Abstract
Circulating tumor cells (CTCs), isolated as a ‘liquid biopsy’, may provide important diagnostic and prognostic information. Therefore, rapid, reliable and unbiased detection of CTCs are required for routine clinical analyses. It was demonstrated that negative enrichment, an epithelial marker-independent technique for isolating CTCs, exhibits a better efficiency in the detection of CTCs compared with positive enrichment techniques that only use specific anti-epithelial cell adhesion molecules. However, negative enrichment techniques incur significant cell loss during the isolation procedure, and as it is a method that uses only one type of antibody, it is inherently biased. The detection procedure and identification of cell types also relies on skilled and experienced technicians. In the present study, the detection sensitivity of using negative enrichment and a previously described unbiased detection method was compared. The results revealed that unbiased detection methods may efficiently detect >90% of cancer cells in blood samples containing CTCs. By contrast, only 40–60% of CTCs were detected by negative enrichment. Additionally, CTCs were identified in >65% of patients with stage I/II lung cancer. This simple yet efficient approach may achieve a high level of sensitivity. It demonstrates a potential for the large-scale clinical implementation of CTC-based diagnostic and prognostic strategies.
Keywords: circulating tumor cells, non-small-cell lung cancer, negative enrichment, unbiased enrichment, cluster of differentiation 45, cytokeratin, A549 cell line
Introduction
Lung malignancies are the most common cause of mortality among patients with cancer. This is mainly due to the late timing and advanced disease stage at presentation (1). Distant metastases are identified in >55% of patients at presentation (1). For patients diagnosed in the early stages, surgery to resect the tumor and nearby lymph nodes is the most common type of treatment. By contrast, patients with late-stage disease often undergo chemotherapy, alone or in combination with radiation therapy, prior or subsequent to surgery (2). In a previous study, Ilie et al (3) revealed that the annual surveillance of circulating tumor cells (CTCs) in patients with chronic obstructive pulmonary disease by computed tomography was able to detect lung nodules 1 to 4 years subsequent to the detection of the CTCs, leading to prompt surgical resection and histopathological diagnosis of early-stage lung cancer. Therefore, a sensitive method that directly detects lung cancer cells in the early stages may improve the treatment efficacy and prognoses of patients.
CTCs originate from primary tumor masses, enter into the peripheral bloodstream and establish distant metastatic lesions in other organs (4). CTCs provide important diagnostic and prognostic information. They are also important for understanding the biology of metastasis (5). CTCs are rarely observed in the blood, and patients with early stage cancer may exhibit as few as 1 CTC per 1 ml peripheral blood (6). As such, detection of CTCs remains challenging. A number of approaches have been described for the detection and isolation of CTCs, and ~40 commercial products are available at present (7–16). The existing methods fall within two categories: Physical and biological methods. Physical methods mainly exploit the differences in size and density between CTCs and blood cells (17,18). They are label-free and present excellent flexibility in the subsequent characterization of CTCs. However, their reliability has been questioned. In a previous study, cancer cells were able to pass through 5 µm wide channels (19). Additionally, cancer cells that have undergone epithelial-mesenchymal transition (EMT) possess small sizes and spindle shapes, and therefore may easily pass through filters leading to false-negative results (20). Approaches based on the bioaffinities between epithelial cell adhesion molecules (EpCAM) on cell membranes and cognate antibody-coated magnetic beads yield high specificities (21). As EpCAM is not a universal marker that may be used to isolate all CTCs in various types of cancer, these methods are inherently flawed (22,23). Anti-cluster of differentiation (CD45 antibody-coated magnetic beads have been used to deplete the majority of leucocytes and negatively enrich CTCs. This method has demonstrated a relatively high sensitivity of 80–90% (24). However, CTCs and leucocytes may conglutinate into lumps, or CTCs may nonspecifically endocytose these beads, causing a loss of CTCs (25). An unbiased detection method has been developed (11,26), in which blood samples are not subjected to either positive or negative enrichment. Instead, blood samples are directly lysed to remove the red blood cells. The remaining nucleated cells were bound onto substrates, fixed, stained using fluorescence-labeled antibodies and subjected to examination using high-throughput automated microscopy. High levels of sensitivity have been demonstrated for negative enrichment and this unbiased detection method. However, a direct comparison between negative enrichment and the unbiased detection method has not been performed. The correlation between CTC counts and tumor stages is also inconclusive.
