binding to receptors on the surface of tumor cells, directly inhibiting the tumorigenic properties of IGF-II in an IGF-II-dependent manner (18,26). In addition, a number of studies have reported that it may also inhibit growth and modulate cell migration and apoptosis in an IGF-II-independent manner via interactions with several plasma, extracellular matrix and cell surface molecules (18,24,26,27).
However, to date, there have been no studies on the expression information and prognostic importance of IGFBP-6 in primary gastric adenocarcinoma. Therefore, the expression level of IGFBP-6 was assessed using reverse transcription-quantitative polymerase chain reaction (RT-qPCR), western blotting and immunohistochemical (IHC) staining in the present study. Additionally, the association between IGFBP-6, clinicopathological features and prognostic significance was investigated in gastric adenocarcinoma patients.
Patients and methods
Approval and consent
The Research Ethics Committee of the Sun Yat-sen University Cancer Center (SYSUCC; Guangzhou, Guangdong, China) approved the present study, and each patient provided written informed consent prior to the study.
Patients and tissue specimens
This study retrospectively analyzed clinicopathological information from 263 gastric adenocarcinoma patients who underwent surgery between October 2003 and October 2005 at SYSUCC. The patients were selected according to the following criteria: i) Histopathologically proven gastric adenocarcinoma; ii) received gastrectomy with D1/D2 lymphadenectomy; iii) complete follow-up information; iv) no chemotherapy or radiotherapy administered prior to surgery; v) no synchronous malignancies, remnant or recurrent gastric cancer; and vi) no perioperative mortality. Every patient underwent radical gastrectomy with D2 lymphadenectomy, which was performed by by experienced surgeons using standard procedures in line with the Japanese Gastric Cancer Association guidelines (28). The study consisted of 181 men and 82 women, with a median age of 60 years (range, 17–85 years). The histological classification was graded based on the World Health Organization classification criteria (29). Tumor-Node-Metastasis (TNM) staging was estimated according to the Union for International Cancer Control (UICC) seventh edition (30).
Fresh gastric cancer tumor tissue and matched normal tissue were obtained from a cohort of 48 consecutive gastric cancer patients who underwent gastrectomy at SYSUCC between February 2004 and February 2005. The postoperative tissue samples were immersed in RNAlater (Ambion; Thermo Fisher Scientific, Inc., Waltham, MA, USA) and were stored at −80°C until RNA extraction. The tumor tissue and matched normal tissue (obtained >2 cm away from the tumor edge) were confirmed pathologically.
RNA extraction and RT-qPCR
The total RNA of the specimens was extracted according to the manufacturer's instructions using TRIzol Reagent (Invitrogen; Thermo Fisher Scientific, Inc.). To avoid DNA contamination, RNAse-free DNAse I was added. RNA quality and quantity were determined using a NanoDrop spectrophotometer (Thermo Fisher Scientific, Inc.). First-strand cDNA was synthesized by reverse transcription using M-MLV reverse transcriptase (Promega Corporation, Madison, WI, USA) and 2µg total RNA. The cDNA was then subjected to RT-qPCR to evaluate the relative expression levels of Image result for glyceraldehyde 3-phosphate dehydrogenase (GAPDH; as an internal control) and IGFBP-6. The Applied Biosystems ABI 7900HT machine was used to measure the binding of SYBR Green I to double-stranded DNA in gene specific amplification. The 15-µl reactions contained cDNA, SYBR Green master mix (Invitrogen; Thermo Fisher Scientific, Inc.) and each pair of oligonucleotide primers. The sequences of the sense and antisense primers were as follows: IGFBP6 forward, 5′-TGGAGCTGTCATCACTCAAC-3′ and reverse, 5′-GCCAACACCAACACTCTTTC-3′; and GAPDH forward, 5′-CTCCTCCTGTTCGACAGTCAGC-3′ and reverse, 5′-CCCAATACGACCAAATCCGTT-3′. The PCR cycling conditions were as follows: 95°C for 10 min, and 45 cycles of melting at 95°C for 30 sec, followed by 60°C for 1 min. The relative quantity and regression curves of all specimens were calculated by the instrument's software (SDS 2.0). GAPDH was selected as the internal control gene for normalizing the relative expression of the target gene to the geometric mean. qPCR was performed in triplicate in 96-well plates and repeated twice separately. The normalized final data were analyzed using the 2−∆∆Cq method (31).
Western blotting
The tumor and non-tumor tissues from the gastric cancer patients were homogenized in radioimmunoprecipitation assay lysis buffer. Following centrifugation (4,000 × g) at 4°C for 15 min, the total proteins were collected and the protein concentrations were determined. The protein samples, in equal amounts of 30 µg, were transferred to polyvinylidene fluoride membranes through 12% SDS-PAGE gels. Next, the membranes were incubated with IGFBP-6 polyclonal antibody (catalog no. ab109765; 1:1,000 dilution) or anti-β-actin antibody (catalog no. ab16039; 1:2,000 dilution) (both Abcam, Cambridge, UK), overnight at 4°C, after immersion in 5% skimmed milk for 60 min to block non-specific binding sites. Subsequent to being washed, the membranes were incubated with horseradish peroxidase-conjugated secondary antibody (catalog no. ab6759; 1:5,000 dilution; Abcam) for 2 h at room temperature, followed by washing again and analysis using an electrochemiluminescence system (Pierce; Thermo Fisher Scientific, Inc.).
