Abstract
In clinical practice ionizing radiation (IR) is primarily applied to cancer treatment in the form of fractionated dose (FD) irradiation. Despite this fact, a substantially higher amount of current knowledge in the field of radiobiology comes from in vitro studies based on the cellular response to single dose (SD) irradiation. In addition, intrinsic and acquired resistance to IR remains an issue in clinical practice, leading to radiotherapy treatment failure. Numerous previous studies suggest that an improved understanding of the molecular processes involved in the radiation-induced DNA damage response to FD irradiation could improve the effectiveness of radiotherapy. Therefore, the present study examined the differential expression of genes and microRNA (miRNA) in murine Lewis lung cancer (LLC)1 cells exposed to SD or FD irradiation. The results of the present study indicated that the gene and miRNA expression profiles of LLC1 cells exposed to irradiation were dose delivery type-dependent. Data analysis also revealed that mRNAs may be regulated by miRNAs in a radiation-dependent manner, suggesting that these mRNAs and miRNAs are the potential targets in the cellular response to SD or FD irradiation. However, LLC1 tumors after FD irradiation exhibited no significant changes in the expression of selected genes and miRNAs observed in the irradiated cells in vitro, suggesting that experimental in vitro conditions, particularly the tumor microenvironment, should be considered in detail to promote the development of efficient radiotherapy approaches. Nevertheless, the present study highlights the primary signaling pathways involved in the response of murine cancer cells to irradiation. Data presented in the present study can be applied to improve the outcome and development of radiotherapy in preclinical animal model settings.
Keywords: fractionated irradiation, radiation response, genome-wide expression analysis, gene and microRNA expression, Lewis lung carcinoma, syngeneic tumor model
Introduction
Radiotherapy (RT) remains one of the most common types of therapy used alone or in combination with other therapeutics to treat cancer. In clinical practice, ~50% of all cancer patients receive radiotherapy at some point during treatment. Currently, to allow repair and recovery of radiation-induced damage to normal tissue cells, radiotherapy is administered in fractions of ~2 Gy every 24 h, 5 days/week for ≤7 weeks. However, the majority of the knowledge in the field of radiobiology comes from single dose (SD) irradiation research (1). Therefore, a detailed investigation of the molecular processes mediating the cellular response to fractionated dose (FD) irradiation is required to improve the efficiency of radiotherapy.
There is an emerging body of knowledge on the comprehensive molecular mechanisms underlying the cellular response to FD irradiation and the mechanisms associated with resistance to RT. Previous studies have shown that treatment with multiple fractions of irradiation produces a different gene expression signature in several cancer cell lines compared with SD irradiation (2,3). For instance, exposure to 10 Gy delivered as fractionated irradiation results in increased changes in differential gene expression in prostate cancer PC3 and DU145 cells (4). In addition, as demonstrated by gene expression profiles, exposure to FD irradiation can induce a significantly different microRNA (miRNA/miR) expression profile compared with SD (5,6). miRNAs perform an important role in the regulation of the expression of genes involved in the cellular response to radiation-induced DNA damage (7). Previous studies have reported that the modulation of miRNA expression levels in cancer cells can alter their sensitivity to irradiation (8–10). Therefore, the integration of gene and miRNA signatures of radiosensitivity could lead to a reliable strategy for predicting radiation-induced cellular responses. Furthermore, the silencing of radiation-induced miRNAs could be implemented in direct antitumor therapies to improve the response of tumor cells to RT.
Several previous studies using a gene expression microarray approach indicated expression of a different set of genes in several human cancer cell xenografts following exposure to irradiation compared with cells irradiated in vitro, suggesting that the tumor microenvironment may affect the outcome of irradiation (2,11). The LLC1 cell line was established from the lung of a C57BL mouse bearing a primary Lewis lung carcinoma tumor. This cell line is highly tumorigenic and immunologically compatible with the murine immune system, unlike widely used human cancer cell xenograft models. Consequently, the LLC1 cell line is primarily used in syngeneic animal models to evaluate the efficacy of anticancer treatment in vivo (12). The present study analyzed global gene and miRNA expression changes in LLC1 cells exposed to SD of 2 or 10 Gy irradiation and FD of 5×2 Gy irradiation.
Materials and methods
Cell culture and maintenance
The LLC1 mouse Lewis lung carcinoma cell line was obtained from the American Type Culture Collection (Manassas, VA, USA). Cells were cultured at 37°C in a humidified atmosphere containing 5% CO2 with Dulbecco's modified Eagle's medium (DMEM; Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum (Thermo Fisher Scientific, Inc.), 2 mM L-glutamine (Thermo Fisher Scientific, Inc.), 100 U/ml penicillin (Carl Roth GmbH Co., KG, Karlsruhe, Germany) and 0.1 mg/ml streptomycin (Carl Roth GmbH Co., KG).
Animals and tumor model
C57BL/6 female mice (Vilnius University, Vilnius, Lithuania) were maintained at a constant temperature (22±1°C), relative humidity (55±10%) and photoperiod (12 h light/dark cycle) in the Open Access Centre at the National Cancer Institute of Lithuania (Vilnius, Lithuania). All animal procedures were performed in accordance with the guidelines established by State Food and Veterinary Service Animal Care and Use Committee (Vilnius, Lithuania) that approved the current study (approval no. 0190). Two female mice at 10–12 weeks of age and 19–22 g body weight were injected subcutaneously with Lewis lung carcinoma LLC1 cells (1×106 cells suspended in DMEM medium) into their right groins. Animals were sedated with ketamine hydrochloride alone (0.1 mg/g body mass; ROTEXMEDICA GmbH, Trittau, Germany) by injection of 0.1–0.2 ml/animal solution in sterile normal saline (B. Braun Melsungen AG, Melsungen, Germany) into the caudal thigh muscles and sacrificed by cervical dislocation, and their tumors were excised, homogenized and resuspended in normal saline 10 days following the implantation. Mice in each experimental group containing 6 female mice were injected with 0.2 ml of the obtained suspension into their right groin. Tumors were allowed to reach a volume of 400–600 mm3 prior to irradiation. Tumor volumes were measured with vernier calipers and calculated according to the following formula: Tumor volume=(length × width × height of tumor) × π/6.
Cell and tumor irradiation
LLC1 cells and tumors were exposed to a SD of 2–10 Gy or a FD course of 2 Gy daily for ≤5 days using a Varian 6MV Clinac 600 C/D linear accelerator X-ray system (Varian Medical Systems, Inc., Palo Alto, CA, USA) at room temperature. The dose rate was ~3 Gy/min. Prior to irradiation, animals were sedated with ketamine hydrochloride alone (0.1 mg/g body mass) by injection of 0.1–0.2 ml/animal solution in sterile normal saline into the caudal thigh muscles and placed in a customized harness that allowed the groin to be exposed to irradiation, whereas the rest of the body was shielded by lead. In all of the experiments separate controls of non-irradiated mice tumors were used for SD or FD regimens.
Clonogenic survival assay
LLC1 cells were plated in 6-well plates 24 h prior to irradiation (500–10,000 cells/well) and treated with SD of up to 10 Gy or FD of 2 Gy of ionizing radiation (IR) daily for ≤5 days. In total, 8 days subsequent to irradiation LLC1 cell colonies (>50 cells/colony) were stained with crystal violet and counted manually. Clonogenic survival was evaluated as described previously (13). The mean cell survival fraction from 3 independent experiments was used to represent survival at each irradiation dose.
Total RNA and miRNA extraction
Total RNA enriched with small noncoding RNAs was isolated using the mirVana RNA isolation kit (Thermo Fisher Scientific, Inc.) according to manufacturer's protocol. For total RNA extraction, LLC1 cells were plated into 25 cm2 cell culture flasks for RNA isolation (0.7×106 or 0.1×106 cells/flask for the SD and FD irradiation regimens, respectively). Subsequently, ~1×106 LLC1 cells were harvested 4 h following SD (2 or 10 Gy) or FD (5×2 Gy) irradiation and were used for total RNA extraction. Following the same experimental design, untreated or irradiated animals were sacrificed, tumors excised and 100 mg of mouse tumor tissue was used for total RNA extraction. The quantity and quality of RNA were measured using a NanoDrop 2000c Spectrophotometer (Thermo Fisher Scientific, Inc.) and Agilent 2100 Bioanalyzer (Agilent Technologies, Inc., Santa Clara, CA, USA).
