Abstract
Recent studies have demonstrated that deregulated microRNA (miRNA/miR) expression has a profound impact on biological and pathological processes; abnormal miR-1469 expression was detected in several human malignancies. In the present study, the clinicopathological and prognostic significance of miR-1469 was assessed in 129 patients with esophageal squamous cell cancer (ESCC) who successfully underwent esophagectomy and esophagogastrostomy. Low miR-1469 expression was identified to be significantly associated with tumor invasion depth (P=0.026), lymph node metastasis status (P<0.001) and pathological tumor stage (P<0.001). Survival analysis demonstrated that patients with low miR-1469 expression had significantly poorer disease-free survival (DFS) (18.2 vs. 43.2%; P=0.004) and overall survival (29.1 vs. 47.3%; P=0.029) 5 years following surgery compared with patients with high miR-1469 expression. Univariate survival analysis demonstrated that low miR-1469 expression significantly predicted unfavorable 5-year DFS among patients with N1-3 disease (7.1 vs. 31.8%; P=0.043). The results from the present study indicate that miR-1469 expression could be used in the clinic to predict ESCC progression and prognosis. This will aid in the identification of high-risk patients with ESCC that require more aggressive therapeutic interventions.
Keywords: miR-1469, progression, relapse, prognosis, esophageal squamous cell cancer
Introduction
Esophageal cancer occurs worldwide, and Eastern Asia has one of the highest incidence rates (1). Esophageal squamous cell cancer (ESCC), the predominant histological type, is the most common subtype of esophageal cancer in Eastern Asia, including China (2). Owing to the aggressive invasion, early metastasis, therapeutic resistance and other malignant biological behaviors of ESCC, the prognosis is poor, despite numerous efforts to develop effective therapeutic interventions, such as early detection, advanced surgical techniques and chemoradiotherapy (3). Therefore, it is important to identify novel molecular biomarkers to allow for the identification of high-risk patients that require prompt and tailored treatment (4). However, efforts to identify specific molecules associated with ESCC progression and prognosis have thus far been unsuccessful (5), to the best of our knowledge.
MicroRNAs (miRNAs/miRs) are small, endogenous non-coding RNAs that are highly conserved across a variety of eukaryotic species (6,7). Since the first report to highlight the roles of miRNAs in chronic lymphocytic leukemia (8), several studies have demonstrated that the deregulation of miRNA expression has a profound impact on human biological and pathological processes through a number of molecular mechanisms (9–11). Abnormal miR-1469 expression has been reported in several human malignancies (12–15). However, the role miR-1469 serves in ESCC has not yet been reported, to the best of our knowledge. In the present study, the expression of miR-1469 in ESCC tissues, and the value of miR-1469 in predicting disease progression, relapse and prognosis, was investigated.
Materials and methods
Patients and follow-up
The present study recruited patients who underwent the surgical resection of a primary ESCC between September 2009 and June 2010 at Shandong Provincial Hospital Affiliated to Shandong University and Jinan Central Hospital Affiliated to Shandong University (Jinan, China). The 129 patients included in the present study had not received preoperative chemotherapy and/or radiotherapy, had undergone R0 resection and esophagogastrostomy without perioperative death, and also had complete clinicopathological and follow-up data. Clinicopathological data for the recruited patients is illustrated in Table I.
Table I.
Association between of miR-1469 expression and the clinicopathological characteristics of patients with esophageal squamous cell cancer.