Immunohistochemistry
All specimens from the patients were processed with formalin-fixation and paraffin-embedding, followed by the formation of 2-μm thick slides, which were subsequently deparaffinized and rehydrated. Subsequently, the sections were soaked in ethylenediaminetetraacetic acid (pH 8.0), followed by heating for antigen retrieval. After blocking the endogenous peroxidase with hydrogen peroxide solution, the sections were soaked in IGFBP-6 antibody (catalog no. SC-6007; Santa Cruz Biotechnology, Inc., Dallas, TX, USA) at a dilution of 1:500, overnight at 4°C. The sections were then washed and immersed with secondary antibody (OriGene Technologies, Inc., Beijing, China) for 30 min at room temperature. Finally, the reaction signals were visualized with diaminobenzidine tetrahydrochloride, and all of the sections were counterstained with hematoxylin.
Semi-quantitative methods
Two experienced pathologists who were blinded to the clinical parameters evaluated these specimens. Deeper discussion was required to reach a consensus when faced with discrepancies. The total IGFBP-6 immunostaining score was estimated based on the positive cell percentage (the percentage of tumor cells positively staining versus the total cells) and the staining intensity, as described previously (32–34). The positive cell percentages were identified as follows: <5% (negative), score of 0; 5–25% (sporadic), score of 1; 25–50% (focal), score of 2; and >50% (diffuse), score of 3. The staining intensities of no staining (−), weakly-stained (+), moderately-stained (++) and strongly-stained (+++) were scored as 0, 1, 2 and 3, respectively. The total IGFBP-6 IHC expression score was the product of the aforementioned factors, which ranged from 0 to 9. IGFBP-6 expression was grouped into low expression (scores of 0–3) and high expression (scores of 4–9).
Statistical analysis
Associations between the clinicopathological features and IGFBP-6 expression were evaluated with the χ2 test and Pearson's index. The overall survival curves and survival differences were analyzed by the Kaplan-Meier method and log-rank test. Multivariate analysis was assessed using the Cox proportional-hazard regression model. The predictive ability of the two models including and excluding IGFBP-6 expression and other prognostic clinicopathological variables were compared based on Akaike information criterion (AIC) and Harrell's concordance index (c-index). The minimum AIC value indicates the optimum predictive prognosis model with the least clinicopathological data loss and inefficiency. Harrell's c-index was an index to evaluate the predictive accuracy, with a value much closer to 1.0 implying higher accuracy. All data were analyzed statistically using the SPSS 18.0 software package (SPSS, Inc., Chicago, IL, USA), and statistical significance was considered to be indicated by P<0.05 in a two-tailed test.
Results
RT-qPCR of IGFBP-6 mRNA expression
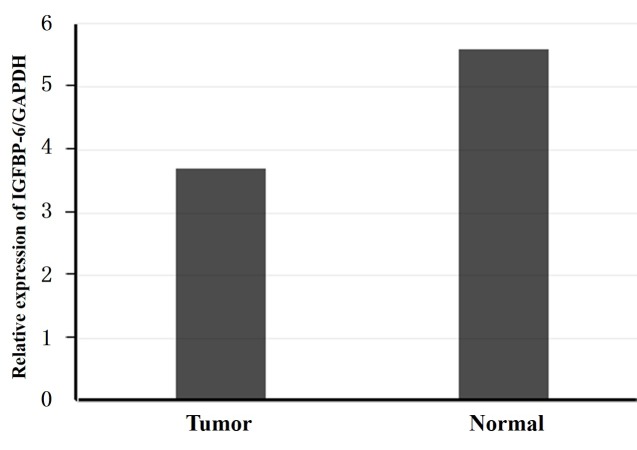
The IGFBP-6 transcriptional levels were detected by RT-qPCR in 48 pairs of post-operative samples, including tumor tissue and normal tissue, from gastric cancer patients. The IGFBP-6 mRNA levels were significantly decreased in the tumor tissue of 36 (75.0%) patients compared with those in the normal tissue (P=0.0073; Fig. 1).
Figure 1.
RT-qPCR analysis of IGFBP-6 expression in gastric cancer patients. Relative expression of IGFBP6 in gastric cancer tumor tissues compared with adjacent non-tumor tissues, as assessed by RT-qPCR (n=48; P=0.0073). RT-qPCR, reverse transcription-quantitative polymerase chain reaction; IGFBP-6, insulin-like growth factor binding protein 6; GAPDH, glyceraldehyde 3-phosphate dehydrogenase.
Western blotting of IGFBP-6 expression
The IGFBP-6 protein expression level in gastric cancer tissue specimens was detected by western blotting. The results showed IGFBP-6 expression was decreased in the tumor tissue compared with the matched normal tissue in 33 (68.8%) of the 48 patients (P=0.016; Fig. 2). These findings were consistent with those of the RT-qPCR.
Figure 2.
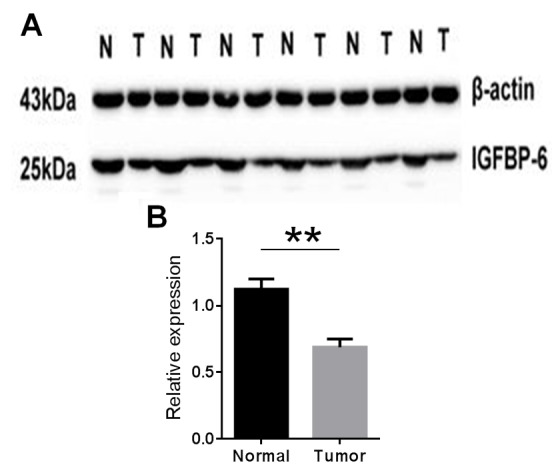
(A and B) Western blotting analysis of IGFBP6 expression in gastric cancer patients. Relative expression of IGFBP6 in gastric cancer tumor tissues compared with adjacent non-tumor tissues (n=41), as assessed by western blotting (n=48; **P=0.016). IGFBP-6, insulin-like growth factor binding protein 6; T, tumor tissues; N, non-tumor tissues.