Gene expression microarrays
RNA sample preparation, labeling and hybridization were performed using the kits described below according to the manufacturer's protocol. Briefly, 1 µg of total RNA was used for cDNA synthesis and amplification using the MessageAmp aRNA Amplification kit (Thermo Fisher Scientific, Inc.). Subsequently, 825 ng of cDNA was labeled with Cy3/Cy5 using the Arcturus® Turbo Labeling™ Cy®3/Cy®5 kit (Thermo Fisher Scientific, Inc.). The cDNA was then hybridized to the Mouse Whole Genome 4×44k Oligonucleotide Microarray (Agilent Technologies, Inc.) using a HS 400 Hybridization station (Tecan Group, Ltd., Männedorf, Switzerland). A total of 3 independent replicates for each sample were run. Microarray slides were scanned using the LS Reloaded laser scanner (Tecan Group Ltd.). Microarray image analysis and data generated were analyzed using ImaGene software (version 9.0; BioDiscovery, El Segundo, CA, USA) and GeneSpring GX software (version 11.5; Agilent Technologies, Inc.). Raw extracted gene expression data were normalized through Loess regression analysis to account for variation. Genes that exhibited a significant (P<0.05) fold-change in expression of >1.5 were defined as differentially expressed in LLC1 cells between the untreated and irradiated groups. The microarray design and data are available from the Gene Expression Omnibus (GEO) database (accession no. GSE84108; ncbi.nlm.nih.gov/geo) (14).
miRNA expression microarrays
miRNA labeling was performed using the miRNA Complete Labeling and Hyb kit (Agilent Technologies, Inc.) according to manufacturer's protocol. Briefly, 100 ng of total RNA was dephosphorylated and directly labeled with Cy3. Samples were dried out and resuspended in Hi-RPM Hybridization Buffer (Agilent Technologies, Inc.), containing a GE Blocking Agent (Agilent Technologies, Inc.) and denaturized by heating for 5 min at 100°C. In a further step, samples were hybridized to Mouse miRNA 8×15K Microarrays (Agilent Technologies, Inc.) containing probes for 627 mouse miRNAs from the Sanger database version 12 (15) for 20 h at 55°C in a rotating hybridization oven. A total of 3 independent replicates for each sample were used. Slides were then washed 3 times in PBS and scanned with the SureScan Microarray Scanner (Agilent Technologies, Inc.). Microarray images were extracted using Feature Extraction software (version 10.7.3.1; Agilent Technologies, Inc.). To normalize raw probe values, experimental samples were normalized to the mean of all samples using GeneSpring GX software (version 11.5; Agilent Technologies, Inc.). miRNAs that exhibited a significant (P<0.05) fold-change in expression of >2 were defined as differentially expressed in LLC1 cells between the untreated and irradiated groups. Microarray data are available at the GEO database (accession no. GSE84109).
Enrichment analysis
Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis of gene expression data was performed using the WEB-based GEne SeT AnaLysis Toolkit, as described previously (16). P-values were calculated using the hypergeometric test and adjusted using the Benjamini and Hochberg procedure. Functional KEGG pathway categories associated with ≥5 genes were considered as significantly enriched (P<0.05) in differentially expressed genes. In silico miRNA target analysis was performed with Diana Tools using the microT-CDS algorithm, as described previously (17,18).
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) for the evaluation of the mRNA expression
To validate differential gene expression changes, the RevertAid RT kit (Thermo Fisher Scientific, Inc.) was used for cDNA synthesis according to manufacturer's protocol. Briefly, 1 µg of total RNA was added to a 20 µl RT reaction containing 5 µM random hexamer primers, 1 µM deoxynucleotide (dNTP) mix, 20 units RNase inhibitor and 20 units reverse transcriptase. Thermocycling conditions were as follows: 25°C for 5 min; 42°C for 60 min; 70°C for 5 min.
RT-qPCR was performed using a MasterCycler RealPlex4 RT-PCR system (Eppendorf, Hamburg, Germany.) and 2X Kapa SYBR Fast qPCR Master mix (Kapa Biosystems, Inc., Wilmington, MA, USA) according to manufacturer's protocol. All reactions were performed in a 10 µl reaction volume containing 5 µl 2X Kapa SYBR Fast qPCR Master mix, 1 µl 10 ng/µl cDNA, 0.2 µl 10 µM forward and reverse primer mixture and 3.8 µl nuclease-free water. Thermocycling conditions were as follows: 95°C for 3 min; and 40 cycles of 3 sec at 95°C and 30 sec at 60°C. The relative changes in gene expression were evaluated using the 2−∆∆Cq method as described previously (19). For the normalization of the expression data, hypoxanthine phosphoribosyltransferase 1 (Hprt1) was used as a reference gene. Each experiment was repeated at least 3 times. RT controls were used for determination of genomic DNA contamination.
The sequences of the primers were as follows: Hprt1 forward (F), 5′-CCTAAGATGAGCGCAAGTTGAA-3′ and reverse (R), 5′-CCACAGGACTAGAACACCTGCTAA-3′; p21 F, 5′-CCAGGCCAAGATGGTGTCTT-3′ and R, 5′-TGAGAAAGGATCAGCCATTGC-3′; cyclin G1 (Ccng1) F, 5′-ACAACTGACTCTCAGAAACTGC-3′ and R, 5′-CATTATCATGGGCCGACTCAAT-3′; thrombospondin 2 (Thbs2) F, 5′-CTGGGCATAGGGCCAAGAG-3′ and R, 5′-GCTTGACAATCCTGTTGAGATCA-3′; BTG anti-proliferation factor 2 (Btg2) F, 5′-GGACGCACTGACCGATCATTA-3′ and R, 5′-GATACAGCGATAGCCAGAACC-3′.
RT-qPCR for the evaluation of the miRNA expression
To validate differential changes in miRNA expression, the RevertAid RT kit (Thermo Fisher Scientific, Inc.) was used for cDNA synthesis as described previously (20). Briefly, 0.2 µg of total RNA was added to a 20 µl RT reaction containing 1 µM specific RT primer, 1 µM dNTP mix, 20 units RNase inhibitor and 20 units reverse transcriptase. Thermocycling conditions were as follows: 25°C for 20 min; 37°C for 60 min; and 70°C for 10 min. The sequences of the specific RT primers were as follows: SnoRNA-135,5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCTTCAG-3′; miR-34b-3p, 5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACGATGGC-3′; miR-34c-5p, 5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACGCAATC-3′; miR-186-5p, 5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAGCCCA-3′; and miR-145a-5p, 5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAGGGAT-3′.
RT-qPCR was performed using the Eco™ RT-PCR system (Illumina, San Diego, CA, USA) and 2X Kapa SYBR Fast qPCR Master mix (Kapa Biosystems, Inc.) according to manufacturer's protocol. All reactions were performed in a 10 µl reaction volume containing 5 µl 2X Kapa SYBR Fast qPCR Master mix, 1 µl 5 ng/µl cDNA, 0.2 µl forward and reverse primer mixture (10 µM) and 3.8 µl nuclease-free water. Thermocycling conditions were as follows: 95°C for 3 min; 3 cycles of 15 sec at 95°C, 1 min at 55°C and 30 sec at 60°C; and 32 cycles of 10 sec at 95°C and 30 sec at 60°C. The relative changes in miRNA expression were evaluated using the 2−∆∆Cq method (17). For the normalization of the expression data, SnoRNA-135 was used as a reference gene. The sequences of primers used for the amplification were as follows: SnoRNA-135 forward, 5′-GTAGTGGTGAGCCTATGGTTTT-3′; miR-34b-3p forward, 5′-CGGCGAATCACTAACTCCACT-3′; miR-34c-5p forward, 5′-GGCGAGGCAGTGTAGTTAGCT-3′; miR-186-5 forward, 5′-GGCGCAAAGAATTCTCCTTT-3′; miR-145a-5p forward, 5′-CGGTCCAGTTTTCCCAGGA-3′; and a reverse primer, 5′-GTGCAGGGTCCGAGGT-3′.
Statistical analysis
Data were analyzed using GraphPad Prism software (version 6.0; GraphPad Software, La Jolla, CA, USA). A Student's t-test was performed to statistically compare differences between the untreated and irradiated groups. P<0.05 was considered to indicate a statistically significant difference. All experiments were independently repeated ≥3 times.