| miR-1469 expression (no. of patients) | ||||
|---|---|---|---|---|
| Clinicopathological characteristic | No. of patients | Low | High | P-valuea |
| Age (years) | 0.829 | |||
| <60 | 53 | 22 | 31 | |
| ≥60 | 76 | 33 | 43 | |
| Gender | 0.124 | |||
| Male | 102 | 47 | 55 | |
| Female | 27 | 8 | 19 | |
| Tumor length (cm) | 0.224 | |||
| <5 | 85 | 33 | 52 | |
| ≥5 | 44 | 22 | 22 | |
| Tumor differentiation | 0.082 | |||
| High | 31 | 8 | 23 | |
| Moderate | 63 | 29 | 34 | |
| Low | 35 | 18 | 17 | |
| Tumor invasion depth | 0.026 | |||
| T1 | 23 | 6 | 17 | |
| T2 | 40 | 13 | 27 | |
| T3 | 51 | 26 | 25 | |
| T4 | 15 | 10 | 5 | |
| Lymph node metastasis stage | <0.001 | |||
| N0 | 65 | 13 | 52 | |
| N1 | 29 | 19 | 10 | |
| N2 | 23 | 14 | 9 | |
| N3 | 12 | 9 | 3 | |
| Pathological tumor stage | <0.001 | |||
| I | 18 | 3 | 15 | |
| II | 58 | 17 | 41 | |
| III | 53 | 35 | 18 | |
χ2 test.
Tumor samples were collected immediately following surgical resection, snap-frozen in liquid nitrogen and then stored at −80°C until required for the extraction of total RNA. The histopathologic diagnosis of ESCC was verified based on sections stained with hematoxylin and eosin, according to the classification system of the World Health Organization (16). Tumor-node-metastasis (TNM) staging was determined based on the 7th classification guidelines of the American Joint Committee on Cancer (17). Written informed consent was obtained from each patient, and the study design was approved by the Institutional Review Boards of Shandong Provincial Hospital Affiliated to Shandong University and Jinan Central Hospital Affiliated to Shandong University, (Shandong, China).
All patients were followed up at 3–6 month intervals, using a number of medical procedures, including physical examination, upper gastrointestinal barium meal examination, ultrasound examination, computed tomography and gastroscopy, until patient mortality (all causes) or the end of the study period (June 2015). The median follow-up period was 35 months, with a range of 6–68 months.
Real-time quantitative reverse transcription-polymerase chain reaction (RT-qPCR)
Total RNA was extracted from each frozen tissue sample using TRIzol (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) according to the manufacturer's instructions. The purity and concentration of all RNA samples was measured using a spectrophotometer (NanoDrop; Thermo Fisher Scientific, Wilmington, DE, USA).
cDNA was reverse transcribed from 100 ng RNA using SYBR® Premix Ex Taq™ (Takara Biotechnology Co., Ltd., Dailan, China) in a final volume of 20 µl, according to the manufacturer's instructions. The RT conditions used are as follows: 37°C for 60 min, 85°C for 5 sec and 4°C for 60 min. Subsequently, quantification of miR-1469 expression was performed by RT-qPCR using SYBR® Premix Ex Taq™ II on a MX3000P instrument (Agilent Technologies, Inc., Santa Clara, CA, USA). The qPCR consisted of 2 µl (10 ng) cDNA, 12.5 µl SYBR Premix Ex Taq II, 0.5 µl Dye II, 2 µl 5 µM forward primer, 1 µl 10 µM Uni-miR RT-qPCR primer and 7 µl of ddH2O. The qPCR conditions used were as follows: 95°C for 30 sec, 45 cycles of 95°C for 5 sec and 57°C for 34 sec. The sequences of the primers are as follows: miR-1469 forward, 5′-GGGCTGAGCTGAAGCAGTG-3′, and reverse, 5′-CAGTGCGTGTCGTGGAGT-3′, U6 forward, 5′-GCTTCGGCAGCACATATACTAAAAT-3′, and reverse primer, 5′-CGCTTCACGAATTTGCGTGTCAT-3′. The small nuclear RNA U6, which is highly conserved and universally expressed, was used as the endogenous reference. Relative changes of miR-1469 expression were calculated using the comparative threshold cycle (2−ΔΔCq) method (18) and normalized to U6 expression. All RT-qPCR reactions were performed in triplicate. High or low miR-1469 expression was defined based on the median expression cut-off value, which was calculated separately for each cohort (19).