Correlation between IGFBP-6 expression and clinicopathological characteristics
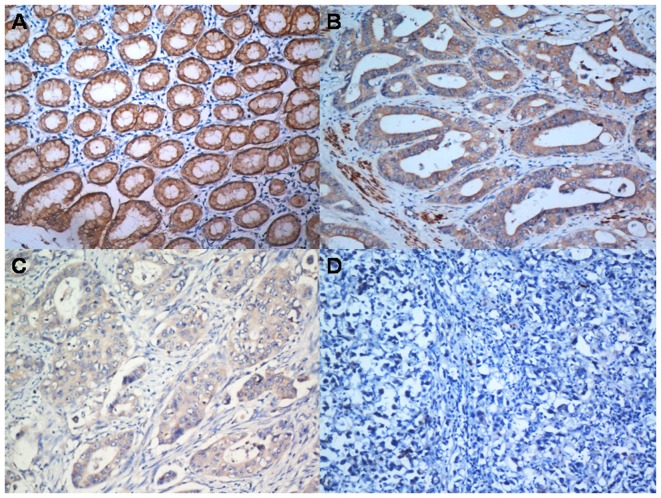
To further determine the role of IGFBP-6, the expression level of IGFBP-6 protein in paraffin-embedded tissue slides, including tumor tissue and matched normal tissue, were examined by immunohistochemistry in 263 gastric cancer patients (Fig. 3). IGFBP-6 protein was localized positively in the cytoplasm upon IHC staining.
Figure 3.
IGFBP6 protein expression in gastric cancer surgical specimens, as assessed by immunohistochemistry. (A) Strong IGFBP6 staining (+++) was observed in the non-cancerous gastric mucosa. (B) IGFBP6 staining (++) in well-differentiated gastric cancer. (C) IGFBP6 staining (+) in poorly-differentiated gastric cancer. (D) Negative IGFBP6 staining (−) in mucinous carcinoma. IGFBP-6, insulin-like growth factor binding protein 6.
As aforementioned, all patients were categorized into two groups according to the IGFBP-6 expression levels. The correlations between IGFBP-6 expression and the clinicopathological features are listed in Table I. IGFBP-6 expression was significantly decreased in poorly-differentiated (G3) carcinomas compared with well-differentiated (G1) and moderately-differentiated (G2) carcinomas (P=0.001). There were significant associations between the reduced expression of IGFBP-6 and larger tumor size (P<0.001) and the presence of a palliative gastrectomy (P=0.015). Reduced IGFBP-6 expression was also significantly associated with an increased depth of tumor invasion (T3/4a/4b; P<0.001) and lymph node metastasis (P<0.001). Additionally, the majority of the patients in stage III (72.4%) and stage IV (80.7%) exhibited a reduced IGFBP-6 expression level (P<0.001).
Table I.
Association between IGFBP-6 expression and the clinicopathological features of patients with gastric cancer.
| IGFBP-6 expression | ||||
|---|---|---|---|---|
| Variables | Patients, n | Low (n=170) | High (n=93) | P-value |
| Age, years | 0.912 | |||
| ≤60 | 160 | 103 | 57 | |
| >60 | 103 | 67 | 36 | |
| Gender | 0.096 | |||
| Male | 181 | 111 | 70 | |
| Female | 82 | 59 | 23 | |
| Tumor size, cm | <0.001a | |||
| ≤5.0 | 179 | 102 | 77 | |
| >5.0 | 84 | 68 | 16 | |
| Histological grade | 0.001a | |||
| Well-differentiated (G1)/moderately-differentiated (G2) | 70 | 34 | 36 | |
| Poorly-differentiated (G3) | 193 | 136 | 57 | |
| Location | 0.228 | |||
| Proximal | 150 | 104 | 46 | |
| Distant | 105 | 59 | 46 | |
| Total | 8 | 7 | 1 | |
| Radical resection | 0.015a | |||
| Yes | 228 | 141 | 87 | |
| No | 35 | 29 | 6 | |
| Tumor invasion (T) | <0.001a | |||
| T1+T2 | 51 | 14 | 37 | |
| T3+T4a/T4b | 212 | 156 | 56 | |
| Nodal status (N) | <0.001a | |||
| No | 69 | 31 | 38 | |
| Yes | 194 | 139 | 55 | |
| Metastasis status (M) | 0.070 | |||
| M0 | 237 | 149 | 88 | |
| M1 | 26 | 21 | 5 | |
| TNM staging | <0.001a | |||
| I | 29 | 1 | 28 | |
| II | 63 | 43 | 20 | |
| III | 145 | 105 | 40 | |
| IV | 26 | 21 | 5 | |
P<0.05. IGFBP-6, insulin-like growth factor binding protein 6.
Correlation between IGFBP-6 expression and patient survival
The median survival time and overall survival rate of all 263 patients were 58 months (3–87 months) and 56.0%, respectively. Compared with the IGFBP-6 low expression group, the overall survival rate (78.8 vs. 43.4%) and the 5-year overall survival rate (78.8 vs. 47.4%) in the high expression group were significantly improved (both P<0.001; Fig. 4).
Figure 4.