The aim of the present study was to compare the detection sensitivity between these two approaches using blood samples mixed with cancer cell lines. In the negative enrichment group, anti-CD45 coated magnetic beads, group M, and CD45 depletion cocktail, group C, were selected to isolate and detect cancer cells. In the other group, group U, the unbiased detection method was performed. The results demonstrated that the unbiased detection method exhibited improved sensitivity but also possessed significant cost advantages compared with the negative enrichment group. Additionally, this method was used to study the clinical significance of CTCs in non-small-cell lung cancer stages, and detected CTCs in >65% patients with stage I/II lung cancer. It was concluded that the unbiased detection method has the potential to provide a cost-effective technological solution for large-scale clinical implementation.
Materials and methods
Ethics statement
The present study was approved by the institutional review board and ethical committee of Zhongda Hospital (Nanjing, China). The data were analyzed anonymously and written informed consent from was obtained from the participants.
Collection and processing of blood samples of patients
A total of 83 lung cancer patients and 151 non-cancerous controls were recruited for the present study. The clinicopathological characteristics of the patients are listed in Table I. The blood samples were collected at the Affiliated Zhongda Hospital (Nanjing, China) and Nanjing Jinling Hospital (Nanjing, China) between July 2013 and June 2014. To avoid epithelial cell contamination by the needle when it pierced the skin, the first tube with 2 ml of peripheral blood was discarded and only the second tube with 3 ml of blood was assayed. To avoid significant degradation of CD45, blood samples were processed 3 h subsequent to collection in normal EDTA-treated tubes. No patient underwent preoperative radiotherapy, chemotherapy or any other treatment. The cancer of each patient was staged according to AJCC Lung Cancer Staging, 7th edition (2009). The clinical data including sex, age and TNM stages were obtained from medical records. The nucleated cells were collected and seeded onto substrates following the same protocol described below. The cells were incubated with the mixture of anti-CD45 (1:400; catalog no. 11–0459-42; eBioscience, Inc., San Diego, CA, USA) and anti-pan cytokeratin (1:500; catalog no. 50-9003-82; CK; eBioscience, Inc.) at 4°C for 5 h, and subsequently stained with DAPI. Only CK+/CD45−/DAPI+ cells were identified as CTCs.
Table I.
Patient demographics.
| Characteristic | Lung cancer patients | Healthy controls |
|---|---|---|
| Total, n | 83 | 107 |
| Sex, n | 56/27a | 60/47a |
| Age, yearsb | 66 (25–82) | 56 (21–78) |
| Cancer stage, n | ||
| Stage I | 13 | – |
| Stage II | 10 | – |
| Stage III | 33 | – |
| Stage IV | 27 | – |
Male/female.
Data are presented as the median and range.
Cell culture and preparation of blood samples
Green fluorescent protein (GFP)-transfected adenocarcinomic human alveolar basal epithelial A549 cell lines (American Type Culture Collection, Manassas, VA, USA) were cultured in Dulbecco's modified Eagle's medium (Corning Incorporated, Corning, NY, USA) supplemented with 10% fetal bovine serum (FBS; Corning Incorporated), 100 U/ml penicillin (Hyclone; GE Healthcare Life Sciences, Logan, UT, USA) and 100 µg/ml streptomycin (Hyclone; GE Healthcare Life Sciences). Cells were placed in T-25 tissue culture flasks. Flasks were placed into an incubator maintained at 5% CO2 at 37°C and 10% humidity. The GFP expression rate of the A549 cells was calculated based on fluorescence expression and was ~99%. Triplicate 6 ml human peripheral blood samples were collected from 10 healthy volunteers using Vacutainers (BD Biosciences, Franklin Lakes, NJ, USA). A total of 6,000 A549 cells were mixed into 6 ml healthy blood, and the 6 ml sample was equally split into M, C and U groups.