Results
Clonogenic cell survival
Clonogenic survival analysis revealed that LLC-1 cells were more sensitive to SD irradiation compared with FD irradiation (data not shown). The surviving fractions of LLC1 cells following 2–10 Gy SD irradiation were 62.2±4.1–1.1±0.51% compared with non-irradiated cells. LLC1 cell survival decreased to 19.81±4.65% following 5×2 Gy FD irradiation compared with non-irradiated cells.
Global mRNA expression changes
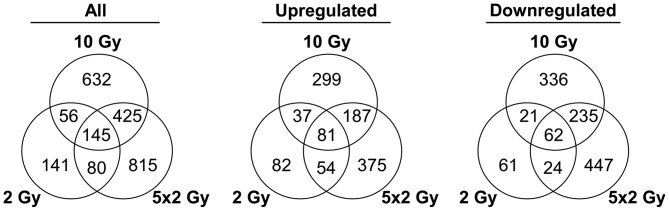
Genome wide gene expression microarray data analysis revealed that a total of 2,294 genes were differentially expressed (fold-change >1.5; P<0.05) in LLC1 cells 4 h following treatment with 2 Gy SD (SD2), 10 Gy (SD10) or 5×2 Gy FD irradiation compared with the untreated cells (Fig. 1). The amount of differentially expressed genes following irradiation was dose delivery-dependent. The exposure of LLC1 cells to SD2 resulted in the differential expression of 422 genes. By contrast, the expression of 1,258 and 1,465 genes was significantly altered following exposure to SD10 and FD irradiation, respectively. The ratio of upregulated and downregulated genes was similar following all irradiation regimens. Microarray data analysis also revealed that 145 differently expressed genes were common between all irradiation regimens.
Figure 1.
Venn diagrams demonstrating the number of genes and microRNAs differentially expressed (fold-change ≥1.5; P<0.05) in LLC1 cells following single dose (2 or 10 Gy) or fractionated dose (5×2 Gy) irradiation.
KEGG pathway enrichment analysis
In order to elucidate which pathways were significantly affected by irradiation treatment, genes identified to be differentially expressed (fold-change >1.5; P<0.05) following SD or FD were grouped into functional KEGG pathway categories (Table I). KEGG pathway analysis revealed that the ‘cell cycle’ and ‘p53 signaling pathway’ categories were the most significantly altered following all irradiation regimens, and the ‘DNA replication’ and ‘apoptosis’ categories were also significantly altered subsequent to SD10 and FD irradiation regimens. Genes associated with ‘Pathways in cancer’ were the most significantly enriched among all KEGG categories following SD10 and FD irradiation. Furthermore, subsequent to exposure to SD10 or FD the second most significantly altered functional categories were related to DNA repair (‘mismatch repair’, ‘nucleotide excision repair’ and ‘base excision repair’) and the immune response (‘cytokine-cytokine receptor interaction’, ‘hepatitis C’, ‘chemokine signaling pathway’, ‘B cell receptor signaling pathway’, ‘Janus kinase-signal transducer and activator of transcription signaling pathway’ and ‘Toll-like receptor signaling pathway’). The pathway enrichment data also revealed that SD10 and FD irradiation significantly altered the expression of genes involved in the ‘mitogen activated protein kinase (MAPK’, ‘tumor growth factor-β’, ‘vascular endothelial growth factor’, ‘wingless-type MMTV integration site family’ and ‘insulin’ signaling pathways.
Table I.
Kyoto Encyclopedia of Genes and Genomes pathway enrichment categories for genes differentially expressed in LLC1 cells following single dose (2 or 10 Gy) or fractionated dose (5×2 Gy) irradiation.
| 2 Gy | 10 Gy | 5×2 Gy | ||||
|---|---|---|---|---|---|---|
| Category | No. of genes | P-value | No. of genes | P-value | No. of genes | P-value |
| Pathways in cancer | 11 | 0.0005 | 38 | 1.76×10−16 | 53 | 3.50×10−27 |
| Cell cycle | 5 | 0.0069 | 25 | 2.88×10−16 | 28 | 3.48×10−18 |
| p53 signaling pathway | 10 | 2.37×10−09 | 15 | 2.07×10−10 | 24 | 2.46×10−20 |
| MAPK signaling pathway | 0 | NS | 20 | 2.73×10−06 | 29 | 8.91×10−11 |
| Cytokine-cytokine receptor interaction | 7 | 0.0066 | 19 | 2.84×10−06 | 26 | 1.35×10−09 |
| DNA replication | 0 | NS | 8 | 2.09×10−06 | 10 | 3.89×10−08 |
| TGF-β signaling pathway | 3 | NS | 14 | 1.92×10−08 | 14 | 6.72×10−08 |
| Apoptosis | 6 | 0.0007 | 16 | 3.41×10−10 | 17 | 1.40×10−10 |
| VEGF signaling pathway | 0 | NS | 9 | 7.35×10−05 | 12 | 8.71×10−07 |
| Hepatitis C | 0 | NS | 16 | 1.63×10−07 | 19 | 4.13×10−09 |
| Mismatch repair | 0 | NS | 7 | 1.10×10−06 | 8 | 1.20×10−07 |
| Nucleotide excision repair | 0 | NS | 7 | 7.45×10−05 | 12 | 1.72×10−09 |
| Wnt signaling pathway | 0 | NS | 9 | 0.0076 | 15 | 1.24×10−05 |
| Chemokine signaling pathway | 0 | NS | 15 | 2.07×10−05 | 16 | 2.50×10−05 |
| B cell receptor signaling pathway | 5 | 0.0022 | 11 | 1.96×10−06 | 10 | 3.51×10−05 |
| Base excision repair | 2 | NS | 8 | 3.42×10−06 | 7 | 7.77×10−05 |
| Jak-STAT signaling pathway | 6 | 0.0035 | 14 | 1.13×10−05 | 14 | 4.64×10−05 |
| Insulin signaling pathway | 4 | NS | 17 | 2.63×10−08 | 17 | 1.20×10−07 |
| RIG-I-like receptor signaling pathway | 0 | NS | 2 | NS | 6 | 0.01 |
| Toll-like receptor signaling pathway | 0 | NS | 7 | 0.008 | 9 | 0.0013 |
| Homologous recombination | 0 | NS | 5 | 0.0006 | 5 | 0.0009 |
No significance; MAPK, mitogen activated protein kinase; TGF, transforming growth factor; VEGF, vascular endothelial growth factor; Wnt, wingless-type MMTV integration site family; Jak, Janus kinase; STAT, signal transducer and activator of transcription.
Heat map analysis
Radiation-induced changes in the expression of individual genes from the p53, cell cycle, apoptosis and immune response-associated KEGG pathway categories, which were the most significantly altered in LLC1 cells following SD and FD irradiation, were color coded to demonstrate the expression patterns of genes within each category following exposure to SD and FD irradiation protocols (Fig. 2). Typically, the heat maps demonstrated that the differential expression of genes peaked in cells exposed to SD10 or FD irradiation. In addition, the extent of certain differentially expressed genes was different in cells irradiated with FD compared with SD.
Figure 2.