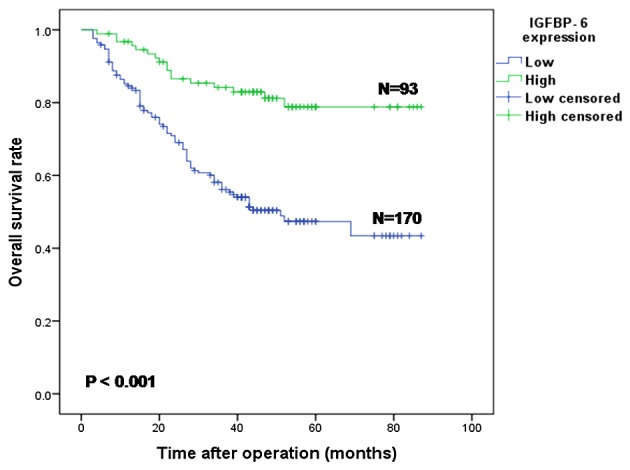
Kaplan-Meier survival curves of gastric cancer patients (n=263) after gastrectomy. The survival rate of the patients in the IGFBP6-low group (n=170) was significantly lower than that of the patients in the IGFBP6-high group (n=93) (P<0.001). IGFBP-6, insulin-like growth factor binding protein 6.
Univariate and multivariate analyses
The associations between IGFBP-6 expression and other clinicopathological features and the prognosis of gastric cancer patients were examined by univariate and multivariate analyses. According to the results of the univariate analysis, the following 9 factors were significantly associated with overall survival: Age (P=0.012), tumor location (P=0.001), tumor size (P<0.001), histological grade (P=0.002), radical resection (P<0.001), IGFBP-6 expression (P<0.001), T stage (P<0.001), N stage (P<0.001) and M stage (P<0.001) based on the UICC TNM staging, seventh edition (Table II). Next, two Cox regression models were constructed, which contained the factors of IGFBP-6 expression or no IGFBP-6 expression, in order to realize the role of IGFBP-6 expression in prognostic prediction. Model A (without IGFBP-6 expression) showed that tumor location (P=0.010), tumor size (P=0.001), histological grade (P=0.012), tumor infiltration degree (P=0.018), lymph node metastases (P<0.001) and distant metastases (P=0.008) were independent prognostic predictors in gastric cancer patients (Table II). Model B (with IGFBP-6 expression) identified that tumor location (P=0.008), tumor size (P=0.007), IGFBP-6 expression (P=0.014), tumor infiltration degree (P=0.046), lymph node metastases (P<0.001) and distant metastases (P=0.004) were independent prognostic prediction factors in gastric cancer patients (Table II). Compared with the IGFBP-6 high expression group, the relative risk of mortality was 2.104 times higher in the IGFBP-6 low expression group (hazard ratio, 2.104; 95% confidence interval, 1.162–3.807; P=0.014). Furthermore, AIC and Harrell's c-index were used to evaluate the predictive accuracy of the two prognostic models. The results showed that the model including IGFBP-6 expression (AIC, 924.881; c-index, 0.878) possessed superior prognostic value compared with the model excluding IGFBP-6 expression (AIC, 947.164; c-index, 0.825).
Table II.
Univariate and multivariate survival analyses of clinicopathological variables for 263 gastric carcinoma patients.
| A, Model without IGFBP6 expression | ||||||
|---|---|---|---|---|---|---|
| Univariate analysis | Multivariate analysis | |||||
| Variables | HR | 95% CI | P-value | HR | 95% CI | P-value |
| Gender (male vs. female) | 1.513 | 0.994–2.303 | 0.054 | |||
| Age, years (≥60 vs. <60) | 1.674 | 1.120–2.504 | 0.012a | 1.437 | 0.953–2.166 | 0.083 |
| Location (distal/proximal/total) | 0.475 | 0.311–0.725 | 0.001a | 0.574 | 0.375–0.878 | 0.010a |
| Size, cm (>5 vs. ≤5) | 2.392 | 1.595–3.588 | <0.001a | 1.985 | 1.302–3.025 | 0.001a |
| Histological grade (G3 vs. G2/G1) | 2.549 | 1.420–4.577 | 0.002a | 2.142 | 1.186–3.867 | 0.012a |
| Radical resection (no vs. yes) | 4.288 | 2.694–6.824 | <0.001a | 1.139 | 0.454–2.859 | 0.781 |
| T (T4/T3 vs. T2/T1) | 5.812 | 2.359–14.317 | <0.001a | 3.130 | 1.217–8.047 | 0.018a |
| N (yes vs. no) | 4.095 | 2.123–7.899 | <0.001a | 3.940 | 2.002–7.753 | <0.001a |
| M (M1 vs. M0) | 5.218 | 3.145–8.656 | <0.001a | 3.887 | 1.432–10.554 | 0.008a |
| B, Model containing IGFBP6 expression | ||||||
| Univariate analysis | Multivariate analysis | |||||
| Variables | HR | 95% CI | P-value | HR | 95% CI | P-value |
| Gender (male vs. female) | 1.513 | 0.994–2.303 | 0.054 | |||
| Age, years (≥60 vs. <60) | 1.674 | 1.120–2.504 | 0.012a | 1.457 | 0.965–2.198 | 0.073 |
| Location (distal/proximal/total) | 0.475 | 0.311–0.725 | 0.001a | 0.562 | 0.367–0.860 | 0.008a |
| Size, cm (>5 vs. ≤5) | 2.392 | 1.595–3.588 | <0.001a | 1.795 | 1.170–2.753 | 0.007a |
| Histological grade (G3 vs. G2/G1) | 2.549 | 1.420–4.577 | 0.002a | 1.819 | 0.996–3.321 | 0.052 |
| Radical resection (no vs. yes) | 4.288 | 2.694–6.824 | <0.001a | 1.023 | 0.407–2.574 | 0.961 |
| IGFBP-6 expression (low vs. high) | 3.949 | 2.237–6.971 | <0.001a | 2.104 | 1.162–3.807 | 0.014a |
| T (T4/T3 vs. T2/T1) | 5.812 | 2.359–14.317 | <0.001a | 2.637 | 1.018–6.831 | 0.046a |
| N (yes vs. no) | 4.095 | 2.123–7.899 | <0.001a | 3.772 | 1.914–7.435 | <0.001a |
| M (M1 vs. M0) | 5.218 | 3.145–8.656 | <0.001a | 4.359 | 1.600–11.876 | 0.004a |
P<0.05. T, tumor; N, node; M, metastasis; IGFBP-6, insulin-like growth factor binding protein 6.
