Abstract
Acute myeloid leukemia (AML) is a heterogeneous malignant disorder derived from the myeloid hematopoietic cells that accounts for ~80% of all adult acute leukemia. Numerous studies have shown that drug resistance not only exists against conventional chemotherapeutic drugs, but also limits the efficacy of new biological agents. Therefore, it is important to identify the mechanisms behind chemoresistance and seek therapeutic strategies to enhance efficacy in AML chemotherapy. MicroRNA (miR)-217 has been recognized as a tumor suppressor that is downregulated in various types of cancer, however the mechanisms behind the expression and function of miR-217 in AML have not yet been recognized. The expression of miR-217 was determined by quantitative polymerase chain reaction (qPCR). Following transfection with miR-217 mimics, an MTT assay, chemosensitivity assay, cell apoptosis assay and western blot analysis were performed in AML cell lines. Functional assays were also performed to explore the effects of endogenous Kirsten rat sarcoma viral oncogene homolog (KRAS) in AML. The results revealed that miR-217 was downregulated in patients with AML. Overexpression of miR-217 inhibited cellular proliferation and enhanced cell chemosensitivity to doxorubicin by the cell apoptosis pathway in AML cells. A dual-luciferase reporter assay demonstrated that KRAS was a direct target gene of miR-217 in vitro. qPCR and western blot analysis revealed that miR-217 negatively regulated KRAS protein expression, but had no impact on KRAS mRNA expression. Knockdown of KRAS expression markedly suppressed AML cellular proliferation, and enhanced cell chemosensitivity to doxorubicin via the cell apoptosis pathway. These findings indicate that miR-217 functions as a tumor suppressor in AML by directly targeting KRAS. Therefore, miR-217-based therapeutic strategies may provide a novel strategy for the enhancement of efficacy in the treatment of AML.
Keywords: acute myeloid leukemia, Kirsten rat sarcoma viral oncogene homolog, microRNA-217, apoptosis, chemosensitivity
Introduction
Leukemia is a cancer of the white blood cells, and can be classified as acute myeloid leukemia (AML), acute lymphoblastic leukemia or chronic lymphoblastic leukemia (1). AML is the most common type of acute leukemia, and accounts for ~80% of all cases of acute leukemia in adults (2). AML is a clonal disorder caused by rapid proliferation, accumulation and differentiation arrest of hematopoietic progenitors in the bone marrow and blood (3). It was predicted that in 2015, there would be 20,830 new cases and 10,460 mortalities due to AML in the United States (4).
AML can be divided into 3 risk-based categories based on cytogenetic information: Favorable, intermediate and poor, with a 5-year overall survival rate of 55, 24–42 and 11%, respectively (5). Currently, the main treatment for patients with AML is cytarabine/anthracycline-based chemotherapy. However, the majority of patients cannot be cured by this approach (6). Furthermore, the majority of AML patients will relapse, with the major cause of relapse and therapeutic failure in AML being resistance to chemotherapy (7). Therefore, it is important to further investigate the molecular mechanisms behind chemoresistance and develop effective new therapeutic strategies for the enhancement of efficacy in AML chemotherapy.
MicroRNAs (miRs) are highly conserved, non-protein-coding, single-stranded small RNAs (~22 nucleotides in length) that post-transcriptionally regulate gene expression in cancerous and non-cancerous cells (8). miRs typically decrease the expression level of target mRNA by interacting preferentially with the 3′ untranslated region (3′UTR) of target mRNA, resulting in translation repression or degradation (9,10). miRs act as either oncogenes or tumor suppressors in the development and progression of human carcinogenesis (11). Certain miRs that are upregulated in cancer directly target oncogenes, and function in a proliferative and anti-apoptotic manner. Conversely, miRs that are downregulated in certain types of cancer function as tumor suppressors and inhibit cancer initiation and progression (12–14). Studies have revealed that miRs control various key cellular physiological and pathological processes, including the cell cycle, cellular proliferation, apoptosis, differentiation and development, and are involved in several human diseases, including cancer (15,16). Additionally, miRs have been demonstrated to perform important roles in the regulation of chemoresistance (17,18). Therefore, miRs may be investigated as targets for anticancer drug resistance in AML.
Abnormal expression of miR-217 has been demonstrated in numerous human malignancies, however the mechanisms behind the expression and function of miR-217 in AML have not yet been recognized. The present study demonstrated that miR-217 was downregulated in patients with AML compared with healthy controls. In addition, upregulation of miR-217 suppressed cell proliferation and enhanced the chemosensitivity of AML cells to doxorubicin (DOX) through the cell apoptosis pathway. In addition, the Kirsten rat sarcoma viral oncogene homolog (KRAS) was identified as a direct target of miR-217. The present findings have therapeutic implications and may be explored for implications for the treatment of AML.
Materials and methods
Clinical specimens
In the present study, samples of bone marrow from 62 patients with AML were collected at Tongji Hospital (Wuhan, China). A total of 25 healthy subjects were also used in this study as the control group. The diagnosis of AML was made based on standard diagnostic methods, including morphological assessment and cytochemical studies of bone marrow smears. No patients received anti-leukemic therapy during bone marrow aspiration. The present study was approved by the Ethics Committee of Tongji Hospital and informed written consent was also obtained from all patients in the AML and control groups of this study.
Cell culture
Human leukemia HL-60 and K562 cell lines, were obtained from the American Type Culture Collection (Manassas, VA, USA). Cells were cultured in RPMI-1640 medium supplemented with 10% fetal bovine serum, 100 U/ml penicillin, 100 mg/ml streptomycin and 2 mM L-glutamine (All from Gibco; Thermo Fisher Scientific, Inc.) in a humidified air atmosphere of 5% CO2 at 37°C.