Statistical analysis
All statistical analyses were performed using SPSS software (version 18.0; SPSS Inc., Chicago, IL, USA). A χ2 test was used to determine the association between miR-1469 expression levels and clinicopathological characteristics. The Kaplan-Meier method was used to calculate survival curves and the log-rank test for the univariate analysis was performed to determine the statistical significance of differences between the survival curves of patient subgroups. Disease-free survival (DFS) was calculated from the date of surgery to the date of tumor recurrence or the last follow-up. Overall survival (OS) was calculated from the date of surgery to the date of death or the last follow-up. Multivariate analysis was performed using the Cox proportional hazards regression model to identify independent prognostic factors for predicting survival. P<0.05 was considered to indicate a statistically significant difference.
Results
Association between miR-1469 expression and clinicopathological characteristics
The expression levels of miR-1469 in each sample were determined by RT-qPCR, and compared with clinicopathological characteristics using a χ2 test (Table I). Low miR-1469 expression was significantly associated with tumor invasion depth (26.1% in T1, 32.5% in T2, 51.0% in T3 and 66.7% in T4 cases; P=0.026), lymph node metastases (20.0% in N0, 65.5% in N1, 60.9% in N2, and 75.0% in N3 cases; P<0.001) and pathological stage (16.7% in I, 29.3% in II and 66.0% in III cases; P<0.001). However, no association was identified between miR-1469 expression and patient age or gender. Additionally, no association was identified between miR-1469 expression and tumor length or differentiation.
Association between miR-1469 expression and tumor relapse
The location and time of tumor relapse was recorded during the follow-up period. A total of 87 patients suffered tumor relapse and the 5-year DFS rate was 32.6%. Of the 87 patients, 45 (51.7%) had low tumor miR-1469 expression and 42 cases (48.3%) had high tumor miR-1469 expression (P=0.003; data not shown). A univariate analysis demonstrated that the 5-year DFS was significantly decreased for patients with low tumor miR-1469 expression than for those with high tumor miR-1469 expression (18.2 vs. 43.2%; P=0.004; Fig. 1A). In addition, multivariate analysis demonstrated that only lymph node metastasis status retained its significance as an independent predictor for unfavorable DFS [P=0.031; 95% confidence interval (CI), 1.035–2.089; Table II].