Pretreatment of samples in respective groups
In group M, 2 ml peripheral blood was mixed with 28 ml cold red blood cell (RBC) lysis buffer, containing 15.5 mM NH4Cl, 0.1 mM EDTA and 1 mM NaHCO3, for 10 min at 20°C. All nucleated cells were resuspended in PBS and subsequently incubated with anti-CD45 monoclonal antibody-coated magnetic beads at 37°C for 30 min, followed by the separation of the magnetic beads using a magnetic stand (Promega Corporation, Madison, WI, USA). Following centrifugation at 800 × g for 5 min at room temperature, the supernatants were discarded, and the isolated cells were resuspended in PBS. In group C, 2 ml blood was mixed with 100 µl RosetteSep Human CD45 Depletion Cocktail (STEMCELL Technologies, Inc., Vancouver, Canada) and incubated for 20 min at room temperature. The sample was diluted with an equal volume of PBS containing 2% FBS and was layered onto 4 ml Ficoll-Paque PLUS solution (GE Healthcare Life Sciences, Shanghai, China). The mixture was centrifuged at 1,200 × g for 20 min at room temperature. The enriched cells were collected and washed with PBS twice. In group U, blood samples were treated only with RBC lysis buffer. All nucleated cells were washed with PBS twice, centrifuged at 400 × g for 5 min at room temperature and resuspended in PBS. All samples were added to poly-l-lysine (PLL)-coated polystyrene culture dishes (Corning Incorporated, Corning, NY, USA), and incubated for 2 h at 37°C. The typical total cell density on the polystyrene (PS) surface was ~1,000 cells/mm2.
Cell staining
Cell samples from each group were fixed with 4% paraformaldehyde for 10 min on ice and permeabilized with ice-cold pure methanol for 10 min. All samples were stained with 0.1 µg/ml DAPI for 10 min at room temperature. The plates were analyzed using Operetta (version 1.0; PerkinElmer, Inc., Waltham, MA, USA) high-content imaging system.
Quantification and statistical analysis
All images were analyzed by Harmony software (version 6.4; PerkinElmer, inc.). The number of A549 cells with green fluorescence was automatically counted (version 10.8; IN CELL Analyzer; GE Healthcare Life Sciences, Chalfont, UK). The detection sensitivity of each group was calculated using the following equation: Sensitivity=(N)/(2,000×3), where N was the total number of captured CTCs in the three groups, and 2,000 was the number of A549 cells in 2 ml blood. Data were statistically analyzed using paired two-tailed Student's t-test. Clinical samples were imaged and analyzed. The number of CTCs bearing red fluorescence was automatically counted (version 10.8; IN CELL Analyzer; GE Healthcare Life Sciences). P<0.05 was considered to indicate a statistically significant difference.