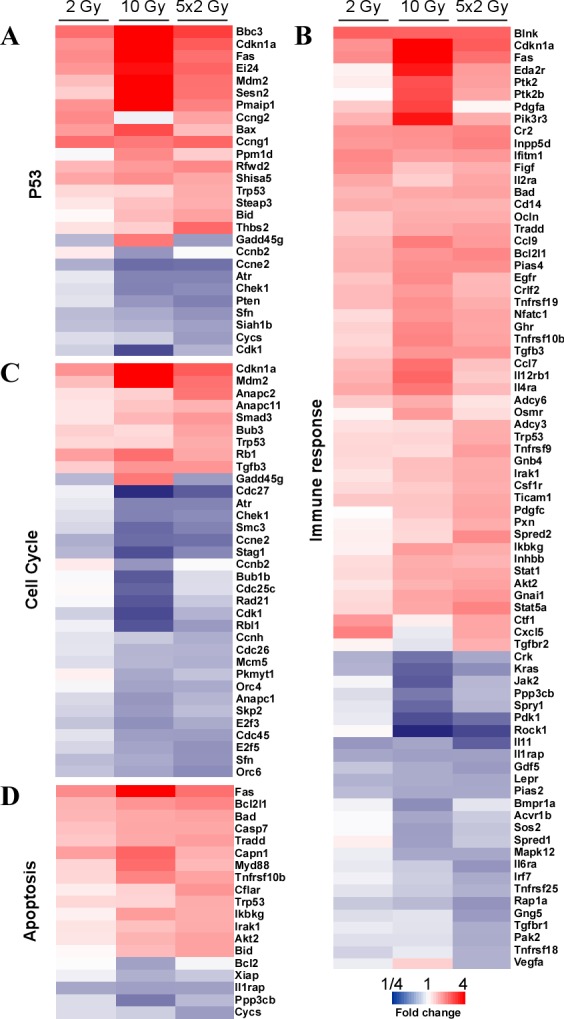
Heat maps of differentially expressed genes according to their Kyoto Encyclopedia of Genes and Genomes pathway category in LLC1 cells following single dose (2 or 10 Gy) or fractionated dose (5×2 Gy) irradiation. (A) p53 signaling pathway. (B) Immune response. (C) Cell cycle regulation. (D) Apoptosis.
The microarray data indicated that a total of 27 genes involved in the p53 signaling pathway were significantly altered in LLC1 cells exposed to all irradiation regimens (Fig. 2A). Fig. 2B depicts a total of 77 genes associated with immune response regulation that were differentially expressed in LLC1 cells following irradiation treatment. This subset of the heat map reveals that the expression of 51 genes was upregulated and 26 genes were downregulated. The expression of chemokines ccl7 and ccl9 peaked in response to 10 Gy, whereas FD induced the expression of cxcl5. The irradiation regimens also induced the expression of tumor necrosis factor-associated cytokines tnfrsf10b and tnfrsf19, which peaked following 10 Gy irradiation. FD also significantly altered the expression of tnfrsf9, tnfrsf18 and tnfrsf25. The expression of cytokines, including figf, vegfa pdgfc, ctf1 and il11, was significantly altered in LLC1 cells in response to FD irradiation. The irradiation regimens induced the expression of transcription factors Nfatc1 and Stat1, which peaked following SD10. In addition, FD significantly induced the expression of Stat5a. Exposure to radiation also altered the expression of a total of 34 genes involved in cell cycle regulation (Fig. 2C). The majority of the differentially expressed genes were downregulated, whereas the expression of only 9 genes was upregulated in this category. Heat map analysis also demonstrated that 19 apoptosis related genes were differentially expressed in cells following irradiation (Fig. 2D). The expression of a total of 14 genes was upregulated in this category, including proapoptotic Fas, Bad, Bid, Casp7, Trp53, Tradd, Thfrsf10b, and anti-apoptotic Bcl2l1 and Cfalr genes. In addition, the expression of 5 apoptosis-associated genes was downregulated in cells following to exposure to irradiation in this group, including Bcl2 and Xiap peaked following SD10.
Global miRNA expression changes
The miRNA microarray data revealed that a total of 18 miRNAs were differentially expressed (>2-fold; P<0.05) in LLC1 cells exposed to all irradiation protocols (Table II). The expression of 2 miRNAs, miR-34c-5p and miR-145a-3p, was significantly altered by all irradiation protocols, whereas miR-34c-3p and miR-34b-3p were upregulated following exposure to SD10 and FD. Data in Table II also revealed that the highest number of miRNAs was differently altered in LLC1 cells following exposure to FD, resulting in deregulated expression of 7 unique miRNAs. The expression of miR-186-5p, miR-145a-5p, miR-129-5p, miR-192-5p, miR-129-2-3p and miR-30c-5p was upregulated, and miR-105 was downregulated, in LLC1 cells following the FD regimen.
Table II.
Relative expression of differentially expressed miRNAs in LLC1 cells following single dose (2 or 10 Gy) or fractionated dose (5×2 Gy) irradiation.
| Relative expression (irradiation dose) | ||||
|---|---|---|---|---|
| miRNA | miRbase ID no. | 2 Gy | 10 Gy | 5×2 Gy |
| miR-34c-5p | MIMAT0000381 | 2.19a | 2.79a | 5.30a |
| miR-145a-3p | MIMAT0004534 | −2.46a | −2.08a | −2.94a |
| miR-878-5p | MIMAT0004932 | −2.20a | −2.74a | −2.43 |
| miR-126a-5p | MIMAT0000137 | −2.16a | −1.6 | −2.15 |
| miR-338-5p | MIMAT0004647 | −2.03a | 1.10 | −1.89 |
| miR-26b-3p | MIMAT0004630 | −1.31 | −2.32a | −1.33 |
| miR-136-5p | MIMAT0000148 | −1.28 | 2.17a | −1.43 |
| miR-466a-5p | MIMAT0004759 | −1.73 | −2.58a | −3.28 |
| miR-710 | MIMAT0003500 | −1.71 | −2.45a | −3.11 |
| miR-34b-3p | MIMAT0004581 | 2.18 | 4.07a | 14.78a |
| miR-34c-3p | MIMAT0004580 | 2.32 | 4.77a | 24.93a |
| miR-30c-5p | MIMAT0000514 | −1.10 | 1.11 | 5.50a |
| miR-105 | MIMAT0004856 | −1.54 | −1.31 | −3.65a |
| miR-129-5p | MIMAT0000209 | 1.27 | 2.07 | 4.26a |
| miR-129-2-3p | MIMAT0000544 | −1.09 | 1.35 | 8.10a |
| miR-145a-5p | MIMAT0000157 | 1.12 | 1.12 | 6.99a |
| miR-186-5p | MIMAT0000215 | 1.46 | 1.55 | 3.55a |
| miR-192-5p | MIMAT0000517 | 1.55 | 1.36 | 2.62a |
Relative miRNA expression >2-fold and P<0.05 compared with the expression levels in untreated cells.
miRNA target filter analysis
In order to determine functions of the 18 miRNA significantly altered following exposure to SD and FD irradiation in the post-transcriptional regulation of gene expression, the present study identified 6,343 individual target genes potentially regulated by these miRNA using in silico miRNA target analysis. Subsequently, negative associations between all differently expressed genes and miRNAs associated with cell cycle regulation, the p53 signaling pathway, apoptosis and the immune response were identified, indicating a potential miRNA-mRNA connection in these processes.
The miRNAs showing inverse associations with differently expressed target genes involved in selected pathways are shown in Table III. A negative association was identified between the differential expression of 6 miRNAs and 11 mRNAs in the cell cycle, p53 signaling pathway and apoptosis KEGG categories. miRNA target analysis also revealed that 20 differentially expressed genes from the immune response category were inversely associated with the differential expression of 12 miRNAs.
Table III.
miRNA target filter analysis of differentially expressed target genes and miRNAs from the cell cycle, p53, apoptosis and immune response categories that demonstrated an inverse association in LLC1 cells exposed to single dose (10 Gy) or fractionated dose (5×2 Gy) irradiation.
| 10 Gy | 5×2 Gy | |||
|---|---|---|---|---|
| Category | miRNA | Target gene | miRNA | Target gene |
| Cell cycle | miR-34c-5p↑ | E2f3↓; E2f5↓; Ccne2↓ | miR-30c-5p↑ | Ccne2↓; Stag1↓; Orc4↓; Skp2↓ |
| miR-34c-5p↑ | Ccne2↓; E2f3↓ | |||
| miR-129-5p↑ | Stag1↓; Orc4↓ | |||
| miR-145a-5p↑ | Orc4↓ | |||
| miR-186-5p↑ | Cdc27↓; Stag1↓ | |||
| p53 signaling pathway | miR-34c-5p↑ | Ccne2↓ | miR-30c-5p↑ | Ccne2↓ |
| miR-34c-5p↑ | Ccne2↓ | |||
| miR-129-5p↑ | Pten↓ | |||
| miR-145a-3p↓ | Pmaip1↑; Sesn2↑ | |||
| Apoptosis | miR-30c-5p↑ | Ppp3cb↓ | ||
| Immune response | miR-34b-3p↑ | Spred1↓ | miR-30c-5p↑ | Lepr↓; Kras↓; Ppp3cb↓ |
| miR-34c-3p↑ | Spred1↓ | miR-34c-3p↑ | Gng5↓ | |
| miR-34c-5p↑ | Pdk1↓ | miR-34c-5p↑ | Pdk1↓ | |
| miR-136-5p↓ | Eda2r↑ | miR-129-5p↑ | Il6ra↓; Rock1↓ | |
| miR-145a-3p↓ | Cr2↑; Inpp5d↑ | miR-186-5p↑ | Vegfa↓; Pias2↓ | |
| miR-466a-5p↓ | Eda2r↑; Egfr↑; Inhbb↑ | miR-192-5p↑ | Crk↓; Pias2↓ | |
| miR-710↓ | Stat1↑; Pik3r3↑ | miR-105↓ | Tgfbr2↑; Stat1↑ | |
| miR-145a-3p↓ | Tgfbr2↑; Cr2↑; Inpp5d↑; Ticam1↑ | |||
An upwards and downwards pointing arrow indicates increased and decreased expression, respectively. miRNA/miR, microRNA.