Figure 1.
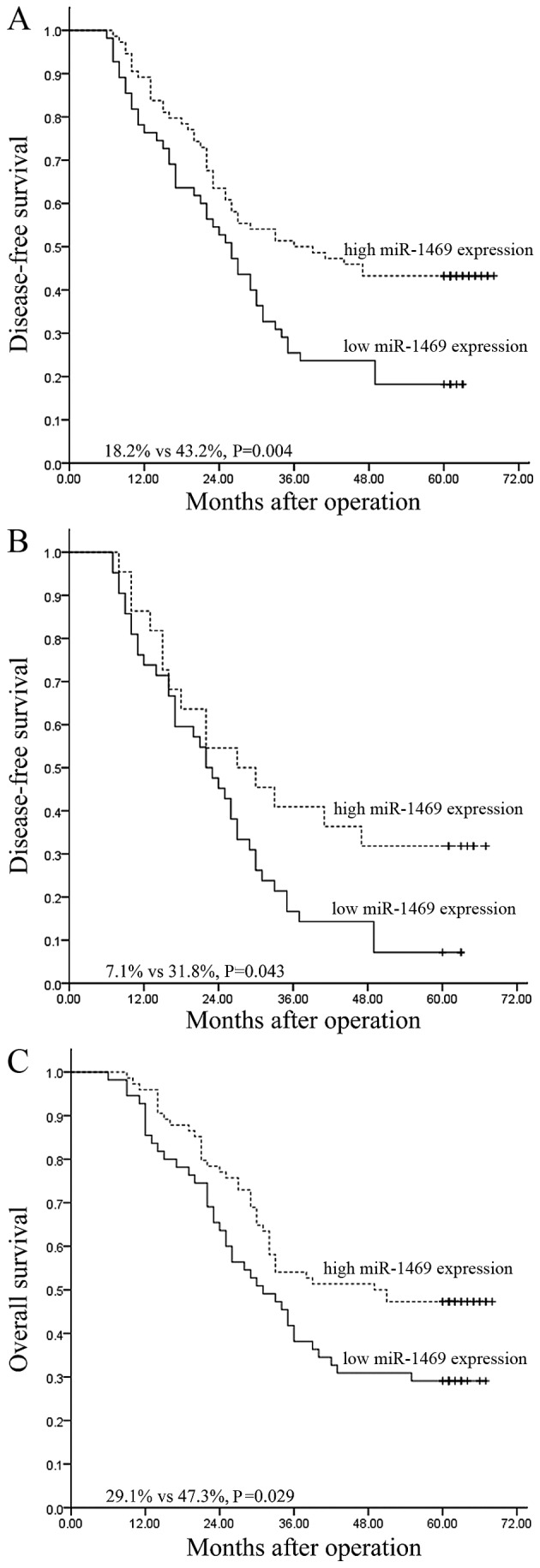
Kaplan-Meier survival curves stratified according to miR-1469 expression levels. The survival curves represent (A) the DFS of all patients, (B) the DFS of patients with N1-3 disease and (C) the overall survival of all patients. DFS, disease-free survival.
Table II.
Results of univariate and multivariate analyses for the association between clinicopathological characteristics and survival in patients with esophageal squamous cell cancer.
| Disease-free survival | Overall survival | |||||
|---|---|---|---|---|---|---|
| Multivariate analysis | Multivariate analysis | |||||
| Clinicopathological characteristic | Univariate analysis P-value | P-value | 95.0% CI | Univariate analysis P-value | P-value | 95.0% CI |
| Age | 0.598 | 0.352 | 0.516–1.516 | 0.809 | 0.539 | 0.534–1.534 |
| Gender | 0.802 | 0.761 | 0.636–1.636 | 0.847 | 0.783 | 0.518–1.518 |
| Tumor length | 0.665 | 0.947 | 0.614–1.614 | 0.617 | 0.935 | 0.621–1.621 |
| Tumor differentiation | 0.095 | 0.512 | 0.803–1.803 | 0.280 | 0.812 | 0.741–1.741 |
| Tumor invasion depth | 0.033 | 0.512 | 0.785–1.785 | 0.009 | 0.361 | 0.815–1.815 |
| Lymph node metastasis stage | <0.001 | 0.031 | 1.035–2.035 | <0.001 | 0.020 | 1.070–2.070 |
| Pathological tumor stage | <0.001 | 0.742 | 0.552–2.552 | <0.001 | 0.582 | 0.582–2.582 |
| miR-1469 expression | 0.004 | 0.322 | 0.488–1.488 | 0.029 | 0.977 | 0.595–1.595 |
CI, confidence interval.
The predictive significance of miR-1469 expression in selective patient subgroups was stratified according to lymph node metastasis status and pathological stage, respectively. Univariate analysis demonstrated that the 5-year DFS of patients with low tumor miR-1469 expression was significantly lower compared with that of the remaining cases with high miR-1469 expression among N1-3 patients (7.1 vs. 31.8%; P=0.043; Fig. 1B). However, this was not the case for patients with N0 (53.8 vs. 48.1%; P=0.751), pathological stage I–II (40.0 vs. 48.2%; P=0.582) or pathological stage III (5.7 vs. 27.8%; P=0.128) tumors with low miR-1469 tumor expression compared with the corresponding groups with high miR-1469 tumor expression (data not shown).