Results and Discussion
Comparison of detection sensitivity
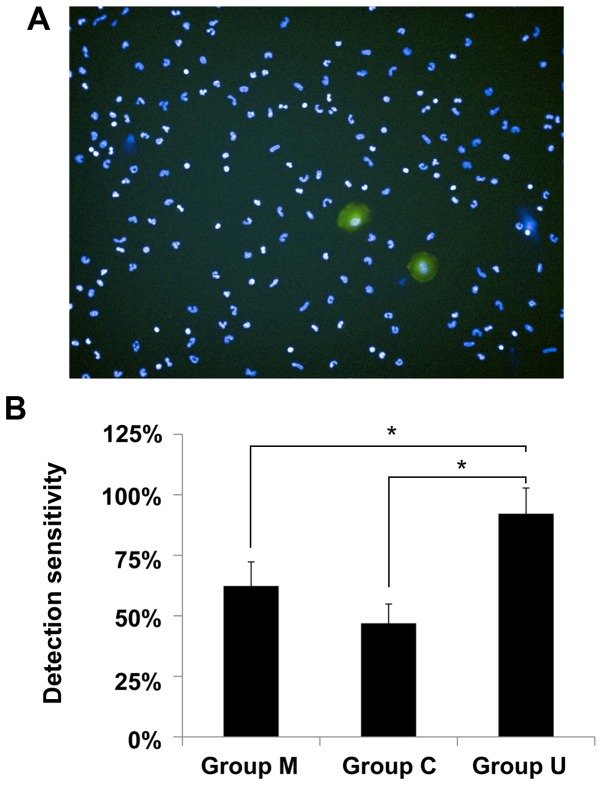
The number of A549 cells in three groups was investigated. A representative image captured by Operetta high-content imaging system is illustrated in Fig. 1A. Nucleated cells, including leucocytes and A549 cells, were stained with DAPI, and all nuclei exhibited blue fluorescence. The A549 cells bearing GFP demonstrated strong green fluorescence. In previous studies, an average of 94±2% binding efficiency at the 2 h time point was observed for the PLL-treated PS substrate, and notably, cell retention on the PLL-treated PS was revealed to be 96±3% (11). The high binding efficiency, high retention, small standard deviation and automatic counting in this detection platform provided good reliability for the present study.
Figure 1.
Comparison of sensitivities between negative enrichment and unbiased detection methods. (A) A representative image of A549 cells mixed with a blood sample. Magnification, ×20. (B) The quantified data of sensitivities in each group. *P<0.01.
In group M, the detection sensitivity of magnetic beads-based negative enrichment was 62.3±9.9% (Fig. 1B). In previous studies, SW620 cells were mixed into blood and negatively enriched using magnetic beads, and 57–94% of cells were recovered (24). It is noted that in the present study, only 100 SW620 cells were mixed into the blood samples for the negative enrichment analysis of the SW620 cells. The relatively low number may cause accidental errors, and therefore the recovered efficiency demonstrated a large standard deviation. By contrast, 6,000 A549 cells were mixed into the blood samples in the present study, ensuring sufficient data was obtained for the statistical analyses. The results of the present study fall into the range 57–94%. The loss of A549 cells may be attributed to non-specifically endocytosed magnetic beads. Alternatively, magnetic beads may adhere to the surface of the A549 cells.
In group C, the detection sensitivity was the lowest, 46.9±8% (Fig. 1B). The isolation of CTCs is simple and target cells may be collected within 1 h. The RosetteSep antibody cocktail crosslinks CD45+ leucocytes to erythrocytes, forming immunorosettes. This increases the density of the leucocytes, meaning they pellet along with the free erythrocytes when centrifuged over a buoyant density medium. If correct protocol is followed, the desired cells are not labeled with antibody and are easily collected as a highly enriched population at the interface between the plasma and the Ficoll-Paque PLUS solution. However, antibodies are often subject to high levels of off-target cross-reactivity. Additionally, cancer cells and particularly cell clusters, may physically stick amongst the complexes. Therefore, the sensitivity of this detection method may be unreliable. Conversely, this strategy produces samples with a high concentration of cancer cells, and therefore may facilitate subsequent molecular analyses if sufficient cancer cells are recovered.
The highest detection sensitivity was achieved in group U: The detection sensitivity was 92.2±10.6% (Fig. 1B). In this group, the cancer cells mixed with the blood samples were only treated with RBC lysis buffer followed by PBS washing and surface seeding. The cells did not experience any immuno-enrichment procedure, which typically causes a certain level of cell loss. A paired t-test was performed, and a significant difference in the detection sensitivity was observed between groups M, C and U (P<0.01).