Microarray data validation
To validate the microarray data, the present study selected 4 upregulated genes and miRNAs for RT-qPCR analysis (Table IV). The results indicated that the expression of genes involved in the p53 signaling pathway, including Btg2, cyclin Ccng1, p21 and Thbs2, were significantly upregulated in LLC1 cells following irradiation compared with the untreated cells. RT-qPCR analysis also revealed that miR-34b-3p and miR-34c-5p were significantly upregulated in LLC1 cells following exposure of SD10 and FD, whereas miR-186-5p and miR-145a-5p were significantly upregulated following FD, compared with untreated cells. RT-qPCR analysis validated the gene and miRNA microarray data.
Table IV.
Validation of gene and miRNA microarray data by RT-qPCR.
| A, Gene | ||
|---|---|---|
| Irradiation dose | ||
| Data | 10 Gy | 5×2 Gy |
| Btg2 | ||
| RT-qPCR | 5.09±0.34 | 4.51±0.73 |
| Microarrays | 5.38±1.55 | 2.74±0.09 |
| Ccng1 | ||
| RT-qPCR | 4.02±0.57 | 3.2±0.47 |
| Microarrays | 2.06±0.47 | 2.30±0.45 |
| P21 | ||
| RT-qPCR | 2.64±0.06 | 2.6±0.49 |
| Microarrays | 5.96±1.28 | 2.43±0.16 |
| Thbs2 | ||
| RT-qPCR | 1.30±0.10 | 2.76±0.67 |
| Microarrays | 1.29±0.18 | 2.26±0.11 |
| B, miRNA | ||
| Irradiation dose | ||
| Data | 10 Gy | 5×2 Gy |
| miR-34b-3p | ||
| RT-qPCR | 2.76±0.48 | 3.6±0.58 |
| Microarrays | 4.07±1.53 | 14.78±4.75 |
| miR-34c-5p | ||
| RT-qPCR | 2.67±0.66 | 2.32±0.34 |
| Microarrays | 2.79±0.33 | 5.3±0.90 |
| miR-186-5p | ||
| RT-qPCR | 1.62±0.48 | 2.28±0.06 |
| Microarrays | 1.55±0.59 | 3.55±0.81 |
| miR-145a-5p | ||
| RT-qPCR | 1.41±0.46 | 3.44±0.49 |
| Microarrays | 1.12±0.88 | 6.99±1.41 |
miRNA/miR, microRNA; RT-qPCR, reverse transcription-quantitative polymerase chain reaction.
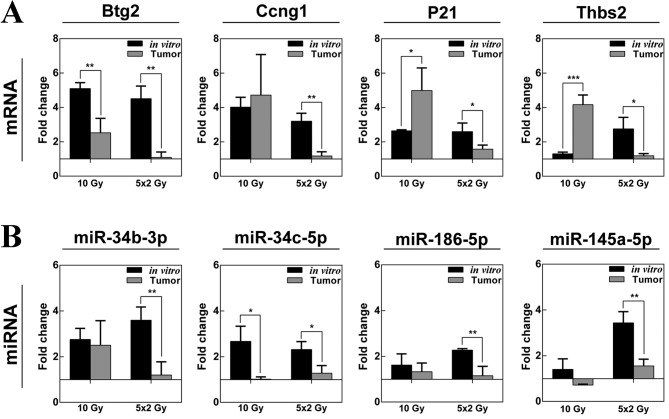
The present study compared the difference in expression of selected genes and miRNAs in LLC1 cells grown in vitro and LLC1 tumors in vivo following SD10 and FD irradiation (Fig. 3A and B). RT-qPCR analysis revealed that the expression of btg2, ccng1 and p21 was upregulated in tumors and LLC1 cells following SD treatment with 10 Gy. Thbs2 was also upregulated in vivo following irradiation of 10 Gy, whereas the expression of thbs2 was only significantly altered in vitro following FD. Notably, the expression of the selected genes was not significantly altered in LLC tumors following the FD irradiation regimen, which is in contrast to the results identified in LLC1 cells. Additionally, RT-qPCR analysis indicated no significant changes in the expression of the selected miRNAs in LLC1 tumors following irradiation (Fig. 3B).
Figure 3.
Validation of microarray gene and miRNA expression data through quantitative polymerase chain reaction analysis. Graphs showing the fold-change of selected (A) genes (Btg2, Ccng1, p21 and Thbs2) and (B) miRNAs (miR-34b-3p, miR-34c-5p, miR186-5p and miR-145a-5p) in LLC1 cells in vitro and in mouse LLC1 xenograft tumors following exposure to a single dose (10 Gy) or fractionated dose (5×2 Gy) radiation compared with the expression levels in untreated cells. Results are presented as the mean ± standard deviation (n=3). *P<0.05; **P<0.01; ***P<0.001.
Discussion
The present study investigated the changes in gene and miRNA expression signatures following SD2, SD10 and 5×2 Gy FD irradiation in mouse lung carcinoma LLC1 cells and syngeneic LLC1 tumors. The obtained data revealed that the gene expression profiles of LLC1 cells were irradiation dose delivery-dependent. In addition, the present study demonstrated through KEGG pathway enrichment analysis that the p53 signaling, cell cycle, apoptosis and immune response pathways were the most significantly altered functional categories in LLC1 cells following irradiation. The extent of differential expression was also irradiation dose delivery-dependent. The results of the miRNA microarray indicated that FD irradiation induced a significantly different miRNA expression pattern compared with SD irradiation. Furthermore, miRNA target filter analysis revealed a significant association between mRNA and miRNA expression signatures in LLC1 cells following exposure to radiation. However, RT-qPCR analysis demonstrated that LLC1 tumors exhibited no significant change in the expression of selected genes and miRNAs following a FD irradiation regimen.
The microarray data revealed that FD irradiation induced differential expression in the highest number of genes. In addition, a total of 145 genes were commonly expressed between all irradiation regimens, demonstrating a significantly different gene expression pattern in LLC1 cells following exposure to SD or FD radiation. These results are supported by previous studies (2,4,5) that also indicated that different gene expression profiles in human breast and prostate carcinoma and normal endothelium cell lines were irradiation dose delivery-dependent. In addition, Palayoor et al (5) have previously identified potential therapeutic targets from investigated the response of human prostate carcinoma cells exposed to FD radiation (3). The results of previous studies and the present study suggest that FD irradiation could be a relevant approach to identifying genes and molecular pathways that are clinically important for the improving the efficacy of radiotherapy.