Association between miR-1469 expression and prognosis
Of the 129 patients, 78 succumbed to mortality within the follow-up period and the 5-year OS rate was 39.5% (data not shown). Univariate analysis revealed that low miR-1469 expression was associated with an unfavorable 5-year OS compared with high miR-1469 expression (29.1 vs. 47.3%; P=0.029; Fig. 1C). Multivariate analysis revealed that only lymph node metastasis retained its significance as an independent predictor for unfavorable OS (P=0.020; 95% CI, 1.070–2.202; Table II).
A further stratified survival analysis demonstrated that miR-1469 expression was not associated with an unfavorable 5-year OS for subgroups of patients grouped according to lymph node metastasis status (N0, 69.2 vs. 51.9%; P=0.353; N1-3, 16.7 vs. 36.4%; P=0.145) and pathological stage (I–II, 65.0 vs. 53.6%, P=0.566; III, 8.6 vs. 27.8%; P=0.223) (data not shown).
Discussion
Esophageal cancer is one of the most aggressive and deadly malignancies of the digestive tract, with patients typically presenting with bulky tumors and metastasis at the time of clinical diagnosis. The prognosis of patients with esophageal cancer remains poor and only a subset of patients respond well to treatment (4,20). The current TNM classification cannot accurately predict clinical outcome for patients with ESCC (21), even for patients with identical clinicopathological risk factors. Clearly distinguishing between subgroups of patients who have a high or low risk of relapse, treatment resistance and metastasis remains a challenge (22), therefore there is an urgent need to identify biomarkers that can be used to identify patients that require more aggressive postoperative treatment.
miRNAs are deregulated in numerous human cancers (23,24). Previous studies have demonstrated that miRNAs are reliable and cost-effective biomarkers that can be utilized in the diagnosis and assessment of the progression, therapeutic response and prognosis of a wide spectrum of malignant tumors, including esophageal cancer (7,25–27). A better understanding of the miRNAs associated with particular cancers would provide valuable insights for treatment. Previous studies have demonstrated that miR-1469 may function as an oncogene (12,13) or a tumor suppressor gene (14,15) in malignant tumors originating from different tissues. However, to the best of our knowledge, the effects of miR-1469 expression on the progression and clinical outcome of human ESCC has not yet been reported.
In the present study, the expression of miR-1469 in tumor samples from 129 ESCC patients was analyzed by RT-qPCR and compared with clinicopathological characteristic and survival data. Low miR-1469 expression was significantly associated with invasion depth, lymph node metastasis status and pathological stage, suggesting that miR-1469 acts as a tumor suppressor. Low expression of miR-1469 was significantly associated with a poor prognosis; a univariate analysis revealed a correlation between low miR-1469 expression and a poor 5-year DFS and OS. A multivariate analysis demonstrated that only lymph node metastasis status could predict tumor relapse and prognosis independently of other clinicopathological parameters.
To explore whether miR-1469 expression levels could be employed as a predictor to improve the current risk stratification for ESCC based on the conventional staging system, a further stratified survival analysis split by lymph node metastasis status and pathological stage was performed. The results demonstrated that low miR-1469 expression only significantly predicted an unfavorable 5-year DFS among the subgroup of patients with N1-3 disease. Although statistically significant differences were not detected, low miR-1469 expression was associated with a poorer DFS and OS in the subgroup of patients with pathologic stage III disease. Therefore, the data from the present study indicate that miR-1469 expression can predict the clinical outcome of ESCC, particularly in patients with advanced disease. Measuring miR-1469 expression levels in patients with ESCC may allow clinicians to more accurately identify high-risk patients who would benefit from more aggressive therapeutic interventions.
Previous studies have identified an association between dysregulated miRNA expression and cancer diagnosis (28,29). In addition, it has been demonstrated that miRNAs modulate numerous signaling pathways and regulate the expression of thousands of genes (7,30). The current gold standard of cancer diagnosis is a pathological diagnosis, due to the heterogeneous nature of tumors. Future studies will need to be performed to ascertain whether miR-1469 can be reliably used as biomarker for the diagnosis of ESCC.