The respective detection sensitivities of two diagnostic tests among three groups were compared, and it was observed that an unbiased detection was superior to negative enrichment. Additionally, an unbiased detection method does not require expensive leucocyte depletion reagents, and therefore offers an affordable option for clinical CTC examination. In the analysis of blood samples from patients with lung cancer, the unbiased detection method was used.
Detection of CTCs in patients samples
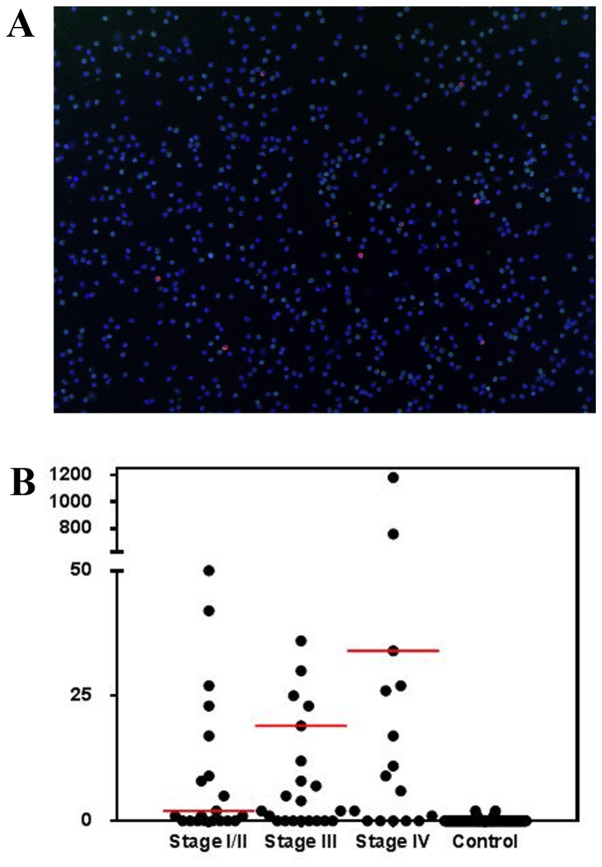
A representative image of detected CTCs in the blood samples of patients is presented in Fig. 2A. Using the unbiased detection method, the overall diagnostic rate of CTCs in the patient population was 74.7%, with a median of 17 CTCs/ml, a range of 0–1,182 CTCs/ml and a mean of 83.3±180.8 CTCs/ml. Of the 23 patients with stage I/II disease, 65.2% exhibited detectable CTCs. A total of 75.8% of patients with stage III and 81.5% of patients with stage IV disease exhibited detectable CTCs. No significant difference (P=0.12) was observed in CTC count between stages III (range 0–561) and IV (range 0–1,182) disease (Fig. 2B). The mean CTC count in patients with stage III and IV disease was significantly increased compared with that of patients with stage I/II disease (range 0–92; P<0.05; Table II). Due to the sensitivity constraints in current positive or negative enrichment-based CTC detection methods, there are few published data about CTC prevalence rates or the correlation of CTCs with disease progression and their usability for clinical staging. Marrinucci et al (26) demonstrated that there was no correlation between CTC counts for all stages of lung cancer. However, in another study, CTCs in gastrointestinal cancer patients were revealed to be a useful tool for judging tumor stage (27). In the present study, a correlation between CTCs and stage of cancer was not observed, although lower CTCs counts were identified in patients with stage I/II disease. It was hypothesized that the lack of correlation may be due to small patient groups. It should be noted that vimentin and/or N-cadherin and other EMT markers were not analyzed in the present study. Therefore, the potential mesenchymal CTCs were not counted. This limitation will be addressed in future studies. However, the results of the present study provide a preliminary indication that the unbiased detection method is able to demonstrate high detection sensitivity, which may be useful for addressing EMT detection in future studies.