The present study demonstrated that the most significantly altered functional categories in LLC1 cells following all irradiation regimens were cell cycle regulation and the p53 signaling pathways. In addition, the most significant pathway enrichment was identified in cells exposed to FD. The transcription factor p53 serves an essential role in the cellular response to IR-induced DNA damage (21). The activation of p53 results in temporary cell cycle arrest to facilitate the repaired of damaged DNA, in addition to apoptosis if the damage cannot be repaired (22,23). The present study identified that the majority of cell cycle-associated genes were downregulated in LLC1 cells following SD and FD irradiation regimens, including genes promoting G1/S and G2/M transition. Notably, the expression of the kinases Atr and Chek1, which are implicated in S-phase DNA damage checkpoint arrest (24), were also downregulated in LLC1 cells following irradiation. In addition, irradiation resulted in a significant upregulation of p21, which is a master regulator of cell cycle checkpoint progression or arrest (25). However, the deregulation of gene expression was more robust in cells exposed to FD. These included differentially expressed genes associated with the progression of DNA replication and mitosis. For example, the expression of anaphase-promoting complex (APC/C) encoding genes, including Cdc26, Cdc27, Anapc2 and Anapc11, was altered in cells exposed to FD radiation. In addition, FD irradiation deregulated the expression of Bub1 and Bub3, which are involved in the spindle assembly checkpoint and the regulation of APC/C catalytic activity (26). Furthermore, the irradiated LLC1 cells demonstrated significantly upregulated expression of genes involved in the regulation of apoptosis. However, microarray data indicated that the deregulation of gene expression associated with pro-apoptotic processes peaked in cells exposed to SD10. The expression levels of anti-apoptotic genes, including Bcl2l1 and Akt2, were increased in cells following the exposure to FD compared with SD. These data indicate that the survival of LLC1 cells is significantly higher following exposure to FD compared with SD10. The pro-survival effect of the radiation-induced DNA damage response of LLC1 cells treated with FD is likely dependent on the cumulative effect of the differential expression of genes.
The present study revealed that the expression of genes involved in the immune response was significantly altered in LLC1 cells following all irradiation regimens. Despite this, the number of differentially expressed genes was similar in cells exposed to SD10 and FD. However, the set of specific differentially expressed inflammatory genes was significantly different following the different irradiation protocols. This is in accordance with the results of previous studies, which identified a distinct expression profile of immune response genes between cells exposed to SD or FD (3,5). Microarray analysis performed in the current study also demonstrated that genes from the immune response category, including genes encoding chemokines, cytokines, cytokine receptors and tumor necrosis factors, were differently expressed in the irradiated LLC1 cells compared with non-irradiated cells. The results of the present study are consistent with previous studies demonstrating that RT can promote the immune recognition of tumor cells by increasing the expression of antigen-presenting molecules, pro-inflammatory cytokines and the release of ‘Damage-associated molecular signals’, leading to the attraction of immune cells to the irradiated tumor site (27–29). Additionally, the expression of transcription factors, including Nfatc1 and Stat1, were upregulated in cells exposed to SD and FD radiation. Members of the Stat family have been demonstrated to activate the transcription of genes involved in cancer cell survival, proliferation and angiogenesis (30). Furthermore, Stat1 is considered to serve an important role in regulating the expression of interferon-stimulated genes (ISGs) (31). The transactivation of ISGs by Stat1 can be induced as a part of the cellular response to IR and can lead to increased radioresistance (32). In the current study, treatment with FD radiation elevated the expression of Stat5a, suggesting that other Stat family members may be involved in resistance to radiotherapy. This is further supported by a previous study that demonstrated an association between the expression of Stat5a and radiosensitivity in head and neck squamous cell carcinoma cells (33). These findings suggest that the radiation-induced immune response in irradiated LLC1 cells may contribute to tumor development in a dose delivery-dependent manner. Radiation-induced alterations in the expression of inflammatory genes are considered to be pro-immunogenic, highlighting the potential of combining RT with immunotherapy for the treatment of cancer (34,35). Preclinical data also indicates that the promotion of antitumor immune response is irradiation delivery type-dependent, since RT delivered as a SD is not sufficient to induce antitumor immunity (36–38). Together these findings indicate possible directions for the development of more efficient anticancer irradiation treatment strategies, based on exploiting the pro-survival and immunogenic tumor signaling pathway alterations during FD irradiation.
It has previously been demonstrated that exposure to SD radiation results in the differential expression of miRNAs in various cancer and normal cells (39). In the present study, the expression of a total of 18 miRNAs was significantly altered in LLC1 cells exposed to IR. Microarray analysis also identified that the expression of miR-34 cluster miRNAs, including miR-34b and miR34c, was significantly upregulated in cells exposed to SD or FD radiation. Previous studies have demonstrated that members of the miR-34 cluster are regulated by p53 and involved in the regulation of cell cycle arrest, proliferation inhibition and apoptosis (40). miR-34 cluster miRNAs have also been identified to be upregulated in different human cancer cells exposed to IR (41,42). These observations suggest that miR-34 cluster miRNAs serve an important role in the response of LLC1 cells to IR. The microarray data analysis demonstrated that FD irradiation induced the most robust deregulation of miRNA expression, indicating that the expression of miRNA is also altered in an irradiation delivery-dependent manner. Similar results were identified in previous studies, which demonstrated a high degree of alteration in the expression of miRNAs in prostate cancer and endothelium cells following FD (5,6). In addition, Leung et al (43) reported that only a small number of miRNAs differentially expressed in breast cancer cells exposed to SD or FD were the same, suggesting that FD induces a distinct miRNA signature compared with SD radiation.
To further extend the understanding of the roles miRNAs serve in the cellular response to IR, the present study performed miRNA target filter analysis to identify potential functional associations between differentially expressed mRNAs and miRNAs in LLC1 cells exposed to radiation. However, the identification of regulatory miRNAs and their target mRNAs remains a major challenge since a single miRNA may regulate multiple mRNAs and vice versa. In addition, statistical methods which are able to identify these miRNA-controlled regulations may result in thousands of putative miRNA-mRNA pairs, leading to an inability to extract a biologically relevant understanding of the collective function of differentially expressed miRNAs (44). Therefore, the present study investigated the negative association between the expression of miRNAs and genes associated with p53, cell cycle regulation, apoptosis and immune response pathways, which were shown to be the most prominently altered in LLC1 cells following SD and FD irradiation.
It has been shown that the transcription factor p53 performs an important role in the regulation of the transcription of several miRNAs, which in turn control the expression of p53-regulated genes that mediate cell cycle arrest and apoptosis (40,45). The present miRNA target analysis revealed an inverse association between the expression of miR-34c and E2f3, E2f5 and Ccne2, suggesting that the upregulation of miR-34c could be associated with G1 phase regulation in LLC1 cells exposed to radiation. This is also supported by previous studies that have highlighted the role of miR-34c in the induction of G1 and G2/M cell cycle arrest (46,47). In addition, Li et al (48) have demonstrated that the expression of E2F3 is reduced following the upregulation of miR-34c in endometrial carcinoma cells, indicating that E2F3 could be a target of miR-34c. The present study identified a negative association between several miRNAs and genes that were differentially expressed in cells exposed to FD radiation. For example, the upregulation of miR-30c was associated with the downregulation of Ccne2, Stag1, Orc4 and Skp2. In addition, the expression of Stag1 and Orc4 was inversely associated with the expression of miR-129, miR-145a and miR-186. These observations suggest that these miRNAs perform important roles in the cell cycle arrest response to radiation in LLC1 cells in a dose delivery-dependent manner. However, no significant association was observed between differentially expressed miRNAs and genes involved in apoptosis, with the exception of a negative association between the expression of miR-30c and Ppp3cb. Nevertheless, several miRNAs that were differentially expressed subsequent to irradiation have previously been demonstrated to be associated with the regulation of apoptosis, indicating that they may serve a similar role in irradiated LLC1 cells. For example, the overexpression of miR-129 was identified to promote the death of irradiated breast cancer cells by targeting high mobility group box 1 for degradation (49). In addition, miR-30c was demonstrated to serve an important role in the radiation-induced hematopoietic cell damage response (50).
There is emerging evidence that miRNAs are involved in the radiation-induced regulation of inflammatory responses (5–7). miRNA target filter analysis revealed that the majority of miRNAs differentially expressed in LLC1 cells exposed to SD and FD radiation were inversely associated with several genes associated with the immune response, highlighting the role of miRNA in the inflammatory response to irradiation. Target filter analysis also indicated that the regulation of miRNAs and inflammatory response-associated genes in LLC1 cells treated with fractionated irradiation was significantly different. In addition, the upregulation of Stat1 was associated with the downregulation of miR-710 in cells exposed to SD and with the downregulation of miR-105 in cells exposed to FD. The present study demonstrated that treatment with FD also upregulated the expression of miR-145-5p, which has previously been identified to target Stat1 (51), indicating that certain differentially expressed genes could be regulated by distinct miRNAs in cells exposed to SD or FD radiation. These data suggest that the regulation of the immune response by miRNAs in irradiated LLC1 cells may be irradiation dose delivery-dependent.