Discussion
It has been shown that IGF-I and IGF-II are involved in cancer development, acting as strong mitogenic agents in different cell types (35–38). Certain studies have shown that IGF-II is an autocrine growth factor in a number of tumors; IGF-II was overexpressed in numerous tumors, including those in ovarian cancer, functioning by binding to the IGF-I receptor (IGF-IR) and insulin receptor (IR-A) (39). Meanwhile, malignant cells that predominantly express IR-A have been shown to overexpress IGF-II (40–43). As IGF-II could bind to IGF-IR and IR-A, it has been hypothesized that when IGF-II binds to these receptors, the signals of mitosis and antiapoptosis may be activated. In certain regional tissues with receptors and IGF-II overexpression, the effect of IGF-II is thus substantially amplified (39). A powerful autocrine or paracrine growth stimulatory cycle based on all these key factors is supposed to contribute to the development of cancer (44). Consistent results from in vitro studies have suggested that IGF-II is likely to increase the cellular ability of migration and invasion by acting on IR-A (43). These findings indicate that IGF-II is critical in the tumorigenesis, progression and prognosis of numerous malignancies.
As a relatively specific inhibitor of IGF-II (20–26), IGFBP-6 could interfere with the process of cell proliferation, cell growth, cell differentiation and cell adhesion in numerous types of cells (18). However, IGFBP-6 not only inhibits IGF-II activity directly, but also develops IGF-II intrinsic functions, such as cell growth suppression and apoptosis stimulation (19,25,27,31). Previous studies demonstrated that IGFBP-6 overexpression in rhabdomyosarcoma significantly reduced tumor growth in nude mice (24,45) and stimulated the programmed cell death of cancer cells (46). IGFBP-6 was reported to inhibit IGF-II-induced proliferation in colon cancer cells, but not IGF-I-induced proliferation (47). These in vivo and in vitro findings hinted that IGFBP-6 suppresses tumor growth. A number of studies have demonstrated that the development and progression of a tumor is a multi-factor and multi-step process that involves various cytokines, proteins and their corresponding receptors, proteases and chemokines, which are affected by the tumor microenvironment and genetic and epigenetic factors (48,49). IGFBP-6 has been shown to regulate the function of IGFs, particularly IGF-II, which is involved in tumor progression. Therefore, it is plausible that the loss of IGFBP-6 expression causes cell dedifferentiation and colony formation, which accelerate metastasis and tumor invasion. However, to date, IGFBP-6 expression information and its prognostic significance in primary gastric adenocarcinoma patients have not been evaluated.
The expression of IGFBP-6 at the gene and protein levels was analyzed using three different methods in the present study. First, IGFBP-6 expression at the mRNA and protein levels in post-operative gastric adenocarcinoma samples was investigated by RT-qPCR and western blotting. The results revealed that IGFBP-6 expression was decreased in tumor tissue compared with that in matched normal tissue at the mRNA (P=0.0073) and protein (P=0.016) levels. These preliminary data coincide with the aforementioned hypothesis that lower expression of IGFBP-6 may play a vital role in gastric tumorigenesis and development, and function as a tumor suppressor.
Furthermore, immunohistochemistry confirmed the expression of IGFBP-6 in the tumor tissues of the gastric adenocarcinoma patients. IGFBP-6 was expressed in the cytoplasm, and the staining intensity varied among the different tissues. The results demonstrated that low IGFBP-6 expression was associated with a larger tumor size, poorly-differentiated adenocarcinoma (G3), whole stomach cancer, radical resection, deeper depth of tumor infiltration (T3 and T4a/b) and lymph node metastasis. Low IGFBP-6 expression was more likely to be detected in stage III (73%) and stage IV (81%) patients, findings that were consistent with the experimental results of RT-qPCR and western blotting. These findings indicated that IGFBP-6 could be viewed as a potential suppressor participating in the occurrence and development of gastric cancer.
According to the results from the Kaplan-Meier survival analysis, patients with higher expression levels of IGFBP-6 experienced significantly improved survival outcomes compared with those with lower expression levels of IGFBP-6. Univariate analysis demonstrated that decreased IGFBP-6 expression was significantly associated with the OS rate and the 5-year OS rate in gastric cancer patients. Multivariate analysis showed that IGFBP-6 expression (P=0.014), combined with certain other conventional clinicopathological factors, including tumor location (P=0.008), tumor size (P=0.007), tumor differentiated degree (P=0.046), lymph node metastasis status (P<0.001) and distant metastasis status (P=0.004), were independent prognostic factors. Furthermore, the AIC and c-index values manifested that the prognosis model that included IGFBP-6 expression (AIC, 924.881; c-index, 0.878) possessed a stronger predictive ability than the model excluding expression (AIC, 947.164; c-index, 0.825) in this study. These results indicated that the decreased expression of IGFBP-6 may be useful to identify an unfavorable prognosis in gastric cancer patients and may consequently be viewed as a potentially novel prognostic factor. Additionally, these results indicated that the incorporation of certain gene expression levels, such as those of IGFBP-6, in traditional clinicopathological prognostic models may be of benefit to better predict the survival outcomes of patients.