Cell transfection
Mature miR-217 mimics, negative control (NC), KRAS small interfering (si) RNA, NC siRNA, and luciferase reporter plasmids were obtained from Shanghai GenePharma Co., Ltd. (Shanghai, China). To evaluate the functions of miR-217 and KRAS in AML cells, cells were transfected with miR-217 mimics or NC, and KRAS siRNA or NC siRNA using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. Subsequent to transfection at 37°C for 6 h, cell culture medium was replaced with RPMI-1640 medium containing 10% FBS and 2 mM L-glutamine.
RNA isolation, reverse transcription and quantitative polymerase chain reaction (qPCR)
Bone marrow mononuclear cells from bone marrow aspirates were isolated using Ficoll-Hypaque density gradient centrifugation (400 × g for 30 min at 20°C followed by 100 × g for 10 min at 20°C) (Ficoll, Pharmacia LKB Biotechnology, Piscataway, NY, USA). Total RNA was extracted from bone marrow mononuclear cells using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. Single-stranded cDNA for the miR analysis was synthesized by reverse-transcription using PrimeScripts RT reagent kit (Takara Biotechnology Co., Ltd., Dalian, China) according to the manufacturer's protocol. qPCR was performed using a SYBR premix Ex Taq kit (Takara Biotechnology Co., Ltd.) on an Applied Biosystems 7300 Real-time PCR system (Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. The thermocycling conditions for qPCR were as follows: 95°C for 30 sec; 40 cycles of 95°C for 5 sec; and 60°C for 30 sec. The relative expression level was calculated using the 2−ΔΔCq method (19). U6 small nuclear RNA and GAPDH were used as an internal control. The primer sequences used were as follows: miR-217 forward, 5′-TACTCAACTCACTACTGCATCAGGA-3′ and reverse, 5′-TATGGTTGTTCTGCTCTCTGTGTC-3′; and U6 forward, 5′-GCTTCGGCAGCACATATACTAAA-3′ and reverse, 5′-GCTTCACGAATTTGCGTGTCAT-3′. KRAS forward, 5′-GACTCTGAAGATGTACCTATGGTCCTA-3′ and reverse, 5′-CATCATCAACACCCTGTCTTGTC-3′; and GAPDH forward, 5′-ATAGCACAGCCTGGATAGCAACGTAC-3′ and reverse, 5′-CACCTTCTACAATGAGCTGCGTGTG-3′. Each sample was analyzed in triplicate.
MTT assay
Subsequent to transfection for 24 h, transfected cells were harvested and seeded into 96-well culture plates at a density of 3×104 cells. Following the incubation at 37°C for various time periods (24–96 h), an MTT assay (5 mg/ml, Sigma-Aldrich; Merck Millipore, St. Louis, MO, USA) was performed according to the manufacturer's protocol. Briefly, 20 µl MTT assay solution was added into each well. Subsequent to a 4 h incubation at 37°C, the 96-well plate was centrifuged at 100 × g for 5 min at room temperature, and the purple colored precipitate of formazan was dissolved in 200 µl dimethyl sulfoxide. Subsequent to being slowly spun at 37°C for 15 min, the absorbance at 490 nm wavelength was detected using an automatic multi-well spectrophotometer (Bio-Rad Laboratories, Inc., Hercules, CA, USA). All experiments were performed in triplicate. Each experiment was repeated at least 3 times.
Chemosensitivity assay
Following transfection for 48 h, transfected cells were harvested and seeded into 96-well culture plates at a density of 5×104 cells. Cells were treated with DOX (Sigma-Aldrich; Merck Millipore) at various concentrations (32–1048576 ng/ml). Subsequent to incubation at 37°C for 48 h, a chemosensitivity assay was performed using the MTT assay as described previously. The dose-response curve was charted at different concentrations. Each concentration was analyzed in triplicate. Each experiment was repeated at least 3 times.
Cell apoptosis assay
Following transfection for 48 h, transfected cells were harvested and seeded into 6-well plates at a density of 2×106 cells. Cells were then treated with DOX at 32 ng/ml. Subsequent to incubation for 48 h at 37°C, cells were collected and washed with PBS (Gibco; Thermo Fisher Scientific). Subsequently, cells were centrifuged at 100 × g for 5 min at room temperature, all PBS was carefully removed and the cells were fixed in 80% ice-cold ethanol in PBS. Subsequently, cells were re-suspended in 1X binding buffer to a concentration of 1×104 cells/µl. Then cells were treated with 5 µl of Annexin V-fluorescein isothiocyanate and 10 µl of propidium iodide (PI) and incubated for 15 min at room temperature in dark. Cells were then analyzed with flow cytometry (BD FACS Calibur; BD Biosciences, Franklin Lakes, NJ, USA). Apoptotic cells were recognized by a high Annexin V fluorescence signal combined with a low PI signal and analyzed using CellQuest version 5.1 (BD Biosciences).
Western blot analysis
Subsequent to transfection for 72 h, transfected cells were lysed with RIPA lysis buffer (Beyotime Institute of Biotechnology, Haimen, China). Protein concentration was measured using a BCA assay kit (Pierce; Thermo Fisher Scientific, Inc.). Equal amounts of protein (20 µg) were then separated by 10% SDS-PAGE and transferred to polyvinylidene difluoride membranes (Merck Millipore). Subsequent to blocking with 5% non-fat dry milk in TBS saline containing 0.05% Tween-20 (Beyotime Institute of Biotechnology), membranes were incubated at 4°C overnight with mouse anti-human KRAS (cat. no. ab157255) or β-actin (cat. no. ab8226) monoclonal antibody (both 1:1,000 dilution; Abcam, Cambridge, MA, USA), followed by incubation with horseradish peroxidase conjugated secondary antibody (1:3,000 dilution; cat. no. ab6789; Abcam) at room temperature for 2 h. Bands were then visualized with an enhanced chemiluminescence detection system (Pierce; Thermo Fisher Scientific, Inc.). β-actin was used as an internal control.