Figure 2.
Samples from patients with various stages of lung cancer were examined using the unbiased detection method. (A) A typical image of CTCs detected by a high-content imaging system (version 10.8; IN CELL Analyzer). Magnification, ×4. Green light, anti-cluster of differentiation 45; red light, anti-pan cytokeratin; blue, DAPI. (B) The distribution of CTC counts in patients with lung cancer according to tumor stage. The red lines illustrate the median. CTC, circulating tumor cell.
Table II.
Results of CTC detection in various lung cancer stages and healthy controls.
| Detectable CTCs | ||||
|---|---|---|---|---|
| Stage | Detection, n (%) | Range | Mean | Median |
| I/II | 15 (65.2) | 0–92 | 17.9 | 2 |
| III | 25 (75.8) | 0–561 | 67.9 | 19 |
| IV | 22 (81.5) | 0-1,182 | 157.9 | 34 |
| Healthy control | 6 (5.6) | 0–2 | 0.1 | 0 |
CTC, circulating tumor cell.
When analyzing the numbers of captured CTCs, anthropogenic factors, including artificial determination of cell type, adjustment of fluorescence exposure time and adjustment of image contrast, were minimized as much as possible. Once all images were captured, the software automatically tiled and stitched them, followed by signal processing and analysis. The background noise and detectable fluorescent signal of each cell in each fluorescence channel were first quantified, and subsequently the value of background noise was deducted. The difference was subsequently compared to the critical threshold value developed by the PMR Institute (unpublished data, the average fluorescence intensity of white blood cells). The shape and size of the nucleus was also quantified, as according to cytomorphology, cells exhibiting a large nucleus and an irregular shape may be considered to be abnormal. Therefore, 5 independent parameters were used to determine cell types in the data processing protocol. Depending on the number of markers used to characterize the CTCs, the automated detection and analysis procedures may be completed as quickly as 1 h, giving a total isolation and analysis time of ~7 h, from blood lysis to report, with minimum manipulation. Therefore, this methodology possesses great potential towards enabling an unbiased routine clinical analysis of CTCs. Once clinically validated, it may be applied in any clinical center equipped with an automated microscopy facility, at a fraction of the cost of current CTC isolation technologies.
In summary, the present study suggests that an unbiased detection method may sensitively detect CTCs in patients with early stage lung cancer, and this method demonstrates significantly improved sensitivity compared with negative enrichment analysis methods. The automatic detection and analysis of the unbiased detection method limits human error, and therefore may offer reliable and objective results. Additional improvements to the imaging systems used may be able to detect and analyze ~5 markers, including epithelial and mesenchymal markers. This platform is simple, efficient and low cost. It may be a powerful tool in routine clinical analysis of tumors.
Acknowledgements
The present study was supported by the Colleges and Universities Graduate Student Scientific Research Innovation Project of Jiangsu Province, China (grant no. CXZZ12_0120). The authors would like to thank PerMed Biomedical Company (Beijing, China) and PMR Institute (Shanghai, China) for providing high-content imaging system and analysis software.