The results of the present study demonstrate that the expression of genes and miRNAs is different in LLC1 tumors and LLC1 cells in vitro following exposure to SD10 or FD irradiation. Despite the fact that treatment with SD10 increased the expression of genes involved in the p53 signaling pathway in LLC1 tumors, no significant change in the expression of selected genes and miRNAs were observed in vivo following the exposure to FD radiation, suggesting that the radiation-induced changes in gene and miRNA expression may be modulated by the tumor microenvironment. Similar results of different cellular response to irradiation under the different microenvironment conditions were obtained by previous studies, which have applied different strategies to validate in vitro data. Camphausen et al (11) reported that glioblastoma U87 and U251 cells exposed to 6 Gy SD radiation in vivo exhibited a different set of differentially expressed genes compared with cells grown in vitro. In addition, Tsai et al (2) demonstrated that prostate cancer DU145 cell xenografts exhibited a different profile of genes induced by SD and FD compared with the same cells exposed to irradiation in vitro, indicating that a 10 Gy exposure in vivo could only reach an effect of up to 3 Gy exposure under in vitro growth conditions. These data suggest that investigations into the effects of gene and miRNA expression will require more biologically relevant experimental conditions.
In conclusion, the present study indicates that the gene and miRNA expression profiles in LLC1 cells exposed to radiation are dose delivery type-dependent. In addition, data analysis revealed that the altered expression of miRNAs and targeted mRNAs may affect radiation-induced DNA damage response pathways differently in LLC1 cells exposed to SD and FD irradiation. The results of the present study may be applied to improve the outcome of radiotherapy. However, experimental in vitro conditions, including the tumor microenvironment, should be considered in more detail in further investigations.
Acknowledgements
The present study was supported by the European Social Fund under the National Integrated Programme of Biotechnology and Biopharmacy (grant no. VP1-3.1-SMM-08-K01-005).
Glossary
Abbreviations
- RT
radiotherapy
- IR
ionizing radiation
- SD
single dose
- FD
fractionated dose
References
- 1.Lawrence YR, Vikram B, Dignam JJ, Chakravarti A, Machtay M, Freidlin B, Takebe N, Curran WJ, Jr, Bentzen SM, Okunieff P, et al. NCI-RTOG translational program strategic guidelines for the early-stage development of radiosensitizers. J Natl Cancer Inst. 2013;105:11–24. doi: 10.1093/jnci/djs472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tsai MH, Cook JA, Chandramouli GV, DeGraff W, Yan H, Zhao S, Coleman CN, Mitchell JB, Chuang EY. Gene expression profiling of breast, prostate, and glioma cells following single versus fractionated doses of radiation. Cancer Res. 2007;67:3845–3852. doi: 10.1158/0008-5472.CAN-06-4250. [DOI] [PubMed] [Google Scholar]
- 3.John-Aryankalayil M, Palayoor ST, Cerna D, Simone CB, II, Falduto MT, Magnuson SR, Coleman CN. Fractionated radiation therapy can induce a molecular profile for therapeutic targeting. Radiat Res. 2010;174:446–458. doi: 10.1667/RR2105.1. [DOI] [PubMed] [Google Scholar]
- 4.Simone CB, II, John-Aryankalayil M, Palayoor ST, Makinde AY, Cerna D, Falduto MT, Magnuson SR, Coleman CN. mRNA expression profiles for prostate cancer following fractionated irradiation are influenced by p53 status. Transl Oncol. 2013;6:573–585. doi: 10.1593/tlo.13241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Palayoor ST, John-Aryankalayil M, Makinde AY, Falduto MT, Magnuson SR, Coleman CN. Differential expression of stress and immune response pathway transcripts and miRNAs in normal human endothelial cells subjected to fractionated or single-dose radiation. Mol Cancer Res. 2014;12:1002–1015. doi: 10.1158/1541-7786.MCR-13-0623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.John-Aryankalayil M, Palayoor ST, Makinde AY, Cerna D, Simone CB, II, Falduto MT, Magnuson SR, Coleman CN. Fractionated radiation alters oncomir and tumor suppressor miRNAs in human prostate cancer cells. Radiat Res. 2012;178:105–117. doi: 10.1667/RR2703.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao L, Lu X, Cao Y. MicroRNA and signal transduction pathways in tumor radiation response. Cell Signal. 2013;25:1625–1634. doi: 10.1016/j.cellsig.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gong P, Zhang T, He D, Hsieh JT. MicroRNA-145 modulates tumor sensitivity to radiation in prostate cancer. Radiat Res. 2015;184:630–638. doi: 10.1667/RR14185.1. [DOI] [PubMed] [Google Scholar]
- 9.Weidhaas JB, Babar I, Nallur SM, Trang P, Roush S, Boehm M, Gillespie E, Slack FJ. MicroRNAs as potential agents to alter resistance to cytotoxic anticancer therapy. Cancer Res. 2007;67:11111–11116. doi: 10.1158/0008-5472.CAN-07-2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Balça-Silva J, Neves Sousa S, Gonçalves AC, Abrantes AM, Casalta-Lopes J, Botelho MF, Sarmento-Ribeiro AB, Silva HC. Effect of miR-34b overexpression on the radiosensitivity of non-small cell lung cancer cell lines. Anticancer Res. 2012;32:1603–1609. [PubMed] [Google Scholar]
- 11.Camphausen K, Purow B, Sproull M, Scott T, Ozawa T, Deen DF, Tofilon PJ. Orthotopic growth of human glioma cells quantitatively and qualitatively influences radiation-induced changes in gene expression. Cancer Res. 2005;65:10389–10393. doi: 10.1158/0008-5472.CAN-05-1904. [DOI] [PubMed] [Google Scholar]
- 12.Pawlik A, Delmar P, Bosse S, Sainz L, Petat C, Pietu G, Thierry D, Tronik-Le Roux D. Changes in transcriptome after in vivo exposure to ionising radiation reveal a highly specialised liver response. Int J Radiat Biol. 2009;85:656–671. doi: 10.1080/09553000903020024. [DOI] [PubMed] [Google Scholar]
- 13.Buch K, Peters T, Nawroth T, Sänger M, Schmidberger H, Langguth P. Determination of cell survival after irradiation via clonogenic assay versus multiple MTT Assay-a comparative study. Radiat Oncol. 2012;7:1. doi: 10.1186/1748-717X-7-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Holko M, et al. NCBI GEO: Archive for functional genomics data sets-update. Nucleic Acids Res. 2013;41:D991–D995. doi: 10.1093/nar/gks1193. (Database issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang J, Duncan D, Shi Z, Zhang B. WEB-based GEne SeT AnaLysis Toolkit (WebGestalt): Update 2013. Nucleic Acids Res. 2013;41:W77–W83. doi: 10.1093/nar/gkt439. (Web Server issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Paraskevopoulou MD, Georgakilas G, Kostoulas N, Vlachos IS, Vergoulis T, Reczko M, Filippidis C, Dalamagas T, Hatzigeorgiou AG. DIANA-microT web server v5.0: Service integration into miRNA functional analysis workflows. Nucleic Acids Res. 2013;41:W169–W173. doi: 10.1093/nar/gkt393. (Web Server issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vlachos IS, Kostoulas N, Vergoulis T, Georgakilas G, Reczko M, Maragkakis M, Paraskevopoulou MD, Prionidis K, Dalamagas T, Hatzigeorgiou AG. DIANA miRPath v.2.0: Investigating the combinatorial effect of microRNAs in pathways. Nucleic Acids Res. 2012;40:W498–W504. doi: 10.1093/nar/gks494. (Web Server issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative pcr and the 2(−Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 20.Butkytė S, Čiupas L, Jakubauskienė E, Vilys L, Mocevicius P, Kanopka A, Vilkaitis G. Splicing-dependent expression of microRNAs of mirtron origin in human digestive and excretory system cancer cells. Clin Epigenetics. 2016;8:33. doi: 10.1186/s13148-016-0200-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rashi-Elkeles S, Elkon R, Shavit S, Lerenthal Y, Linhart C, Kupershtein A, Amariglio N, Rechavi G, Shamir R, Shiloh Y. Transcriptional modulation induced by ionizing radiation: p53 remains a central player. Mol Oncol. 2011;5:336–348. doi: 10.1016/j.molonc.2011.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fei P, El-Deiry WS. P53 and radiation responses. Oncogene. 2003;22:5774–5783. doi: 10.1038/sj.onc.1206677. [DOI] [PubMed] [Google Scholar]