Dual-luciferase reporter assay
A dual-luciferase reporter assay was performed to explore whether KRAS was a direct target of miR-217. Cells were transfected with miR-217 mimics or NC, and co-transfected with PGL3-KRAS-3′UTR wild type (Wt) or PGL3-KRAS-3′UTR mutant (Mut) using Lipofectamine® 2000. Following incubation at 37°C for 48 h, a Dual-Luciferase Reporter assay (Promega Corporation, Madison, WI) was performed to detect Firefly and Renilla luciferase activity according to the manufacturer's protocol. Renilla luciferase activity was measured as an internal control. Each experiment was repeated at least 3 times.
Statistical analysis
Data are presented as the mean ± standard deviation and compared using the Student's t-test or one-way analysis of variance with SPSS software (version 13.0; SPSS, Inc., Chicago, IL, USA). Double-tailed P<0.05 was considered to indicate a statistically significant difference.
Results
miR-217 was downregulated in AML
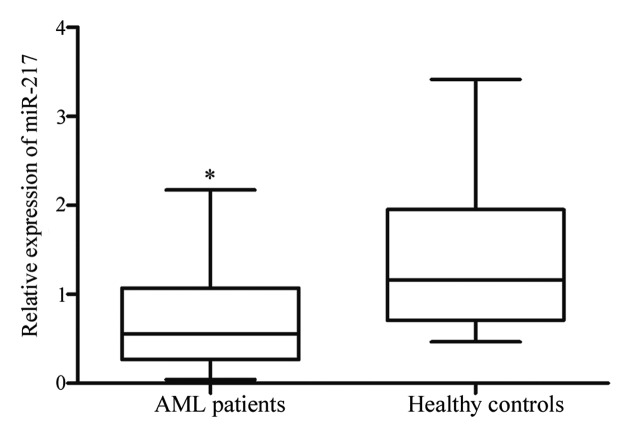
miR-217 expression was measured in patients with AML and healthy controls using qPCR. As shown in Fig. 1, miR-217 was significantly lower in patients with AML compared with healthy controls (P=0.001). The results indicate that miR-217 may perform an important role in AML.
Figure 1.
Expression levels of miR-217 in patients with AML and healthy controls. miR-217 was significantly downregulated in AML patients compared with those of healthy controls. Data are presented as box-and-whisker plots. *P<0.05 compared with their respective controls. miR, microRNA; AML, acute myeloid leukemia.
miR-217 was markedly upregulated in HL-60 and K562 cells following transfection with miR-217 mimics
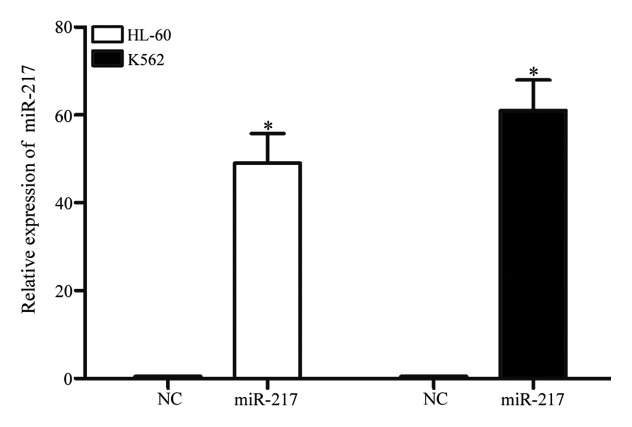
To investigate the functions of miR-217 on AML cells, miR-217 mimics or NC was transfected into HL-60 and K562 cells. qPCR was performed to evaluate transfection efficiency. As demonstrated in Fig. 2, miR-217 was markedly upregulated in HL-60 and K562 (both P<0.001) cells transfected with miR-217 mimics, in comparison with cells transfected with NC.
Figure 2.
Following transfection with miR-217 mimics, miR-217 was upregulated in human leukemia HL-60 and K562 cell lines, when compared with cells transfected with NC. Data are presented as the mean ± standard deviation. *P<0.05 compared with their respective controls. miR, microRNA; NC, normal control.
miR-217 decreased cell proliferation in HL-60 and K562 cells
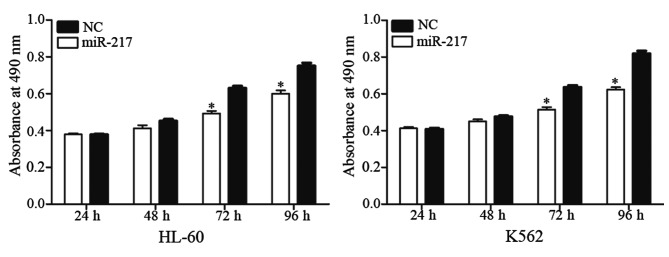
An MTT assay was performed to explore the effect of miR-217 on cell proliferation. As shown in Fig. 3, enforced miR-217 expression resulted in growth inhibition relative to NC in HL-60 (P=0.023) and K562 (P=0.015) cell lines. These results indicate that miR-217 was a negative regulator of AML cell proliferation.