Although the possible association between the decreased expression of IGFBP-6 and other cellular factors, and the exact mechanism of the tumor suppression function of IGFBP-6 have not been further investigated in the present study, the comparative quantity of post-operative samples from gastric cancer patients who underwent uniformly standard treatment enhances the value of the present results. All the data are conducive to understanding gastric cancer in more depth.
In conclusion, the data from the present study demonstrated that the decreased expression of IGFBP-6 predicts poor clinical outcomes in gastric cancer patients. Nevertheless, the exact molecular mechanisms and relative signal channel of IGFBP-6 involved in gastric cancer must be investigated further in future studies. A greater and more thorough comprehension of the functional mechanism of IGFBP-6 in the genesis and development of tumors is beneficial to improving clinical outcomes and developing more effective treatment strategies in gastric cancer patients.
Acknowledgements
This study was supported by grants from the Natural Science Foundation of Guangdong Province (2015A030313089), the Medical Science and Technology Research Foundation of Guangdong Province (B2014160), the Major Program of Collaborative Innovation of Guangzhou (201508030042) and the Science and Technology Project of Fujian Province (2014J01284).
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics, 2009. CA Cancer J Clin. 2009;59:225–249. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 3.Hartgrink HH, Jansen EP, van Grieken NC, van de Velde CJ. Gastric cancer. Lancet. 2009;374:477–490. doi: 10.1016/S0140-6736(09)60617-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nobili S, Bruno L, Landini I, Napoli C, Bechi P, Tonelli F, Rubio CA, Mini E, Nesi G. Genomic and genetic alterations influence the progression of gastric cancer. World J Gastroenterol. 2011;17:290–299. doi: 10.3748/wjg.v17.i3.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen CN, Lin JJ, Chen JJ, Lee PH, Yang CY, Kuo ML, Chang KJ, Hsieh FJ. Gene expression profile predicts patient survival of gastric cancer after surgical resection. J Clin Oncol. 2005;23:7286–7295. doi: 10.1200/JCO.2004.00.2253. [DOI] [PubMed] [Google Scholar]
- 6.Yasui W, Oue N, Aung PP, Matsumura S, Shutoh M, Nakayama H. Molecular-pathological prognostic factors of gastric cancer: A review. Gastric Cancer. 2005;8:86–94. doi: 10.1007/s10120-005-0320-0. [DOI] [PubMed] [Google Scholar]
- 7.Lee HS, Cho SB, Lee HE, Kim MA, Kim JH, Park DJ, Kim JH, Yang HK, Lee BL, Kim WH. Protein expression profiling and molecular classification of gastric cancer by the tissue array method. Clin Cancer Res. 2007;13:4154–4163. doi: 10.1158/1078-0432.CCR-07-0173. [DOI] [PubMed] [Google Scholar]
- 8.Oue N, Hamai Y, Mitani Y, Matsumura S, Oshimo Y, Aung PP, Kuraoka K, Nakayama H, Yasui W. Gene expression profile of gastric carcinoma: Identification of genes and tags potentially involved in invasion, metastasis, and carcinogenesis by serial analysis of gene expression. Cancer Res. 2004;64:2397–2405. doi: 10.1158/0008-5472.CAN-03-3514. [DOI] [PubMed] [Google Scholar]
- 9.Hippo Y, Taniguchi H, Tsutsumi S, Machida N, Chong JM, Fukayama M, Kodama T, Aburatani H. Global gene expression analysis of gastric cancer by oligonucleotide microarrays. Cancer Res. 2002;62:233–240. [PubMed] [Google Scholar]
- 10.Yasui W, Oue N, Sentani K, Sakamoto N, Motoshita J. Transcriptome dissection of gastric cancer: Identification of novel diagnostic and therapeutic targets from pathology specimens. Pathol Int. 2009;59:121–136. doi: 10.1111/j.1440-1827.2009.02329.x. [DOI] [PubMed] [Google Scholar]
- 11.Iosef C, Vilk G, Gkourasas T, Lee KJ, Chen BP, Fu P, Bach LA, Lajoie G, Gupta MB, Li SS, Han VK. Insulin-like growth factor binding protein-6 (IGFBP-6) interacts with DNA-end binding protein Ku80 to regulate cell fate. Cell Signal. 2010;22:1033–1043. doi: 10.1016/j.cellsig.2010.02.006. [DOI] [PubMed] [Google Scholar]
- 12.Minniti CP, Luan D, O'Grady C, Rosenfeld RG, Oh Y, Helman LJ. Insulin-like growth factor II overexpression in myoblasts induces phenotypic changes typical of the malignant phenotype. Cell Growth Differ. 1995;6:263–269. [PubMed] [Google Scholar]
- 13.Osborne CK, Coronado EB, Kitten LJ, Arteaga CI, Fuqua SA, Ramasharma K, Marshall M, Li CH. Insulin-likegrowth factor-II (IGF-II): A potential autocrine/paracrine growth factor forhuman breast cancer acting via the IGF-I receptor. Mol Endocrinol. 1989;3:1701–1709. doi: 10.1210/mend-3-11-1701. [DOI] [PubMed] [Google Scholar]