References
- 1.Crinò L, Weder W, van Meerbeeck J, Felip E. ESMO Guidelines Working Group: Early stage and locally advanced (non-metastatic) non-small-cell lung cancer: ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2010;21(Suppl 5):v103–v115. doi: 10.1093/annonc/mdq207. [DOI] [PubMed] [Google Scholar]
- 2.Egner JR. AJcc cancer staging manual. JAMA. 2010;304:1726–1727. doi: 10.1001/jama.2010.1525. [DOI] [Google Scholar]
- 3.Ilie M, Hofman V, Long-Mira E, Selva E, Vignaud JM, Padovani B, Mouroux J, Marquette CH, Hofman P. ‘Sentinel’ circulating tumor cells allow early diagnosis of lung cancer in patients with chronic obstructive pulmonary disease. PLoS One. 2014;9:e111597. doi: 10.1371/journal.pone.0111597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang L, Asghar W, Demirci U, Wan Y. Nanostructured substrates for isolation of circulating tumor cells. Nano Today. 2013;8:374–387. doi: 10.1016/j.nantod.2013.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yu M, Stott S, Toner M, Maheswaran S, Haber DA. Circulating tumor cells: Approaches to isolation and characterization. J Cell Biol. 2011;192:373–382. doi: 10.1083/jcb.201010021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zheng Q, Iqbal SM, Wan Y. Cell detachment: Post-isolation challenges. Biotechnol Adv. 2013;31:1664–1675. doi: 10.1016/j.biotechadv.2013.08.013. [DOI] [PubMed] [Google Scholar]
- 7.Wan Y, Kim YT, Li N, Cho SK, Bachoo R, Ellington AD, Iqbal SM. Surface-Immobilized aptamers for cancer cell isolation and microscopic cytology. Cancer Res. 2010;70:9371–9380. doi: 10.1158/0008-5472.CAN-10-0568. [DOI] [PubMed] [Google Scholar]
- 8.Wan Y, Liu Y, Allen PB, Asghar W, Mahmood MA, Tan J, Duhon H, Kim YT, Ellington AD, Iqbal SM. Capture, isolation and release of cancer cells with aptamer-functionalized glass bead array. Lab Chip. 2012;12:4693–4701. doi: 10.1039/c2lc21251j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wan Y, Mahmood MA, Li N, Allen PB, Kim YT, Bachoo R, Ellington AD, Iqbal SM. Nanotextured substrates with immobilized aptamers for cancer cell isolation and cytology. Cancer. 2012;118:1145–1154. doi: 10.1002/cncr.26349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wan Y, Tan J, Asghar W, Kim YT, Liu Y, Iqbal SM. Velocity effect on aptamer-based circulating tumor cell isolation in microfluidic devices. J Phys Chem B. 2011;115:13891–13896. doi: 10.1021/jp205511m. [DOI] [PubMed] [Google Scholar]
- 11.Wan Y, Winter M, Delalat B, Hardingham JE, Grover PK, Wrin J, Voelcker NH, Price TJ, Thierry B. Nanostructured polystyrene well plates allow unbiased high-throughput characterization of circulating tumor cells. ACS Appl Mater Interfaces. 2014;6:20828–20836. doi: 10.1021/am505201s. [DOI] [PubMed] [Google Scholar]
- 12.Wang S, Wan Y, Liu Y. Effects of nanopillar array diameter and spacing on cancer cell capture and cell behaviors. Nanoscale. 2014;6:12482–12489. doi: 10.1039/C4NR02854F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Asghar W, Kim YT, Ilyas A, Sankaran J, Wan Y, Iqbal SM. Synthesis of nano-textured biocompatible scaffolds from chicken eggshells. Nanotechnology. 2012;23:475601. doi: 10.1088/0957-4484/23/47/475601. [DOI] [PubMed] [Google Scholar]
- 14.Asghar W, Wan Y, Ilyas A, Bachoo R, Kim YT, Iqbal SM. Electrical fingerprinting, 3D profiling and detection of tumor cells with solid-state micropores. Lab Chip. 2012;12:2345–2352. doi: 10.1039/c2lc21012f. [DOI] [PubMed] [Google Scholar]