Figure 3.
miR-217 inhibited acute myeloid leukemia cell proliferation. The MTT assay revealed that upregulation of miR-217 significantly suppressed cell proliferation in human leukemia HL-60 and K562 cell lines. Data are presented as the mean ± standard deviation. *P<0.05 compared with their respective controls. miR, microRNA; NC, normal control.
miR-217 enhanced cell chemosensitivity to DOX in HL-60 and K562 cells
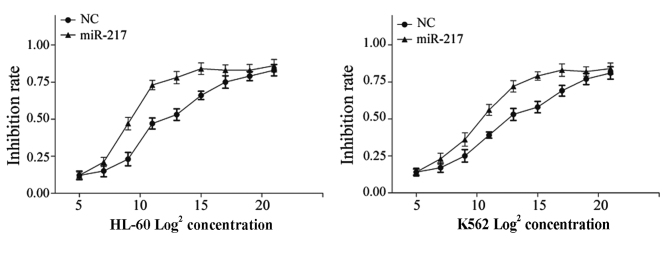
It has previously been demonstrated that miRs perform important roles in the regulation of chemoresistance. Therefore, the present study investigated the effect of miR-217 on cell chemoresistance in AML using a chemosensitivity assay. Following transfection with miR-217 mimics or NC, cells were treated with DOX at various concentrations (32–1048576 ng/ml) for 48 h. As shown in Fig. 4, miR-217 enhanced cell chemosensitivity of HL-60 (P=0.012) and K562 (P=0.018) cells to DOX compared to cells transfected with NC.
Figure 4.
The effect of miR-217 on chemoresistance in acute myeloid leukemia cells was determined by a chemosensitivity assay. miR-217 significantly enhanced cell chemosensitivity of human leukemia HL-60 and K562 cell lines to doxorubicin compared to NC. Data are presented as the mean ± standard deviation. miR, microRNA; NC, normal control.
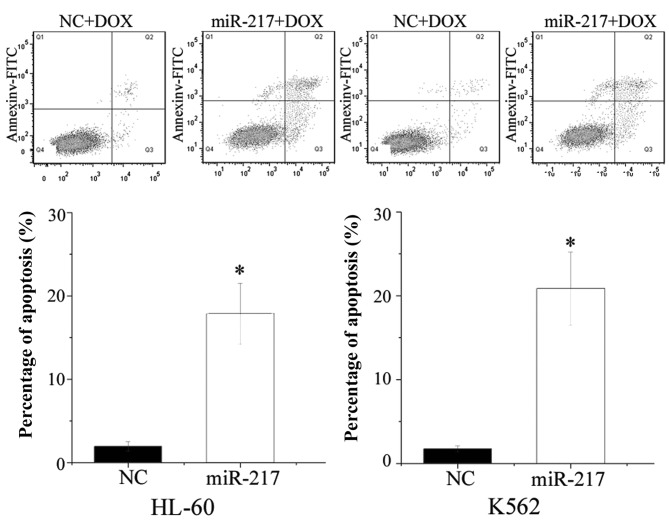
miR-217 enhanced cell apoptosis induced by DOX in HL-60 and K562 cells
The present study performed flow cytometry to evaluate the influence of miR-217 on cell apoptosis induced by DOX. As shown in Fig. 5, ectopic expression of miR-217 enhanced cell apoptosis induced by DOX in HL-60 (P=0.010) and K562 (P=0.005) cells. The present findings are consistent with the change of drug sensitivity, and demonstrate that miR-217-increased cell chemosensitivity was possibly mediated by the cell apoptosis pathway.
Figure 5.
Influence of miR-217 on cell apoptosis induced by DOX as determined by flow cytometry. Upregulation of miR-217 increased cell apoptosis in human leukemia HL-60 and K562 cells induced by DOX. Data are presented as the mean ± standard deviation. *P<0.05 compared with their respective controls. miR, microRNA; DOX, doxorubicin; FITC, fluorescein isothiocyanate; NC, normal control.
KRAS was a direct target gene of miR-217 in vitro
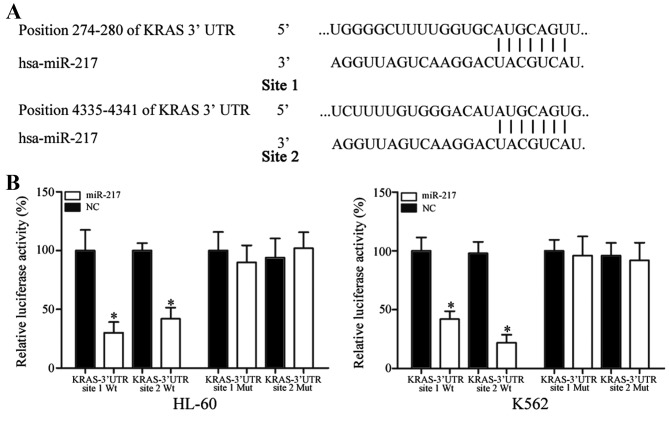
To identify the target of miR-217, bioinformatic algorithms (TargetScan, Whitehead Institute for Biomedical Research, Cambridge, MA, USA) were performed. KRAS was predicted to be a target of miR-217. As shown in Fig. 6A, 2 miR-217 putative binding sites were identified covering the nucleotides 274–280 (site 1) and 4335–4341 (site 2) of KRAS 3′UTR.
Figure 6.
KRAS was a direct target gene of miR-217 in vitro. (A) Two miR-217 putative binding sites were identified covering the nucleotides 274–280 (site 1) and 4,335–4,341 (site 2) of KRAS 3′UTR. (B) Ectopic expression of miR-217 significantly inhibited the PGL3-KRAS-3′UTR site 1 Wt and PGL3-KRAS-3′UTR site 2 Wt luciferase activity, but not the PGL3-KRAS-3′UTR site 1 Mut and PGL3-KRAS-3′UTR site 1 Mut luciferase activity in human leukemia HL-60 and K562 cells. Data are presented as the mean ± standard deviation. *P<0.05 compared with their respective controls. KRAS, Kirsten rat sarcoma viral oncogene homolog; miR, microRNA; Wt, wild type; Mut, mutant; UTR, untranslated region; NC, normal control.