- 14.Thompson MA, Cox AJ, Whitehead RH, Jonas HA. Autocrine regulation of human tumor cell proliferation by insulin-like growth factor II: An in-vitro model. Endocrinology. 1990;126:3033–3042. doi: 10.1210/endo-126-6-3033. [DOI] [PubMed] [Google Scholar]
- 15.Christofori G, Naik P, Hanahan D. A second signal supplied by insulin-like growth factor II in oncogene-induced tumorigenesis. Nature. 1994;369:414–418. doi: 10.1038/369414a0. [DOI] [PubMed] [Google Scholar]
- 16.Cullen KJ, Allison A, Martire I, Ellis M, Singer C. Insulin-like growth factor expression in breast cancer epithelium and stroma. Breast Cancer Res Treat. 1992;22:21–29. doi: 10.1007/BF01833330. [DOI] [PubMed] [Google Scholar]
- 17.Ho MN, Delgado CH, Owens GA, Steller MA. Insulin-like growth factor-II participates in the biphasic effect of a gonadotropin-releasing hormone agonist on ovarian cancer cell growth. Fertil Steril. 1997;67:870–876. doi: 10.1016/S0015-0282(97)81399-4. [DOI] [PubMed] [Google Scholar]
- 18.Firth SM, Baxter RC. Cellular actions of the insulin-like growth factor binding proteins. Endocr Rev. 2002;23:824–854. doi: 10.1210/er.2001-0033. [DOI] [PubMed] [Google Scholar]
- 19.Kiefer MC, Schmid C, Waldvogel M, Schläpfer I, Futo E, Masiarz FR, Green K, Barr PJ, Zapf J. Characterization of recombinant human insulin-like growth factor binding proteins 4, 5, and 6 produced in yeast. J Biol Chem. 1992;267:12692–12699. [PubMed] [Google Scholar]
- 20.Bach LA. IGFBP-6 five years on; not so ‘forgotten’? Growth Horm IGF Res. 2005;15:185–192. doi: 10.1016/j.ghir.2005.04.001. [DOI] [PubMed] [Google Scholar]
- 21.Bach LA, Hsieh S, Brown AL, Rechler MM. Recombinant human insulin-like growth factor (IGF)-binding protein-6 inhibits IGF-II-induced differentiation of L6A1 myoblasts. Endocrinology. 1994;135:2168–2176. doi: 10.1210/endo.135.5.7525263. [DOI] [PubMed] [Google Scholar]
- 22.Dake BL, Boes M, Bach LA, Bar RS. Effect of an insulin-like growth factor binding protein fusion protein on thymidine incorporation in neuroblastoma and rhabdomyosarcoma cell lines. Endocrinology. 2004;145:3369–3374. doi: 10.1210/en.2003-1667. [DOI] [PubMed] [Google Scholar]
- 23.Gallicchio MA, Kaun C, Wojta J, Binder B, Bach LA. Urokinase type plasminogen activator receptor is involved in insulin-like growth factor-induced migration of rhabdomyosarcoma cells in vitro. J Cell Physiol. 2003;197:131–138. doi: 10.1002/jcp.10352. [DOI] [PubMed] [Google Scholar]
- 24.Gallicchio MA, Kneen M, Hall C, Scott AM, Bach LA. Overexpression of insulin-like growth factor binding protein-6 inhibits rhabdomyosarcoma growth in vivo. Int J Cancer. 2001;94:645–651. doi: 10.1002/ijc.1519. [DOI] [PubMed] [Google Scholar]
- 25.Kim EJ, Kang YH, Schaffer BS, Bach LA, MacDonald RG, Park JH. Inhibition of Caco-2 cell proliferation by all-trans retinoic acid: Role of insulin-like growth factor binding protein-6. J Cell Physiol. 2001;190:92–100. doi: 10.1002/jcp.10045. [DOI] [PubMed] [Google Scholar]
- 26.Mohan S, Baylink DJ. IGF-binding proteins are multifunctional and act via IGF-dependent and -independent mechanisms. J Endocrinol. 2002;175:19–31. doi: 10.1677/joe.0.1750019. [DOI] [PubMed] [Google Scholar]
- 27.Sueoka N, Lee HY, Wiehle S, Cristiano RJ, Fang B, Ji L, Roth JA, Hong WK, Cohen P, Kurie JM. Insulin-like growth factor binding protein-6 activates programmed cell death in non-small cell lung cancer cells. Oncogene. 2000;19:4432–4436. doi: 10.1038/sj.onc.1203813. [DOI] [PubMed] [Google Scholar]
- 28.Sano T. Evaluation of the gastric cancer treatment guidelines of the Japanese Gastric Cancer Association. Gan To Kagaku Ryoho. 2010;37:582–586. (In Japanese) [PubMed] [Google Scholar]
- 29.Berlth F, Bollschweiler E, Drebber U, Hoelscher AH, Moenig S. Pathohistological classification systems in gastric cancer: Diagnostic relevance and prognostic value. World J Gastroenterol. 2014;20:5679–5684. doi: 10.3748/wjg.v20.i19.5679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wittekind C. 2010 TNM system: On the 7th edition of TNM classification of malignant tumors. Pathologe. 2010;31:331–332. doi: 10.1007/s00292-010-1349-3. (In German) [DOI] [PubMed] [Google Scholar]