In conclusion, to the best of our knowledge, the data in the present study provides the first evidence for the role of miR-1469 in predicting disease progression and post-surgical clinical outcomes in patients with ESCC. These data are consistent with previous reports evaluating the tumor suppressor roles of miR-1469 in human cancer (14,15). However, multicenter and large-scale studies are required to verify this data prior to the clinical application of miR-1469, in addition to in vitro and in vivo functional studies to identify the target genes and signaling pathways of miR-1469. The results of these studies may allow for the identification and selection of high-risk patients that would benefit from specific treatments following surgery, therefore improving clinical outcomes.
Glossary
Abbreviations
- ESCC
esophageal squamous cell cancer
- TNM
tumor-node-metastasis
- DFS
disease-free survival
- OS
overall survival
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Chen W, He Y, Zheng R, Zhang S, Zeng H, Zou X, He J. Esophageal cancer incidence and mortality in China, 2009. J Thorac Dis. 2013;5:19–26. doi: 10.3978/j.issn.2072-1439.2013.01.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pennathur A, Gibson MK, Jobe BA, Luketich JD. Oesophageal carcinoma. Lancet. 2013;381:400–412. doi: 10.1016/S0140-6736(12)60643-6. [DOI] [PubMed] [Google Scholar]
- 4.Kim T, Grobmyer SR, Smith R, Ben-David K, Ang D, Vogel SB, Hochwald SN. Esophageal cancer-the five year survivors. J Surg Oncol. 2011;103:179–183. doi: 10.1002/jso.21784. [DOI] [PubMed] [Google Scholar]
- 5.Hong L, Han Y, Zhang H, Fan D. Prognostic markers in esophageal cancer: From basic research to clinical use. Expert Rev Gastroenterol Hepatol. 2015;9:887–889. doi: 10.1586/17474124.2015.1041507. [DOI] [PubMed] [Google Scholar]
- 6.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-Y. [DOI] [PubMed] [Google Scholar]
- 7.Naidu S, Magee P, Garofalo M. MiRNA-based therapeutic intervention of cancer. J Hematol Oncol. 2015;8:68. doi: 10.1186/s13045-015-0162-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, et al. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci USA. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10:704–714. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Di Leva G, Garofalo M, Croce CM. MicroRNAs in cancer. Annu Rev Pathol. 2014;9:287–314. doi: 10.1146/annurev-pathol-012513-104715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ohtsuka M, Ling H, Doki Y, Mori M, Calin GA. MicroRNA processing and human cancer. J Clin Med. 2015;4:1651–1667. doi: 10.3390/jcm4081651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fix LN, Shah M, Efferth T, Farwell MA, Zhang B. MicroRNA expression profile of MCF-7 human breast cancer cells and the effect of green tea polyphenon-60. Cancer Genomics Proteomics. 2010;7:261–277. [PubMed] [Google Scholar]
- 13.White NM, Khella HW, Grigull J, Adzovic S, Youssef YM, Honey RJ, Stewart R, Pace KT, Bjarnason GA, Jewett MA, et al. miRNA profiling in metastatic renal cell carcinoma reveals a tumour-suppressor effect for miR-215. Br J Cancer. 2011;105:1741–1749. doi: 10.1038/bjc.2011.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang B, Jing C, Wang J, Guo X, Chen Y, Xu R, Peng L, Liu J, Li L. Identification of microRNAs associated with lymphangiogenesis in human gastric cancer. Clin Transl Oncol. 2014;16:374–379. doi: 10.1007/s12094-013-1081-6. [DOI] [PubMed] [Google Scholar]