In addition, a dual-luciferase reporter assay was performed to explore whether KRAS was a direct target of miR-217. As shown in Fig. 6B, miR-217 led to a significant decrease of pGL3-KRAS-3′UTR site 1 Wt and pGL3-KRAS-3′UTR site 2 Wt luciferase activity in HL-60 (site 1, P=0.020; site 2, P=0.032) and K562 (site 1, P=0.035; site 2, P=0.014) cells compared with NC cells. Mutation of the two miR-217 binding sites (site 1 Mut or site 2 Mut) restored normal luciferase activity of KRAS-3′UTR in HL-60 and K562 cells. These results indicate that KRAS was a direct target gene of miR-217 in vitro.
miR-217 negatively regulated KRAS protein expression at the post-transcriptional level
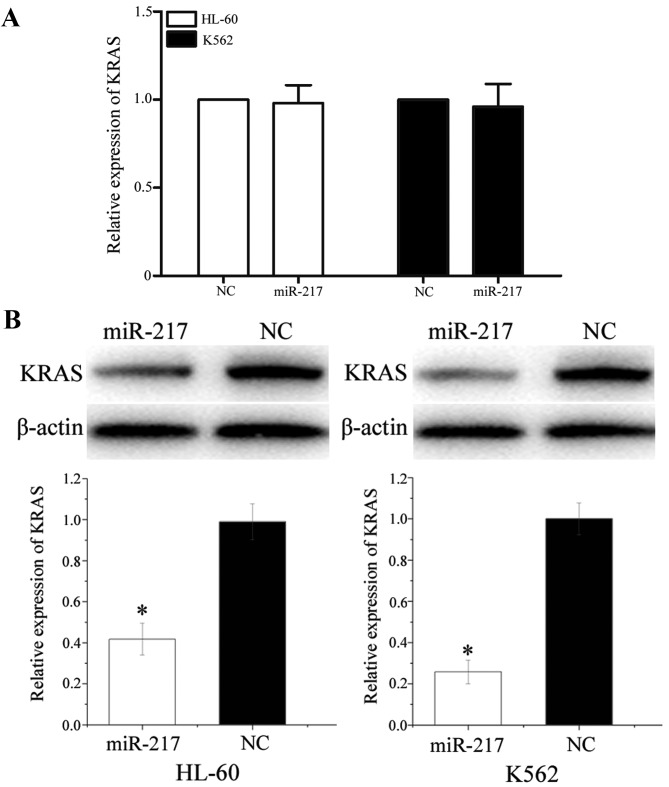
To determine the association between miR-217 and KRAS at the mRNA and protein levels, miR-217 mimics or NC was transfected into HL-60 and K562 cells. The expression of KRAS at mRNA level was detected using qPCR. The expression of KRAS at protein level was monitored using western blot analysis. As shown in Fig. 7A, KRAS mRNA levels were not significantly altered during these treatments (P>0.05). However, western blot analysis revealed that compared to NC, the expression of KRAS was significantly downregulated in HL-60 (P=0.022) and K562 (P=0.016) cells transfected with miR-217 mimics (Fig. 7B). These results indicated that miR-217 did not affect KRAS mRNA stability, but decreased KRAS expression at the post-transcriptional level.
Figure 7.
miR-217 negatively regulated KRAS expression at the post-transcriptional level. (A) No differences were found in KRAS mRNA levels between human leukemia HL-60 and K562 cells transfected with miR-217 and NC. Data are presented as mean ± standard deviation. (B) Human leukemia HL-60 and K562 cells transfected with miR-217 showed a decreased KRAS expression at protein level compared with cells transfected with NC. Data are presented as mean ± standard deviation. β-actin was used as a control in the western blot analysis. *P<0.05 compared with their respective controls. KRAS, Kirsten rat sarcoma viral oncogene homolog; miR, microRNA; NC, normal control.
KRAS was involved in miR-217-induced effects in HL-60 and K562 cells
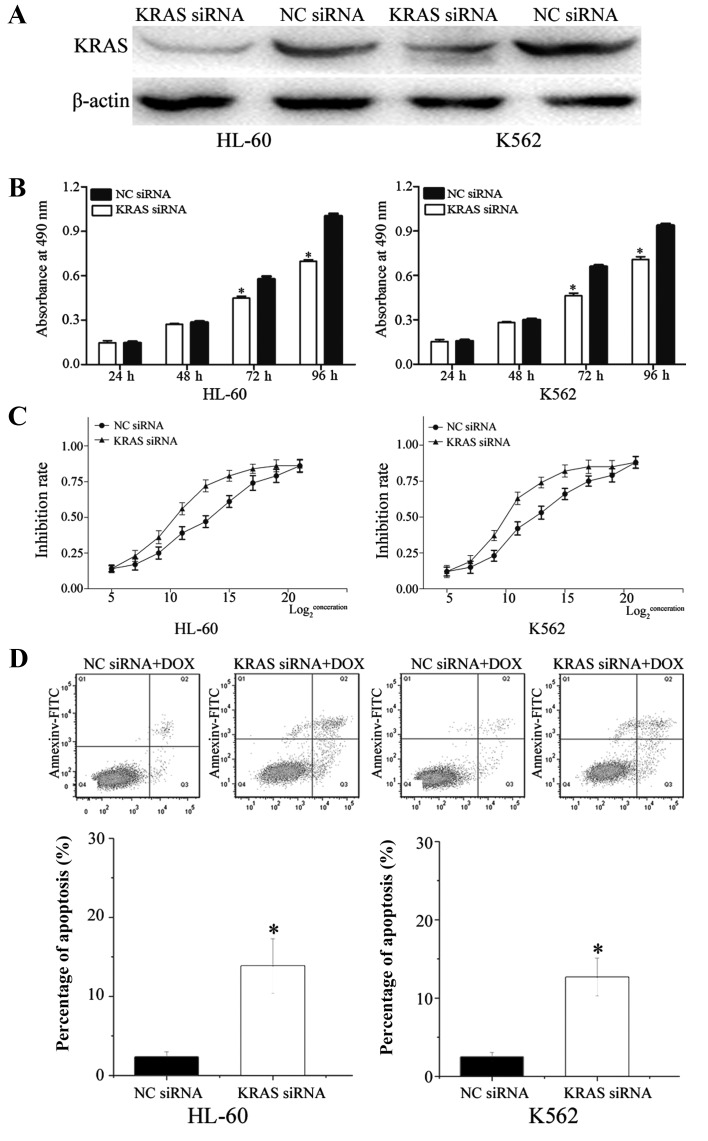
To determine whether KRAS serves as a critical mediator of the effects of miR-217 in AML cells, KRAS siRNA and NC siRNA were transfected into HL-60 and K562 cells. Following transfection for 72 h, western blot analysis demonstrated that KRAS was downregulated in HL-60 (P=0.010) and K562 (P=0.015) cells transfected with KRAS siRNA (Fig. 8A).