- 15.Wang L, Zheng Q, Zhang Q, Xu H, Tong J, Zhu C, Wan Y. Detection of single tumor cell resistance with aptamer biochip. Oncol Lett. 2012;4:935–940. doi: 10.3892/ol.2012.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mahmood MA, Wan Y, Islam M, Ali W, Hanif M, Kim YT, Iqbal SM. Micro+nanotexturing of substrates to enhance ligand-assisted cancer cell isolation. Nanotechnology. 2014;25:475102. doi: 10.1088/0957-4484/25/47/475102. [DOI] [PubMed] [Google Scholar]
- 17.Farace F, Massard C, Vimond N, Drusch F, Jacques N, Billiot F, Laplanche A, Chauchereau A, Lacroix L, Planchard D, et al. A direct comparison of CellSearch and ISET for circulating tumour-cell detection in patients with metastatic carcinomas. Br J Cancer. 2011;105:847–853. doi: 10.1038/bjc.2011.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim MS, Sim TS, Kim YJ, Kim SS, Jeong H, Park JM, Moon HS, Kim SI, Gurel O, Lee SS, et al. SSA-MOA: A novel CTC isolation platform using selective size amplification (SSA) and a multi-obstacle architecture (MOA) filter. Lab Chip. 2012;12:2874–2880. doi: 10.1039/c2lc40065k. [DOI] [PubMed] [Google Scholar]
- 19.Wan Y, Tamuly D, Allen PB, Kim YT, Bachoo R, Ellington AD, Iqbal SM. Proliferation and migration of tumor cells in tapered channels. Biomed Microdevices. 2013;15:635–643. doi: 10.1007/s10544-012-9721-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sarrió D, Rodriguez-Pinilla SM, Hardisson D, Cano A, Moreno-Bueno G, Palacios J. Epithelial-mesenchymal transition in breast cancer relates to the basal-like phenotype. Cancer Res. 2008;68:989–997. doi: 10.1158/0008-5472.CAN-07-2017. [DOI] [PubMed] [Google Scholar]
- 21.Nagrath S, Sequist LV, Maheswaran S, Bell DW, Irimia D, Ulkus L, Smith MR, Kwak EL, Digumarthy S, Muzikansky A, et al. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature. 2007;450:1235–1239. doi: 10.1038/nature06385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Green JV, Murthy SK. Microfluidic enrichment of a target cell type from a heterogenous suspension by adhesion-based negative selection. Lab Chip. 2009;9:2245–2248. doi: 10.1039/b906768j. [DOI] [PubMed] [Google Scholar]
- 23.Liu Z, Jiang M, Zhao J, Ju H. Circulating tumor cells in perioperative esophageal cancer patients: Quantitative assay system and potential clinical utility. Clin Cancer Res. 2007;13:2992–2997. doi: 10.1158/1078-0432.CCR-06-2072. [DOI] [PubMed] [Google Scholar]
- 24.Liu Z, Fusi A, Klopocki E, Schmittel A, Tinhofer I, Nonnenmacher A, Keilholz U. Negative enrichment by immunomagnetic nanobeads for unbiased characterization of circulating tumor cells from peripheral blood of cancer patients. J Transl Med. 2011;9:70. doi: 10.1186/1479-5876-9-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ozkumur E, Shah AM, Ciciliano JC, Emmink BL, Miyamoto DT, Brachtel E, Yu M, Chen PI, Morgan B, Trautwein J, et al. Inertial focusing for tumor antigen-dependent and -independent sorting of rare circulating tumor cells. Sci Transl Med. 2013;5:179ra47. doi: 10.1126/scitranslmed.3005616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marrinucci D, Bethel K, Kolatkar A, Luttgen MS, Malchiodi M, Baehring F, Voigt K, Lazar D, Nieva J, Bazhenova L, et al. Fluid biopsy in patients with metastatic prostate, pancreatic and breast cancers. Phys Biol. 2012;9:016003. doi: 10.1088/1478-3975/9/1/016003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hiraiwa K, Takeuchi H, Hasegawa H, Saikawa Y, Suda K, Ando T, Kumagai K, Irino T, Yoshikawa T, Matsuda S, et al. Clinical significance of circulating tumor cells in blood from patients with gastrointestinal cancers. Ann Surg Oncol. 2008;15:3092–3100. doi: 10.1245/s10434-008-0122-9. [DOI] [PubMed] [Google Scholar]