- 23.Mirzayans R, Andrais B, Scott A, Murray D. New insights into p53 signaling and cancer cell response to DNA damage: Implications for cancer therapy. J Biomed Biotechnol. 2012;2012:170325. doi: 10.1155/2012/170325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Willis N, Rhind N. Regulation of DNA replication by the S-phase DNA damage checkpoint. Cell Division. 2009;4:13. doi: 10.1186/1747-1028-4-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cazzalini O, Scovassi AI, Savio M, Stivala LA, Prosperi E. Multiple roles of the cell cycle inhibitor p21CDKN1A in the DNA damage response. Mutat Res. 2010;704:12–20. doi: 10.1016/j.mrrev.2010.01.009. [DOI] [PubMed] [Google Scholar]
- 26.Shepperd LA, Meadows JC, Sochaj AM, Lancaster TC, Zou J, Buttrick GJ, Rappsilber J, Hardwick KG, Millar JB. Phosphodependent recruitment of Bub1 and Bub3 to Spc7/KNL1 by Mph1 kinase maintains the spindle checkpoint. Curr Biol. 2012;22:891–899. doi: 10.1016/j.cub.2012.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Formenti SC, Demaria S. Combining radiotherapy and cancer immunotherapy: A paradigm shift. J Natl Cancer Inst. 2013;105:256–265. doi: 10.1093/jnci/djs629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Derer A, Deloch L, Rubner Y, Fietkau R, Frey B, Gaipl US. Radio-immunotherapy-induced immunogenic cancer cells as basis for induction of systemic anti-tumor immune responses-pre-clinical evidence and ongoing clinical applications. Front Immunol. 2015;6:505. doi: 10.3389/fimmu.2015.00505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wattenberg MM, Fahim A, Ahmed MM, Hodge JW. Unlocking the combination: Potentiation of radiation-induced antitumor responses with immunotherapy. Radiat Res. 2014;182:126–138. doi: 10.1667/RR13374.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.O'Shea JJ, Schwartz DM, Villarino AV, Gadina M, McInnes IB, Laurence A. The JAK-STAT pathway: Impact on human disease and therapeutic intervention. Annu Rev Med. 2015;66:311–328. doi: 10.1146/annurev-med-051113-024537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Au-Yeung N, Mandhana R, Horvath CM. Transcriptional regulation by STAT1 and STAT2 in the interferon JAK-STAT pathway. JAKSTAT. 2013;2:e23931. doi: 10.4161/jkst.23931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cheon H, Holvey-Bates EG, Schoggins JW, Forster S, Hertzog P, Imanaka N, Rice CM, Jackson MW, Junk DJ, Stark GR. IFNβ-dependent increases in STAT1, STAT2, and IRF9 mediate resistance to viruses and DNA damage. EMBO J. 2013;32:2751–2763. doi: 10.1038/emboj.2013.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stegeman H, Kaanders JH, Verheijen MM, Peeters WJ, Wheeler DL, Iida M, Grénman R, van der Kogel AJ, Span PN, Bussink J. Combining radiotherapy with MEK1/2, STAT5 or STAT6 inhibition reduces survival of head and neck cancer lines. Mol Cancer. 2013;12:133. doi: 10.1186/1476-4598-12-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kalbasi A, June CH, Haas N, Vapiwala N. Radiation and immunotherapy: A synergistic combination. J Clin Invest. 2013;123:2756–2763. doi: 10.1172/JCI69219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Frey B, Rubner Y, Kulzer L, Werthmöller N, Weiss EM, Fietkau R, Gaipl US. Antitumor immune responses induced by ionizing irradiation and further immune stimulation. Cancer Immunol Immunother. 2014;63:29–36. doi: 10.1007/s00262-013-1474-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dewan MZ, Galloway AE, Kawashima N, Dewyngaert JK, Babb JS, Formenti SC, Demaria S. Fractionated but not single-dose radiotherapy induces an immune-mediated abscopal effect when combined with anti-CTLA-4 antibody. Clin Cancer Res. 2009;15:5379–5388. doi: 10.1158/1078-0432.CCR-09-0265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schaue D, Ratikan JA, Iwamoto KS, McBride WH. Maximizing tumor immunity with fractionated radiation. Int J Radiat Oncol Biol Phys. 2012;83:1306–1310. doi: 10.1016/j.ijrobp.2011.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kulzer L, Rubner Y, Deloch L, Allgäuer A, Frey B, Fietkau R, Dörrie J, Schaft N, Gaipl US. Norm- and hypo-fractionated radiotherapy is capable of activating human dendritic cells. J Immunotoxicol. 2014;11:328–336. doi: 10.3109/1547691X.2014.880533. [DOI] [PubMed] [Google Scholar]
- 39.Metheetrairut C, Slack FJ. MicroRNAs in the ionizing radiation response and in radiotherapy. Curr Opin Genet Dev. 2013;23:12–19. doi: 10.1016/j.gde.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hermeking H. p53 Enters the MicroRNA World. Cancer Cell. 2007;12:414–418. doi: 10.1016/j.ccr.2007.10.028. [DOI] [PubMed] [Google Scholar]
- 41.Girardi C, De Pittà C, Casara S, Sales G, Lanfranchi G, Celotti L, Mognato M. Analysis of miRNA and mRNA expression profiles highlights alterations in ionizing radiation response of human lymphocytes under modeled microgravity. PLoS One. 2012;7:e31293. doi: 10.1371/journal.pone.0031293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Josson S, Sung SY, Lao K, Chung LW, Johnstone PA. Radiation modulation of microRNA in prostate cancer cell lines. Prostate. 2008;68:1599–1606. doi: 10.1002/pros.20827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Leung CM, Chen TW, Li SC, Ho MR, Hu LY, Liu WS, Wu TT, Hsu PC, Chang HT, Tsai KW. MicroRNA expression profiles in human breast cancer cells after multifraction and single-dose radiation treatment. Oncol Rep. 2014;31:2147–2156. doi: 10.3892/or.2014.3089. [DOI] [PubMed] [Google Scholar]
- 44.Jayaswal V, Lutherborrow M, Ma DD, Yang Hwa Y. Identification of microRNAs with regulatory potential using a matched microRNA-mRNA time-course data. Nucleic Acids Res. 2009;37:e60. doi: 10.1093/nar/gkp153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hermeking H. MicroRNAs in the p53 network: Micromanagement of tumour suppression. Nat Rev Cancer. 2012;12:613–626. doi: 10.1038/nrc3318. [DOI] [PubMed] [Google Scholar]
- 46.Cannell IG, Kong YW, Johnston SJ, Chen ML, Collins HM, Dobbyn HC, Elia A, Kress TR, Dickens M, Clemens MJ, et al. p38 MAPK/MK2-mediated induction of miR-34c following DNA damage prevents Myc-dependent DNA replication. Proc Natl Acad Sci USA. 2010;107:5375–5380. doi: 10.1073/pnas.0910015107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Achari C, Winslow S, Ceder Y, Larsson C. Expression of miR-34c induces G2/M cell cycle arrest in breast cancer cells. BMC Cancer. 2014;14:538. doi: 10.1186/1471-2407-14-538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li F, Chen H, Huang Y, Zhang Q, Xue J, Liu Z, Zheng F. miR-34c plays a role of tumor suppressor in HEC1-B cells by targeting E2F3 protein. Oncol Rep. 2015;33:3069–3074. doi: 10.3892/or.2015.3894. [DOI] [PubMed] [Google Scholar]
- 49.Luo J, Chen J, He L. mir-129-5p attenuates irradiation-induced autophagy and decreases radioresistance of breast cancer cells by targeting HMGB1. Med Sci Monit. 2015;21:4122–4129. doi: 10.12659/MSM.896661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li XH, Ha CT, Fu D, Xiao M. Micro-RNA30c negatively regulates REDD1 expression in human hematopoietic and osteoblast cells after gamma-irradiation. PLoS One. 2012;7:e48700. doi: 10.1371/journal.pone.0048700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gregersen LH, Jacobsen AB, Frankel LB, Wen J, Krogh A, Lund AH. MicroRNA-145 targets in colon cancer cells. PLoS One. 2010;5:e8836. doi: 10.1371/journal.pone.0008836. [DOI] [PMC free article] [PubMed] [Google Scholar]