Figure 8.
Effects of KRAS in human leukemia HL-60 and K562 cells. (A) Western blot analysis shows that KRAS expression at protein level was downregulated in human leukemia HL-60 and K562 cells transfected with KRAS siRNA, in comparison with cells transfected with NC siRNA. β-actin acted as a control. (B) MTT assay revealed that cells transfected with KRAS siRNA showed significantly inhibited cell proliferation. Data are presented as mean ± standard deviation. (C) Chemosensitivity assays revealed that the KRAS siRNA group showed markedly enhanced cell chemosensitivity of human leukemia HL-60 and K562 cells to doxorubicin compared to cells transfected with NC siRNA. Data are presented as mean ± standard deviation. (D) Inhibition of KRAS enhanced HL-60 and K562 cells apoptosis induced by DOX. Data are presented as mean ± standard deviation.*P<0.05 compared with their respective controls. KRAS, Kirsten rat sarcoma viral oncogene homolog; siRNA, small interfering RNA; NC, normal control; FITC, fluorescein isothiocyanate; DOX, doxorubicin.
In the MTT assay, the KRAS siRNA group significantly inhibited cell growth (HL-60, P=0.008; K562, P=0.020) compared with the NC siRNA group (Fig. 8B). In addition, chemosensitivity assays revealed that the KRAS siRNA group exhibited markedly enhanced cell chemosensitivity of HL-60 (P=0.024) and K562 (P=0.026) cells to DOX compared with cells transfected with NC siRNA (Fig. 8C). Additionally, the apoptosis assay verified that knockdown of KRAS increased HL-60 (P=0.017) and K562 (P=0.030) cell apoptosis induced via DOX (Fig. 8D). These data indicate that effects of KRAS siRNA were similar to those induced by miR-217 in AML cells, suggesting KRAS as a functional target of miR-217 in AML.
Discussion
Abnormal expression of miRs has been verified to contribute to carcinogenesis and the development of various types of tumor (20). However, the role of miRs in AML needs additional investigation. The present study demonstrated that miR-217 was significantly downregulated in AML. In addition, ectopic expression of miR-217 suppressed cellular proliferation and enhanced the chemosensitivity of AML cells to DOX through the cellular apoptosis pathway. To the best of our knowledge, this is the first study to explore the expression and functions of miR-217 in AML.
The abnormal expression of miR-217 has been demonstrated in a number of human malignancies. For example, in human osteosarcoma, the expression level of miR-217 was decreased in tumor tissues and cell lines. Decreased miR-217 expression was closely correlated with large tumor size, positive distant metastasis, advanced clinical stage and shorter overall survival (21,22). These findings suggest that in osteosarcoma, miR-217 may be involved in the initiation and progression of cancer, and could be investigated as a prognostic biomarker in the future (23). miR-217 was also found to be downregulated in colorectal cancer tissues compared with corresponding noncancerous tissues. The low expression level of miR-217 was notably correlated with tumor differentiation and shorter overall survival for patients with colorectal cancer (24). Additionally, miR-217 was found to be downregulated in lung (25), hepatocellular (26) and clear cell renal cell carcinoma (27) and pancreatic ductal adenocarcinoma (28). However, in breast cancer, miR-217 was upregulated in tumor tissues in comparison with normal breast tissues. High levels of miR-217 were significantly associated with high histological grade, the triple negative subtype and advanced tumor stage (29).
These conflicting studies indicate that miR expression levels vary in different types of tumor, and that is has tissue specificity. In addition, in gastric cancer, upregulation of miR-217 significantly suppressed cell growth and invasion through negative regulation of Glypican-5 (23). miR-217 has been verified as a tumor suppressor in osteosarcoma via blockade of the WAS protein family member 3 and Wnt5a (21,22,30). In colorectal cancer, enforced miR-217 expression targeted astrocyte-elevated gene-1 to decrease cell growth, colony formation, migration and invasion by promoting apoptosis and G0/G1 phase arrest (24). In hepatocellular carcinoma, endogenous miR-217 expression inhibited cell invasion by directly targeting E2F transcription factor 3 (26). In clear cell renal cell carcinoma, ectopic expression of miR-217 suppressed cell proliferation, migration and invasion (27). These studies indicate that miR-217 mainly functions as a tumor suppressor in several types of cancer. However, miR-217 also functions as an oncogene in human breast cancer by targeting the Dachshund homolog 1 to enhance cell proliferation (29). These ambivalent results suggest that the functions of miR-217 in cancers are tissue-type dependent. In addition, miR-217 enhanced the sensitivity of lung cancer cells to cisplatin (25). The present study revealed that miR-217 decreased cell proliferation and enhanced chemosensitivity of AML cells to DOX via the cell apoptosis pathway. The present results therefore provide support for the use of DOX in combination with miR-217 as a treatment therapy for patients with AML.
In addition, an important molecular link between miR-217 and KRAS was demonstrated in the present study. miR-217 targeted KRAS to function as a tumor suppressor in AML. KRAS mainly functions as a critical on-off switch in signaling networks that transfer extracellular signals to the nucleus, and connect multiple upstream signals to various downstream signaling pathways (31). These signaling pathways contribute to cellular differentiation, proliferation, survival, cell cycle, apoptosis, migration and invasion (32,33). Therefore, it is important to pay close attention to KRAS as a potential targeted therapy for inhibition in cancer.