- 31.Livak and Schmittgen: Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCt method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 32.Wang W, Lv L, Pan K, Zhang Y, Zhao JJ, Chen JG, Chen YB, Li YQ, Wang QJ, He J, et al. Reduced expression of transcription factor AP-2α is associated with gastric adenocarcinoma prognosis. PLoS One. 2011;6:e24897. doi: 10.1371/journal.pone.0024897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li YF, Wang DD, Zhao BW, Wang W, Yuan SQ, Huang CY, Chen YM, Zheng Y, Keshari RP, et al. Poor prognosis of gastric adenocarcinoma with decreased expression of AHRR. PLoS One. 2012;7:e43555. doi: 10.1371/journal.pone.0043555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang CY, Chen YM, Zhao JJ, Chen YB, Jiang SS, Yan SM, Zhao BW, Pan K, Wang DD, Lv L, et al. Decreased expression of transcription elongation factor A-like 7 is associated with gastric adenocarcinoma prognosis. PLoS One. 2013;8:e54671. doi: 10.1371/journal.pone.0054671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.El-Badry OM, Romanus JA, Helman LJ, Cooper MJ, Rechler MM, Israel MA. Autonomous growth of a human neuroblastoma cell line is mediated by insulin-like growth factor II. J Clin Invest. 1989;84:829–839. doi: 10.1172/JCI114243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stewart CE, Rotwein P. Growth, differentiation, and survival: Multiple physiological functions for insulin-like growth factors. Physiol Rev. 1996;76:1005–1026. doi: 10.1152/physrev.1996.76.4.1005. [DOI] [PubMed] [Google Scholar]
- 37.Toretsky JA, Helman LJ. Involvement of IGF-II in human cancer. J Endocrinol. 1996;149:367–372. doi: 10.1677/joe.0.1490367. [DOI] [PubMed] [Google Scholar]
- 38.LeRoith D, Baserga R, Helman L, Roberts CT., Jr Insulin-like growth factors and cancer. Ann Intern Med. 1995;122:54–59. doi: 10.7326/0003-4819-122-1-199501010-00009. [DOI] [PubMed] [Google Scholar]
- 39.Lu L, Katsaros D, Wiley A, de la Longrais IA Rigault, Risch HA, Puopolo M, Yu H. The relationship of insulin-like growth factor-II, insulin-like growth factor binding protein-3, and estrogen receptor-alpha expression to disease progression in epithelial ovarian cancer. Clin Cancer Res. 2006;12:1208–1214. doi: 10.1158/1078-0432.CCR-05-1801. [DOI] [PubMed] [Google Scholar]
- 40.Denley A, Wallace JC, Cosgrove LJ, Forbes BE. The insulin receptor isoform exon 11-(IR-A) in cancer and other diseases: A review. Horm Metab Res. 2003;35:778–785. doi: 10.1055/s-2004-814157. [DOI] [PubMed] [Google Scholar]
- 41.Frasca F, Pandini G, Scalia P, Sciacca L, Mineo R, Costantino A, Goldfine ID, Belfiore A, Vigneri R. Insulin receptor isoform A, a newly recognized, high-affinity insulin-like growth factor II receptor in fetal and cancer cells. Mol Cell Biol. 1999;19:3278–3288. doi: 10.1128/MCB.19.5.3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kalli KR, Falowo OI, Bale LK, Zschunke MA, Roche PC, Conover CA. Functional insulin receptors on human epithelial ovarian carcinoma cells: Implications for IGF-II mitogenic signaling. Endocrinology. 2002;143:3259–3267. doi: 10.1210/en.2001-211408. [DOI] [PubMed] [Google Scholar]
- 43.Sciacca L, Mineo R, Pandini G, Murabito A, Vigneri R, Belfiore A. In IGF-I receptor-deficient leiomyosarcoma cells autocrine IGF-II induces cell invasion and protection from apoptosis via the insulin receptor isoform A. Oncogene. 2002;21:8240–8250. doi: 10.1038/sj.onc.1206058. [DOI] [PubMed] [Google Scholar]
- 44.Vella V, Pandini G, Sciacca L, Mineo R, Vigneri R, Pezzino V, Belfiore A. A novel autocrine loop involving IGF-II and the insulin receptor isoform-A stimulates growth of thyroid cancer. J Clin Endocrinol Metab. 2002;87:245–254. doi: 10.1210/jcem.87.1.8142. [DOI] [PubMed] [Google Scholar]
- 45.Bach LA. Insulin-like growth factor binding protein-6: The ‘forgotten’ binding protein? Horm Metab Res. 1999;31:226–234. doi: 10.1055/s-2007-978723. [DOI] [PubMed] [Google Scholar]
- 46.Kuo YS, Tang YB, Lu TY, Wu HC, Lin CT. IGFBP-6 plays a role as an oncosuppressor gene in NPC pathogenesis through regulating EGR-1 expression. J Pathol. 2010;222:299–309. doi: 10.1002/path.2735. [DOI] [PubMed] [Google Scholar]
- 47.Leng SL, Leeding KS, Whitehead RH, Bach LA. Insulin-like growth factor (IGF)-binding protein-6 inhibits IGF-II-induced but not basal proliferation and adhesion of LIM 1215 colon cancer cells. Mol Cell Endocrinol. 2001;174:121–127. doi: 10.1016/S0303-7207(00)00444-5. [DOI] [PubMed] [Google Scholar]
- 48.Saeki N, Ono H, Sakamoto H, Yoshida T. Genetic factors related to gastric cancer susceptibility identified using a genome-wide association study. Cancer Sci. 2013;104:1–8. doi: 10.1111/cas.12042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nagini S. Carcinoma of the stomach: A review of epidemiology, pathogenesis, molecular genetics and chemoprevention. World J Gastrointest Oncol. 2012;4:156–169. doi: 10.4251/wjgo.v4.i7.156. [DOI] [PMC free article] [PubMed] [Google Scholar]