- 15.Xu C, Zhang L, Li H, Liu Z, Duan L, Lu C. MiRNA-1469 promotes lung cancer cells apoptosis through targeting STAT5a. Am J Cancer Res. 2015;5:1180–1189. [PMC free article] [PubMed] [Google Scholar]
- 16.Hamilton SR, Aaltonen LA. Pathology and genetics of tumours of the digestive system, World Health Organization classification of tumours. IARC Press; Lyon: 2000. pp. 10–25. [Google Scholar]
- 17.Edge SF, Byrd DR, Compton CC, Fritz AG, Greene FL, Trotti A. AJCC Cancer Staging Manual. 7th edition. Springer; New York, NY: 2010. Esophagus and esophagogastric junction; pp. 103–111. [Google Scholar]
- 18.Zhou H, Tang K, Xiao H, Zeng J, Guan W, Guo X, Xu H, Ye Z. A panel of eight-miRNA signature as a potential biomarker for predicting survival in bladder cancer. J Exp Clin Cancer Res. 2015;34:53. doi: 10.1186/s13046-015-0167-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mathé EA, Nguyen GH, Bowman ED, Zhao Y, Budhu A, Schetter AJ, Braun R, Reimers M, Kumamoto K, Hughes D, et al. MicroRNA expression in squamous cell carcinoma and adenocarcinoma of the esophagus: Associations with survival. Clin Cancer Res. 2009;15:6192–6200. doi: 10.1158/1078-0432.CCR-09-1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Enzinger PC, Mayer RJ. Esophageal Cancer. N Engl J Med. 2003;349:2241–2252. doi: 10.1056/NEJMra035010. [DOI] [PubMed] [Google Scholar]
- 21.Li SH, Tian H, Yue WM, Li L, Gao C, Li WJ, Hu WS, Hao B. Metastasis-associated protein 1 nuclear expression is closely associated with tumor progression and angiogenesis in patients with esophageal squamous cell cancer. World J Surg. 2012;36:623–631. doi: 10.1007/s00268-011-1421-z. [DOI] [PubMed] [Google Scholar]
- 22.Guo Y, Chen Z, Zhang L, Zhou F, Shi S, Feng X, Li B, Meng X, Ma X, Luo M, et al. Distinctive microRNA profiles relating to patient survival in esophageal squamous cell carcinoma. Cancer Res. 2008;68:26–33. doi: 10.1158/0008-5472.CAN-06-4418. [DOI] [PubMed] [Google Scholar]
- 23.Pipan V, Zorc M, Kunej T. MicroRNA polymorphisms in cancer: A literature analysis. Cancers (Basel) 2015;7:1806–1814. doi: 10.3390/cancers7030863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin S, Gregory RI. MicroRNA biogenesis pathways in cancer. Nat Rev Cancer. 2015;15:321–333. doi: 10.1038/nrc3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 26.Garzon R, Calin GA, Croce CM. MicroRNAs in Cancer. Annu Rev Med. 2009;60:167–179. doi: 10.1146/annurev.med.59.053006.104707. [DOI] [PubMed] [Google Scholar]
- 27.Raza U, Zhang JD, Sahin O. MicroRNAs: Master regulators of drug resistance, stemness, and metastasis. J Mol Med (Berl) 2014;92:321–336. doi: 10.1007/s00109-014-1129-2. [DOI] [PubMed] [Google Scholar]
- 28.Tricoli JV, Jacobson JW. MicroRNA: Potential for cancer detection, diagnosis, and prognosis. Cancer Res. 2007;67:4553–4555. doi: 10.1158/0008-5472.CAN-07-0563. [DOI] [PubMed] [Google Scholar]
- 29.Kosaka N, Iguchi H, Ochiya T. Circulating microRNA in body fluid: A new potential biomarker for cancer diagnosis and prognosis. Cancer Sci. 2010;101:2087–2092. doi: 10.1111/j.1349-7006.2010.01650.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ye Y, Wang KK, Gu J, Yang H, Lin J, Ajani JA, Wu X. Genetic variations in microRNA-related genes are novel susceptibility loci for esophageal cancer risk. Cancer Prev Res (Phila) 2008;1:460–469. doi: 10.1158/1940-6207.CAPR-08-0135. [DOI] [PMC free article] [PubMed] [Google Scholar]