DOX, a cytotoxic antiproliferative drug, is widely used as cytotoxic agent for chemotherapy in a wide range of cancers, including AML (34,35). It operates by intercalating into the DNA and preventing DNA from being resealed; this stops replication and eventually damages the DNA structure, resulting in the arrest of the cell growth cycle and the cellular apoptosis pathway (36). However, the use of DOX in clinical settings is limited due to the risk of cardiotoxicity and the ability of the cancer cells to develop resistance to the drug (35). Additionally, drug resistance is the major reason for treatment failure. This indicates that intrinsic chemoresistance may have an important role in cancer. The present study verified for the first time that upregulation of miR-217 induced chemosensitivity to DOX through the cellular apoptosis pathway in vitro. Above all, the present results have the potential to lead to the development of novel strategies in treating AML.
In summary, the present study was the first to show that miR-217 was downregulated in AML and contributed to the cellular proliferation. In addition, it was demonstrated that miR-217 enhanced cell chemosensitivity of AML cells to DOX through the cell apoptosis pathway. In the present study, KRAS was identified as a direct target of miR-217. The identification of candidate target genes of miR-217 may aid understanding of the potential mechanisms involved in AML.
Additional studies are required to address whether the potential of miR-217 may be fully realized in AML treatment. If so, miR-217 may be beneficial for treatment of AML.
Acknowledgements
The present study was supported by the Natural Science Foundation of Hubei (grant no. 2012FFB02435) and the central university special funding (Huazhong University of Science and Technology; grant no. 2013QN191).
References
- 1.Das RP, Konkimalla VB, Rath SN, Hansa J, Jagdeb M. Elucidation of the molecular interaction between miRNAs and the HOXA9 gene, involved in acute myeloid leukemia, by the assistance of argonaute protein through a computational approach. Genomics Inform. 2015;13:45–52. doi: 10.5808/GI.2015.13.2.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Estey EH. Acute myeloid leukemia: 2013 update on risk-stratification and management. Am J Hematol. 2013;88:318–327. doi: 10.1002/ajh.23404. [DOI] [PubMed] [Google Scholar]
- 3.Ferrara F, Schiffer CA. Acute myeloid leukaemia in adults. Lancet. 2013;381:484–495. doi: 10.1016/S0140-6736(12)61727-9. [DOI] [PubMed] [Google Scholar]
- 4.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 5.Gregory TK, Wald D, Chen Y, Vermaat JM, Xiong Y, Tse W. Molecular prognostic markers for adult acute myeloid leukemia with normal cytogenetics. J Hematol Oncol. 2009;2:23. doi: 10.1186/1756-8722-2-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burnett A, Wetzler M, Löwenberg B. Therapeutic advances in acute myeloid leukemia. J Clin Oncol. 2011;29:487–494. doi: 10.1200/JCO.2010.30.1820. [DOI] [PubMed] [Google Scholar]
- 7.Stanisic S, Kalaycio M. Treatment of refractory and relapsed acute myelogenous leukemia. Expert Rev Anticancer Ther. 2002;2:287–295. doi: 10.1586/14737140.2.3.287. [DOI] [PubMed] [Google Scholar]
- 8.He L, Hannon GJ. MicroRNAs: Small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1415. [DOI] [PubMed] [Google Scholar]
- 9.Ameres SL, Zamore PD. Diversifying microRNA sequence and function. Nat Rev Mol Cell Biol. 2013;14:475–488. doi: 10.1038/nrm3611. [DOI] [PubMed] [Google Scholar]
- 10.Berindan-Neagoe I, Pdel C Monroig, Pasculli B, Calin GA. MicroRNAome genome: A treasure for cancer diagnosis and therapy. CA Cancer J Clin. 2014;64:311–336. doi: 10.3322/caac.21244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ueda T, Volinia S, Okumura H, Shimizu M, Taccioli C, Rossi S, Alder H, Liu CG, Oue N, Yasui W, et al. Relation between microRNA expression and progression and prognosis of gastric cancer: A microRNA expression analysis. Lancet Oncol. 2010;11:136–146. doi: 10.1016/S1470-2045(09)70343-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Braun J, Hüttelmaier S. Pathogenic mechanisms of deregulated microRNA expression in thyroid carcinomas of follicular origin. Thyroid Res. 2011;4(Suppl 1):S1. doi: 10.1186/1756-6614-4-S1-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang B, Pan X, Cobb GP, Anderson TA. microRNAs as oncogenes and tumor suppressors. Dev Biol. 2007;302:1–12. doi: 10.1016/j.ydbio.2006.08.028. [DOI] [PubMed] [Google Scholar]
- 14.Esquela-Kerscher A, Slack FJ. Oncomirs-microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 15.Bhardwaj A, Singh S, Singh AP. MicroRNA-based cancer therapeutics: Big hope from small RNAs. Mol Cell Pharmacol. 2010;2:213–219. doi: 10.4255/mcpharmacol.10.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ryan BM, Robles AI, Harris CC. Genetic variation in microRNA networks: The implications for cancer research. Nat Rev Cancer. 2010;10:389–402. doi: 10.1038/nrc2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu F, Zhang J, Ji M, Li P, Du Y, Wang H, Zang S, Ma D, Sun X, Ji C. miR-181b increases drug sensitivity in acute myeloid leukemia via targeting HMGB1 and Mcl-1. Int J Oncol. 2014;45:383–392. doi: 10.3892/ijo.2014.2390. [DOI] [PubMed] [Google Scholar]
- 18.Nagano H, Tomimaru Y, Eguchi H, Hama N, Wada H, Kawamoto K, Kobayashi S, Mori M, Doki Y. MicroRNA-29a induces resistance to gemcitabine through the Wnt/β-catenin signaling pathway in pancreatic cancer cells. Int J Oncol. 2013;43:1066–1072. doi: 10.3892/ijo.2013.2037. [DOI] [PubMed] [Google Scholar]
- 19.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 20.Wu D, Niu X, Pan H, Zhang Z, Zhou Y, Qu P, Zhou J. MicroRNA-497 targets hepatoma-derived growth factor and suppresses human prostate cancer cell motility. Mol Med Rep. 2016;13:2287–2292. doi: 10.3892/mmr.2016.4756. [DOI] [PubMed] [Google Scholar]
- 21.Sun B, Yang M, Li M, Wang F. The microRNA-217 functions as a tumor suppressor and is frequently downregulated in human osteosarcoma. Biomed Pharmacother. 2015;71:58–63. doi: 10.1016/j.biopha.2015.02.014. [DOI] [PubMed] [Google Scholar]
- 22.Wei R, Deng Z, Su J. miR-217 targeting Wnt5a in osteosarcoma functions as a potential tumor suppressor. Biomed Pharmacother. 2015;72:158–164. doi: 10.1016/j.biopha.2015.04.012. [DOI] [PubMed] [Google Scholar]
- 23.Wang H, Dong X, Gu X, Qin R, Jia H, Gao J. The MicroRNA-217 functions as a potential tumor suppressor in gastric cancer by targeting GPC5. PLoS One. 2015;10:e0125474. doi: 10.1371/journal.pone.0125474. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 24.Wang B, Shen ZL, Jiang KW, Zhao G, Wang CY, Yan YC, Yang Y, Zhang JZ, Shen C, Gao ZD, et al. MicroRNA-217 functions as a prognosis predictor and inhibits colorectal cancer cell proliferation and invasion via an AEG-1 dependent mechanism. BMC Cancer. 2015;15:437. doi: 10.1186/s12885-015-1438-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guo J, Feng Z, Huang Z, Wang H, Lu W. MicroRNA-217 functions as a tumour suppressor gene and correlates with cell resistance to cisplatin in lung cancer. Mol Cells. 2014;37:664–671. doi: 10.14348/molcells.2014.0121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Su J, Wang Q, Liu Y, Zhong M. miR-217 inhibits invasion of hepatocellular carcinoma cells through direct suppression of E2F3. Mol Cell Biochem. 2014;392:289–296. doi: 10.1007/s11010-014-2039-x. [DOI] [PubMed] [Google Scholar]
- 27.Li H, Zhao J, Zhang JW, Huang QY, Huang JZ, Chi LS, Tang HJ, Liu GQ, Zhu DJ, Ma WM. MicroRNA-217, down-regulated in clear cell renal cell carcinoma and associated with lower survival, suppresses cell proliferation and migration. Neoplasma. 2013;60:511–515. doi: 10.4149/neo_2013_066. [DOI] [PubMed] [Google Scholar]
- 28.Vychytilova-Faltejskova P, Kiss I, Klusova S, Hlavsa J, Prochazka V, Kala Z, Mazanec J, Hausnerova J, Kren L, Hermanova M, et al. MiR-21, miR-34a, miR-198 and miR-217 as diagnostic and prognostic biomarkers for chronic pancreatitis and pancreatic ductal adenocarcinoma. Diagn Pathol. 2015;10:38. doi: 10.1186/s13000-015-0272-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang Q, Yuan Y, Cui J, Xiao T, Jiang D. MiR-217 promotes tumor proliferation in breast cancer via targeting DACH1. J Cancer. 2015;6:184–191. doi: 10.7150/jca.10822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shen L, Wang P, Yang J, Li X. MicroRNA-217 regulates WASF3 expression and suppresses tumor growth and metastasis in osteosarcoma. PLoS One. 2014;9:e109138. doi: 10.1371/journal.pone.0109138. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 31.Zuber J, Tchernitsa OI, Hinzmann B, Schmitz AC, Grips M, Hellriegel M, Sers C, Rosenthal A, Schäfer R. A genome-wide survey of RAS transformation targets. Nat Genet. 2000;24:144–152. doi: 10.1038/72799. [DOI] [PubMed] [Google Scholar]
- 32.Crespo P, León J. Ras proteins in the control of the cell cycle and cell differentiation. Cell Mol Life Sci. 2000;57:1613–1636. doi: 10.1007/PL00000645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu Y, Zhuang Y, Han M, Xu T, Deng K. Ras promotes cell survival by antagonizing both JNK and Hid signals in the Drosophila eye. BMC Dev Biol. 2009;9:53. doi: 10.1186/1471-213X-9-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lagadinou ED, Ziros PG, Tsopra OA, Dimas K, Kokkinou D, Thanopoulou E, Karakantza M, Pantazis P, Spyridonidis A, Zoumbos NC. c-Jun N-terminal kinase activation failure is a new mechanism of anthracycline resistance in acute myeloid leukemia. Leukemia. 2008;22:1899–1908. doi: 10.1038/leu.2008.192. [DOI] [PubMed] [Google Scholar]
- 35.Kweon SH, Song JH, Kim TS. Resveratrol-mediated reversal of doxorubicin resistance in acute myeloid leukemia cells via downregulation of MRP1 expression. Biochem Biophys Res Commun. 2010;395:104–110. doi: 10.1016/j.bbrc.2010.03.147. [DOI] [PubMed] [Google Scholar]
- 36.Tao J, Lu Q, Wu D, Li P, Xu B, Qing W, Wang M, Zhang Z, Zhang W. ΜicroRNA-21 modulates cell proliferation and sensitivity to doxorubicin in bladder cancer cells. Oncol Rep. 2011;25:1721–1729. doi: 10.3892/or.2011.1245. [DOI] [PubMed] [Google Scholar]